Abstract
Although phosphorylation directs serine-arginine (SR) proteins from nuclear storage speckles to the nucleoplasm for splicing function, dephosphorylation paradoxically induces similar movement raising the question of how such chemical modifications are balanced in these essential splicing factors. In this new study we investigated the interaction of protein phosphatase 1 (PP1) with the SR protein SRSF1 to understand the foundation of these opposing effects in the nucleus. We found that RNA recognition motif 1 (RRM1) in SRSF1 binds PP1 and represses its catalytic function through an allosteric mechanism. Disruption of RRM1-PP1 interactions reduces the phosphorylation status of the RS domain in vitro and in cells, re-directing SRSF1 in the nucleus. The data imply that an allosteric SR protein-phosphatase platform balances phosphorylation levels in a “goldilocks” region for the proper subnuclear storage of an SR protein splicing factor.
Keywords: kinase, kinetics, phosphorylation, phosphatase, SR Protein
Graphical abstract
Introduction
The splicing of mRNA transcripts is catalyzed by a nuclear complex known as the spliceosome, a dynamic, macromolecular machine composed of several small nuclear ribonucleoproteins (U1–6 snRNPs) and over 100 auxilliary protein factors 1,2. Spliceosome assembly and its associated chemical reactions that remove noncoding introns and stitch together exons are regulated by the serine-arginine (SR) ¶ proteins 3. SR proteins constitute an essential family of twelve splicing factors (SRSF1–12) that guide intermolecular contacts between precursor mRNA and several early spliceosomal components including the U1 and U2 snRNPs4, 5. Their expression levels influence splice-site identity and, in combination with competing splicing factors (eg., hnRNPs), control alternative exon incorporation into mature mRNA 6. In addition to their role in proteome expansion for normal cell function, SR proteins are often over-expressed in many cancer cells highlighting the relevance of these splicing factors in human disease 7,8.
All SR proteins possess one or two RNA recognition motifs (RRMs) and a signature C-terminal domain rich in arginine-serine repeats (RS domain). Whereas RRMs are globular, RS domains (50–200 residues) have low sequence diversity and are intrinsically disordered 9,10. Although RRMs bind pre-mRNA and are generally well-characterized at the structural level, RS domain structure and function is still not fully understood. RS domains are disordered but they become more rigid upon phosphorylation based on NMR studies9, 11. However, owing to their poor solubility and self-association, no high-resolution structure of a full-length SR protein has been reported. Despite this limitation, biochemical studies have highlighted important functions of the individual domains in SR proteins. For example, RS domains can interact with some RNA sequences, assist in RRM recognition of splice-site junctions and recruit several spliceosomal components through direct RS-RS interactions 12–14 Some studies suggest that, in addition to RS-RS and RRM-RNA interactions, RS domains regulate RRMs through intra- or intersteric contacts (RRM-RS) in the SR protein. For example, the RS domain in the prototype SRSF1 (aka ASF/SF2) has been shown to interact intrasterically with its RRMs, thereby regulating contacts with an RRM from the 70K subunit of U1 snRNP, an important initiating step for assembly of the pre-spliceosome 15.
RS domain phosphorylation is critical for general SR protein activities including splicing function and subcellular trafficking. SR protein kinase-1 (SRPK1), present in both the cytoplasm and nucleus, has been shown to polyphosphorylate the N-terminus (RS1) of the SRSF1 RS domain (Fig. 1A), promoting interactions with an SR-specific transportin (TRN-SR) that transfers it to the nucleus 16. SRSF1 is typically found concentrated in dynamic, membrane-free organelles known as speckles that act as storage areas for SR proteins and other splicing constituents 17. The cdc2-like kinases (CLK1–4), strictly localized to the nucleus, mobilize SRSF1 from speckles to the nucleoplasm by phosphorylation of the C-terminal half of the RS domain (RS2) (Fig. 1A) 18, 19 CLK activity is thought to be important for SRSF1 recruitment to sites of active gene splicing near the periphery of the speckle 20–22. However, either SRPK1 or CLK1 expression can diffuse speckles suggesting that hyper-phosphorylation may serve a general role in mobilizing SR proteins for splicing activities 19,20,23–25 . We showed recently that SRPK1 and CLK1 form a nuclear complex that activates splicing function and plays a synergistic role in controlling RS domain phosphorylation and SR protein release from CLK1 26. Both SRPKs and CLKs are often over-expressed in various cancers emphasizing the relationship between SR protein phosphorylation and splicing as well as the potential for therapeutic intervention through kinase inhibition strategies 27,28 . Indeed, inhibition of SRPKs using the small molecule SRPIN340 induces apoptosis in leukemia cell lines and reduces the splicing of a pro-angiogenic form of VEGF 29–31.
Figure 1. PP1 binding to RRM1 in SRSF1.
A) Domain organization of SRSF1. B-D,F) Interactions of PP1 with wild-type and mutant forms of SRSF1. Pull-down assays were performed using GST-PP1, immobilized on g-agarose resin, and His-tagged SRSF1 (B), SRSF1RATA (C), RRM1 (D), RRM1RATA (D), and SRSF1ΔRS2 (F). E) Fraction bound of PP1 compared to SRSF1 and variants. Integrations are performed using ImageJ. G) Far-UV CD spectra of RRM1 and RRM1RATA. H) RS2 binds specifically to RRM1. Pull-down assays were performed using GST-RS2, immobilized on g-agarose resin, with His-tagged RRM1 and RRM2.
The ability to diffuse nuclear speckles upon SRPK/CLK expression has led to the general principle that increasing RS domain phosphorylation liberates SR proteins from these storage compartments for splicing function. However, this simple model conflicts with other studies regarding the phosphorylation-dependent dynamics of nuclear SR proteins. For example, Mistelli and Spector showed that treatment of permeabilized cells with PP1 does not enhance speckle formation, as expected, but rather causes speckle diffusion 32. Also, the addition of a phosphatase inhibitor to these cells does not diffuse speckles but instead leads to enlarged speckles 32. Another study showed that hypophosphorylated SR proteins are initially localized near nucleoli prior to entry into speckles in daughter nuclei suggesting that lower RS domain phosphorylation levels may not restrict SR proteins to speckles 33. These combined studies suggest that the relationship between RS domain phosphoryl content and SR protein mobility is not strictly linear but instead may be more complex than originally conceived.
Much attention has been duly placed on the role of phosphorylation in regulating SR protein function and the mechanism of action of relevant kinases. We showed that the lengthy Arg-Ser repeat in RS1 of SRSF1 is phosphorylated using a directional (C-to-N-terminal) mechanism that involves dipeptide translocation from a docking groove to the active site 34, 35. CLK1 phosphorylates Arg-Ser dipeptides throughout the entire RS domain along with several critical Ser-Pro dipeptides in RS2 that play a role in SR protein mobilization for splicing function 19. CLK1 lacks a docking groove like SRPK1 but instead possesses a disordered N-terminus that recognizes RS domains 36. In comparison, less is known about the interaction of protein phosphatases with SR proteins and their mechanisms of action but some insights may be garnered from studies on the splicing factor Tra2β1. The Stamm laboratory proposed that Tra2β1 contains a short segment (RVDF) in its RNA binding domain that interacts with the protein phosphatase PP1 37. They noted the conservation of this motif in all known Tra2β1 proteins and its similarity to the classic PP1 binding motif RVxF. Mutation of these sequences reduced PP1 binding affecting the alternative splicing of several genes. These findings are intriguing since this putative docking region is also present in three of the twelve members of the canonical SR protein family. Stamm’s laboratory showed that mutations in RRM1 (RVEF→RATA) not only disrupt PP1 binding to SRSF1 but also affects splice variants of Tra2β1 and SMN2 suggesting that the SRSF1-PP1 complex is important for SR protein-dependent splicing function 37. However, it is still unclear whether SRSF1-PP1 interactions regulate phosphatase activity or SR protein mobilization from nuclear speckles, a key prerequisite for splicing activity.
