Abstract
The transcriptional pathways activated downstream of vascular endothelial growth factor (VEGF) signaling during angiogenesis remain incompletely characterized. By assessing the signals responsible for induction of the Notch ligand delta-like 4 (DLL4) in endothelial cells, we find that activation of the MAPK/ERK pathway mirrors the rapid and dynamic induction of DLL4 transcription and that this pathway is required for DLL4 expression. Furthermore, VEGF/ERK signaling induces phosphorylation and activation of the ETS transcription factor ERG, a prerequisite for DLL4 induction. Transcription of DLL4 coincides with dynamic ERG-dependent recruitment of the transcriptional co-activator p300. Genome-wide gene expression profiling identified a network of VEGF-responsive and ERG-dependent genes, and ERG chromatin immunoprecipitation (ChIP)-seq revealed the presence of conserved ERG-bound putative enhancer elements near these target genes. Functional experiments performed in vitro and in vivo confirm that this network of genes requires ERK, ERG and p300 activity. Finally, genome-editing and transgenic approaches demonstrate that a highly conserved ERG-bound enhancer located upstream of HLX (which encodes a transcription factor implicated in sprouting angiogenesis) is required for its VEGF-mediated induction. Collectively, these findings elucidate a novel transcriptional pathway contributing to VEGF-dependent angiogenesis.
KEY WORDS: Endothelial cell, Transcription, Enhancer, Angiogenesis, Genome editing, ETS factor, Zebrafish, Mouse, Human
Summary: A network of VEGF-dependent angiogenic genes is regulated by the transcription factor ERG, which dynamically recruits the co-activator p300 to VEGF-dependent enhancers in endothelial cells in vitro and in vivo.
INTRODUCTION
The growth of new blood vessels is requisite for tissue repair and homeostasis and contributes to the pathogenesis of several diseases, including cancer and diabetic retinopathy. A comprehensive understanding of the signaling pathways and downstream transcriptional networks that control angiogenesis could be leveraged to identify novel therapeutic targets to either promote or inhibit vascular growth. The central mechanism responsible for the majority of vascular growth is angiogenesis. Angiogenesis is a highly coordinated process that requires the interaction of several intracellular and intercellular signaling pathways that ultimately converge on a network of transcriptional pathways to elicit cellular behaviors (Herbert and Stainier, 2011). Vascular endothelial growth factor (VEGF), one of the central drivers of angiogenesis, is required for blood vessel development during embryogenesis (Carmeliet et al., 1996; Ferrara et al., 1996) and contributes to vascular homeostasis, as well as physiological and pathological postnatal vascular growth (Kim et al., 1993; Lee et al., 2007). VEGF activates a number of signal transduction pathways in endothelial cells (ECs) that modulate cytoskeletal dynamics and gene expression (Olsson et al., 2006), resulting in a suite of angiogenic cell behaviors, including directed, polarized cell migration. Although some of the transcriptional networks involved in VEGF signaling have been identified (Herbert and Stainier, 2011), much remains to be discovered regarding the mechanisms by which VEGF coordinates new vessel growth.
ECs receiving a threshold of VEGF stimulation initiate a signal transduction pathway that culminates in the transcription of the Notch ligand delta-like 4 (DLL4) (Lobov et al., 2007), as well as a network of other angiogenic genes (Liu et al., 2008). Phenotypic changes occur in the VEGF receiving cell, endowing it with ‘tip’ cell characteristics, including acquisition of numerous filopodial projections, increased migratory behavior, and elevated VEGF receptor 2 (VEGFR2; also known as KDR) expression (Blanco and Gerhardt, 2013). DLL4 on the surface of a tip cell binds to, and activates, Notch receptors on adjacent stalk cells. Notch activity in stalk cells induces the transcription of Notch-dependent genes, such as those encoding members of the basic helix-loop-helix transcription factor families HEY and HES, and suppresses filopodia formation and cell migration, while also dampening VEGFR2 expression. Importantly, tip and stalk cell phenotypes are dynamic, and in time a stalk cell can become a tip cell, and vice versa (Jakobsson et al., 2010). Coordinating these dynamic cellular behaviors is essential for an effective angiogenic response. The molecular mechanisms responsible for the maintenance and conversion between these phenotypes are only partially understood, and include oscillations in DLL4 induction in the tip cell (Lobov et al., 2007; Ubezio et al., 2016), as well as tight control of Notch signal duration in neighboring stalk cells (Guarani et al., 2011). How VEGF-regulated transcriptional programs control the dynamic expression of DLL4 and other angiogenic genes during sprouting angiogenesis remains poorly understood.
We previously identified a highly conserved enhancer element located within intron 3 of murine Dll4 that directs expression in arteries and angiogenic vessels (Wythe et al., 2013). Activity of this enhancer in arteries is VEGF responsive, and this is at least in part dependent on ETS transcription factors, including ETS-related gene (ERG) (Wythe et al., 2013). The ETS family of transcription factors play crucial roles in multiple stages of vascular development, including angiogenesis (Randi et al., 2009). More than a dozen ETS factors are expressed in ECs, and several of these [e.g. ETV2, TEL (ETV6), ETS1, ETS2, FLI1, ERG] have been implicated in vascular growth (Liu and Patient, 2008; Pham et al., 2007). ETS factors bind to a consensus 5′-GGA(A/T)-3′ sequence in the genome through a highly conserved ∼85 amino acid ETS domain (Sharrocks, 2001). Several of the family members also contain additional functional domains, such as the pointed (PNT) domain, a docking site for the serine/threonine kinase extracellular regulated kinase-2 (ERK2; MAPK1), which phosphorylates ETS1 and ETS2 in response to mitogen-activated protein kinase (MAPK) activation (Seidel and Graves, 2002). Phosphorylation of ETS1 and ETS2 enhances their activity through the recruitment of the transcriptional co-activator proteins p300 (EP300) and Creb-binding protein (CBP; CREBBP) (Foulds et al., 2004). Modulation of ETS factor activity by signal transduction pathways is not unique to ERK2, as other MAPK signaling pathways, such as p38 (MAPK14) and JNK (MAPK8) have been documented (Wasylyk et al., 1998; Yordy and Muise-Helmericks, 2000).
The specificity of MAPK pathways for particular ETS family members has recently been explored in prostate cancer cells in vitro. Interestingly, of the three MAPKs analyzed (p38, JNK and ERK2), only ERK2 phosphorylates ERG (yet multiple MAPK members act on ETS1/2) (Selvaraj et al., 2015). ERK2 primarily phosphorylates three residues on ERG: S96 (amino terminal to the PNT domain), S215 and S276 (both carboxy terminal to the PNT domain). Crucially, mutation of S215 to alanine, an amino acid refractory to phosphorylation (a so-called ‘phospho mutant’), abolishes ERG function in prostate cancer cells (Selvaraj et al., 2015). Although several studies have implicated ERG as a mediator of EC survival, proliferation, motility and vascular integrity (Birdsey et al., 2008, 2012, 2015; Liu and Patient, 2008; Yuan et al., 2011), whether ERG acts as a hub, integrating signals downstream of VEGF to control these diverse EC behaviors is not known.
Here, we explore the signaling and transcriptional pathways activated downstream of VEGF signaling in ECs. We find that the dynamic induction of MAPK/ERK activity controls DLL4 transcription in human ECs and that MAPK/ERK is required for angiogenesis in zebrafish in vivo. Furthermore, we demonstrate that MAPK/ERK activity leads to phosphorylation of ERG, and that ERG is required for the induction of DLL4 and a network of other angiogenic genes in human, mouse and zebrafish ECs. Mechanistically, we show that ERG recruits p300 to enhancer elements to coordinate angiogenic gene expression. These findings provide new insight into the molecular mechanisms of VEGF-mediated angiogenesis, and suggest that MAPK/ERK activation of ERG/p300 might represent a novel therapeutic target for modulating vascular growth.
RESULTS
Dynamic MAPK/ERK signaling regulates gene induction in response to VEGF stimulation
Dll4 is dynamically expressed in tip cells during sprouting angiogenesis (Hellström et al., 2007; Jakobsson et al., 2010; Suchting et al., 2007; Ubezio et al., 2016). We first delineated the kinetics of VEGF-dependent DLL4 transcription in vitro. We assayed DLL4 unspliced pre-mRNA (as a surrogate of transcription) and mature mRNA transcript levels in VEGF-stimulated serum- and growth factor-starved human microvascular ECs (MVECs) or human umbilical vein ECs (HUVECs). In both cell types, DLL4 transcription responded dynamically to VEGF stimulation, peaking at 15-30 min (15-30′) post addition of VEGF, and returning to baseline levels by 2 h (Fig. S1A; Fig. 1A). The expression of spliced DLL4 mRNA was also transient and dynamic, with expression peaking at 1 h and returning to near baseline levels by 2 h (Fig. S1A; Fig. 1A).
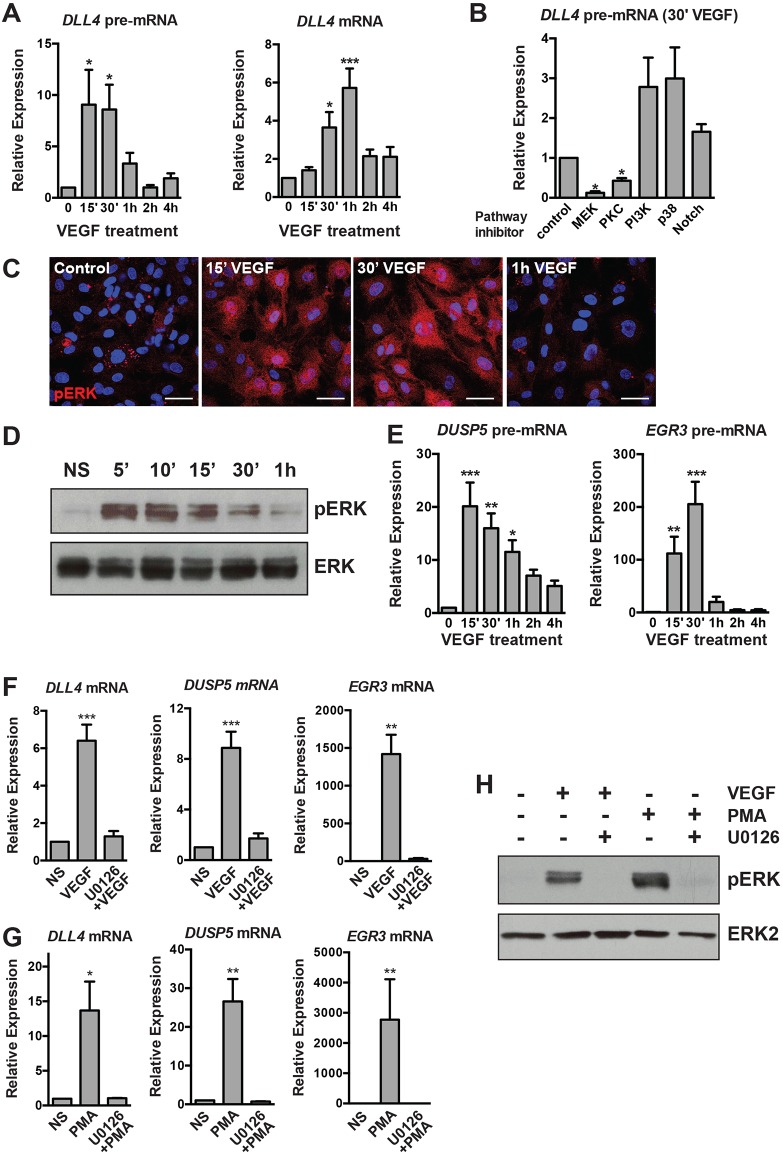
Fig. 1.
Transcriptional activation of DLL4 and other angiogenic genes in response to VEGF stimulation requires active MAPK/ERK signaling. (A) Kinetics of DLL4 transcriptional activation (as measured by qRT-PCR of unspliced DLL4 pre-mRNA) and expression of mature DLL4 mRNA in HUVECs treated with VEGF (n=7). (B) Inhibition of the MAPK pathway (by PKC and MEK inhibitors) ablates induction of DLL4 pre-mRNA (measured 30′ after VEGF treatment by qRT-PCR), whereas Notch inhibition has no effect (n=3). Expression is relative to vehicle-treated, VEGF-stimulated cells. (C) Kinetics of MAPK/ERK activation as detected by pERK (red) immunofluorescence in HUVECs. Blue, DAPI staining. Scale bars: 40 μm. Representative experiment of three. (D) Kinetics of pERK in VEGF-stimulated cells assessed by western blot. Total ERK is included as a loading control. Representative experiment of two. (E) Kinetics of DLL4 pre-mRNA induction are similar to known MAPK/ERK-dependent genes (EGR3, DUSP5) (n=3). (F) Induction of DLL4, DUSP5 and EGR3 by VEGF is abrogated in cells pre-treated with the MEK inhibitor U0126 (n=5). (G) Induction of DLL4, DUSP5 and EGR3 by PMA is abrogated in cells pre-treated with the MEK inhibitor U0126 (n=4). (H) Western blot demonstrating the efficacy of U0126 pre-treatment of VEGF- or PMA-treated cells. pERK is not induced in U0126 pre-treated cells (n=1). NS, non-stimulated.
