Abstract
Objective
To develop a simple, effective, time-saving and low-cost fluorescence protein microarray method for detecting serum alpha-fetoprotein (AFP) in patients with hepatocellular carcinoma (HCC).
Method
Non-contact piezoelectric print techniques were applied to fluorescence protein microarray to reduce the cost of prey antibody. Serum samples from patients with HCC and healthy control subjects were collected and evaluated for the presence of AFP using a novel fluorescence protein microarray. To validate the fluorescence protein microarray, serum samples were tested for AFP using an enzyme-linked immunosorbent assay (ELISA).
Results
A total of 110 serum samples from patients with HCC (n = 65) and healthy control subjects (n = 45) were analysed. When the AFP cut-off value was set at 20 ng/ml, the fluorescence protein microarray had a sensitivity of 91.67% and a specificity of 93.24% for detecting serum AFP. Serum AFP quantified via fluorescence protein microarray had a similar diagnostic performance compared with ELISA in distinguishing patients with HCC from healthy control subjects (area under receiver operating characteristic curve: 0.906 for fluorescence protein microarray; 0.880 for ELISA).
Conclusion
A fluorescence protein microarray method was developed for detecting serum AFP in patients with HCC.
Keywords: Alpha-fetoprotein, hepatocellular carcinoma, fluorescence protein microarray, detecting
Introduction
Hepatocellular carcinoma (HCC) is the second most frequent cause of cancer-related death.1 Treatments with curative intent, such as resection, are feasible at an early stage.2 Despite this, even after complete resection, patients remain at a high risk for disease recurrence, either due to early recurrence of the initial tumour or due to the formation of new lesions (leading to late recurrence).3 In the early stages, liver transplantation has the clearest benefit.4 However, due to the shortage of donor organs, resection and radiofrequency ablation are alternatives.5 HCC is often diagnosed at the late stages when only limited therapeutic options are available; consequently, the survival rate for patients with HCC remains low.6 It is the second global cause of cancer-related mortality, with 746 000 global deaths in 2012.1 In developing countries, more than two-thirds of patients with HCC are diagnosed at the advanced stages.7 To increase the chance of intervention and, more importantly, to improve survival, early detection of subclinical HCC by alpha-fetoprotein (AFP) and/or ultrasonography screening is implemented in many countries. AFP is the most commonly used diagnostic test for HCC surveillance.8–10 Research has shown that high plasma levels of AFP are related to poor prognosis, as well as to the histological grade of malignancy.11
Enzyme-linked immunosorbent assay (ELISA) and fluorescence protein microarray are successfully used in the detection of AFP.12 Our previous work developed a protein microarray assay with horseradish peroxidase (HRP) chemiluminescence for the quantification of AFP in serum from patients with HCC.13 Three types of fluorescence protein microarrays are currently used to study the biochemical activities of proteins: analytical microarrays, functional microarrays, and reverse phase microarrays. Antibody microarrays are the most commonly used analytical microarray.
This study reports the development and application of fluorescence protein microarrays with cyanine 3 (Cy3) to detect AFP in the serum from patients with HCC and healthy control subjects. We have recently updated our methodology to use non-contact piezoelectric print techniques using a microarray spotter (Nano-Plotter™; GeSiM, Radeberg, Germany). The Nano-Plotter™ 2.1 instrument family is ideal for generating high-quality spots by the non-contact micro-dispensing of sub-nanolitre volumes. The versatile ‘drop on demand’ piezo technology allows the placement of very small drops onto various surfaces, but also the mixing of samples into pre-filled cavities to run nano-assays. The application of non-contact piezoelectric print techniques reduces the cost of prey antibody.14
Patients and methods
Study population
The study recruited patients with HCC who underwent liver resection surgery at the Department of Hepatobiliary Surgery, Beijing You’an Hospital, Beijing, China between 1 August 2012 and 31 December 2014. Patients were required to be positive for serum hepatitis B surface antigen and free from chronic hepatitis C infection. HCC diagnosis was histologically confirmed after surgical resection. Healthy control subjects were recruited from volunteers with no reported gastrointestinal or hepatobiliary disease who were undergoing routine health screening at the outpatient department of Beijing You’an Hospital, Beijing, China during the same time period. The study protocol was approved by the Ethics Committee of Beijing You’an Hospital. Written informed consent was obtained from all participants prior to enrolment in the study.