In this new study, we wished to investigate how the interplay of kinases and phosphatases influences the phosphorylation state and mobility of SR proteins in cells. For these investigations we focused on the prototype SR protein SRSF1 which has been extensively studied with regard to its phosphorylation mechanism and role in mRNA splicing 38,39. We found that SRSF1 acts as a platform for both PP1 and the protein kinases SRPK1 and CLK1. Rather than activating the phosphatase, binding of PP1 to one of the RRMs (RRM1) represses activity in an allosteric manner, thereby enhancing the phosphoryl content of the RS domain in SRSF1. Disruption of a putative docking site increases PP1 activity leading to increases in hypo-phosphorylated forms of SRSF1 both in vitro and in cells. Decreased RS domain phosphorylation correlates with both decreased speckle population/size and increased nucleoplasmic levels of SRSF1. These new findings indicate that SRSF1 acts as an allosteric platform that binds and represses PP1 catalysis allowing appropriate, intermediate phosphorylation levels for the correct localization of an SR protein in nuclear speckles.
Results
A Short Sequence in RRM1 Promotes PP1 Binding to SRSF1
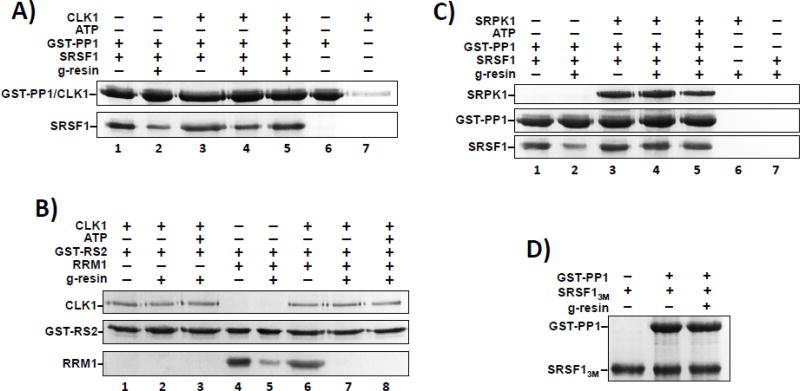
Previous studies identified a putative docking sequence in RRM1 (RVEF) that facilitates interaction of SRSF1 with PP1 (Fig. 1A) 37. We wished to replace this minimal sequence in order to test the role of PP1 binding on SRSF1 dephosphorylation. To characterize these residues in our recombinant protein, we showed that GST-PP1, immobilized on g-agarose resin, interacts directly with His-tagged SRSF1 in pull-down assays (Fig. 1B). In control experiments, we showed that pull down of SRSF1 is dependent on GST-PP1 and that SRSF1 does not interact nonspecifically with the g-agarose resin (Fig. 1B). Either removing RRM1 (SRSF1ΔRRM1) or mutating the putative docking sequences in RRM1 using mutations reported previously 37 (RVEF → RATA; SRSF1RATA) results in proteins that are incapable of binding PP1 (Fig. 1C & Suppl. Fig. S1A). To determine whether the binding epitope for PP1 is restricted to RRM1, we demonstrated that GST-PP1 interacts with the isolated RRM1 domain but not with a construct containing the RATA mutations (RRM1RATA) (Fig. 1D). By integrating the input and pull-down lanes, we noticed that GST-PP1 binds fully to RRM1 but binds less efficiently to the full-length SRSF1 under our assay conditions (Fig. 1E).
Since we showed previously that RS1 interacts specifically with RRM2 40, we speculated that RS2 could influence PP1 association either directly or indirectly. We deleted either the full RS domain or RS2 (residues 220–248) and found that these mutants (SRSF1ΔRS & SRSF1ΔRS2) interacted robustly with GST-PP1 similar to that for RRM1 (Fig. 1E,F & Suppl. Fig. S1B). Since the RVEF region is part of a terminal three-residue β strand, we tested whether mutations could unfold the protein damaging other potential binding sites in RRM1. We ruled out this possibility by showing that the circular dichroism spectra of RRM1 and RRM1RATA are similar (Fig. 1G). We next monitored the structure of the RRMs as a function of temperature and found that RRM1 and RRM1RATA display similar temperature-dependent denaturation curves indicating that the putative docking mutations do not alter overall domain stability (Suppl. Fig. S2). To probe for direct contacts between RS2 and RRM1, we generated a GST-tagged form of RS2 and found that it interacts directly with RRM1 but not with RRM2 (Fig. 1H). In control experiments we confirmed that GST-RS1 interacts robustly with RRM2 but not with RRM1 (Suppl. Fig. S1C). Overall, these studies indicate that RRM1 presents a docking surface or surfaces for PP1 that is subject to regulation by the C-terminal RS domain of SRSF1. The data imply that RS2 likely interacts directly with RRM1, down-regulating PP1 binding.
PP1 Interactions With SRSF1 Are Phosphorylation-Dependent
Since we demonstrated previously that SRPK1-dependent phosphorylation breaks RRM2-RS1 interactions 40, we speculated whether phosphorylation could have similar effects on RRM1-RS2 contacts, thereby enhancing PP1 binding to RRM1. To address this, we pre-treated SRSF1 with CLK1 in the absence and presence of ATP, inhibited the kinase with the CLK1-specific inhibitor TG003 and performed pull-down assays using GST-PP1. We showed that CLK1 binding and phosphorylation enhances PP1 binding to SRSF1 (compare lanes 2 & 5 in Fig. 2A & Suppl. Fig. S3A). Furthermore, CLK1 binding and phosphorylation of GST-RS2 breaks contacts with RRM1 (compare lanes 5, 7 & 8 in Fig. 2B). In control experiments we demonstrated that, unlike SRPK1, CLK1 stays attached to GST-RS2 after phosphorylation (compare lanes 1–3 in Fig. 2B) in keeping with prior reports showing that CLK1 binds with high affinity to both the phosphorylated and unphosphorylated RS domain of SRSF1 26, 41. Overall, these findings indicate that CLK1 phosphorylation of RS2 breaks contacts with RRM1, promoting PP1 binding to the SR protein.
Figure 2. PP1-SRSF1 interactions are phosphorylation dependent.
A) CLK1 enhances PP1 binding to SRSF1. His-tagged SRSF1 and CLK1 are incubated in the absence and presence of ATP and inhibited with TG003 before pull down using GST-PP1 and g-agarose resin. B) CLK1 disrupts RS2 binding to RRM1. His-tagged CLK1 and GST-RS2 are incubated in the absence and presence of ATP and inhibited with TG003 before pull down with and without His-tagged RRM1. C) SRPK1 enhances PP1 binding to SRSF1. His-tagged SRSF1 and SRPK1 are incubated in the absence and presence of ATP and inhibited with SRPIN340 before pull down using GST-PP1 and g-agarose resin. D) Mutations in RRM2 enhance PP1 binding to SRSF1. GST-PP1, immobilized on g-agarose resin, is incubated with His tagged SRSF13M.
Although SRPK1 does not efficiently phosphorylate RS2 42, we wondered whether the kinase could promote PP1 binding by phosphorylating RS1 and indirectly destabilizing RS2-RRM1 interactions. To address this, we pre-treated SRSF1 with SRPK1 in the absence and presence of ATP, inhibited the kinase with the SRPK-specific inhibitor SRPIN340, and performed pull-down assays using GST-PP1. We found that SRPK1 phosphorylation promotes PP1 binding to SRSF1 (compare lanes 2, 4, & 5 in Fig. 2C & Suppl. Fig. S3B). To provide further support that the phosphorylation-dependent disruption of RS1-RRM2 interactions promotes PP1 binding we investigated the interaction of PP1 to a mutant form of SRSF1 lacking three charged residues in RRM2 that bind RS1. We showed in previous studies that RS1 does not interact with RRM2 in this mutant (SRFS13M) and, thus, SRFS13M could mimic SRPK1-dependent phosphorylation and separation of RS1 and RRM2 40. In line with the phosphorylation experiments, we found that SRFS13M binds fully to PP1 suggesting that severing contacts between RS1 and RRM2 enhances PP1-RRM1 binding (Fig. 2D). Overall, these experiments reveal that while RS2 interacts with RRM1, RS domain phosphorylation by either SRPK1 or CLK1 breaks these contacts enhancing PP1 association with SRSF1. Interestingly, severing RS2-RRM1 contacts can occur either directly through CLK1 phosphorylation of RS2 or indirectly through phosphorylation of RS1 by SRPK1.