As VEGF engagement of its principal angiogenic receptor, VEGFR2, can activate multiple downstream signaling pathways, we employed a panel of pharmacological cell signaling inhibitors to define the pathway(s) responsible for the rapid and transient induction of DLL4 transcription. Inhibition of the MAPK/ERK signaling pathway [using inhibitors of either MEK or protein kinase C (PKC)] abrogated DLL4 induction in response to VEGF (Fig. S1B; Fig. 1B). Immunofluorescent staining of VEGF-stimulated HUVECs revealed the presence of phosphorylated ERK (pERK) in both the nucleus and cytoplasm 15-30′ after treatment, with levels returning to baseline after 1 h (Fig. 1C). Measurement of pERK by western blotting mirrored the rapid and dynamic MAPK/ERK activation observed in immunofluorescence experiments, as pERK levels returned to near baseline levels by 1 h after stimulation (Fig. 1D). The kinetics of MAPK/ERK activation therefore parallels that of DLL4 transcription in response to VEGF treatment.
Several VEGF/MAPK/ERK-responsive genes have been characterized, including the immediate early gene early growth response 3 (EGR3) (Liu et al., 2008) and the ERK phosphatase dual specificity phosphatase 5 (DUSP5) (Bellou et al., 2009; Kucharska et al., 2009). The transcriptional induction of EGR3 and DUSP5 (as measured by qRT-PCR analysis of unspliced pre-mRNA) largely mirrored that of MAPK/ERK activation and DLL4 transcription (Fig. 1E). As expected, the induction of DLL4, EGR3 and DUSP5 mRNA was completely inhibited in the presence of the highly selective small molecule MEK inhibitor U0126 (Fig. 1F). In addition, the induction of these genes by VEGF stimulation was attenuated in HUVECs in which ERK1 (MAPK3) and ERK2 (MAPK1) were knocked down by siRNA (Fig. S1C). To determine whether MAPK/ERK activity in the absence of VEGF signaling was sufficient to induce expression of these genes, we treated serum-starved HUVECs with a PKC activator (and therefore an activator of MEK/ERK signaling), phorbol-ester myristate acetate (PMA) (Franklin et al., 1994; Schultz et al., 1997). PMA treatment markedly elevated DLL4, EGR3 and DUSP5 mRNA levels, and this response was blocked by pre-treatment with U0126, demonstrating that the MEK/ERK pathway is necessary and sufficient to activate transcription of a subset of angiogenic genes (Fig. 1G,H).
We further assessed the physiological relevance of MAPK/ERK signaling during sprouting angiogenesis in vivo. In agreement with recent reports (Costa et al., 2016; Shin et al., 2016), pERK was enriched in angiogenic sprouts (i.e. intersomitic vessels) in developing zebrafish embryos, indicative of active MAPK/ERK signaling (Fig. 2A; Fig. S2A). Importantly, inhibition of MAPK/ERK signaling using the MEK inhibitor SL327 completely abrogated the pERK signal throughout the embryo, including in the sprouting vessels (Fig. 2A; Fig. S2A). Inhibition of MEK signaling had a functional effect on angiogenesis, as sprout length (Fig. 2B) and the number of ECs per sprout (Fig. S2B) were decreased, as demonstrated previously (Shin et al., 2016). At this dose, SL327 did not cause developmental delay or necrosis (Fig. S2C). Inhibition of MAPK/ERK signaling diminished the expression of dll4 mRNA, as determined by qRT-PCR (Fig. 2C). Furthermore, time-lapse microscopy using a Notch biosensor revealed that attenuation of MEK activity reduced Notch signaling within the developing vasculature in vivo, and these results were confirmed by static confocal microscopy of a conventional Notch reporter (Fig. 2D,E; Fig. S3; Movies 1 and 2).
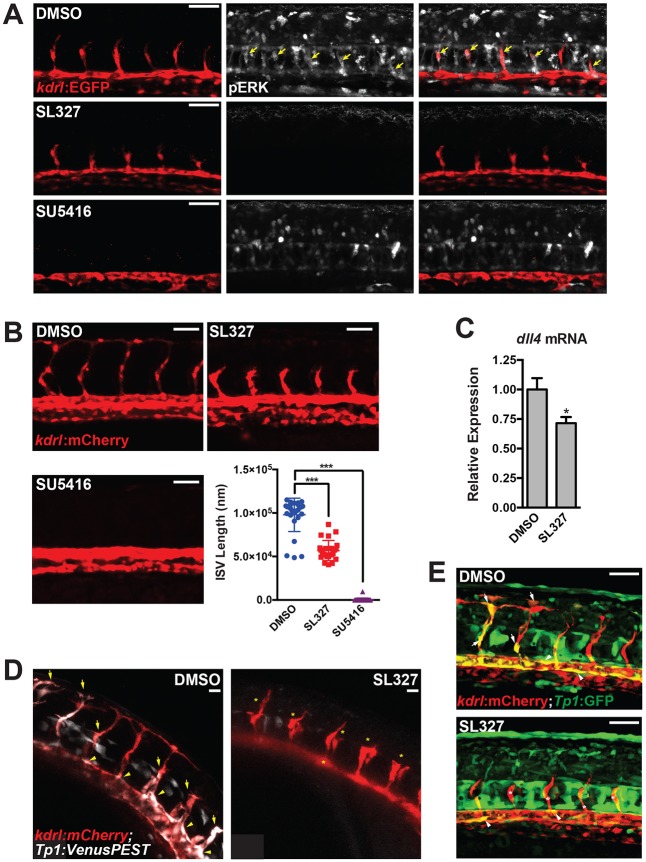
Fig. 2.
Active MAPK/ERK signaling regulates sprouting angiogenesis in zebrafish. (A) pERK staining (white) in embryos treated with vehicle (i.e. DMSO), MEK (i.e. SL327) or VEGFR2 (i.e. SU5416) inhibitors [treated from 18-20 hours post-fertilization (hpf) to ∼24 hpf]. See Fig. S2A for additional images and quantification. Yellow arrows indicate pERK-positive sprouting endothelial cells. (B) Inhibition of MEK activity by SL327 inhibits ISV sprout length. Inhibition of VEGFR2 signaling with SU5416 is included as a positive control. Quantification of ISV length at 28 hpf is shown. (C) dll4 expression in SL327-treated embryos at 28 hpf (treatment initiated at 18-20 hpf) as assessed by qRT-PCR (n=6 individual embryos). (D) Notch activity is reduced in the vasculature of SL327-treated Tg(kdrl:mCherry); Tg(Tp1bglob:Venus-PEST) embryos. Still images from time-lapse microscopy of a representative experiment are shown. Arrows indicate Notch signaling-positive ISVs, arrowheads indicate Notch signaling-positive endothelial cells in the dorsal aorta, asterisks indicate Notch-negative ISVs. See Fig. S3 (for additional still images) and Movies 1 and 2. (E) Similar experiment to that shown in D, but with Tg(kdrl:mCherry); Tg(Tp1bglob:EGFP) embryos. Scale bars: 50 μm (A,B,E); 20 μm (D).
ERG activity is controlled by VEGF/MAPK/ERK signaling
To determine whether ERG is required for the dynamic induction of DLL4 downstream of VEGF, we knocked down ERG using siRNA in HUVECs. ERG knockdown reduced the basal levels of DLL4 and completely abrogated the induction of DLL4 in response to VEGF stimulation (Fig. 3A,B). Furthermore, activation of MAPK/ERK with PMA stimulation failed to elevate DLL4 transcription in ERG knockdown cells, confirming that ERG functions downstream of VEGF and MAPK/ERK (Fig. 3C).
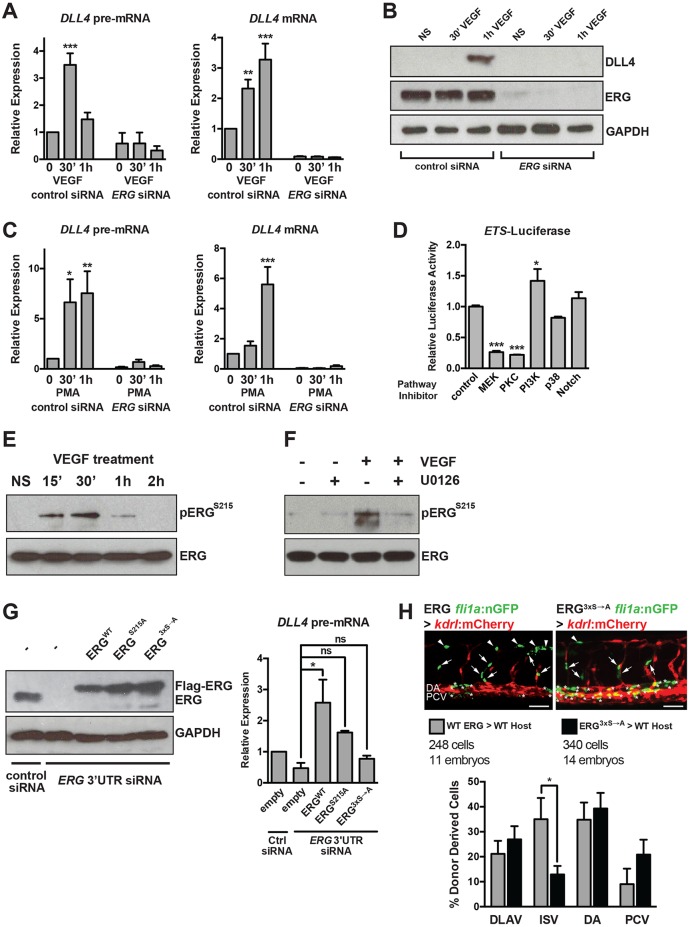
Fig. 3.
VEGF/MAPK signaling stimulates ERG transcriptional activity to induce DLL4 expression. (A) VEGF induction of DLL4 transcription (as assessed by qRT-PCR measurement of DLL4 pre-mRNA) and mature DLL4 mRNA expression in HUVECs requires ERG (n=4). (B) VEGF induction of DLL4 protein expression requires ERG. Representative experiment of three. (C) Induction of DLL4 transcription by PMA, an activator of MAPK/ERK signaling, requires ERG (n=4). (D) ETS activity (as assessed by activity of an 8× concatamer of an ETS element driving luciferase expression) in BAECs is suppressed by MEK or PKC inhibition. Triplicate determinations from a representative experiment of three. (E) ERG is phosphorylated at S215 in response to VEGF stimulation (15-30′) in HUVECs. Representative experiment of two. (F) VEGF-induced phosphorylation of ERG requires MEK activity. Representative experiment of two. (G) ERG was knocked down using siRNAs directed to the 3′ UTR, followed by overexpression of Flag-tagged wild-type (WT) or mutant (S215A or S96A;S215A;S276A, indicated as 3xS→A) ERG. ERG western blot indicates restoration of expression using electroporated constructs (a representative experiment of three is shown). DLL4 expression as assessed by qRT-PCR of pre-mRNA after 1 h of VEGF treatment (n=3). (H) Representative images of transplanted cells from Tg(fli1a:nls-GFP) embryos injected with wild-type or mutant ERG mRNA into Tg(kdrl:mCherry) embryos. Arrows and asterisks indicate endothelial cells that are donor derived. Quantification of cellular position is shown below (n=248 cells from 11 embryos for wild type and n=340 cells from 14 embryos for mutant). Scale bar: 50 µm. DA, dorsal aorta (asterisks); DLAV, dorsal longitudinal anastomotic vessel (arrowheads); ISV, intersomitic vessel (arrows); NS, non-stimulated; PCV, posterior cardinal vein.
To explore further the relationship between ETS factors and MAPK activity, we tested whether MAPK/ERK signaling modulates ETS factor activity by creating a luciferase reporter construct under the control of a concatemer (eight tandem copies) of the ETS-DNA binding site within the intron 3 enhancer of murine Dll4 [identified by Wythe et al. (2013)]. ETS reporter activity in bovine aortic ECs (BAECs) was attenuated by both MEK and PKC inhibition (Fig. 3D). This suggests that ETS factor transactivation is controlled by MAPK/ERK signaling.
Selvaraj et al. recently demonstrated that ERK2 preferentially bound and phosphorylated ERG at serines 96 (S96), 215 (S215) and 276 (S276), and that S215 phosphorylation was required for ERG activity in prostate cancer cells (Selvaraj et al., 2015). We found that S215 was dynamically phosphorylated in ECs in response to VEGF stimulation, with peak phosphorylation occurring at 15-30′, which coincides with increased MAPK/ERK activity following VEGF treatment (Fig. 3E). Pretreatment with a MEK inhibitor abolished S215 phosphorylation (Fig. 3F). To determine the functional importance of ERK-mediated phosphorylation of ERG, we eliminated endogenous ERG using an siRNA directed to the 3′ UTR of ERG and then reintroduced wild-type or phospho-mutant ERG. Expression of wild-type ERG restored DLL4 transcription, whereas expression of ERG containing a mutation of one phosphorylation site (S215A) had less activity, and ERG containing mutations in all three ERK-phosphorylated residues (S96A, S215A, S276A) failed to rescue DLL4 transcription (Fig. 3G). This suggests that ERK phosphorylation is functionally important in dictating ERG activity.
To test further the functional importance of ERG phosphorylation, transplantation experiments were performed in zebrafish. Wild-type or mutant (S96A, S215A, S276A) ERG mRNA was injected into kdrl:nls-EGFP donor embryos, followed by transplantation of these cells into kdrl:mCherry recipient hosts at sphere stage. The location of the donor cells within the trunk vasculature was scored at 28-30 hours post-fertilization (hpf) to determine whether expression of mutant ERG affects the ability of these cells to contribute to angiogenesis. There appeared to be no overt phenotypic consequence following mosaic overexpression of wild-type or mutant ERG. However, the percentage of ERG mutant-expressing cells contributing to intersegmental vessels (ISVs) (but not other vascular structures) was significantly reduced compared with wild-type ERG-expressing cells (Fig. 3H).