Blood collection
Whole blood (1 ml) was collected from each study participant (preoperative blood for patients with HCC). Blood was allowed to clot, centrifuged at 3000 g for 10 min at 4℃ using aLX-300 mini-centrifuge (Kylin-Bell Lab Instruments, Haimen, China), and the resulting serum was stored at –80℃. Prior to use, serum was diluted 1:4 with 0.05% phosphate-buffered saline (PBS)-Tween 20 (pH 7.5) and stored at 4℃ for a maximum of 7 days.
Microarray preparation
Reaction wells were created by attaching slips of hydrophobic paper (10 holes per slip) to aldehyde-coated glass slides. The prey antibody (0.5 mg/ml mouse antihuman AFP monoclonal antibody with 30% glycerol to prevent evaporation; Fapon Biotech Company, Shenzhen, China) was spotted onto the surface of the aldehyde-modified glass slides in triplicate using a Nano-Plotter™ non-contact microarrayer (microarrays 3*2 spots; spot size 400 µm; spot-to-spot distance 800 µm; GeSiM). Each well contained three anti-AFP antibody and three negative control (5% bovine serum albumen) spots. Following spotting, the slides were incubated for 24 h at 4℃ to fully immobilize the proteins. Each slide was then blocked with 200 µl blocking buffer (10% normal goat serum with 0.1% sodium azide) for 2 h at 37℃, washed four times with 0.05% PBS-Tween 20 (pH 7.5) at room temperature (5 s each wash), air dried at room temperature and then stored at 4℃ until use.
AFP quantification
Rabbit antihuman AFP polyclonal antibody (ab128028; Abcam, Cambridge, UK) was labelled with biotin using a biotin (type A) conjugation kit (ab102865; Abcam) according to the manufacturer’s instructions. Serum samples were added to the microarray reaction wells, hybridized for 30 min at 37℃, then washed four times with 0.05% PBS-Tween 20 (pH7.5) at room temperature (5 s each wash). The biotin-labelled rabbit antihuman AFP polyclonal antibody (1:50 dilution) was added to the wells and hybridized for 30 min at 37℃. Slides were washed twice with 0.05% PBS-Tween 20 (pH 7.5) at room temperature (5 s each wash), then incubated with Cy3-labelled streptavidin (1:50 dilution, ab136196; Abcam) for 30 min at 37℃. Slides were washed four times with 0.05% PBS-Tween 20 (pH 7.5) at room temperature (5 s each wash). Slides were scanned using a GenePix® 4000B Microarray Scanner (Axon, Rye Brook, NY, USA). The standard curve of AFP was created by applying different concentrations of AFP antigen (Fapon Biotech Company; 80 ng/ml, 40 ng/ml, 20 ng/ml, 10 ng/ml, 5 ng/ml, 2.5 ng/ml, 1.25 ng/ml, 0.0625 ng/ml, and 0.03125 ng/ml) and 0.05% PBS-Tween 20 as a negative control to the microarray plate coated with the anti-AFP antibody. The reliability and stability of the assay were verified by repeat quantification (10 assays) of the AFP standard curve to calculate within-run and between-run variation.