RRM1-PP1 Contacts Inhibit Phosphatase Activity
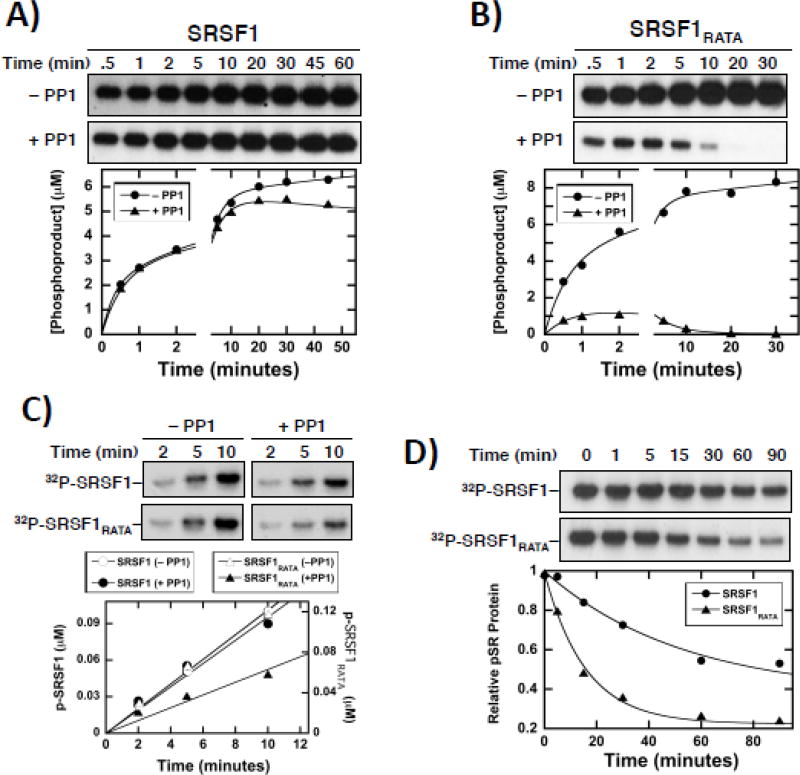
Having shown that PP1 interacts directly with RRM1 in SRSF1, we wished to address whether such docking interactions influence phosphatase activity. For these studies, we purified an active form of PP1 that displays monomeric characteristics based on size exclusion chromatography (Suppl. Fig. S4). We initially measured the phosphorylation of SRSF1 (1µM) under steady-state conditions using SRPK1 (75 nM) in the absence and presence of PP1 (300 nM). In the absence of PP1, SRSF1 is phosphorylated by SRPK1 in a multiphasic progress curve consistent with a multi-site phosphorylation reaction (Fig. 3A). PP1 addition had no effect in the first 10 minutes but elicited some dephosphorylation after 20 minutes (Fig. 3A). We repeated this experiment using SRSF1RATA (1µM) and amounts of SRPK1, PP1 and ATP identical to those in Fig. 3A and found that the phosphatase had significant effects on the progress curve (Fig. 3B). PP1 appeared to display much higher activity using SRSF1RATA as the substrate compared to SRSF1 at fixed SRPK1. This enhanced phosphatase activity is not due to decreases in SRPK1 activity toward SRSF1RATA compared to SRSF1 since the initial velocity for the mutant substrate is approximately 75 % higher than for the wild-type substrate in the absence of PP1 (Fig. 3A,B). For these experiments we used substoichiometric amounts of PP1 compared to SRSF1 (PP1<SRSF1) to minimize free PP1. Overall, these results illustrate that removing the putative docking sequence on RRM1 substantially increases the activity of PP1 for the RS domain of SRSF1.
Figure 3. PP1-SRSF1 interactions repress phosphatase activity.
A,B) Phosphorylation of SRSF1 (A) and SRSF1RATA (B) by SRPK1 (75 nM) in the absence and presence of PP1 (300 nM). In the absence of PP1, the initial velocities are 3.6 ± 0.77 µM/min for SRSF1 and 6.3 ± 1.7 µM/min for SRSF1RATA. In the presence of PP1, the initial velocities are 3.8 ± 0.42 µM/min for SRSF1 and 2.3 ± 0.16 µM/min for SRSF1RATA. C) Phosphorylation of SRSF1 and SRSF1RATA by CLK1 (150 nM) in the absence and presence of PP1 (100 nM). In the absence of PP1, the initial velocities are 10 ± 0.20 nM/min for SRSF1 and 12 ± 0.60 nM/min for SRSF1RATA. In the presence of PP1, the initial velocities are 9.5 ± 0.75 nM/min for SRSF1 and 6.0 ± 0.69 nM/min for SRSF1RATA. D) Dephosphorylation of SRPK1-phosphorylated SRSF1 and SRSF1RATA by PP1. Data were fit to single exponential curves with rate constants of 0.019 ± 0.00050 and 064 ± 0.0078 min−1 for SRSF1 and SRSF1RATA
To determine whether the effects of PP1 on SRSF1 versus SRSF1RATA are kinase-specific, we repeated the kinetic experiments using CLK1. Unlike SRPK1, CLK1 lacks a docking groove and instead relies on an RS-like N-terminus that interacts strongly with RS domains in SR proteins 24, 41. CLK1 is less efficient than SRPK1 so we used higher CLK1 (150 nM) and lower PP1 concentrations (100 nM) to monitor SR protein phosphorylation. Under these conditions we measured phosphorylation kinetics using fixed amounts of CLK1 and PP1 and found that while PP1 had no effect on the steady-state phosphorylation of SRSF1, it had higher activity against SRSF1RATA (Fig. 3C). These experiments along with those using SRPK1 were performed under conditions where the kinase was allowed to compete with PP1 during the phosphorylation reaction. We next wished to ask whether RRM1 binding affects PP1 activity after RS domain phosphorylation is complete. To address this, we pre-phosphorylated equal amounts of both SR proteins (80 nM) with SRPK1 (1 µM), inhibited the kinase using SRPIN340 and then dephosphorylated using PP1 (300 nM). We found that SRSF1RATA was dephosphorylated about 3-fold faster than SRSF1 (Fig. 3D). These combined experiments demonstrate that PP1 binding to RRM1 down-regulates phosphatase activity.
RRM1 Is An Allosteric Regulator of PP1
We next wished to explore the mechanism by where RRM1 interactions down-regulate PP1 function. We posited that if RRM1 contacts inhibit PP1 by unfavorably positioning it with respect to the RS domain then the addition of excess RRM1 should dissociate PP1 from SRSF1 enhancing dephosphorylation. We initially measured PP1-dependent dephosphorylation of CLK1-phosphorylated SRSF1 in the absence and presence of excess RRM1. We found that adding RRM1 (1 µM) to PP1 (0.1 µM) had no significant effect on the dephosphorylation rate of 32P-SRSF1 (0.2 µM) (Fig. 4A). Furthermore, we showed that neither RRM1RATA nor RRM2 significantly affects PP1 activity toward 32P-SRSF1 (Fig. 4A). These findings suggest that SRSF1 does not down-regulate PP1 catalysis through positioning of the RS domain in the SRSF1-PP1 complex. Further, it is likely that PP1 has equal access to the RS domain whether it is free in solution or bound to SRSF1. As a control, we showed that RRM1 (not RRM1RATA) dissociates 32P-SRSF1 from GST-PP1 in pull-down assays performed at conditions identical to those in the kinetic assays (Suppl. Fig. S5).
Figure 4. RRM1 allosterically represses PP1 Activity.
A) PP1 activity in the absence and presence of RRM1, RRM1RATA, and RRM2 using 32P-SRSF1 (CLK1 phosphorylated) and PNPP as substrates. Data were collected in triplicate with standard deviations displayed in the error bars. B) Double reciprocal plots of velocity versus PNPP as a function of fixed, varying amounts of RRM1. C) RRM1-dependent inhibition of PP1. Initial velocities are recorded at a function of RRM1 using two fixed concentrations of PNPP. The data were fit to apparent KI values of 410 ± 46 and 350 ± 22 nM at 5 (●) and 50 mM PNPP (▲). D) Effects of 30% sucrose on the initial velocities for p-SRSF1 and p-SRSF1RATA dephosphorylation by PP1.