ERG coordinates dynamic co-activator recruitment to the DLL4 intronic enhancer
p300 is recruited to VEGF-dependent enhancers and is required for regulating the expression of many angiogenic genes (Zhang et al., 2013). As the earliest time-point previously examined was 1 h post-VEGF treatment, we sought to define the dynamic nature of p300 recruitment to the DLL4 enhancer by ChIP assays in VEGF-stimulated HUVECs. ERG recruitment was modestly enhanced by VEGF treatment (Fig. 4A), but there is high basal ERG occupancy at this enhancer (Wythe et al., 2013). Strikingly, p300 was transiently recruited to the intronic DLL4 enhancer 15-30′ after VEGF stimulation, mirroring the robust and transient increase in MEK/ERK activity and ERG phosphorylation (Fig. 4A). Of note, p300 recruitment did not coincide with increased acetylation of K27 of histone H3 (H3K27ac), although acetylation is already high at this region in ECs (Wythe et al., 2013). Importantly, we found that p300 recruitment to the intronic DLL4 enhancer required ERG (Fig. 4B). In addition, co-immunoprecipitation in ECs demonstrated that ERG and p300 physically interacted following VEGF stimulation (Fig. 4C), and that this interaction was lost in cells expressing a phospho-mutant ERG (S96A, S215A, S276A) protein (Fig. 4D). To determine the functional importance of p300 in DLL4 induction, we utilized a small molecule inhibitor of p300 and CBP histone acetyltransferase activity, c646 (Bowers et al., 2010). Inhibition of p300/CBP activity in HUVECs in vitro did not affect basal levels of DLL4, but completely blocked VEGF induction of DLL4 mRNA (Fig. 4E). Furthermore, inhibition of p300/CBP in zebrafish suppressed elongation of intersomitic vessels (Fig. 4F), but did not result in other gross developmental defects (Fig. S2C).
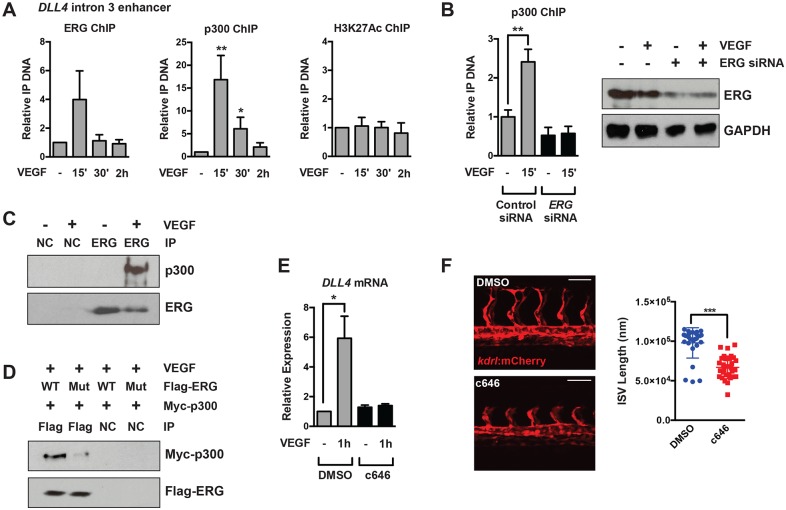
Fig. 4.
p300 is dynamically recruited to the DLL4 enhancer and regulates sprouting angiogenesis. (A) Recruitment of p300 to the DLL4 enhancer located in intron 3 15-30′ after VEGF stimulation, as assessed by ChIP (n=5 for ERG and p300 ChIP, n=3 for H3K27ac ChIP). (B) p300 recruitment in response to VEGF stimulation requires ERG. Shown is a representative experiment of two with triplicate determinations. The extent of ERG knockdown as assessed by western blot is shown to the right. (C) Endogenous p300 and ERG physically interact by co-immunoprecipitation in HUVECs stimulated with VEGF. Shown is a representative experiment of three. (D) Exogenous Myc-p300 interacts with wild-type Flag-ERG in BAECs, but does not interact with phospho-mutant (S96A;S215A;S276A) Flag-ERG by co-immunoprecipitation. Shown is a representative experiment of three. (E) p300 activity is required for DLL4 mRNA induction in HUVECs in response to VEGF stimulation (n=5). c646 is a potent inhibitor of p300/CBP activity. (F) Inhibition of p300 activity suppresses ISV elongation in zebrafish. Quantification is shown to the right. Note: Quantification of the DMSO control is the same as that shown in Fig. 2B, as both inhibitors were used in the same experiment. Scale bars: 50 μm. NC, negative control (V5 antibody).
ERG regulates a network of constitutive and VEGF-inducible genes
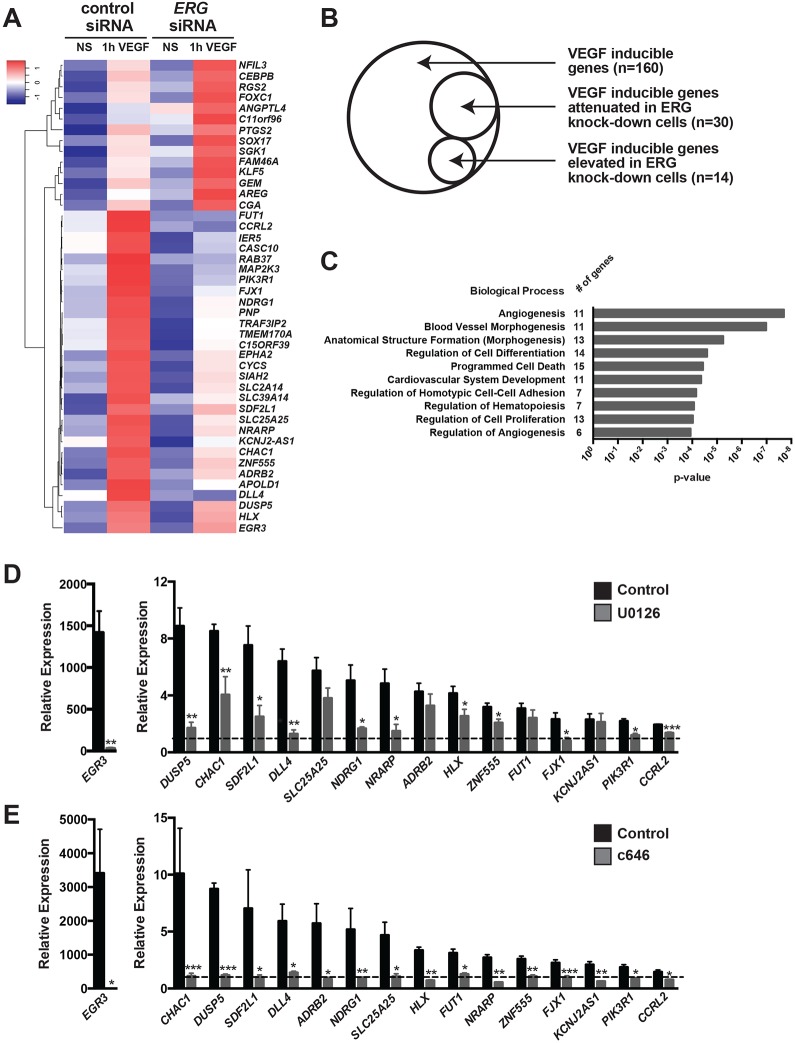
To determine the extent of the genetic network regulated by ERG, we transfected HUVECs with control or ERG siRNAs and performed microarray analysis of gene expression in serum-starved and VEGF-stimulated cells. We focused on transcripts induced at early stages of VEGF stimulation (i.e. within 1 h) to identify genes directly regulated by VEGF/ERK/ERG. Knockdown of ERG resulted in the downregulation of 202 genes, including CLDN5, RASIP1 and ARHGAP28 in serum-starved cells (Fig. S4A), consistent with previous studies (Birdsey et al., 2012; Yuan et al., 2012), and the upregulation of 68 genes. Gene ontology (GO) analysis revealed that the most frequent functional categories altered following loss of ERG were: response to wounding, inflammation, cell migration, cell motility and cell adhesion (Fig. S4B). VEGF treatment increased the expression of 160 genes and downregulated only four genes (Fig. S5A). Analysis of the genes modulated by VEGF revealed GO terms associated with transcriptional regulation and gene expression, cell proliferation and vascular development (Fig. S5B). Of the VEGF-induced genes, 30 (representing ∼19% of all VEGF-inducible genes) were attenuated in ERG knockdown cells, and 14 genes (∼9% of all VEGF-inducible genes) were further elevated in ERG knockdown cells (Fig. 5A,B). GO analysis revealed that these genes (e.g. NRARP, HLX, DUSP5, EGR3 and PIK3R1; Bellou et al., 2009; Herbert et al., 2012; Liu et al., 2008; Nicoli et al., 2012; Phng et al., 2009) are implicated in angiogenesis, blood vessel morphogenesis and development, homotypic cell-cell adhesion, cell proliferation and differentiation (Fig. 5C).
Fig. 5.
A network of VEGF-inducible genes are ERK/ERG/p300 dependent. (A) Hierarchical clustering of microarray analysis identified a group of 30 VEGF-inducible genes that are suppressed in ERG knockdown cells, and a group of 14 VEGF-inducible genes that were further induced in ERG knockdown cells (HUVECs). NS, non-stimulated. (B) Venn diagram depicting the number of VEGF-inducible genes and the subset that are ERG dependent. See Fig. S5 for further details on VEGF-regulated transcripts. (C) Gene Ontology (GO) analysis of ERG/VEGF-dependent genes. Selected representative GO terms are displayed with their associated P-value. The number of genes in each GO category is indicated. The GO terms depicted are: Angiogenesis (GO:0001525), Blood vessel morphogenesis (GO:0048514), Anatomical structure formation (morphogenesis) (GO:0048646), Regulation of cell differentiation (GO:0045595), Programmed cell death (GO:0012501), Cardiovascular system development (GO:0072358), Regulation of homotypic cell-cell adhesion (GO:0034110), Regulation of hematopoiesis (GO:1903706), Regulation of cell proliferation (GO:0042127), Regulation of angiogenesis (GO:0045765). (D) qRT-PCR analysis of the MEK dependency of the VEGF induction of a subset of the genes identified by microarray. HUVECs were pre-treated with DMSO or U0126 (MEK inhibitor) prior to VEGF stimulation. Expression is relative to non-VEGF-stimulated cells (dashed line). Genes are arranged in decreasing order of VEGF induction. The induction of 12 out of 16 genes was found to be MEK dependent (n=4-5). (E) qRT-PCR analysis of the p300 dependency of the VEGF induction of a subset of the genes identified by microarray. HUVECs were pre-treated with DMSO or c646 (p300/CBP inhibitor) prior to VEGF stimulation. Expression is relative to non-VEGF-stimulated cells (dashed line). Genes are arranged in decreasing order of VEGF induction. All 16 VEGF-induced genes were found to be p300 dependent (n=4-5).
Examining the kinetics of a subset of the ERG-dependent, VEGF-induced genes revealed that their transcription is increased transiently by VEGF stimulation, with peak transcription occurring between 15′ and 1 h (Fig. S6). To query the requirement of MAPK/ERK signaling and p300 activity in this response, we measured the induction of these genes in the presence of MEK or p300/CBP inhibitors. We found that 12 of 16 (75%) were MEK dependent, and all required p300/CBP activity (Fig. 5D,E).
To probe the relevance of this pathway in vivo, we assessed the expression of several of the identified genes by in situ hybridization in zebrafish embryos treated with inhibitors of VEGF, MEK or p300/CBP. Importantly, dll4, hlx1 and dusp5 were regulated by this pathway within ISVs (Fig. 6A,B). In addition, flt4, which is regulated by MAPK/ERK signaling during sprouting angiogenesis (Shin et al., 2016), was also dependent on p300/CBP and MEK (Fig. 6A,B). To induce ectopic activation of the MAPK/ERK pathway in a VEGF-independent manner, kdrl:GFP zebrafish embryos were exposed to PMA for 2 h (until 24 hpf). PMA treatment induced the phosphorylation of ERK1/2 in a MEK-dependent manner (Fig. 6C) and led to the induction of dll4, hlx1, dusp5 and egr3 expression in the endothelium, as determined by qRT-PCR from fluorescence-activated cell sorting (FACS)-isolated ECs (Fig. 6D). Importantly, pre-treatment of the embryos with c646 inhibited the PMA-induced induction of dll4, hlx1 and egr3, whereas dusp5 was refractory to c646 inhibition (Fig. 6E).
Fig. 6.
Regulation of VEGF/ERK/ERG/p300-dependent genes in vivo. (A) In situ hybridization using probes for dll4, hlx1, dusp5 and flt4 at 26 hpf. Embryos were treated with inhibitors of MEK (SL327), p300/CBP (c646) or VEGF (SU5416) starting at the 20-somite stage. Expression of each of these genes is MEK, p300 and VEGF dependent. Arrows indicate ISVs expressing the indicated genes. Representative images are shown. (B) Quantification of in situ hybridization experiments. The number of embryos analyzed is indicated. (C) pERK western blot in embryos pre-treated with SL327 for 1 h, followed by addition of PMA for 2 h. Coomassie staining was used to assess loading. Representative experiment of two. (D) qRT-PCR analysis of endothelial or non-endothelial cells isolated from kdrl:GFP embryos exposed to PMA for 2 h (starting at 24 hpf). All of these genes are induced in the endothelium in response to ectopic MEK activation (n=3). (E) qRT-PCR of whole individual embryos that were exposed to DMSO or c646 for 1 h, prior to stimulation with PMA for 2 h at 24 hpf. The induction of dll4, hlx and egr3 by PMA is p300 dependent (n=6).