ELISA
The serum AFP levels were quantified using the human AFP ELISA kit (ab108838; Abcam) according to the manufacturer’s instructions. First, 50 µl of the AFP standard or sample was added to each well. The wells were covered with sealing tape and incubated for 2 h at 37℃. Then, the wells were manually washed five times with 200 µl of 1X Wash Buffer from the kit. Secondly, 50 µl of 1X biotinylated anti-AFP antibody was added to each well and incubated for 1 h at 37℃. The microplate was washed as described above for the protein microarray. Thirdly, 50 µl of 1X SP conjugate was added to each well and incubated for 30 min at 37℃. The microplate was washed as described above for the protein microarray. Fourth, 50 µl of chromogen substrate was added to each well and incubated for 10 min at 37℃ or until the optimal blue colour density was obtained. Finally, 50 µl of stop solution was added to each well. The colour changed from blue to yellow. The absorbance was immediately determined on a microplate reader (Multiskan™ GO Microplate Spectrophotometer; Thermo Fisher Scientific, Waltham, MA, USA) at a wavelength of 450 nm.
Statistical analyses
All statistical analyses were performed using the SPSS® statistical package, version 17.0 (SPSS Inc., Chicago, IL, USA) for Windows®. Data are presented as mean ± SD (coefficient of variation) or median (95% confidence interval [CI]). Between-group comparisons of AFP concentrations were made using Mann–Whitney U-test for fluorescence protein microarray. Spearman’s rank correlation coefficient was used to determine the associations between serum AFP concentrations as determined by fluorescence protein microarray or ELISA. Receiver operating characteristic (ROC) curve analysis was performed to determine the diagnostic performance of fluorescence protein microarray and ELISA. All tests were two-tailed, and P-values < 0.05 were considered statistically significant.
Results
The study recruited patients with HCC (n = 65) who underwent liver resection surgery and who provided blood samples. The mean ± SD age of the patients with HCC was 51.8 ± 5.2 years (35 males/30 females; age range 43–70 years). A total of 45 healthy control subjects (29 males/16 females; mean ± SD age 50.0 ± 4.5 years; age range 46–65 years) were also enrolled in the study. There were no significant between-group differences in age or sex distribution between the two groups.
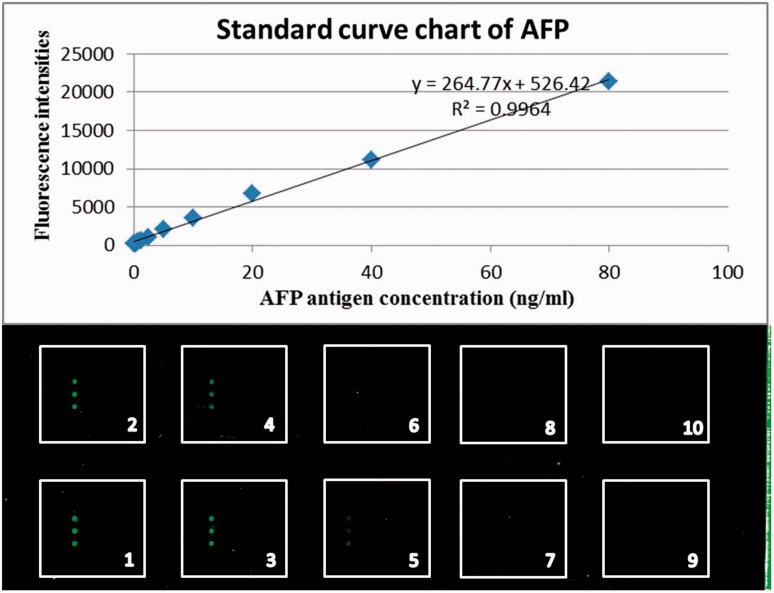
The standard curve of AFP was created by applying different concentrations of AFP antigen (80 ng/ml, 40 ng/ml, 20 ng/ml, 10 ng/ml, 5 ng/ml, 2.5 ng/ml, 1.25 ng/ml, 0.0625 ng/ml, and 0.03125 ng/ml) and 0.05% PBS-Tween 20 as a negative control to the microarray plate coated with the anti-AFP antibody (Figure 1). The reliability and stability of the assay were verified by repeat quantification (10 assays) of the AFP standard curve to calculate within-run and between-run variation. Table 1 presents the within-run and between-run coefficients of variation for the different concentrations of AFP antigen (within-run coefficient of variation, range 4–11; between-run coefficient of variation, range 3–11).
Figure 1.