To determine whether RRM1 could behave as an allosteric regulator (i.e.- bind outside the active site of PP1), we monitored the PP1-dependent dephosphorylation of PNPP in the absence and presence of RRM1 and RRM1RATA. PNPP was chosen as an alternative substrate since it is small and can bind only in the active site and not in the remote docking groove occupied by many physiological regulators of PP1 43. We found that RRM1 (1 µM) significantly inhibited PNPP dephosphorylation whereas RRM1RATA (1 µM) had a minor effect (Fig. 4A). We also showed that RRM2 had no significant influence on the PNPP reaction indicating that the inhibition phenomenon is largely RRM1-specific (Fig. 4A). To evaluate how RRM1 interacts with PP1, we monitored the initial velocity of PP1 as a function of varying PNPP concentrations at fixed amounts of RRM1. Double reciprocal plots show a common intersection point on the 1/PNPP axis, consistent with noncompetitive inhibition (Fig. 4B). These findings are consistent with other reports showing that traditional phosphatase-interacting proteins (e.g., I-1 & CPI-17) also inhibit PP1 noncompetitively 44–46. We next measured the velocity of the PP1 reaction as a function of varying RRM1 to obtain similar KI values of 400 and 350 nM at 5 and 50 mM PNPP, consistent with noncompeittive inhibition (Fig. 4C). This KI value is between 1 and 2 orders of magnitude higher than those for several classic phosphatase-interacting proteins (e.g., CPI-17, I-1, I-2, & Spinophilin) 47–50. Such findings suggest that the binding regions for PP1 on RRM1 may be fewer in number or accessibility compared to the traditional regulators. Taken together, these results indicate that RRM1 binds outside the PP1 active site with a high nanomolar affinity acting as an allosteric repressor of phosphatase catalysis.
Docking Interactions Alter the Dephosphorylation Mechanism
The data presented, thus far, suggest that PP1 binds to RRM1 and that mutation of the docking site releases the phosphatase from SRSF1. We wished to determine whether the dissociation of PP1 results in a fundamental change in the kinetic mechanism consistent with this model. We speculated that PP1 would be limited by dephosphorylation chemistry while bound to SRSF1 owing to its low, observed turnover rates (Fig. 3D) but instead would be limited by substrate diffusion upon mutation of the PP1 docking site. To address this possibility, we studied the dephosphorylation kinetics of 32P-SRSF1 and 32P-SRSF1RATA in the absence and presence of an added viscosogenic agent. We phosphorylated both SR proteins (0.4 µM) using SRPK1 (1 µM), inhibited the reaction with SRPIN340, and added PP1 (200 nM) in the absence and presence of 30 % sucrose. The relative amount of 32P-labeled SR protein was monitored as a function of time (Suppl. Fig. S6) and the initial velocity data were extracted from these plots (Fig. 4D). We found that the viscosogen did not significantly alter the rate for p-SRSF1 consistent with rate limitation by a unimolecular step, most likely the dephosphorylation step. In contrast, the rate for p-SRSF1RATA was considerably reduced (~4-fold) consistent with the idea that diffusion of PP1 limits dephosphorylation of this mutant substrate. The data suggest that PP1 forms a complex with p-SRSF1 dephosphorylating the RS domain in a series of unimolecular steps that are resistant to buffer viscosity. Although PP1 activity appears to extrapolate to zero at infinite RRM1 concentrations (Fig. 4C), it is possible that the phosphatase maintains some activity with RRM1 attached and, thus, acts in a unimolecular fashion in the PP1-SRSF1 complex with no solvent viscosity effect. On the other hand, PP1 may need to briefly dissociate from RRM1 to dephosphorylate the RS domain. Such transient dissociation may not require sufficient movement of PP1 through solvent and, accordingly, may lack a solvent viscosity effect. In contrast, PP1 that is not tethered by the RRM1 docking site owing to mutation must diffuse a longer distance through bulk solvent to bind and dephosphorylate the RS domain in SRFS1RATA, a clear bimolecular event that is sensitive to buffer viscosity. Overall, these findings support a docking model for PP1 on RRM1 that is disrupted through mutation.
PP1 Interactions Regulate SRSF1 Subnuclear Localization
Since RS domain phosphorylation status has been reported to affect SR protein subcellular localization 18,53 , we wished to determine whether disrupting the PP1 docking site in SRSF1 could impact both its phosphorylation state and biological function. To address this possibility we expressed GFP-tagged forms of SRSF1 and SRSF1RATA in HeLa cells and monitored their subcellular localization in fractionation experiments. We found that both GFP-SRSF1 and GFP-SRSF1RATA were largely observed in nuclear fractions of HeLa cell lysates suggesting that removal of the PP1 docking site did not impair nuclear entry of the SR protein (Fig. 5A). Integrity of the nuclear and cytoplasmic fractions were confirmed using specific antibodies for GAPDH and Histone-3. We then imaged HeLa cells that express these constructs using confocal microscopy and found that whereas GFP-SRSF1 mostly resides in large nuclear speckles, cells expressing GFP-SRSF1RATA show comparatively fewer and smaller speckles and greater nucleoplasmic distribution of the splicing factor (Fig. 5B). When considering a larger sample size, speckle areas are statistically smaller with cells expressing GFP-SRSF1RATA compared to those expressing GFP-SRSF1 (Suppl. Fig S7). To determine whether these changes in localization correlate with changes in PP1 interactions, we performed co-immunoprecipitation assays using specific monoclonal antibodies. We found that endogenous PP1 co-immunoprecipitates with GFP-SRSF1 but not with GFP-SRSF1RATA in nuclear lysates (Fig. 5C). In control experiments we showed that endogenous PP1 does not interact with protein A beads in the absence of anti-GFP antibody (Fig. 5C). These findings suggest that disruption of the PP1 binding to RRM1 affects the subnuclear localization of SRSF1 shifting greater amounts of the SR protein from speckles to the nucleoplasm.
Figure 5. Subcellular localization and phosphorylation state of SRSF1 and SRSF1RATA.
A) Cytoplasmic and nuclear fractions of HeLa cells expressing GFP-SRSF1 and GFP-SRSF1RATA. B) Confocal imaging of HeLa cells expressing GFP-SRSF1 and GFP-SRSF1RATA. C) Co-immunopreciptitation of GFP-SRSF1 and GFP-SRSF1RATA with endogenous PP1 in nuclear lysates. D) Treatment of nuclear fractions of HeLa cells expressing GFP-SRSF1 and GFP-SRSF1RATA with CIP. E) Treatment of nuclear fractions of HeLa cells expressing GFP-SRSF1 and GFP-SRSF1RATA with PP1 and run on a Phos-Tag/SDS-PAGE gel. Arrows designate relative differences in two phosphoforms. Corresponding A and B bands are integrated using ImageJ and ratios (A/B) shown at bottom of gel. F) Treatment of HeLa cells expressing GFP-SRSF1 and GFP-SRSF1RATA with 200 nM okadaic acid for 1 hr.
PP1 Contacts Regulate Cellular Phosphorylation of SRSF1
We noticed that both GFP-SRSF1 and GFP-SRSF1RATA appear as doublets in fractionation experiments (Fig. 5A) and wondered whether this could be due to different phosphorylation states that drive changes in speckles. SR proteins are often observed as multiple bands on SDS/PAGE with slower-migrating species reflecting higher phosphorylation states (hyper-phosphorylation) 18,52–54. Interestingly, the ratio of slow- vs. fast-migrating bands is smaller for GFP-SRSF1RATA than GFP-SRSF1 suggesting that the mutant SR protein may be less phosphorylated than the wild-type protein (Fig. 5A,C). To investigate this possibility, we treated nuclear lysates with the protein phosphatase CIP and observed an increase in the mobility of GFP-SRSF1 suggesting that the doublet likely results from variable phosphoforms (Fig. 5D). In comparison, GFP-SRSF1RATA displayed a smaller amount of the slow-migrating form compared to GFP-SRSF1 and CIP treatment removed this band leading to an increase in mobility similar to GFP-SRSF1 (Fig. 5D). We also showed in replicate studies of nuclear lysates that GFP-SRSF1 displays greater relative amounts of the slow- versus fast-migrating species compared to GFP-SRSF1RATA (Suppl. Fig. S8). These findings indicate that the mutant is generally less phosphorylated compared to the wild-type SR protein.