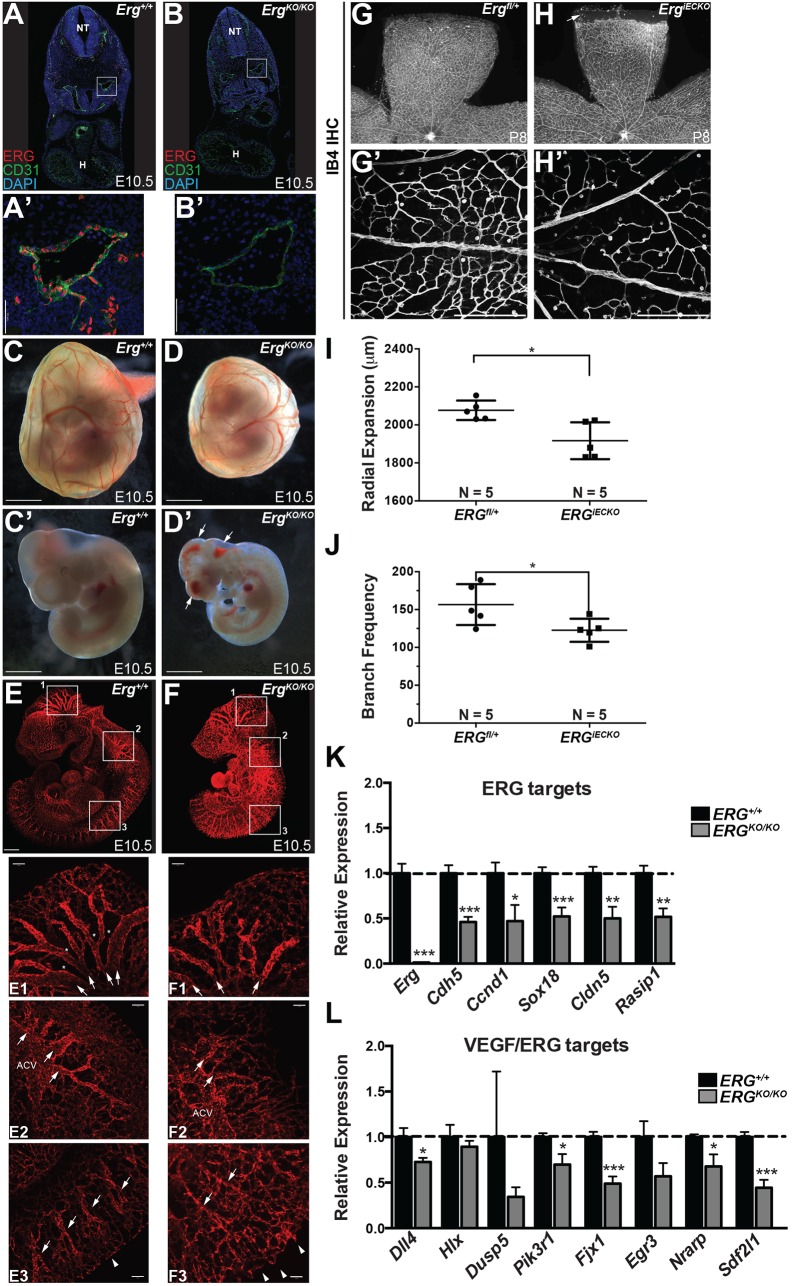
To assess further the role of ERG in angiogenesis in vivo, we generated a novel Erg knockout/lacZ knock-in mouse line. Deletion of Erg resulted in embryonic lethality by embryonic day (E) 11.5-E12.5, similar to previous reports (Birdsey et al., 2015; Vijayaraj et al., 2012) (Fig. S7A-F). Following loss of ERG protein (Fig. 7A-B′), we observed major defects in vascular integrity and angiogenesis during embryogenesis, within both the cranial and the trunk vasculature (Fig. 7C-F). Conditional deletion of Erg (ErgiECKO) using an EC-specific CreERT2 driver [Cdh5(PAC)-CreERT2] (Wang et al., 2010) led to defects in physiological angiogenesis, as determined by examination of angiogenesis within the postnatal retina (Fig. 7G-J; Fig. S7G-K). These data, combined with the embryonic lethality, hemorrhage and reduced angiogenesis all demonstrate a requirement for ERG in physiological angiogenesis.
Fig. 7.
ERG loss of function alters angiogenesis in vivo. (A-B′) Confocal microscopy following staining for ERG and CD31 on mouse embryo cryosections. Magnified views (A′,B′) of the dorsal aorta (boxed areas) reveal loss of ERG, and decreased CD31 (PECAM1), in ErgKO/KO embryos compared with wild-type littermate controls. Nuclei are stained with DAPI (blue). H, heart; NT, neural tube. (C-D′) Representative whole-mount bright-field images of E10.5 Erg+/+ (C,C′) and ErgKO/KO (D,D′) yolk sacs and embryos. Arrows indicate hemorrhage. (E,F) Representative light-sheet microscopy images of endomucin-stained blood vessels in E10.5 Erg+/+ (E) and ErgKO/KO (F) embryos. Boxed areas 1, 2 and 3 are shown below the whole-mount images at a higher magnification. Arrows in box 1 denote remodeled, larger caliber vessels, which are smaller in ErgKO/KO animals, and asterisks denote remodeled areas devoid of vessels, which are reduced in ErgKO/KO embryos compared with Erg+/+. In box 2, the anterior cardinal vein (ACV), although present in the knockouts, showed a decreased diameter and the major large caliber vessels sprouting from it (denoted by arrows) were also smaller and more tortuous. In box 3, the remodeled ISVs are denoted by arrows, and the sprouting front (dorsal-most edge) is denoted by arrowheads. The vascular front appears less uniform in knockouts and the ISVs appear less organized compared with wild-type littermates. (G-H′) Representative images of the total retinal vasculature (G,H) and magnified view of the proximal region (G′,H′) stained with IB4 in Ergfl/+ (G,G′) and ErgiECKO (H,H′) retinas at P8 following tamoxifen administration at P1 and P3. The arrow in H indicates an avascular area in ErgiECKO retina. (I) Quantification of radial expansion of the IB4+ vasculature within the P8 retina (n=5 for both genotypes). (J) Vascular density as determined by quantification of IB4+ branches in the proximal retinal vascular plexus at P8 (n=5 for each genotype). (K,L) ECs were isolated by FACS from Erg+/+ or ErgKO/KO embryos at E10.5. qRT-PCR was performed on the indicated VEGF-independent, ERG-dependent (K) and VEGF-responsive, ERG-dependent (L) genes [n=14 (Erg+/+) and 9 (ErgKO/KO)]. Scale bars: 50 µm (A′,B′); 1000 µm (C-D′); 500 µm (E,F); 100 µm (E1-3,F1-3,G′,H′).
To determine whether the candidate genes identified by our in vitro screen are downstream of Erg in vivo, we isolated ECs from wild-type or Erg mutant mouse embryos and performed qRT-PCR for several of the ERG- or VEGF/ERG-dependent genes (Fig. 7K,L). We found that several ERG-dependent, VEGF-independent genes identified in our screen (e.g. Rasip1, Sox18) or by others [e.g. Cdh5 (Birdsey et al., 2008; Gory et al., 1998) and Cldn5 (Yuan et al., 2012)] were downregulated in Erg loss-of-function embryos (Fig. 7K). Similarly, we observed a significant reduction in a typical VEGF-induced, ERG-dependent transcript, Dll4, in agreement with previous results (Wythe et al., 2013) (Fig. 7L). Additional candidates in this category, which showed robust sensitivity to MAPK and P300 activity in vitro, were substantially downregulated in vivo (e.g. Fjx1, Pik3r1, Sdf2l1, Nrarp). Collectively, these findings demonstrate that a VEGF/MAPK/ERG/p300 cascade is a crucial regulator of angiogenesis in vitro and in vivo.
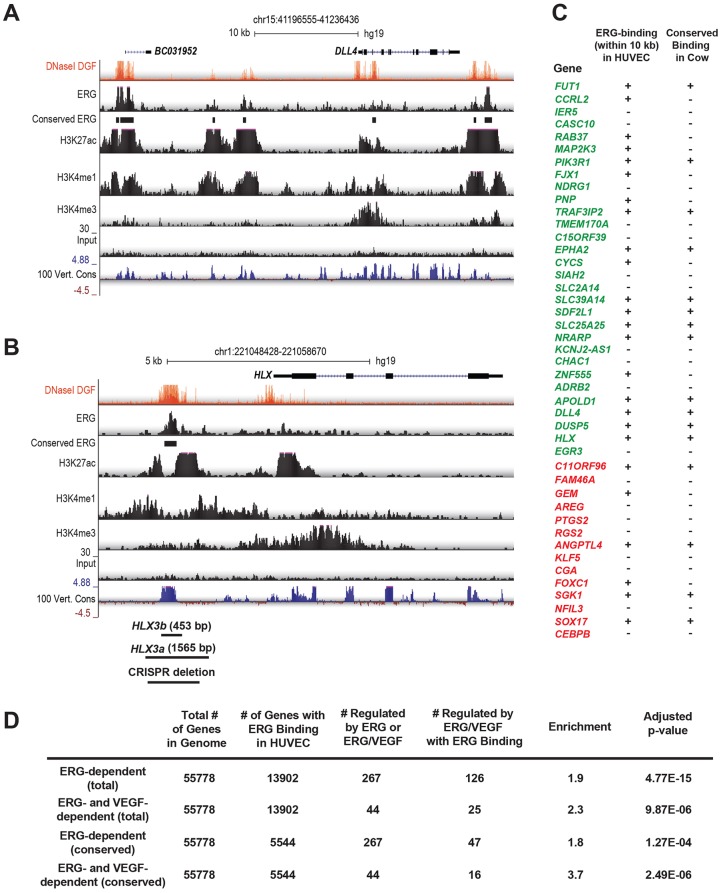
We next sought to identify the enhancers/promoters that ERG might directly act upon to regulate this gene network. We previously found that conserved orthologous transcription factor binding can reveal functional enhancers (Ballester et al., 2014). To identify evolutionarily conserved, epigenetically modified enhancers for further functional analyses, we performed ERG ChIP-seq experiments in both human (HUVECs) and bovine (BAECs) ECs cultured in complete media (i.e. containing VEGF) (Fig. 8). We identified 31,175 ERG ChIP-seq peaks in HUVECs and 34,773 peaks in BAECs, and found that 8337 of the human peaks were conserved in cow (Tables S3 and S4). We also performed H3K27ac ChIP-seq and found that 94% of conserved ERG peaks overlapped H3K27ac-enriched regions, supporting their association with active enhancers. We found that the DLL4 locus contains multiple conserved ERG-bound enhancers, including regions ∼12 kb upstream of the transcriptional start site (TSS) and within intron 3 (Fig. 8A), both of which were previously shown to have arterial-specific activity in vivo (Sacilotto et al., 2013; Wythe et al., 2013). We also identified an ERG-bound enhancer, conserved in cows and humans, ∼3.0 kb upstream of the gene H2.0-like homeobox (HLX) (Fig. 8B; Fig. S8). HLX, a homeobox transcription factor, expression of which is induced by VEGF in vitro (Schweighofer et al., 2009), has been implicated in controlling angiogenic sprouting of human cells in vitro (Prahst et al., 2014; Testori et al., 2011), and in ISV formation in zebrafish (Herbert et al., 2012), as well as yolk sac vascular remodeling in the mouse (Prahst et al., 2014). Further analysis of our ChIP-seq data revealed that the majority of ERG/VEGF-regulated genes (25 of 44) had an ERG ChIP-seq peak within 10 kb of the TSS, and ERG binding was conserved in cow for 16 of these genes (Fig. 8C). This is suggestive of direct regulation of these genes by ERG. Furthermore, ERG binding was significantly enriched near ERG- and ERG/VEGF-regulated genes (Fig. 8D).
Fig. 8.
Identification of ERG-bound enhancers. (A) Visualization of ChIP-seq assessing ERG binding, enhancer modifications (H3K27ac, H3K4me1, DNaseI hypersensitivity) and promoter modifications (H3K3me3) in HUVECs at the DLL4 locus. y-axis denotes reads per million (RPM) sequences. Conservation of ERG binding in BAECs is indicated, as is sequence conservation across 100 vertebrate species (y-axis denotes the magnitude of the conservation score). (B) Visualization of ChIP-seq data and sequence conservation surrounding the human HLX locus, a VEGF- and ERG-regulated gene, revealing a putative enhancer located ∼3 kb upstream of the transcriptional start site (TSS). The locations of the HLX-3a and HLX-3b fragments used in subsequent functional analyses are indicated, as is the region deleted by CRISPR/Cas9-mediated genome editing. (C) The presence of ERG-bound regions within 10 kb of the TSS of ERG/VEGF-dependent genes in HUVECs is indicated. Binding that is conserved in cow (i.e. in BAECs) is indicated. Genes that are induced by ERG are shown in green, whereas those repressed by ERG are shown in red. (D) Analysis of the enrichment of ERG-bound enhancers nearby ERG- and ERG/VEGF-dependent genes, compared with all genes in the genome.