Alpha-fetoprotein (AFP) antigen levels detected and the standard curve chart by the AFP protein microarray. The standard curve of AFP was created by applying different concentrations of AFP antigen (1: 80 ng/ml, 2: 40 ng/ml, 3: 20 ng/ml, 4: 10 ng/ml, 5: 5 ng/ml, 6: 2.5 ng/ml, 7: 1.25 ng/ml, 8: 0.0625 ng/ml, and 9: 0.03125 ng/ml) and 10: 0.05% phosphate-buffered saline-Tween 20 as a negative control to the microarray plate coated with the anti-AFP antibody. The colour version of this figure is available at: http://imr.sagepub.com.
Table 1.
Within-run and between-run variation for fluorescence protein microarrays for the detection of alpha-fetoprotein (AFP) standard.
| AFP standards, ng/ml | Within-run variation | Between-run variation |
|---|---|---|
| 80 | 21045 ± 1322 (6) | 20085 ± 1282 (6) |
| 40 | 11793 ± 1066 (9) | 11605 ± 966 (8) |
| 20 | 6581 ± 246 (4) | 6452 ± 221 (3) |
| 10 | 3650 ± 202(6) | 3720 ± 199 (5) |
| 5 | 2032 ± 124 (6) | 1998 ± 111 (6) |
| 2.5 | 1060 ± 96 (9) | 1090 ± 101 (9) |
| 1.25 | 654 ± 37 (6) | 642 ± 35 (5) |
| 0.625 | 458 ± 31 (7) | 458 ± 31 (7) |
| 0.3125 | 247 ± 27 (11) | 221 ± 25 (11) |
| 0 | 213 ± 18 (8) | 203 ± 18 (9) |
Data were presented as mean ± SD (coefficient of variation) of 10 experiments.
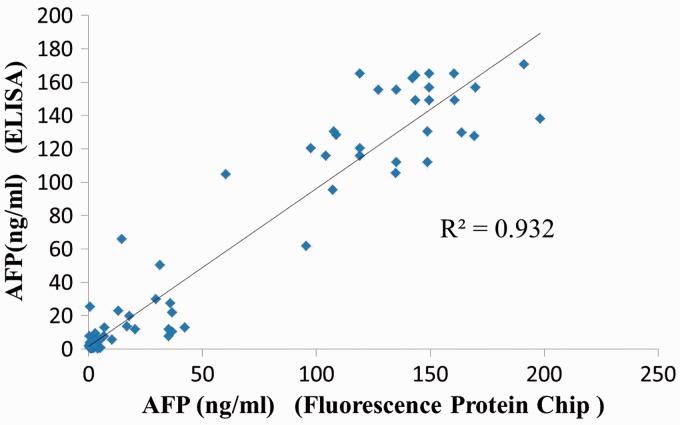
Serum AFP concentrations were significantly higher in patients than in control subjects when quantified via ELISA (mean ± SD: 66.82 ± 65.19 ng/ml versus 2.02 ± 1.43 ng/ml, respectively; P < 0.001) and fluorescence protein microarray (mean ± SD: 66.63 ± 66.51 ng/ml versus 1.68 ± 0.91 ng/ml, respectively; P < 0.001). The results of the fluorescence protein microarray showed that an AFP level ≥ 20 ng/ml was detected in 38 of 65 HCC samples (58.5%) (Table 2). By the ELISA method, an AFP level ≥ 20 ng/ml was detected in 36 of 65 HCC samples (55.4%). When the AFP cut-off value was set at 20 ng/ml, the fluorescence protein microarray had a sensitivity of 91.67% (33/36) and a specificity of 93.24% (69/74) for detecting serum AFP (Table 3). There was a statistically significant positive correlation between AFP concentrations quantified via ELISA and those quantified via fluorescence protein microarray (r = 0.932, P < 0.001; Figure 2).
Table 2.