To attain additional information on this phosphorylation phenomenon, we initially ran nuclear lysates on an SDS/PAGE gel containing Phos-Tag. This chemical additive chelates phosphates inducing slower migration on SDS/PAGE as more phosphates are attached to the protein 55. We found that both SR proteins migrated as a distribution of species between 70 and 100 kDA, consistent with differing phosphorylation states in the cell (Fig. 5E). However, we observed that two bands near 70 kDa show distinct differences in relative population in the two phospho-proteins (Fig. 5E, A & B arrows). The ratio of the A and B bands is approximately 10-fold higher for GFP-SRSF1 than for GFP-SRSF1RATA (Fig. 5D, A/B). For both constructs, treatment of the nuclear lysates with PP1 removed the slow-migrating bands indicating that they are the result of phosphorylation. The dephosphorylated proteins ran near their expected molecular masses of 56 kDa (Fig. 5D). These findings suggest that reduced speckle occupancy of GFP-SRSF1RATA is linked to a lower, altered phosphorylation state. To verify this, we wished to determine whether GFP-SRSF1RATA could be directed back to speckles by inhibiting PP1. Indeed, we found that cells expressing GFP-SRSF1RATA display increases in speckle size and number upon treatment with the PP1 inhibitor okadaic acid (Fig. 5F). In control experiments, treatment of cells expressing wild-type GFP-SRSF1 with okadaic acid did not diffuse speckles but rather increased their size and number consistent with a prior report 32. Taken together, these findings indicate that the disruption of PP1 binding lowers the phosphorylation state of SRSF1, a phenomenon that is accompanied by a shift in speckle population and greater nucleoplasmic distribution.
Discussion
Extensive bio- and immunohistochemical studies suggest that RS domain phosphorylation drives SR proteins from nuclear storage speckles to the nucleoplasm 18, 19, 23, 24 . Such phosphorylation-dependent changes in subnuclear localization are important for the recruitment of SR proteins to the developing spliceosome and for their essential function in mRNA splicing catalysis. However, this simple linear phosphorylation model is challenged by other observations showing that increased phosphatase activity can also mobilize SR proteins in the same manner as kinase expression 32. Such findings raise the question of how a mix of nuclear kinases and phosphatases maintains phosphorylation levels at an ideal level for proper storage in speckles. In the present study we have uncovered a novel, synergistic interplay between PP1 and two splicing kinases (SRPK1 and CLK1) about an SR protein platform that offers a molecular explanation for these seemingly opposing activities.
The catalytic subunits of protein phosphatases are capable of dephosphorylating a wide array of phosphoproteins in vitro. In the cell, specificity is largely achieved through regulatory subunits that bind to the phosphatase, targeting it to its physiological substrate. PP1 appears to be highly versatile in this regard interacting with more than 200 proteins that can restrict the phosphatase to different cellular locations 56–59. In many cases, a channel in PP1 interacts with a conserved docking motif in the regulatory protein defined by the general sequence motif RVxF 60. We demonstrated that the phosphorylation of SRSF1 is regulated through such a motif since mutation in RRM1 is sufficient to break contacts between PP1 and SRSF1 both in vitro and in cells. Given that the RVxF motif lies partially in a 3-residue β strand in RRM1 with the terminal phenylalanine in a random coil followed by a proline, PP1 binding may occur initially though this hydrophobic residue followed by local unfolding of the strand. We showed previously that the final β strand in RRM2 unfolds and binds in the docking groove of SRPK1 suggesting that RRMs may possess flexibility in this region for protein-protein interactions 61. Nonetheless, the lower affinity of RRM1 for PP1 compared to traditional phosphatase regulators may reflect a non-ideal docking region owing to local secondary structure 47–50. Furthermore, although RVxF mutation breaks interactions between SRSF1 and PP1, it is very possible that other residues in RRM1 are involved in PP1 docking. Crystallographic studies revealed additional docking motifs in several regulatory proteins 62–64. Irrespective of the full docking surface, we show that disruption of the PP1-SRSF1 complex through mutation of the RVxF motif represses PP1 catalysis through an allosteric mechanism. Prior structural analyses of PP1 with a peptide containing the RVxF motif from the glycogen targeting subunit GM indicate that the docking groove is remote from the active site allowing for such a long-range mechanism 43. Interestingly, the effects of docking groove occupancy on PP1 activity is context specific since many regulatory proteins (e.g., DARP-32 and I-1) containing the RVxF motif inhibit activity 65 and a short peptide based on GM activates the phosphatase by 3-fold. 66 A full picture of how SRSF1 interacts with PP1 awaits high-resolution structural analyses of the RRM1-PP1 complex. Overall. our new findings illustrate that SRSF1 not only is a substrate for PP1 but also serves as a regulatory subunit for PP1.
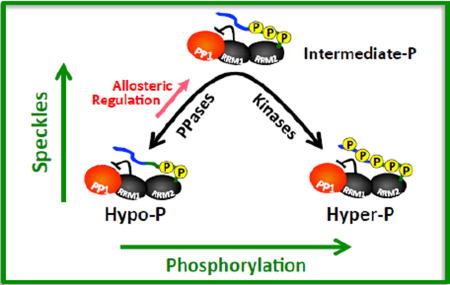
The results from experiments in this study allow us to now assemble a mechanism that describes how kinases and phosphatases regulate SR protein subcellular localization. In this model, RS1 phosphorylation by cytoplasmic SRPKs provides a target for transportin TRN-SR that shuttles SRSF1 into the nucleus (Fig. 6). This translocation process appears to be highly efficient or may require only minimal phosphorylation since we demonstrated that PP1 repression through RRM1 is not required for SRSF1 nuclear localization. In contrast, localization of this splicing factor in the nucleus relies on a delicate balance between kinase and phosphatase activities that maintain an ideal, “goldilocks” amount of RS domain phosphorylation. CLK or SRPK expression disrupts this balance and induces a hyper-phosphorylated SRSF1 that leaves speckles 19, 20, 23–25. While nuclear PP1 is capable of dephosphorylating SRSF1 and also dispersing speckles 32, we now show that this activity is largely repressed through an allosteric mechanism initiated by molecular contacts with RRM1 (Fig. 6). Severing these contacts by mutation disrupts the PP1-SRSF1 complex, increases phosphatase activity and reduces levels of RS domain phosphorylation (hypo-P) that drive SRSF1 into the nucleoplasm. Thus, cells appear to have evolved a mechanism to auto-regulate phosphatase activity so that SRSF1 can reside in storage speckles. Such a mechanism may provide a linchpin for mobilizing SRSF1 and possibly other SR proteins in response to biological signals. Although it is not clear what structural factors control this movement, it is likely that this intermediate phosphorylation state induces an SRSF1 conformation that is ideal for a condensed state in the speckle or facilitates contacts with other speckle components. Overall, this new model can now explain not only how SRPK/CLK expression leads to speckle diffusion but also how PP1 or its activation through RRM1 disconnection leads to the same subnuclear changes.
Figure 6. Regulatory model describing the nonlinear effects of phosphorylation on SRSF1 subnuclear localization.
SRPK1 phosphorylates RS1 allowing TRN-SR-dependent transport of SRSF1 to the nucleus. Speckle localization is regulated by the balanced actions of kinases (e.g., SRPK1 & CLK1) and phosphatases (e.g., PP1).
Materials & Methods
Materials
ATP, Mops, HEPES, Tris, MgCl2, MnCl2, NaCl, EDTA, NP40, Brij 35, glycerol, sucrose, acetic acid, lysozyme, DNAse, RNAse, Phenix imaging film, BSA, Protein G–agarose Ni-resin and liquid scintillant were obtained from Fisher Scientific. γ32P-ATP was obtained from NEN Products. p-Nitrophenyl Phosphate and 10xPMP buffer (500 mM HEPES, 1 M NaCl, 20 mM DTT, 0.1% Brij 35) were purchased from NEB. Protease inhibitor cocktail was obtained from Roche and anti-PP1γ monoclonal antibody was purchased from Thermo, and anti-GFP monoclonal antibody was purchased from Cell Signaling. InstantBlue was purchased from Expedeon, Hybond ECL nitrocellulose blotting membrane was purchased from Amersham.
Expression and purification of recombinant proteins
Human SRPK1, and all forms of His-tagged SRSF1 and PP1γ were expressed and purified from pET19b vectors containing an N-terminal His tag as previously described 41. All forms of GST-SRSF1 and GST-PP1γ were expressed and purified from a pGEX vector as previously described 67. GST-RS2 was further purified using an SP Sepharose column and a linear NaCl gradient (0–1 M) in 50 mM Tris (pH 8). CLK1 virus was transfected and expressed in Hi5 insect cells and CLK1 was purified with a nickel resin and a previously described procedure 36.