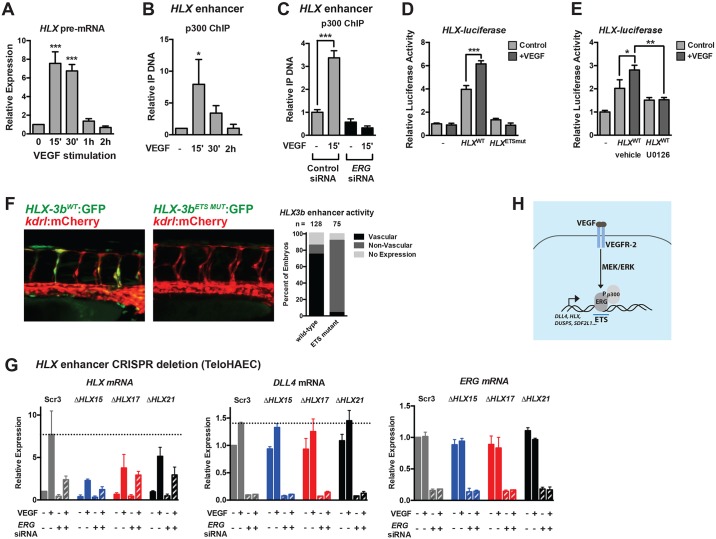
HLX transcription is transiently induced in response to VEGF stimulation, similar to DLL4 (Fig. 9A). We found that p300 was dynamically recruited to this evolutionarily conserved non-coding region (Fig. 9B) in an ERG-dependent manner (Fig. 9C). We cloned this conserved H3K27ac- and ERG-enriched −3 kb 5′ putative regulatory region (HLX-3a, 1565 bp fragment) upstream of a minimal promoter (SV40) driving a luciferase reporter, and found that it was VEGF responsive, and that the basal and VEGF-induced activity of this enhancer required ETS DNA-binding sequences (Fig. 9D). Furthermore, inhibition of MEK activity abrogated the VEGF responsiveness of this regulatory region (Fig. 9E). Although the full HLX-3a regulatory region failed to drive endothelial expression in vivo (data not shown; n=75), refinement of the element to the region bound by ERG (which was highly conserved across vertebrates; Fig. S8) and the 3′ acetylated region (HLX-3b, 435 bp fragment) drove robust EGFP reporter activity in the vasculature of the embryonic zebrafish (Fig. 9F). EGFP reporter expression was preferentially observed in the ECs of the ISVs (which form by angiogenesis) compared with the axial vessels (which form by vasculogenesis), and reporter activity was ETS element dependent (Fig. 9F). To test further the functional importance of this enhancer, we utilized clustered regularly interspaced short palindromic repeats/Cas9 (CRISPR/Cas9) genome editing to delete a portion (1201 bp; see Fig. 8B for schematic) of the H3K27ac-enriched, ERG-bound region upstream of HLX in TeloHAECs, an immortalized human aortic EC line. Several clonal lines (ΔHLX15, ΔHLX17 and ΔHLX21) heterozygous for deletion of this region were generated and confirmed by PCR and DNA sequencing (data not shown). Comparison was made with a clonal line generated following transfection of scrambled control gRNAs (Scr3). Although the basal expression of HLX appeared to be unaffected in the deletion lines, the VEGF-dependent induction of HLX appeared to be diminished (Fig. 9G). In contrast, DLL4 induction was unaffected. Furthermore, knockdown of ERG appeared to attenuate the induction of HLX to a greater extent in the control line compared with the deletion lines, implying that ERG acts through the deleted enhancer region. Collectively, these findings demonstrate the requirement of a highly conserved ERG-bound regulatory element in the VEGF responsiveness of the angiogenic gene HLX.
Fig. 9.
A conserved enhancer upstream of HLX is regulated by ETS factors and is required for VEGF induction. (A) Transcription of HLX (as assessed by qRT-PCR of HLX pre-mRNA) reveals dynamic transcription, peaking at 15-30′ post-VEGF stimulation in HUVECs (n=3). (B) p300 is transiently recruited to a putative HLX enhancer element during VEGF stimulation, as assessed by ChIP assay (n=3). (C) p300 ChIP was performed in control and ERG knockdown cells. Shown are triplicate measures of a representative experiment of two. (D) Luciferase analysis of the HLX enhancer (HLX-3a), demonstrating that it is regulated by VEGF and ETS factors. ETS-binding sites were mutated in the HLX enhancer (HLXETSmut, see Materials and Methods). A representative experiment (of three) with triplicate determinations is shown. (E) Luciferase analysis of the HLX enhancer (HLX-3a), demonstrating that it is regulated by MAPK/ERK activity. A representative experiment (of two) with triplicate determinations is shown. (F) The human HLX enhancer (HLX-3b) is functional in ISVs in zebrafish during sprouting angiogenesis. Activity is lost when the ETS sites in the enhancer are mutated. Shown are representative images of embryos at 42 hpf. Quantification of enhancer activity is shown to the right. (G) CRISPR/Cas9-mediated deletion of a highly conserved enhancer upstream of HLX inhibits VEGF-mediated induction. Shown are a clonal scrambled-control line (Scr3) and three heterozygous deletion lines (ΔHLX15, ΔHLX17, ΔHLX21). Induction of DLL4 is included as a control. Knockdown of ERG affects the induction of HLX in the control line to a greater extent than in the deletion lines. n=2. (H) Schematic of the VEGF/MEK/ERK/ERG/p300 transcriptional pathway identified in this study.
DISCUSSION
Dynamic control of gene expression and the resultant cellular outputs in tip cells and adjacent stalk cells are central to the growth of nascent angiogenic sprouts (Blanco and Gerhardt, 2013; Lobov et al., 2007). The pathways that regulate the temporal VEGF-dependent expression of DLL4 in tip cells are of particular importance considering the central role of this ligand, and its receptor, Notch, in directing tip and stalk cell behaviors (Hellström et al., 2007; Jakobsson et al., 2010; Suchting et al., 2007; Ubezio et al., 2016). Here, we identify one potential mechanism for the transient VEGF-dependent induction of DLL4 transcription. VEGF stimulation initiates a rapid and transient burst of MAPK/ERK activity, with similar kinetics to VEGF induction of DLL4 transcription. Downstream target proteins modulated by ERK kinase activity are also dynamically modified, as illustrated by the transient phosphorylation of ERG at serine 215. Additionally, we find that the co-activator p300 is recruited to angiogenic enhancers in an ERG-dependent manner, with kinetics mirroring ERG phosphorylation. This VEGF/ERK/ERG/p300 transcriptional pathway also dynamically regulates a network of genes shown to positively (e.g. HLX, FJX1, EGR3, APOLD1, ADRB2, EPHA2, FUT1, MAP2K3, NDRG1) and negatively (e.g. DUSP5, NRARP) regulate angiogenesis (Fig. 9H) (Al-Greene et al., 2013; Bellou et al., 2009; Herbert et al., 2012; Iaccarino et al., 2005; Liu et al., 2008; Mirza et al., 2013; Moehler et al., 2008; Phng et al., 2009; Pin et al., 2012; Prahst et al., 2014; Toffoli et al., 2009; Zhou et al., 2011).
The ETS family of transcription factors has previously been implicated as signal-dependent effectors (Wasylyk et al., 1998), but how ETS factors act downstream of VEGF has not been explored in detail. Interestingly, we find that DLL4 induction by VEGF signaling requires MAPK/ERK signaling, as well as ERG expression. Previous studies in cancer cells revealed that phosphorylation of ERG at S96, S215 and S276 is mediated by ERK2 (Selvaraj et al., 2015). We find that VEGF signaling leads to dynamic ERK-dependent phosphorylation of ERG at S215, and that S96, S215 and S276 are required for maximal ERG activity. The ability of ERG to drive expression of VEGF target genes appears to be p300 dependent, as VEGF initiates a physical interaction between ERG and p300, and ERG is required for p300 recruitment to DLL4 and HLX enhancer elements. Furthermore, p300/CBP inhibition abolishes VEGF/ERG-dependent gene expression. Mutation of ERK-phosphorylated residues in ERG prevents its interaction with p300, suggesting a role for ERG phosphorylation in recruitment of p300 to target genes. Furthermore, the termination of p300 recruitment temporally coincides with loss of ERG phosphorylation, implying a functional role for these phosphorylation events. It will be of interest to determine whether all DNA-localized ERG, or only those molecules involved in VEGF signaling output, become phosphorylated in response to VEGF signaling. It will also be of interest to determine how diverse activators of MAPK/ERK signaling, which have distinct effects on angiogenesis (e.g. ANG1/TIE2 (ANGPT1/TEK) signaling), might differentially activate ERG. Answering these questions will be vital for the development of targeted therapeutics to suppress angiogenesis. Because ERG regulates vascular integrity (presumably in cells lacking active ERK), it is possible that ERG functions to maintain vascular stability in an ERK-independent manner, suggesting the possibility of selectively blocking angiogenesis through targeting ERG phosphorylation, while maintaining vascular stability. Furthermore, ERG is known to function as an oncogenic fusion protein (e.g. TMPRSS2-ERG) in prostate cancer (Adamo and Ladomery, 2016). Of note, the amino terminus of ERG (included in many of these fusion proteins) appears to contain the same serine residues phosphorylated by ERK2. It will be of interest to determine how upstream signaling pathways (e.g. activated RAS/MAPK/ERK) influence ERG transcriptional activity in cancer. Perhaps targeting ERG phosphorylation could be of interest to quell ERG oncogenic activity.
Previous studies identified a role for another ETS factor, TEL (ETV6), in the repression, rather than the activation, of DLL4 (Roukens et al., 2010). In this case, TEL bound to the DLL4 promoter under basal conditions to recruit a co-repressor protein, CTBP. Addition of VEGF led to the rapid disassembly of this repressive complex. The kinetics of this repressive TEL/CTBP complex disassembly are comparable to the assembly of the activating ERG/p300 complex that we report here, suggesting that TEL and ERG dynamically control co-activator/co-repressor recruitment. Recently, VEGF has also been shown to stimulate dynamic exchange of co-repressors for co-activators bound to MEF2 transcription factors (Sacilotto et al., 2016), suggesting that several families of transcription factors may coordinate VEGF-dependent sprouting angiogenesis. Indeed, ETS proteins interact with multiple transcription factor families (Carrère et al., 1998; De Val et al., 2008). Of note, we have identified a number of transcription factor binding motifs that are enriched under ERG ChIP-seq peaks in the vicinity of ERG- and ERG/VEGF-dependent genes that might functionally interact with ERG to control gene expression (Table S3). Although a subset of VEGF inducible genes are regulated by ERG, it is equally important to note that many VEGF-dependent genes are ERG independent. This could be attributable to redundancy of ETS factors, but could also imply that additional transcriptional pathways responsible for angiogenic gene regulation remain to be uncovered.
In summary, our study has identified a VEGF/MAPK/ERK/ERG/p300 network that is required for the induction of a subset of VEGF-inducible genes in ECs, allowing us to propose a model for how transient activation of an angiogenic program might be regulated to orchestrate sprouting (Fig. 9H).
MATERIALS AND METHODS
Zebrafish experiments
Zebrafish protocols were approved by the Animal Care Committee at the University Health Network, the University of Texas MD Anderson Cancer Center and Baylor College of Medicine. The following transgenic lines were utilized: Tg(kdrl:mCherry)ci5 (Proulx et al., 2010), Tg(kdrl:GFP)s843 (Jin et al., 2005), Tg(fli1a:nls-EGFP)y7 (Roman et al., 2002), Tg(TP1bglob:EGFP)um14 (Parsons et al., 2009) and Tg(TP1bglob:VenusPEST)S940 (Ninov et al., 2012).
Inhibitor treatments
Embryos were dechorionated, then treated from 18-20 hpf until 26-28 hpf (unless noted otherwise) with the following inhibitors: SU5416 (VEGFR2 inhibitor, 5 μM, LC Laboratories), SL327 (MEK inhibitor, 30 μM, Sigma) or c646 (p300 inhibitor, 3 μM, Sigma), with all inhibitors prepared as 1000× stocks in DMSO, and embryos treated in E3 supplemented with PTU to prevent pigmentation. PMA (Bioshop) was used at a concentration of 1 μM. DMSO (0.1%) was used as a vehicle control. Of note, repeated freeze thaw of c646 stocks diminished efficacy and higher doses of c646 produced serious developmental delay and growth defects (data not shown).
Imaging
See supplementary Materials and Methods for details regarding confocal imaging.
pERK immunofluorescence
Treated Tg(kdrl:GFP)s843 embryos were processed following the protocol of Inoue and Wittbrodt (2011), with the modifications suggested in Le Guen et al. (2014) (see supplementary Materials and Methods for details).
Time-lapse microscopy
Tg(TP1bglob:VenusPEST); Tg(kdrl:mCherry) animals at 18-20 somites were mounted in 1% low-melt agarose on a four-compartment glass-bottom cell culture dish (Cellview, #627975), treated with PTU and tricaine in E3, along with the same concentration indicated above for either DMSO (vehicle control) or SL327 (MEK inhibitor) (see supplementary Materials and Methods for details).
Isolation of ECs by FACS
Embryos were washed with PBS (without calcium/magnesium) and 1 ml of pre-warmed 0.25% Trypsin was added to the embryos. Embryos were incubated at 28°C and gently pipetted up and down every 5 min until digestion was complete. After digestion, 100 μl of fetal bovine serum (FBS; 100%) was added to stop digestion. The cells were spun at 1100 rpm (300 g) for 5 min at 4°C and the supernatant was removed and the cell pellet was re-suspended in 500 μl of FACS solution (450 μl PBS+50 μl 10% bovine serum albumin). Sytox Red (0.5 μl) was added and the samples were incubated at room temperature for 15 min and then passed through a cell sieve. FACS was performed by the UHN Flow Cytometry Facility using a low differential pressure (20 psi). Cells were sorted directly into RLT buffer (Qiagen) for RNA extraction using the RNeasy Micro Kit (Qiagen).