Sensitivity and specificity of serum alpha-fetoprotein (AFP), quantified using fluorescence protein microarray or enzyme-linked immunosorbent assay (ELISA), for the diagnosis of hepatocellular carcinoma (HCC) using a cut-off value of 20 ng/ml AFP.
| Assay | HCC (n = 65) |
Control group (n = 45) |
||
|---|---|---|---|---|
| AFP ≥ 20 ng/ml | AFP<20 ng/ml | AFP ≥ 20 ng/ml | AFP<20 ng/ml | |
| Fluorescence protein microarray | 38 (58.5%) | 27 (41.5%) | 0 | 45 (100.0%) |
| ELISA | 36 (55.4%) | 29 (44.6%) | 0 | 45 (100.0%) |
Data presented as n of patients (%).
Table 3.
Protein microarray and enzyme-linked immunosorbent assay (ELISA) analyses of the alpha-fetoprotein in serum samples.
| ELISA |
Total | |||
|---|---|---|---|---|
| + | – | |||
| Fluorescence protein microarray | + | 33 | 5 | 38 |
| – | 3 | 69 | 72 | |
| Total | 36 | 74 | 110 | |
Data presented as the number of positive or negative AFP in 65 patients with hepatocellular carcinoma and 45 healthy control subjects.
Figure 2.
Scatter plot of serum alpha-fetoprotein (AFP) concentrations in patients with hepatocellular carcinoma and healthy control subjects (n = 110) quantified using fluorescence protein microarray or enzyme-linked immunosorbent assay (ELISA).
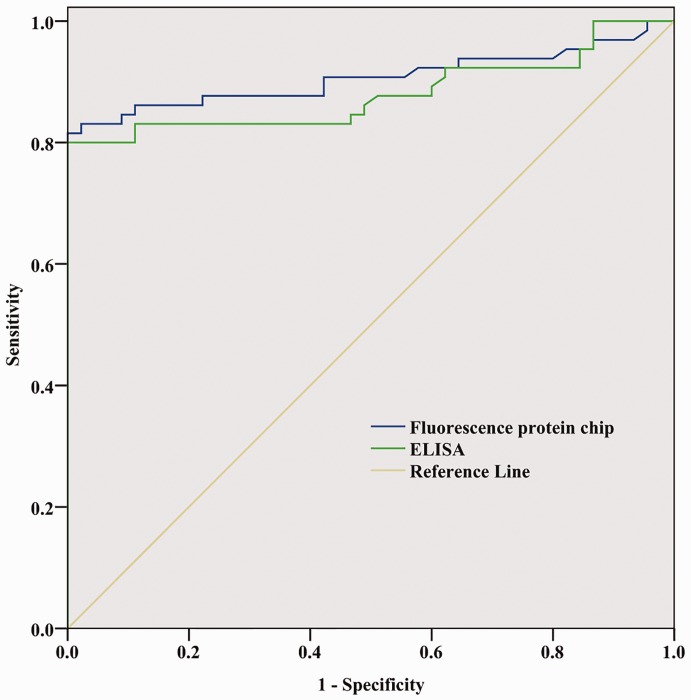
The ROC curve for diagnosis of HCC is shown in Figure 3. Serum AFP quantified via fluorescence protein microarray had a similar diagnostic performance to ELISA in distinguishing patients with HCC from healthy controls (area under ROC curve: 0.906 [95% CI 0.847, 0.966] for fluorescence protein microarray; and 0.880 [95% CI 0.814, 0.946] for ELISA). Fluorescence protein microarray can reliably be used in clinical applications. Meanwhile, compared with ELISA, the method has the advantages of saving time and lower costs (Table 4).
Figure 3.
Receiver operating characteristic curve of serum alpha-fetoprotein (AFP) quantification using fluorescence protein microarray or enzyme-linked immunosorbent assay (ELISA) for the diagnosis of hepatocellular carcinoma in patients with hepatocellular carcinoma and healthy control subjects (n = 110).
Table 4.