Pull-Down Assays
GST-tagged proteins (10 µM) were incubated with His-tagged proteins (10 µM) in binding buffer (0.1% NP40 (Nonidet P40), 20 mM Tris/HCl (pH 7.5) and 75 mM NaCl) in a total volume of 40 µL for 30 min before incubating with 25 µl of g–agarose for 30 min at room temperature. Where phosphorylation is performed, His-tagged SRSF1 protein was incubated with His-tagged kinase (4 µM) in the absence and presence of 100 µM ATP with 10 mM Mg2+ , mM Tris/HCl (pH 7.5), and 75 mM NaCl at 37°C for 30 minutes, followed by a 30 min incubation with GST-PP1γ (10 µM) in the absence of Mn2+ and in the presence of the CLK specific inhibitor TG003 (0.1 mM) or the SRPK1 specific inhibitor SRPIN340 (1 mM) where indicated, followed by a further incubation with 25 µl of g–agarose for 30 min at room temperature. In all cases, the resin was washed 4X with 200 µl of binding buffer, and the bound proteins were eluted with SDS quench buffer and boiled for 5 min. Retained protein was resolved by SDS-PAGE (12% or 18% gel) and visualized by Instant Blue Coomassie stain and integrations performed by ImageJ where measured.
Phosphorylation/Dephosphorylation Reactions
All forms of SRSF1 were phosphorylated by SRPK or CLK1 and dephosphorylated by PP1γ. All reactions were carried out in the presence of 100 mM Mops (pH 7.4), 1× PMP, 1 mM Mn2+, 10 mM Mg2+ and 5 mg/ml BSA at 37°C, using either 50 µM 32P-ATP for SRPK1 or 100 µM 32P-ATP for CLK1 both with a specific activity of 4000–8000 cpm/pmol. PP1γ was either incubated during or after the phosphorylation reactions as indicated. For viscosity experiments, reactions were carried out in the absence and presence of 30% Sucrose as described above. All reactions were carried out in a total volume of 10 µl and quenched with 10 µl of SDS/PAGE loading buffer at various time points. Phosphorylated SR proteins were separated from unreacted 32P-ATP by SDS-PAGE (12% gel), cut from the dried gel and quantified on the 32P channel in liquid scintillant.
PNPP Phosphatase Assay
The p-Nitrophenyl Phosphate (PNPP) phosphatase assay was carried out in a nanodrop spectrophotometer in a volume of 750 µL using a 1 mL plastic cuvette with the absorption monitored at 405 nm. PP1γ (20 nM) is incubated in the presence of 1× PMP buffer, 1 mM MnCl2, 100 mM Mops (pH 7.5), and initial velocities were monitored as a function of either varying PNPP concentration at fixed RRM, or varying RRM concentration at fixed PNPP.
Circular Dichroism Spectroscopy & Temperature Melting
Both RRM1 and RRM1RATA samples were diluted to a concentration of 113 µM in a buffer containing 100 mM Mops, 20 mM Tris and 300 mM NaCl at pH 6. Each sample was placed in a 0.1 cm quartz cuvette and the far-UV spectra was collected (at 25 °C) from 260 to 200 nm in 1 nm increments, with a 5 s averaging time at each wavelength on an Aviv 215 CD spectrometer. Spectra were smoothed using the same number of derivatives with the instrument software. The temperature melts were collected at 226 nm over the range of 25 – 95 °C at 1 degree increments, with an equilibration time of 30 seconds between measurements.
Cell Fractionation Studies
HeLa cells were harvested and lysed in 10 mM HEPES (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT, 0.05% Triton, and protease inhibitor cocktail. The lysates were centrifuged at 228 × g for 10 min to pellet nucleus, and the supernatant was retained as the cytoplasmic fraction. The pellet was washed with lysis buffer and re-suspended in 200 µl of 0.25 mM sucrose and 10 mM MgCl2. The nuclear suspension was layered on 0.88 mM sucrose and 0.5 mM MgCl2 and spun at 2,800 × g to obtain a nuclear pellet. The pellet was re-suspended in 1X RIPA buffer, spun at 2,800 × g, and the supernatant was retained as nuclear fraction.
Immunoprecipitation Experiments
HeLa cell nuclear lysates (200 µL) were pre-cleared with Protein A beads and incubated overnight in the cold room with 25 µL Protein A beads with 2 µg rabbit anti-GFP and the beads were spun at 2,052 × g and washed with 200 µL lysis buffer followed by the addition of 2× SDS loading buffer. The heated slurry was run on a 12% SDS-PAGE followed by immunoblot analysis. GFP-SRSF1 was immunoprecipitated from nuclear lysates (100 µL) using 2 µg GFP antibody and the co-immunoprecipitation of PP1γ was probed using anti-PP1γ antibody. Samples were visualized on either SDS-PAGE, or PhosTag-PAGE (16%) followed by western blot analysis.
Confocal Imaging Experiments
For live cell confocal imaging, HeLa cells were plated on 2.5-cm2 MatTek poly-D-lysine plates and transfected with GFP-SRSF1 constructs (2 µg) for 24 hr. The cells were washed with PBS and transfected HeLa cells were analyzed using an Olympus FV1000 as described previously 19 .Image analysis and quantitation of relative speckle areas were carried out using Image J software. At least 12 cells per condition were analyzed. Unpaired T-test with Welch’s correction was used to determine statistical significance.
Supplementary Material
Research Highlights.
Both phosphorylation and dephosphorylation release SR proteins from nuclear speckles.
A docking site on the SR protein SRSF1 binds PP1 in a phosphorylation-dependent manner.
PP1 binding to SRSF1 represses phosphatase activity through an allosteric mechanism.
Disruption of PP1 binding reduces phosphorylation and diffuses SRSF1 from nuclear speckles.
Subnuclear localization of SRSF1 in speckles is maintained by an SRSF1-PP1 platform.
Acknowledgments
This work was supported by NIH grants GM67969 and GM95828. We would like to thank Dr. Nicolas Villanueva for help with purifying GST-RS2.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: CLK1, cdc2-like kinase 1; PNPP, p-nitrophenyl phosphate; PP1, protein phosphatase 1; RRM, RNA recognition motif; RS domain, domain rich in arginine-serine dipeptide repeats; SR protein, splicing factor containing arginine-serine dipeptide repeats; SRPK1, serine-arginine-specific protein kinase 1; SRSF1, SR protein splicing factor 1 (aka ASF/SF2).