In situ hybridization
Experiments were performed as described previously (Wythe et al., 2011). Riboprobes, with the exception of flt4, were amplified by PCR with primers containing SP6 and T7 overhangs and sense and digoxigenin-labeled antisense probes were synthesized from the PCR template. The hlx1 template was provided by Dr Saulius Sumanas (Cincinnati Children's Medical Center, OH, USA), dll4 in pGEM-T was from Dr Jiandong Liu (University of North Carolina, NC, USA), dusp5 was from GE Dharmacon (Clone ID: 4199935), and flt4 was from Dr Jeffrey Essner (Iowa State University, IA, USA) (digested with EcoRI, transcribed with T7). See supplementary Materials and Methods for primers used.
Enhancer transgenesis
pTol2 enhancer injections were performed as previously published (Wythe et al., 2013). Briefly, an injection mixture consisting of 100 ng of DNA, 125 ng of Tol2 transposase mRNA (Kawakami et al., 2004), 1 µl 0.8% Phenol Red/0.1 M KCl, pH 7.0, and ddH2O in 10 µl total volume was combined and 1 nl injected directly into the cell of one-cell stage Tg(kdrl:mCherry)ci5 zebrafish embryos. Embryos were then maintained at 28.5°C and scored at 42 hpf for enhancer activity within and outside of the vasculature (mCherry+).
Transplantation experiments
Sense-strand-capped mRNA was transcribed using the mMessage mMachine kit (Ambion) from either a pCS2-3xFlag-ERG or pCS2-3xFlag-ERG-3xS→A template (see ‘Cloning’ in supplementary Materials and Methods for details). mRNA (5 µl of 125 ng/µl) was mixed with 1 µl of 2 mg/ml Alexa 647 10,000 MW, Anionic, Fixable Dextran (Life Technologies, D22914) as a lineage tracer, then 1 nl of this mixture was injected into one-cell stage Tg(fli1a:nls-EGFP)y7 donor embryos. Dividers were pulled 1-2 h later for Tg(kdrl:mCherry)ci5 host embryos. Animals were dechorionated on agarose dishes using pronase in E2 media supplemented with penicillin and streptomycin, as suggested by Westerfield (2007). When donor embryos reached approximately sphere stage to 30% epiboly (about 4 hpf), they were transferred to agarose wells (Adaptive Science Tools, PT-1) in E2 plus antibiotics, and 20-40 cells from the lateral margin were transferred from donor to host embryos (at a similar location). Embryos were reared at 28.5°C in 1.5% agarose dishes in E2 supplemented with antibiotics until their fixation in 4% paraformaldehyde (PFA) at 28-30 hpf. See supplementary Materials and Methods for details on imaging. The percentage contribution of donor-derived cells contributing to a location in the host trunk vasculature was quantified as the number of donor-derived cells within the structure divided by the total number of donor-derived cells within the entire vasculature in the region of interest.
Erg murine knockout experiments
Generation of Erg knockout mice was carried out by the KOMP consortium (project ID: 48771; Ergtm1a(KOMP)Wtsi). Cryopreserved sperm were received and in vitro fertilization was performed at the Genetically Engineered Mouse (GEM) core at Baylor College of Medicine. In this Ergtm1a allele, insertion of a splice acceptor-IRES-lacZ-stop, human beta actin promoter driving Neomycin between exon 5 and 6 acts as a null mutation (i.e. a gene trap) (Fig. S7A). For these studies, a global Erg null allele (ErglacZΔNEO/+, ErgKO/+ or Ergtm1b) was generated by Cre-mediated removal of the hBact::Neo cassette by crossing Ergtm1a/+ male mice to females harboring a Tg(ACTB::Cre) (MGI#2176050) (Lewandoski et al., 1997) driver (see Fig. S7A). To generate a conditional allele (Ergflox/+ or Ergtm1c), an Ergtm1a/+ male was crossed to a Tg(ATCB::FlpE) female (MGI#2448985) (Rodríguez et al., 2000) for Flp-mediated removal of the FRT-flanked promoterless lacZ gene trap. The resulting animals, with two loxP sites flanking exon 6, were incrossed to generate Ergflox/flox animals for postnatal studies (see below). For further details regarding PCR genotyping, see Table S1.
Immunohistochemistry
Timed matings between heterozygous mutant animals (ErgKO/+) mice were conducted, and noon of the day a vaginal plug was detected was considered day 0.5. Embryos were collected at E10.5, and the yolk sac was used for PCR genotyping of the embryos. Embryos were fixed overnight in 4% PFA then subjected to whole-mount IHC using an endomucin antibody (eBioscience, V.7C7) (1:200); biotinylated goat anti-rat secondary antibody (Vector, BA-9401) (1:250), ABC Elite Kit (Vector Labs, PK-6100) and Alexa 488 Tyramide (Thermo Fisher Scientific, T20948) and imaged by light-sheet microscopy, as previously published (Wang et al., 2016). For frozen sections, embryos were fixed in fresh 4% PFA at 4°C for 20 min, washed in PBS, cryoprotected in 30% sucrose overnight at 4°C, equilibrated in optimal cutting temperature compound (Sakura, #25608) in peel-away molds (Electron Microscopy Sciences, #701081), solidified on dry ice, then stored overnight at −80°C. For additional detail, see supplementary Materials and Methods.
X-gal staining
Whole-mount embryos were processed as detailed previously (Wythe et al., 2013). See supplementary Materials and Methods for further details.
Analysis of FACS-isolated ECs
Embryos were collected at E10.5 (their yolk sac was removed for PCR genotyping) and dissociated to single cells by a 20-min incubation at 37°C in collagenase type I (1 mg/ml) (ThermoFisher Scientific, #17100017). See supplementary Materials and Methods for details. Next, 25,000 live ECs per embryo were collected into 350 µl RLT Buffer (Qiagen) for downstream processing. Total RNA was isolated from sorted cells using the RNAeasy Mini Kit (Qiagen, #74104). RNA was eluted in 30 µl RNAse-free H2O, and 20 µl of purified RNA was reverse transcribed to cDNA with 5 µl of SuperScript VILO Master Mix (Thermo Fisher Scientific, #11755050). The resulting cDNA was diluted 1:10 with RNAse-free dH2O, and 4 µl was used for each qPCR reaction (15 µl total reaction volume). qPCR reactions were performed in technical triplicate using iTaq Universal SYBR Green Supermix (Bio-Rad, #1725124) and run on a ViiA 7 Real-Time PCR System (Applied Biosystems). Relative abundance of mRNA transcripts was calculated by normalization to Gapdh using the ΔΔCt method (Livak and Schmittgen, 2001). All embryonic RNA samples used for analysis had a deviation no greater than two Cts (range 16-18) for Gapdh. For a list of all primers used, see Table S2.
Analysis of retinal vasculature
Ergflox/flox (aka Erg tm1c/tm1c) females were crossed to ErgKO/+ (aka Erg tm1a/+); Cdh5(PAC)CreERT2 (Wang et al., 2010) males and 30 µl of tamoxifen was administered by subcutaneous injection at a concentration of 10 mg/ml (≈30 µg total per mouse) to their progeny at postnatal days (P) 1 and 3. See supplementary Materials and Methods for details regarding quantification of vascular branching and radial expansion in the retina.
Cell culture
The following cells were utilized for experiments: human umbilical vein endothelial cells (HUVECs, ScienCell), telomerase-immortalized aortic endothelial cells (TeloHAECs, ATCC), dermal microvascular endothelial cells (MVECs, Life Technologies) and bovine aortic endothelial cells (BAECs, Lonza). Cells were cultured according to manufacturer's recommendations. ECs were serum-starved in basal EC media (ScienCell) containing 0.1% FBS and no growth factors for at least 6 h (typically overnight) prior to stimulation with VEGF or PMA. Pathway inhibitors were added 1 h prior to stimulation with VEGF-165 (50 ng/ml, recombinant human protein, R&D Systems or Thermo Fisher Scientific), except for c646, which was added 20 min prior to stimulation. The following inhibitors were used: U0126 (MEK inhibitor, 20 μM, InvivoGen), GF109203X (PKC inhibitor, 5 μM, Tocris Bioscience), LY294002 (PI3K inhibitor, 10 μM, Cell Signaling), SB203580 (p38 inhibitor, 10 μM, Tocris Bioscience), DAPT (γ-secretase inhibitor, 38.5 μM, Sigma) and c646 (p300/CBP inhibitor, 5 μM, Sigma). All drugs were dissolved in DMSO, and comparison was made with vehicle (i.e. DMSO, 0.1%) treated controls. PMA was from BioShop and was used at a concentration of 100 nM.
siRNA experiments
HUVECs were transfected at 30-50% confluency with 40 nM siRNA (Silencer Select, Thermo) targeting the coding region of ERG (assay ID: s4813), ERK1 (MAPK3) (assay ID: S230180) or ERK2 (MAPK1) (assay ID: S11138) using RNAiMax (Invitrogen), and cellular assays were performed 48-72 h later. Western blotting and qRT-PCR were used to assess ERG, ERK1 and ERK2 knockdown. Comparison was made with cells transfected with 40 nM Silencer Select negative control #1. For ERG rescue experiments, an independent siRNA recognizing the 3′ untranslated region of ERG (40 nM, Silencer Select, custom synthesis) was utilized. After 48-72 h, cells were electroporated using the P5 Primary Cell 4D Nucleofector kit and a Lonza 4D Nucleofector; ∼0.5×106 cells were electroporated with 2.5 μg of pCS2 control or pCS2-Flag-ERG expression constructs (wild type and phospho-mutants; see ‘Cloning’ in supplementary Materials and Methods), and 0.2 μg of pmaxGFP (to assess electroporation efficiency). After 18 h, cells were serum-starved for 6 h prior to VEGF stimulation (1 h) and harvested for RNA/protein analyses.
Cloning
For details regarding cloning of ETS concatamer reporter constructs, HLX enhancer reporter constructs and wild-type and mutant ERG expression constructs, please see the supplementary Materials and Methods.
Luciferase experiments
For luciferase assays, BAECs (80% confluent) were transfected with 0.5 µg of luciferase construct (ETS reporter, HLX enhancer, see ‘Cloning’ in supplementary Materials and Methods) and 0.1 µg of pRenilla construct using Lipofectamine 2000 (Invitrogen) (2 µl per 1 µg of plasmid) in 12-well dishes. Cells were treated with cellular signaling pathway inhibitors for 18 h (as above). In some experiments, VEGF (50 ng/ml) was added to OptiMEM medium for 18 h. After 24 h, dual luciferase (Renilla and firefly) was measured using a GloMax20/20 Luminometer (Promega).
Western blotting and co-immunoprecipitation
Western blotting was performed as before (Fish et al., 2011) using the following antibodies: anti-pERK1/2 (Thr202/Tyr204, rabbit polyclonal, Cell Signaling, #9101; 1:1000), anti-ERK2 (mouse monoclonal, Santa Cruz, D-2; 1:500), anti-ERK1/2 (rabbit monoclonal, Cell Signaling, clone 137F5; 1:500), anti-DLL4 (rabbit polyclonal, Cell Signaling, #2589; 1:1000), anti-GAPDH (mouse monoclonal, Santa Cruz, #0411; 1:500), anti-ERG (rabbit polyclonal, Santa Cruz, C-20 or mouse monoclonal antibody, BioCare Medical, 9FY; 1:1000), anti-pERGS215 [rabbit polyclonal, a kind gift from Peter Hollenhorst, Indiana University, IN, USA (Selvaraj et al., 2015); 1:500], anti-p300 (rabbit polyclonal, Santa Cruz, C-20; 1:200). All antibodies have been previously validated. See supplementary Materials and Methods for details regarding pERG western blots and co-immunoprecipitation experiments.
Immunofluorescence on cultured cells
HUVECs were plated on Permanox eight-well chamber slides. Following stimulation with VEGF, cells were fixed with 4% PFA followed by permeabilization with 0.25% Triton X-100. Staining with anti-pERK (rabbit polyclonal, Cell Signaling, #9101, 1:500) was performed overnight at 4°C, followed by addition of a secondary antibody (anti-rabbit Alexa Fluor647, Cell Signaling #4414). Slides were mounted using Vectashield mounting medium with DAPI (Vector Labs H-1200) and imaged using an Olympus FV1000 confocal microscope.
RNA isolation, reverse transcription and quantitative PCR
RNA was isolated from cells and zebrafish using Trizol and reverse transcription was performed using a high-capacity cDNA reverse transcription kit (Applied Biosystems). qRT-PCR was performed using a Roche Lightcycler 480 with LC 480 SYBR Green I Master Mix (Roche). Data were normalized to TATA-box binding protein (TBP) or Gapdh using the ΔΔCt method. For further details regarding primer sequences, see Table S2.
Gene expression array
HUVECs were transfected with control or ERG siRNA and after 48 h the cells were serum-starved overnight and cells were then left unstimulated or were treated with 50 ng/ml VEGF for 1 h. RNA was isolated from four independent experiments using Trizol and analyzed on Agilent microarray, performed at the Princess Margaret Genomics Centre. See supplementary Materials and Methods for details regarding microarray processing and analysis.
Gene ontology analysis
Differentially expressed genes were submitted to the Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics resource (https://david-d.ncifcrf.gov/home.jsp) to be classified into gene ontology (GO) annotation groups (Ashburner et al., 2000; Huang et al., 2009). Fisher's exact test was applied to identify significant GO categories. Select representative GO categories are included in figures.