Comparison of the time involved and cost of running the protein microarray and enzyme-linked immunosorbent assay (ELISA) analyses of the alpha-fetoprotein in human serum samples.
| Assay | Assay duration, h | Volume of serum required, µl | Cost per patient, ¥ | Volume of prey antibody used, nl |
|---|---|---|---|---|
| Fluorescence protein microarray | 1.5–2.0 | 15 | 1.5 | 9–12 |
| ELISA | > 3.0 | 50 | 5–10 | 100 000 |
Discussion
Hepatocellular carcinoma is the second most frequent cause of cancer-related death.1 It usually develops in the setting of chronic liver disease, particularly in patients with chronic hepatitis B and C.15 Hepatocellular carcinoma is the leading cause of death among patients with cirrhosis.15 In our geographical area, hepatitis B virus is the causative agent for in 75–80% of patients.16
Alpha-fetoprotein is a glycoprotein consisting of 591 amino acids and a carbohydrate moiety.17 It is a major plasma protein produced by the yolk sac and the liver during fetal development.18 It is thought to be the fetal form of serum albumin, and it can also be used as a biomarker to detect a subset of tumours in non-pregnant women, men, and children.18 A level above 500 ng/ml of AFP in adults can be indicative of hepatocellular carcinoma, germ cell tumours, and metastatic cancers of the liver.19,20
Microarray technology is a powerful tool for high throughput assays of protein expression, protein–protein interactions and enzyme activity.21 It has become an effective diagnostic tool and antibody microarrays have extensive applications, including in the evaluation of tumour progression and the detection of toxins and bacteria.22
The cut-off value for optimal diagnostic performance for serum AFP quantification has been shown to be 20 ng/ml (sensitivity 41–65%, specificity 80–94%).23 Therefore, the present study used a cut-off value of 20 ng/ml. All serum samples, including those from subjects with HCC and healthy controls, were detected by the fluorescence protein microarray and ELISA. AFP levels greater than 20 ng/ml were not obtained from the healthy serum samples, which indicates that the false positive rate of the protein microarray presented was very low or even zero.
The fluorescence protein microarray developed in this present study has advantages compared with a chemiluminescence protein microarray, which we previously developed for the quantification of AFP in serum from patients with HCC.10 Our previous work applied HRP and chemiluminescence to protein microarray technology for the quantitative detection of AFP in human serum. The HRP chemiluminescent reaction is based on the catalysed oxidation of luminol by peroxide, producing light. In the presence of hydrogen peroxide, HRP converts luminol to an excited intermediate dianion. This dianion emits light on return to its ground state. Many factors affect enzyme activity such as substrate, inhibitors, pH and temperature.24 In this present study, Cy3 was used to label streptavidin and applied to the fluorescence detection techniques. Slides were scanned using a GenePix® 4000B Microarray Scanner without any substrate.
The present study used the following instruments and methods to enhance the stability and sensitivity of the measurement of serum AFP concentration. Protein microarray technology was combined with fluorescence analysis technology,25 in order to build a fluorescence protein microarray method for the quantitative detection of serum AFP. In this present study, the sensitivity of the diagnosis of HCC using the fluorescence protein microarray was similar to that of the ELISA method. In order to enhance the detection sensitivity, this present study used the biotin-avidin system, which is based on the high binding affinity between biotin and avidin. Most proteins, enzymes, and antibodies can be rapidly and stably linked to biotin.26 Avidin consists of four identical subunits, so an avidin molecule has four binding sites for biotin, which creates a multi-stage amplification effect.27 There is a strong affinity between biotin and avidin, and the reaction between them is free from outside interference, and they bind to each other with a high degree of specificity and stability. The biotin-avidin system is widely used in the fields of biology, molecular biology, and biochemistry.28
This present study used a GenePix® 4000B Microarray Scanner to detect the fluorescence intensity, the level of which on the spots was determined using the GenePix® software. Printing is one of the important technologies used to make protein microarrays. Previous protein microarrays have described the use of a contact microarray printer for microarray production. In this present study, a non-contact piezoelectric printer was used, which leads to increased uniformity on the array and it can significantly reduce the amount of antibodies used.14
The fluorescence protein microarray was performed in 1.5–2 h in the present study (excluding the time taken to manufacture the microarray), whereas the ELISA kit used in this experiment required more than 3 h. Fluorescence protein microarray assays are relatively simple to perform and do not require highly skilled technicians. Fluorescence protein microarrays have a number of advantages over ELISA. For example, the small size of the microarray reduces the quantity of prey antibodies, coated antibodies, antigens and other materials required compared with the ELISA. In the present study, the fluorescence protein microarray used 9–12 nl of prey antibody, 15 µl of diluted biotin-labelled rabbit antihuman polyclonal antibody and 15 µl of Cy3-labelled streptavidin for spotting on each well. In contrast, 50 µl of mouse antihuman AFP monoclonal antibody and 50 µl of diluted biotin-labelled rabbit antihuman polyclonal antibody were needed for the ELISA. The amount of serum required for the protein microarray was also greatly reduced; only 15 µl of serum was needed for each well on the protein microarray, whereas 50 µl of serum was needed for the ELISA. Consequently, the fluorescence protein microarray was more economical than the ELISA.