References
- 1.Muller S, Wolpensinger B, Angenitzki M, Engel A, Sperling J, Sperling R. A supraspliceosome model for large nuclear ribonucleoprotein particles based on mass determinations by scanning transmission electron microscopy. J. Mol. Biol. 1998;283:38394. doi: 10.1006/jmbi.1998.2078. [DOI] [PubMed] [Google Scholar]
- 2.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 3.Stojdl DF, Bell JC. SR protein kinases: the splice of life. Biochem. Cell Biol. 1999;77:293–8. [PubMed] [Google Scholar]
- 4.Mermoud JE, Cohen PT, Lamond AI. Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. EMBO J. 1994;13:5679–88. doi: 10.1002/j.1460-2075.1994.tb06906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao SH, Manley JL. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–44. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Z, Fu XD. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma. 2013;122:191–207. doi: 10.1007/s00412-013-0407-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gout S, Brambilla E, Boudria A, Drissi R, Lantuejoul S, Gazzeri S, Eymin B. Abnormal expression of the pre-mRNA splicing regulators SRSF1, SRSF2, SRPK1 and SRPK2 in non small cell lung carcinoma. PLoS One. 2012;7:e46539. doi: 10.1371/journal.pone.0046539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 2007;14:185–93. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiang S, Gapsys V, Kim HY, Bessonov S, Hsiao HH, Mohlmann S, Klaukien V, Ficner R, Becker S, Urlaub H, Luhrmann R, de Groot B, Zweckstetter M. Phosphorylation drives a dynamic switch in serine/arginine-rich proteins. Structure. 2013;21:2162–74. doi: 10.1016/j.str.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 10.Haynes C, Iakoucheva LM. Serine/arginine-rich splicing factors belong to a class of intrinsically disordered proteins. Nucleic Acids Res. 2006;34:305–12. doi: 10.1093/nar/gkj424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamelberg D, Shen T, McCammon JA. A proposed signaling motif for nuclear import in mRNA processing via the formation of arginine claw. Proc. Natl. Acad. Sci. U. S. A. 2007;104:14947–51. doi: 10.1073/pnas.0703151104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen H, Kan JL, Green MR. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol. Cell. 2004;13:367–76. doi: 10.1016/s1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 13.Staknis D, Reed R. SR proteins promote the first specific recognition of Pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol. Cell. Biol. 1994;14:7670–82. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuo P, Maniatis T. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 1996;10:135668. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]
- 15.Cho S, Hoang A, Sinha R, Zhong XY, Fu XD, Krainer AR, Ghosh G. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc. Natl. Acad. Sci. U. S. A. 2011;108:8233–8. doi: 10.1073/pnas.1017700108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai MC, Lin RI, Tarn WY. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10154–9. doi: 10.1073/pnas.181354098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 2011:3. doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ngo JC, Chakrabarti S, Ding JH, Velazquez-Dones A, Nolen B, Aubol BE, Adams JA, Fu XD, Ghosh G. Interplay between SRPK and Clk/Sty kinases in phosphorylation of the splicing factor ASF/SF2 is regulated by a docking motif in ASF/SF2. Mol. Cell. 2005;20:77–89. doi: 10.1016/j.molcel.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 19.Keshwani MM, Aubol BE, Fattet L, Ma CT, Qiu J, Jennings PA, Fu XD, Adams JA. Conserved proline-directed phosphorylation regulates SR protein conformation and splicing function. Biochem. J. 2015;466:311–22. doi: 10.1042/BJ20141373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sacco-Bubulya P, Spector DL. Disassembly of interchromatin granule clusters alters the coordination of transcription and pre-mRNA splicing. J. Cell Biol. 2002;156:425–36. doi: 10.1083/jcb.200107017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xing Y, Johnson CV, Moen PT, Jr, McNeil JA, Lawrence J. Nonrandom gene organization: structural arrangements of specific pre-mRNA transcription and splicing with SC-35 domains. J. Cell Biol. 1995;131:1635–47. doi: 10.1083/jcb.131.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J. Cell Biol. 1993;122:283–93. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gui JF, Lane WS, Fu XD. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature. 1994;369:678–82. doi: 10.1038/369678a0. [DOI] [PubMed] [Google Scholar]
- 24.Colwill K, Pawson T, Andrews B, Prasad J, Manley JL, Bell JC, Duncan PI. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 1996;15:265–75. [PMC free article] [PubMed] [Google Scholar]
- 25.Colwill K, Feng LL, Yeakley JM, Gish GD, Caceres JF, Pawson T, Fu XD. SRPK1 and Clk/Sty protein kinases show distinct substrate specificities for serine/arginine-rich splicing factors. J. Biol. Chem. 1996;271:24569–75. doi: 10.1074/jbc.271.40.24569. [DOI] [PubMed] [Google Scholar]
- 26.Aubol BE, Wu G, Keshwani MM, Movassat M, Fattet L, Hertel KJ, Fu XD, Adams JA. Release of SR Proteins from CLK1 by SRPK1: A Symbiotic Kinase System for Phosphorylation Control of Pre-mRNA Splicing. Mol. Cell. 2016;63:218–28. doi: 10.1016/j.molcel.2016.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayes GM, Carrigan PE, Miller LJ. Serine-arginine protein kinase 1 overexpression is associated with tumorigenic imbalance in mitogen-activated protein kinase pathways in breast, colonic, and pancreatic carcinomas. Cancer Res. 2007;67:2072–80. doi: 10.1158/0008-5472.CAN-06-2969. [DOI] [PubMed] [Google Scholar]
- 28.Yoshida T, Kim JH, Carver K, Su Y, Weremowicz S, Mulvey L, Yamamoto S, Brennan C, Mei S, Long H, Yao J, Polyak K. CLK2 Is an Oncogenic Kinase and Splicing Regulator in Breast Cancer. Cancer Res. 2015;75:1516–26. doi: 10.1158/0008-5472.CAN-14-2443. [DOI] [PubMed] [Google Scholar]
- 29.Siqueira RP, Barbosa Ede A, Poleto MD, Righetto GL, Seraphim TV, Salgado RL, Ferreira JG, Barros MV, de Oliveira LL, Laranjeira AB, Almeida MR, Junior AS, Fietto JL, Kobarg J, de Oliveira EB, Teixeira RR, Borges JC, Yunes JA, Bressan GC. Potential Antileukemia Effect and Structural Analyses of SRPK Inhibition by N-(2-(Piperidin-1-yl)-5-(Trifluoromethyl)Phenyl)Isonicotinamide (SRPIN340) PLoS One. 2015;10:e0134882. doi: 10.1371/journal.pone.0134882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gammons MV, Dick AD, Harper SJ, Bates DO. SRPK1 inhibition modulates VEGF splicing to reduce pathological neovascularization in a rat model of retinopathy of prematurity. Invest. Ophthalmol. Vis. Sci. 2013;54:5797–806. doi: 10.1167/iovs.13-11634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gammons MV, Fedorov O, Ivison D, Du C, Clark T, Hopkins C, Hagiwara M, Dick AD, Cox R, Harper SJ, Hancox JC, Knapp S, Bates DO. Topical antiangiogenic SRPK1 inhibitors reduce choroidal neovascularization in rodent models of exudative AMD. Invest. Ophthalmol. Vis. Sci. 2013;54:6052–62. doi: 10.1167/iovs.13-12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Misteli T, Spector DL. Serine/threonine phosphatase 1 modulates the subnuclear distribution of pre-mRNA splicing factors. Mol. Biol. Cell. 1996;7:1559–72. doi: 10.1091/mbc.7.10.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bubulya PA, Prasanth KV, Deerinck TJ, Gerlich D, Beaudouin J, Ellisman MH, Ellenberg J, Spector DL. Hypophosphorylated SR splicing factors transiently localize around active nucleolar organizing regions in telophase daughter nuclei. J. Cell Biol. 2004;167:51–63. doi: 10.1083/jcb.200404120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma CT, Velazquez-Dones A, Hagopian JC, Ghosh G, Fu XD, Adams JA. Ordered multi-site phosphorylation of the splicing factor ASF/SF2 by SRPK1. J. Mol. Biol. 2008;376:55–68. doi: 10.1016/j.jmb.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 35.Aubol BE, Chakrabarti S, Ngo J, Shaffer J, Nolen B, Fu XD, Ghosh G, Adams JA. Processive phosphorylation of alternative splicing factor/splicing factor 2. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12601–6. doi: 10.1073/pnas.1635129100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keshwani MM, Hailey KL, Aubol BE, Fattet L, McGlone ML, Jennings PA, Adams JA. Nuclear protein kinase CLK1 uses a non-traditional docking mechanism to select physiological substrates. Biochem. J. 2015;472:329–38. doi: 10.1042/BJ20150903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novoyatleva T, Heinrich B, Tang Y, Benderska N, Butchbach ME, Lorson CL, Lorson MA, Ben-Dov C, Fehlbaum P, Bracco L, Burghes AH, Bollen M, Stamm S. Protein phosphatase 1 binds to the RNA recognition motif of several splicing factors and regulates alternative pre-mRNA processing. Hum. Mol. Genet. 2008;17:52–70. doi: 10.1093/hmg/ddm284. [DOI] [PubMed] [Google Scholar]