ChIP-qPCR experiments
ChIP was performed as before (Wythe et al., 2013), using the Imprint ChIP kit (Sigma) or the Magna ChIP A/G kit (Millipore). HUVECs were serum-starved overnight prior to stimulation with 50 ng/ml VEGF for 15′, 30′ or 2 h. Following fixation and shearing of chromatin, immunoprecipitation was performed overnight at 4°C using 1 μg (or 1 μl) of antibodies to ERG (rabbit polyclonal, Santa Cruz, C-20), p300 (rabbit polyclonal, Santa Cruz, C-20) or H3K27ac (rabbit polyclonal, Abcam, ab4729). Mouse IgG (Sigma) was used as a non-specific background control. qPCR was performed using primers that amplified the DLL4 intron 3 enhancer or an enhancer upstream of HLX (see Table S2 for primer sequences). IP DNA was calculated by subtracting the IgG value from the specific antibody value and dividing by a diluted input sample. In some experiments, control or ERG siRNAs were transfected into HUVECs 48 h prior to stimulation with VEGF (15′), followed by p300 ChIP.
ChIP-seq experiments and analysis
Primary HUVECs and BAEC cells were grown in supplier-recommended EC Growth Media (ScienCell) and cultured at 37°C in a 5%-CO2 humidified incubator. Approximately 20 million cells were used for the ERG and ∼3 million cells for the H3K27ac ChIPs. ChIP experiments were conducted as previously described (Ballester et al., 2014). Antibodies used for ChIP were mouse anti-H3K27ac (Millipore, 05-1334 monoclonal) and rabbit anti-ERG 1/2/3 (Santa Cruz Biotechnology, sc353 polyclonal). Two replicates were performed. See supplementary Materials and Methods for details regarding processing and analysis of ChIP-seq experiments.
CRISPR/Cas9-mediated HLX enhancer deletion
The HLX enhancer region to be targeted for deletion was defined by H3K27ac ChIP-seq enhancer marks (∼1200 bp). The MIT CRISPR/Cas9 design tool (http://crispr.mit.edu/) was used to generate gRNAs targeting the 5′ and 3′ boundaries of the HLX enhancer. Two scrambled sequence gRNAs were used as controls. For further details, see the supplementary Materials and Methods.
Statistical analyses
Unless otherwise stated, all experiments were performed a minimum of three times and data represent the mean±s.e.m. Statistical analyses were performed using a Student's t-test (for two groups) or ANOVA (for more than two groups), followed by the Newman–Keuls post-hoc test. P<0.05 was considered statistically significant. In all figures, *P<0.05, **P<0.01 and ***P<0.001.
Acknowledgements
The authors thank the Optical Imaging and Vital Microscopy (OIVM) Core at BCM for support with imaging and data processing; Dr Neil I. Bower (The University of Queensland, Dr Benjamin M. Hogan laboratory) for advice and suggestions regarding pERK staining; Dr Peter Hollenhorst for providing an antibody to phosphorylated ERG and for advice on ERG immunoprecipitation; Dr Jiandong Liu and Ms Nicole Fleming (UNC Chapel Hill) for suggestions on transplantation experiments; and Mrs Karen Berman de Ruiz for excellent mouse husbandry care and colony management.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.E.F., J.D.W.; Methodology: J.E.F., M.C.G., L.T.D., L.A., G.T.E., M.D.W., J.D.W.; Software: L.A., A.M.-R., M.D.W.; Validation: J.E.F., M.C.G., N.K., Z.C., S.V., H.S.C., M.K., L.A., M.-S.N., E.B., O.E.R., A.M.-R., M.D.W., J.D.W.; Formal analysis: J.E.F., M.C.G., L.T.D., N.K., Z.C., S.V., H.S.C., M.K., L.A., M.-S.N., E.B., A.M.H., A.M.R., O.E.R., A.M.-R., M.D.W., J.D.W.; Investigation: J.E.F., M.C.G., L.T.D., N.K., Z.C., S.V., H.S.C., M.K., L.A., M.-S.N., E.B., A.M.H., A.M.R., O.E.R., G.T.E., A.M.-R., M.D.W., J.D.W.; Resources: J.E.F., G.T.E., M.D.W., J.D.W.; Data curation: J.E.F., M.C.G., L.T.D., L.A., A.M.-R., M.D.W., J.D.W.; Writing - original draft: J.E.F., J.D.W.; Writing - review & editing: J.E.F., M.C.G., O.E.R., G.T.E., A.M.-R., M.D.W., J.D.W.; Visualization: J.E.F., M.C.G., E.B., A.M.H., A.M.R., O.E.R., G.T.E., J.D.W.; Supervision: J.E.F., G.T.E., M.D.W., J.D.W.; Project administration: J.E.F., G.T.E., J.D.W.; Funding acquisition: J.E.F., G.T.E., M.D.W., J.D.W.
Funding
J.E.F. was funded by an operating grant from the Canadian Institutes of Health Research (CIHR) (MOP-119506), and a Leaders Opportunity Fund/Canada Foundation for Innovation equipment grant (26422). J.E.F. and M.D.W. were supported by Early Researcher Awards from the Ontario Ministry of Research and Innovation, Canada Research Chairs from CIHR, and a Team Project Award from the University of Toronto's Medicine by Design initiative, which receives funding from the Canada First Research Excellence Fund. L.T.D. was supported by a post-doctoral fellowship from the Toronto General Hospital Research Institute, N.K. was supported by a Canada Graduate Studentship from the Natural Sciences and Engineering Research Council of Canada. M.S.N. was supported by a Vascular Network Scholar Award (CIHR funded). H.S.C. was supported by an Ontario Graduate Student award. ChIP-seq and bioinformatics work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) (436194-2013 to M.D.W.). L.A. was supported by an Alexander Graham Bell Canada Graduate Scholarship; and A.M.-R. was supported by a CONACYT fellowship (203853) and a CIHR STAGE fellowship. A.M.-R.'s laboratory is supported by a CONACYT grant (269449) and a Programa de Apoyo a Proyectos de investigación e innovación tecnológica - Universidad Nacional Autónoma de México (PAPIIT-UNAM) grant (IA206517). G.T.E. was supported by a Cancer Prevention and Research Institute of Texas (CPRIT) First-Time Tenure-Track Faculty Recruitment Award (RR140077). This project was supported by the Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from the National Institutes of Health (CA125123 and RR024574). A.M.H. was supported by a Molecular Physiology of the Cardiovascular System Training Grant, The National Institutes of Health (T32HL07676). J.D.W. was supported by institutional startup funds from the CVRI at Baylor College of Medicine, the Caroline Wiess Law Fund, the Curtis Hankamer Basic Research Fund, and the ARCO Foundation Young Teacher-Investigator Award. Work within the Wythe lab is supported by the American Heart Association (16GRNT31330023). Deposited in PMC for release after 12 months.
Data availability
All microarray and ChIP-seq data have been made publicly available. Microarray data were submitted to ArrayExpress (accession number: E-MTAB-5207). ERG and H3K27ac ChIP-seq data from HUVEC and ERG ChIP-seq data from BAEC were submitted to ArrayExpress (accession number: E-MTAB-5148). We also utilized HUVEC H3K4me3 ChIP-seq data (pooled signal from biological replicates, ENCODE accession numbers: ENCFF000BTS, ENCFF000BTL) and HUVEC H3K4me1 ChIP-seq data (pooled signal from biological replicates, ENCODE accession numbers: ENCFF000BTD, ENCFF000BSY, ENCFF000BSX) from the Encyclopedia of DNA Elements Consortium (ENCODE Project Consortium, 2012). Vertebrate conservation across 100 genomes was extracted from UCSC Genome Browser.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.146050.supplemental
References
- Adamo P. and Ladomery M. R. (2016). The oncogene ERG: a key factor in prostate cancer. Oncogene 35, 403-414. 10.1038/onc.2015.109 [DOI] [PubMed] [Google Scholar]
- Al-Greene N. T., Means A. L., Lu P., Jiang A., Schmidt C. R., Chakravarthy A. B., Merchant N. B., Washington M. K., Zhang B., Shyr Y. et al. (2013). Four jointed box 1 promotes angiogenesis and is associated with poor patient survival in colorectal carcinoma. PLoS ONE 8, e69660 10.1371/journal.pone.0069660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T. et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25-29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester B., Medina-Rivera A., Schmidt D., Gonzàlez-Porta M., Carlucci M., Chen X., Chessman K., Faure A. J., Funnell A. P., Goncalves A. et al. (2014). Multi-species, multi-transcription factor binding highlights conserved control of tissue-specific biological pathways. Elife 3, e02626 10.7554/eLife.02626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellou S., Hink M. A., Bagli E., Panopoulou E., Bastiaens P. I. H., Murphy C. and Fotsis T. (2009). VEGF autoregulates its proliferative and migratory ERK1/2 and p38 cascades by enhancing the expression of DUSP1 and DUSP5 phosphatases in endothelial cells. Am. J. Physiol. Cell Physiol. 297, C1477-C1489. 10.1152/ajpcell.00058.2009 [DOI] [PubMed] [Google Scholar]
- Birdsey G. M., Dryden N. H., Amsellem V., Gebhardt F., Sahnan K., Haskard D. O., Dejana E., Mason J. C. and Randi A. M. (2008). Transcription factor Erg regulates angiogenesis and endothelial apoptosis through VE-cadherin. Blood 111, 3498-3506. 10.1182/blood-2007-08-105346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdsey G. M., Dryden N. H., Shah A. V., Hannah R., Hall M. D., Haskard D. O., Parsons M., Mason J. C., Zvelebil M., Gottgens B. et al. (2012). The transcription factor Erg regulates expression of histone deacetylase 6 and multiple pathways involved in endothelial cell migration and angiogenesis. Blood 119, 894-903. 10.1182/blood-2011-04-350025 [DOI] [PubMed] [Google Scholar]
- Birdsey G. M., Shah A. V., Dufton N., Reynolds L. E., Osuna Almagro L., Yang Y., Aspalter I. M., Khan S. T., Mason J. C., Dejana E. et al. (2015). The endothelial transcription factor ERG promotes vascular stability and growth through Wnt/beta-catenin signaling. Dev. Cell 32, 82-96. 10.1016/j.devcel.2014.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco R. and Gerhardt H. (2013). VEGF and Notch in tip and stalk cell selection. Cold Spring Harb. Perspect. Med. 3, a006569 10.1101/cshperspect.a006569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers E. M., Yan G., Mukherjee C., Orry A., Wang L., Holbert M. A., Crump N. T., Hazzalin C. A., Liszczak G., Yuan H. et al. (2010). Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chem. Biol. 17, 471-482. 10.1016/j.chembiol.2010.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P., Ferreira V., Breier G., Pollefeyt S., Kieckens L., Gertsenstein M., Fahrig M., Vandenhoeck A., Harpal K., Eberhardt C. et al. (1996). Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature 380, 435-439. 10.1038/380435a0 [DOI] [PubMed] [Google Scholar]
- Carrère S., Verger A., Flourens A., Stehelin D. and Duterque-Coquillaud M. (1998). Erg proteins, transcription factors of the Ets family, form homo, heterodimers and ternary complexes via two distinct domains. Oncogene 16, 3261-3268. 10.1038/sj.onc.1201868 [DOI] [PubMed] [Google Scholar]
- Costa G., Harrington K. I., Lovegrove H. E., Page D. J., Chakravartula S., Bentley K. and Herbert S. P. (2016). Asymmetric division coordinates collective cell migration in angiogenesis. Nat. Cell Biol. 18, 1292-1301. 10.1038/ncb3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Val S., Chi N. C., Meadows S. M., Minovitsky S., Anderson J. P., Harris I. S., Ehlers M. L., Agarwal P., Visel A., Xu S.-M. et al. (2008). Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell 135, 1053-1064. 10.1016/j.cell.2008.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57-74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara N., Carver-Moore K., Chen H., Dowd M., Lu L., O'Shea K. S., Powell-Braxton L., Hillan K. J. and Moore M. W. (1996). Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature 380, 439-442. 10.1038/380439a0 [DOI] [PubMed] [Google Scholar]
- Fish J. E., Wythe J. D., Xiao T., Bruneau B. G., Stainier D. Y. R., Srivastava D. and Woo S. (2011). A Slit/miR-218/Robo regulatory loop is required during heart tube formation in zebrafish. Development 138, 1409-1419. 10.1242/dev.060046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulds C. E., Nelson M. L., Blaszczak A. G. and Graves B. J. (2004). Ras/mitogen-activated protein kinase signaling activates Ets-1 and Ets-2 by CBP/p300 recruitment. Mol. Cell. Biol. 24, 10954-10964. 10.1128/MCB.24.24.10954-10964.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin R. A., Tordai A., Patel H., Gardner A. M., Johnson G. L. and Gelfand E. W. (1994). Ligation of the T cell receptor complex results in activation of the Ras/Raf-1/MEK/MAPK cascade in human T lymphocytes. J. Clin. Invest. 93, 2134-2140. 10.1172/JCI117209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gory S., Dalmon J., Prandini M. H., Kortulewski T., de Launoit Y. and Huber P. (1998). Requirement of a GT box (Sp1 site) and two Ets binding sites for vascular endothelial cadherin gene transcription. J. Biol. Chem. 273, 6750-6755. 10.1074/jbc.273.12.6750 [DOI] [PubMed] [Google Scholar]
- Guarani V., Deflorian G., Franco C. A., Krüger M., Phng L.-K., Bentley K., Toussaint L., Dequiedt F., Mostoslavsky R., Schmidt M. H. H. et al. (2011). Acetylation-dependent regulation of endothelial Notch signalling by the SIRT1 deacetylase. Nature 473, 234-238. 10.1038/nature09917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellström M., Phng L.-K., Hofmann J. J., Wallgard E., Coultas L., Lindblom P., Alva J., Nilsson A.-K., Karlsson L., Gaiano N. et al. (2007). Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature 445, 776-780. 10.1038/nature05571 [DOI] [PubMed] [Google Scholar]