In conclusion, this present study successfully developed a fluorescence protein microarray for the detection of serum AFP in patients with HCC. The fluorescence protein microarray demonstrated a similar diagnostic performance compared with an ELISA in distinguishing patients with HCC from healthy control subjects. Meanwhile, it has several advantages over the ELISA, including specificity, rapidity, ease of use, time-saving and lower cost.
Declaration of conflicting interests
The authors declare that there are no conflicts of interest.
Funding
This work was supported by the Ministry of Science and Technology of the People’s Republic of China (grant no. 2012DFA30850). This work was also partially supported by the Beijing Municipal Science & Technology Commission (grant no. D131100005313004), Beijing You’an Hospital (grant no. BJYAH-2013-1-001), Key Laboratory Project of Capital Medical University Foundation (grant no. 2013GYGA03), Beijing Municipal health system high level personnel training programme (grant no. 2013-3-074), Beijing Municipal Education Commission Project (grant no. KM201510025020) and the Collaborative Innovation Centre of Infectious Diseases.
References
- 1.Park JW, Chen M, Colombo M, et al. Global patterns of hepatocellular carcinoma management from diagnosis to death: the BRIDGE Study. Liver Int 2015; 35: 2155–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson AB, Abrams TA, Ben-Josef E, et al. NCCN clinical practice guidelines in oncology: hepatobiliary cancers. J Natl Compr Canc Netw 2009; 7: 350–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poon RT, Fan ST, Ng IO, et al. Different risk factors and prognosis for early and late intrahepatic recurrence after resection of hepatocellular carcinoma. Cancer 2000; 89: 500–507. [PubMed] [Google Scholar]
- 4.Mazzaferro V, Regalia E, Doci R, et al. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. N Engl J Med 1996; 334: 693–699. [DOI] [PubMed] [Google Scholar]
- 5.Roayaie S, Jibara G, Tabrizian P, et al. The role of hepatic resection in the treatment of hepatocellular cancer. Hepatology 2015; 62: 440–451. [DOI] [PubMed] [Google Scholar]
- 6.Llovet JM, Brú C, Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis 1999; 19: 329–338. [DOI] [PubMed] [Google Scholar]
- 7.Stravitz RT, Heuman DM, Chand N, et al. Surveillance for hepatocellular carcinoma in patients with cirrhosis improves outcome. Am J Med 2008; 121: 119–126. [DOI] [PubMed] [Google Scholar]
- 8.Lok AS, Sterling RK, Everhart JE, et al. Des-gamma-carboxy prothrombin and alpha fetoprotein as biomarker for the early detection of hepatocellular carcinoma. Gastroenterology 2010; 138: 493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuen MF, Cheng CC, Lauder IJ, et al. Early detection of hepatocellular carcinoma increases the chance of treatment: Hong Kong experience. Hepatology 2000; 31: 330–335. [DOI] [PubMed] [Google Scholar]
- 10.Bruix J, Sherman M. Management of hepatocellular carcinoma. Hepatology 2005; 42: 1208–1236. [DOI] [PubMed] [Google Scholar]