- 38.Ghosh G, Adams JA. Phosphorylation mechanism and structure of serine-arginine protein kinases. FEBS J. 2011;278:587–97. doi: 10.1111/j.1742-4658.2010.07992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Das S, Krainer AR. Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol. Cancer Res. 2014;12:1195–204. doi: 10.1158/1541-7786.MCR-14-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serrano P, Aubol BE, Keshwani MM, Forli S, Ma CT, Dutta SK, Geralt M, Wuthrich K, Adams JA. Directional Phosphorylation and Nuclear Transport of the Splicing Factor SRSF1 Is Regulated by an RNA Recognition Motif. J. Mol. Biol. 2016;428:243045. doi: 10.1016/j.jmb.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aubol BE, Plocinik RM, Keshwani MM, McGlone ML, Hagopian JC, Ghosh G, Fu XD, Adams JA. N-terminus of the protein kinase CLK1 induces SR protein hyperphosphorylation. Biochem. J. 2014;462:143–52. doi: 10.1042/BJ20140494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma CT, Hagopian JC, Ghosh G, Fu XD, Adams JA. Regiospecific phosphorylation control of the SR protein ASF/SF2 by SRPK1. J. Mol. Biol. 2009;390:618–34. doi: 10.1016/j.jmb.2009.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Egloff MP, Johnson DF, Moorhead G, Cohen PT, Cohen P, Barford D. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 1997;16:1876–87. doi: 10.1093/emboj/16.8.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Senba S, Eto M, Yazawa M. Identification of trimeric myosin phosphatase (PP1M) as a target for a novel PKC-potentiated protein phosphatase-1 inhibitory protein (CPI17) in porcine aorta smooth muscle. J. Biochem. 1999;125:354–62. doi: 10.1093/oxfordjournals.jbchem.a022294. [DOI] [PubMed] [Google Scholar]
- 45.Foulkes JG, Strada SJ, Henderson PJ, Cohen P. A kinetic analysis of the effects of inhibitor-1 and inhibitor-2 on the activity of protein phosphatase-1. Eur. J. Biochem. 1983;132:309–13. doi: 10.1111/j.1432-1033.1983.tb07363.x. [DOI] [PubMed] [Google Scholar]
- 46.Nimmo GA, Cohen P. The regulation of glycogen metabolism. Phosphorylation of inhibitor-1 from rabbit skeletal muscle, and its interaction with protein phosphatases-III and -II. Eur. J. Biochem. 1978;87:353–65. doi: 10.1111/j.1432-1033.1978.tb12384.x. [DOI] [PubMed] [Google Scholar]
- 47.Eto M, Ohmori T, Suzuki M, Furuya K, Morita F. A novel protein phosphatase-1 inhibitory protein potentiated by protein kinase C. Isolation from porcine aorta media and characterization. J. Biochem. 1995;118:1104–7. doi: 10.1093/oxfordjournals.jbchem.a124993. [DOI] [PubMed] [Google Scholar]
- 48.Gibbons JA, Weiser DC, Shenolikar S. Importance of a surface hydrophobic pocket on protein phosphatase-1 catalytic subunit in recognizing cellular regulators. J. Biol. Chem. 2005;280:15903–11. doi: 10.1074/jbc.M500871200. [DOI] [PubMed] [Google Scholar]
- 49.Ragusa MJ, Dancheck B, Critton DA, Nairn AC, Page R, Peti W. Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites. Nat. Struct. Mol. Biol. 2010;17:459–64. doi: 10.1038/nsmb.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Picking WD, Kudlicki W, Kramer G, Hardesty B, Vandenheede JR, Merlevede W, Park IK, DePaoli-Roach A. Fluorescence studies on the interaction of inhibitor 2 and okadaic acid with the catalytic subunit of type 1 phosphoprotein phosphatases. Biochemistry. 1991;30:10280–7. doi: 10.1021/bi00106a028. [DOI] [PubMed] [Google Scholar]
- 51.Koizumi J, Okamoto Y, Onogi H, Mayeda A, Krainer AR, Hagiwara M. The subcellular localization of SF2/ASF is regulated by direct interaction with SR protein kinases (SRPKs) J. Biol. Chem. 1999;274:11125–31. doi: 10.1074/jbc.274.16.11125. [DOI] [PubMed] [Google Scholar]
- 52.Gui JF, Tronchere H, Chandler SD, Fu XD. Purification and characterization of a kinase specific for the serine- and arginine-rich pre-mRNA splicing factors. Proc. Natl. Acad. Sci. U. S. A. 1994;91:10824–8. doi: 10.1073/pnas.91.23.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prasad J, Colwill K, Pawson T, Manley JL. The protein kinase Clk/Sty directly modulates SR protein activity: both hyper- and hypophosphorylation inhibit splicing. Mol. Cell. Biol. 1999;19:6991–7000. doi: 10.1128/mcb.19.10.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Velazquez-Dones A, Hagopian JC, Ma CT, Zhong XY, Zhou H, Ghosh G, Fu XD, Adams JA. Mass spectrometric and kinetic analysis of ASF/SF2 phosphorylation by SRPK1 and Clk/Sty. J. Biol. Chem. 2005;280:41761–8. doi: 10.1074/jbc.M504156200. [DOI] [PubMed] [Google Scholar]
- 55.Yu FX, Zhang Y, Park HW, Jewell JL, Chen Q, Deng Y, Pan D, Taylor SS, Lai ZC, Guan KL. Protein kinase A activates the Hippo pathway to modulate cell proliferation and differentiation. Genes Dev. 2013;27:1223–32. doi: 10.1101/gad.219402.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heroes E, Lesage B, Gornemann J, Beullens M, Van Meervelt L, Bollen M. The PP1 binding code: a molecular-lego strategy that governs specificity. FEBS J. 2013;280:584–95. doi: 10.1111/j.1742-4658.2012.08547.x. [DOI] [PubMed] [Google Scholar]
- 57.Hendrickx A, Beullens M, Ceulemans H, Den Abt T, Van Eynde A, Nicolaescu E, Lesage B, Bollen M. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem. Biol. 2009;16:365–71. doi: 10.1016/j.chembiol.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 58.Esteves SL, Korrodi-Gregorio L, Cotrim CZ, van Kleeff PJ, Domingues SC, da Cruz e Silva OA, Fardilha M, da Cruz e Silva EF. Protein phosphatase 1gamma isoforms linked interactions in the brain. J. Mol. Neurosci. 2013;50:179–97. doi: 10.1007/s12031-012-9902-6. [DOI] [PubMed] [Google Scholar]
- 59.Bollen M, Peti W, Ragusa MJ, Beullens M. The extended PP1 toolkit: designed to create specificity. Trends Biochem. Sci. 2010;35:450–8. doi: 10.1016/j.tibs.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barford D, Das AK, Egloff MP. The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu. Rev. Biophys. Biomol. Struct. 1998;27:133–64. doi: 10.1146/annurev.biophys.27.1.133. [DOI] [PubMed] [Google Scholar]
- 61.Ngo JC, Giang K, Chakrabarti S, Ma CT, Huynh N, Hagopian JC, Dorrestein PC, Fu XD, Adams JA, Ghosh G. A sliding docking interaction is essential for sequential and processive phosphorylation of an SR protein by SRPK1. Mol. Cell. 2008;29:56376. doi: 10.1016/j.molcel.2007.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hurley TD, Yang J, Zhang L, Goodwin KD, Zou Q, Cortese M, Dunker AK, DePaoli-Roach AA. Structural basis for regulation of protein phosphatase 1 by inhibitor-2. J. Biol. Chem. 2007;282:28874–83. doi: 10.1074/jbc.M703472200. [DOI] [PubMed] [Google Scholar]
- 63.Terrak M, Kerff F, Langsetmo K, Tao T, Dominguez R. Structural basis of protein phosphatase 1 regulation. Nature. 2004;429:780–4. doi: 10.1038/nature02582. [DOI] [PubMed] [Google Scholar]
- 64.O’Connell N, Nichols SR, Heroes E, Beullens M, Bollen M, Peti W, Page R. The molecular basis for substrate specificity of the nuclear NIPP1:PP1 holoenzyme. Structure. 2012;20:1746–56. doi: 10.1016/j.str.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cohen P. The role of protein phosphorylation in human health and disease. The Sir Hans Krebs Medal Lecture. Eur. J. Biochem. 2001;268:5001–10. doi: 10.1046/j.0014-2956.2001.02473.x. [DOI] [PubMed] [Google Scholar]
- 66.Tappan E, Chamberlin AR. Activation of protein phosphatase 1 by a small molecule designed to bind to the enzyme’s regulatory site. Chem. Biol. 2008;15:167–74. doi: 10.1016/j.chembiol.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 67.Ma CT, Ghosh G, Fu XD, Adams JA. Mechanism of dephosphorylation of the SR protein ASF/SF2 by protein phosphatase 1. J. Mol. Biol. 2010;403:386–404. doi: 10.1016/j.jmb.2010.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.