- 11.Johnson PJ, Melia WM, Palmer MK, et al. Relationship between serum alpha-foetoprotein, cirrhosis, and survival in hepatocellular carcinoma. Br J Cancer 1981; 44: 502–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu QL, Yan XH, Yin XM, et al. Electrochemical enzyme-linked immunosorbent assay (ELISA) for α-fetoprotein based on glucose detection with multienzyme-nanoparticle amplification. Molecules 2013; 18: 12675–12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y, Zhang Y, Lin D, et al. Protein microarray with horseradish peroxidase chemiluminescence for quantification of serum α-fetoprotein. J Int Med Res 2015; 43: 639–647. [DOI] [PubMed] [Google Scholar]
- 14.Pilobello KT, Agrawal P, Rouse R, et al. Advances in lectin microarray technology: optimized protocols for piezoelectric print conditions. Curr Protoc Chem Biol 2013; 5: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet 2012; 379: 1245–1255. [DOI] [PubMed] [Google Scholar]
- 16.Yuen MF, Poon RT, Lai CL, et al. A randomized placebo-controlled study of long-acting octreotide for the treatment of advanced hepatocellular carcinoma. Hepatology 2002; 36: 687–691. [DOI] [PubMed] [Google Scholar]
- 17.Pucci P, Siciliano R, Malorni A, et al. Human alpha-fetoprotein primary structure: a mass spectrometric study. Biochemistry 1991; 30: 5061–5066. [DOI] [PubMed] [Google Scholar]
- 18.Abelev GI. Alpha-fetoprotein in ontogenesis and its association with malignant tumors. Adv Cancer Res 1971; 14: 295–358. [DOI] [PubMed] [Google Scholar]
- 19.Liao SH, Chen JH, Su YK, et al. Assaying biomarkers via real-time measurements of the effective relaxation time of biofunctionalized magnetic nanoparticles associated with biotargets. Nanomaterials 2015; 17: 1–7. [Google Scholar]
- 20.Ertle JM, Heider D, Wichert M, et al. A combination of α-fetoprotein and des-γ-carboxyprothrombin is superior in detection of hepatocellular carcinoma. Digestion 2013; 87: 121–131. [DOI] [PubMed] [Google Scholar]
- 21.Singh A, Sau AK. Tissue microarray: a powerful and rapidly evolving tool for high-throughput analysis of clinical specimens. Int J Case Rep Images 2010; 1: 1–6. [Google Scholar]
- 22.Kijanka G, IpCho S, Baars S, et al. Rapid characterization of binding specificity and cross-reactivity of antibodies using recombinant human protein arrays. J Immunol Methods 2009; 340: 132–137. [DOI] [PubMed] [Google Scholar]
- 23.Lian W, Wu D, Lim DV, et al. Sensitive detection of multiplex toxins using antibody microarray. Anal Biochem 2010; 401: 271–279. [DOI] [PubMed] [Google Scholar]
- 24.Patrovský V. Hydrogen peroxide determination with luminol and a new catalyst. Talanta 1976; 23: 553–554. [DOI] [PubMed] [Google Scholar]
- 25.Zhang ZY, Mei XG. The application and development of modern fluorescence immunoassay. Letters in Biotechnology 2006; 4: 677–680. [Google Scholar]
- 26.Wilchek M, Bayer EA. The avidin-biotin complex in immunology. Immunol Today 1984; 5: 39–43. [DOI] [PubMed] [Google Scholar]
- 27.Paganelli G, Magnani P, Zito F, et al. Three-step monoclonal antibody tumor targeting in carcinoembryonic antigen-positive patients. Cancer Res 1991; 51: 5960–5966. [PubMed] [Google Scholar]
- 28.Gan Z, Marquardt RR. Colorimetric competitive inhibition method for the quantitation of avidin, streptavidin and biotin. J Biochem Biophys Methods 1999; 39: 1–6. [DOI] [PubMed] [Google Scholar]