- Herbert S. P. and Stainier D. Y. R. (2011). Molecular control of endothelial cell behaviour during blood vessel morphogenesis. Nat. Rev. Mol. Cell Biol. 12, 551-564. 10.1038/nrm3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert S. P., Cheung J. Y. M. and Stainier D. Y. R. (2012). Determination of endothelial stalk versus tip cell potential during angiogenesis by H2.0-like homeobox-1. Curr. Biol. 22, 1789-1794. 10.1016/j.cub.2012.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T. and Lempicki R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44-57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Iaccarino G., Ciccarelli M., Sorriento D., Galasso G., Campanile A., Santulli G., Cipolletta E., Cerullo V., Cimini V., Altobelli G. G. et al. (2005). Ischemic neoangiogenesis enhanced by beta2-adrenergic receptor overexpression: a novel role for the endothelial adrenergic system. Circ. Res. 97, 1182-1189. 10.1161/01.RES.0000191541.06788.bb [DOI] [PubMed] [Google Scholar]
- Inoue D. and Wittbrodt J. (2011). One for all--a highly efficient and versatile method for fluorescent immunostaining in fish embryos. PLoS ONE 6, e19713 10.1371/journal.pone.0019713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson L., Franco C. A., Bentley K., Collins R. T., Ponsioen B., Aspalter I. M., Rosewell I., Busse M., Thurston G., Medvinsky A. et al. (2010). Endothelial cells dynamically compete for the tip cell position during angiogenic sprouting. Nat. Cell Biol. 12, 943-953. 10.1038/ncb2103 [DOI] [PubMed] [Google Scholar]
- Jin S.-W., Beis D., Mitchell T., Chen J. N. and Stainier D. Y. (2005). Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development 132, 5199-5209. 10.1242/dev.02087 [DOI] [PubMed] [Google Scholar]
- Kawakami K., Takeda H., Kawakami N., Kobayashi M., Matsuda N. and Mishina M. (2004). A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev. Cell 7, 133-144. 10.1016/j.devcel.2004.06.005 [DOI] [PubMed] [Google Scholar]
- Kim K. J., Li B., Winer J., Armanini M., Gillett N., Phillips H. S. and Ferrara N. (1993). Inhibition of vascular endothelial growth factor-induced angiogenesis suppresses tumour growth in vivo. Nature 362, 841-844. 10.1038/362841a0 [DOI] [PubMed] [Google Scholar]
- Kucharska A., Rushworth L. K., Staples C., Morrice N. A. and Keyse S. M. (2009). Regulation of the inducible nuclear dual-specificity phosphatase DUSP5 by ERK MAPK. Cell. Signal. 21, 1794-1805. 10.1016/j.cellsig.2009.07.015 [DOI] [PubMed] [Google Scholar]
- Le Guen L., Karpanen T., Schulte D., Harris N. C., Koltowska K., Roukens G., Bower N. I., van Impel A., Stacker S. A., Achen M. G. et al. (2014). Ccbe1 regulates Vegfc-mediated induction of Vegfr3 signaling during embryonic lymphangiogenesis. Development 141, 1239-1249. 10.1242/dev.100495 [DOI] [PubMed] [Google Scholar]
- Lee S., Chen T. T., Barber C. L., Jordan M. C., Murdock J., Desai S., Ferrara N., Nagy A., Roos K. P. and Iruela-Arispe M. L. (2007). Autocrine VEGF signaling is required for vascular homeostasis. Cell 130, 691-703. 10.1016/j.cell.2007.06.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandoski M., Meyers E. N. and Martin G. R. (1997). Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb. Symp. Quant. Biol. 62, 159-168. 10.1101/SQB.1997.062.01.021 [DOI] [PubMed] [Google Scholar]
- Liu F. and Patient R. (2008). Genome-wide analysis of the zebrafish ETS family identifies three genes required for hemangioblast differentiation or angiogenesis. Circ. Res. 103, 1147-1154. 10.1161/CIRCRESAHA.108.179713 [DOI] [PubMed] [Google Scholar]
- Liu D., Evans I., Britton G. and Zachary I. (2008). The zinc-finger transcription factor, early growth response 3, mediates VEGF-induced angiogenesis. Oncogene 27, 2989-2998. 10.1038/sj.onc.1210959 [DOI] [PubMed] [Google Scholar]
- Livak K. J. and Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402-408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lobov I. B., Renard R. A., Papadopoulos N., Gale N. W., Thurston G., Yancopoulos G. D. and Wiegand S. J. (2007). Delta-like ligand 4 (Dll4) is induced by VEGF as a negative regulator of angiogenic sprouting. Proc. Natl. Acad. Sci. USA 104, 3219-3224. 10.1073/pnas.0611206104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza M. A., Capozzi L. A., Xu Y., McCullough L. D. and Liu F. (2013). Knockout of vascular early response gene worsens chronic stroke outcomes in neonatal mice. Brain Res. Bull. 98, 111-121. 10.1016/j.brainresbull.2013.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehler T. M., Sauer S., Witzel M., Andrulis M., Garcia-Vallejo J. J., Grobholz R., Willhauck-Fleckenstein M., Greiner A., Goldschmidt H. and Schwartz-Albiez R. (2008). Involvement of alpha 1-2-fucosyltransferase I (FUT1) and surface-expressed Lewis(y) (CD174) in first endothelial cell-cell contacts during angiogenesis. J. Cell. Physiol. 215, 27-36. 10.1002/jcp.21285 [DOI] [PubMed] [Google Scholar]
- Nicoli S., Knyphausen C.-P., Zhu L. J., Lakshmanan A. and Lawson N. D. (2012). miR-221 is required for endothelial tip cell behaviors during vascular development. Dev. Cell 22, 418-429. 10.1016/j.devcel.2012.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninov N., Borius M. and Stainier D. Y. R. (2012). Different levels of Notch signaling regulate quiescence, renewal and differentiation in pancreatic endocrine progenitors. Development 139, 1557-1567. 10.1242/dev.076000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A.-K., Dimberg A., Kreuger J. and Claesson-Welsh L. (2006). VEGF receptor signalling - in control of vascular function. Nat. Rev. Mol. Cell Biol. 7, 359-371. 10.1038/nrm1911 [DOI] [PubMed] [Google Scholar]
- Parsons M. J., Pisharath H., Yusuff S., Moore J. C., Siekmann A. F., Lawson N. and Leach S. D. (2009). Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech. Dev. 126, 898-912. 10.1016/j.mod.2009.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham V. N., Lawson N. D., Mugford J. W., Dye L., Castranova D., Lo B. and Weinstein B. M. (2007). Combinatorial function of ETS transcription factors in the developing vasculature. Dev. Biol. 303, 772-783. 10.1016/j.ydbio.2006.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phng L.-K., Potente M., Leslie J. D., Babbage J., Nyqvist D., Lobov I., Ondr J. K., Rao S., Lang R. A., Thurston G. et al. (2009). Nrarp coordinates endothelial Notch and Wnt signaling to control vessel density in angiogenesis. Dev. Cell 16, 70-82. 10.1016/j.devcel.2008.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin A.-L., Houle F., Guillonneau M., Paquet E. R., Simard M. J. and Huot J. (2012). miR-20a represses endothelial cell migration by targeting MKK3 and inhibiting p38 MAP kinase activation in response to VEGF. Angiogenesis 15, 593-608. 10.1007/s10456-012-9283-z [DOI] [PubMed] [Google Scholar]
- Prahst C., Kasaai B., Moraes F., Jahnsen E. D., Larrivee B., Villegas D., Pardanaud L., Pibouin-Fragner L., Zhang F., Zaun H. C. et al. (2014). The H2.0-like homeobox transcription factor modulates yolk sac vascular remodeling in mouse embryos. Arterioscler. Thromb. Vasc. Biol. 34, 1468-1476. 10.1161/ATVBAHA.114.303626 [DOI] [PubMed] [Google Scholar]
- Proulx K., Lu A. and Sumanas S. (2010). Cranial vasculature in zebrafish forms by angioblast cluster-derived angiogenesis. Dev. Biol. 348, 34-46. 10.1016/j.ydbio.2010.08.036 [DOI] [PubMed] [Google Scholar]
- Randi A. M., Sperone A., Dryden N. H. and Birdsey G. M. (2009). Regulation of angiogenesis by ETS transcription factors. Biochem. Soc. Trans. 37, 1248-1253. 10.1042/BST0371248 [DOI] [PubMed] [Google Scholar]
- Rodríguez C. I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A. F. and Dymecki S. M. (2000). High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25, 139-140. 10.1038/75973 [DOI] [PubMed] [Google Scholar]
- Roman B. L., Pham V. N., Lawson N. D., Kulik M., Childs S., Lekven A. C., Garrity D. M., Moon R. T., Fishman M. C., Lechleider R. J. et al. (2002). Disruption of acvrl1 increases endothelial cell number in zebrafish cranial vessels. Development 129, 3009-3019. [DOI] [PubMed] [Google Scholar]
- Roukens M. G., Alloul-Ramdhani M., Baan B., Kobayashi K., Peterson-Maduro J., van Dam H., Schulte-Merker S. and Baker D. A. (2010). Control of endothelial sprouting by a Tel-CtBP complex. Nat. Cell Biol. 12, 933-942. 10.1038/ncb2096 [DOI] [PubMed] [Google Scholar]
- Sacilotto N., Monteiro R., Fritzsche M., Becker P. W., Sanchez-Del-Campo L., Liu K., Pinheiro P., Ratnayaka I., Davies B., Goding C. R. et al. (2013). Analysis of Dll4 regulation reveals a combinatorial role for Sox and Notch in arterial development. Proc. Natl. Acad. Sci. USA 110, 11893-11898. 10.1073/pnas.1300805110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacilotto N., Chouliaras K. M., Nikitenko L. L., Lu Y. W., Fritzsche M., Wallace M. D., Nornes S., García-Moreno F., Payne S., Bridges E. et al. (2016). MEF2 transcription factors are key regulators of sprouting angiogenesis. Genes Dev. 30, 2297-2309. 10.1101/gad.290619.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz H., Engel K. and Gaestel M. (1997). PMA-induced activation of the p42/44ERK- and p38RK-MAP kinase cascades in HL-60 cells is PKC dependent but not essential for differentiation to the macrophage-like phenotype. J. Cell. Physiol. 173, 310-318. [DOI] [PubMed] [Google Scholar]
- Schweighofer B., Testori J., Sturtzel C., Sattler S., Mayer H., Wagner O., Bilban M. and Hofer E. (2009). The VEGF-induced transcriptional response comprises gene clusters at the crossroad of angiogenesis and inflammation. Thromb. Haemost. 102, 544-554. 10.1160/th08-12-0830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel J. J. and Graves B. J. (2002). An ERK2 docking site in the Pointed domain distinguishes a subset of ETS transcription factors. Genes Dev. 16, 127-137. 10.1101/gad.950902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj N., Kedage V. and Hollenhorst P. C. (2015). Comparison of MAPK specificity across the ETS transcription factor family identifies a high-affinity ERK interaction required for ERG function in prostate cells. Cell Commun. Signal. 13, 12 10.1186/s12964-015-0089-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrocks A. D. (2001). The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2, 827-837. 10.1038/35099076 [DOI] [PubMed] [Google Scholar]
- Shin M., Beane T., Quillien A., Male I., Zhu L. J. and Lawson N. D. (2016). Vegfa signals through ERK to promote angiogenesis, but not artery differentiation. Development. 143, 3796-3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchting S., Freitas C., le Noble F., Benedito R., Breant C., Duarte A. and Eichmann A. (2007). The Notch ligand Delta-like 4 negatively regulates endothelial tip cell formation and vessel branching. Proc. Natl. Acad. Sci. USA 104, 3225-3230. 10.1073/pnas.0611177104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testori J., Schweighofer B., Helfrich I., Sturtzel C., Lipnik K., Gesierich S., Nasarre P., Hofer-Warbinek R., Bilban M., Augustin H. G. et al. (2011). The VEGF-regulated transcription factor HLX controls the expression of guidance cues and negatively regulates sprouting of endothelial cells. Blood 117, 2735-2744. 10.1182/blood-2010-07-293209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffoli S., Delaive E., Dieu M., Feron O., Raes M. and Michiels C. (2009). NDRG1 and CRK-I/II are regulators of endothelial cell migration under Intermittent Hypoxia. Angiogenesis 12, 339-354. 10.1007/s10456-009-9156-2 [DOI] [PubMed] [Google Scholar]
- Ubezio B., Blanco R. A., Geudens I., Stanchi F., Mathivet T., Jones M. L., Ragab A., Bentley K. and Gerhardt H. (2016). Synchronization of endothelial Dll4-Notch dynamics switch blood vessels from branching to expansion. Elife 5, e12167 10.7554/eLife.12167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayaraj P., Le Bras A., Mitchell N., Kondo M., Juliao S., Wasserman M., Beeler D., Spokes K., Aird W. C., Baldwin H. S. et al. (2012). Erg is a crucial regulator of endocardial-mesenchymal transformation during cardiac valve morphogenesis. Development 139, 3973-3985. 10.1242/dev.081596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Nakayama M., Pitulescu M. E., Schmidt T. S., Bochenek M. L., Sakakibara A., Adams S., Davy A., Deutsch U., Lüthi U. et al. (2010). Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature 465, 483-486. 10.1038/nature09002 [DOI] [PubMed] [Google Scholar]
- Wang J., Xiao Y., Hsu C.-W., Martinez-Traverso I. M., Zhang M., Bai Y., Ishii M., Maxson R. E., Olson E. N., Dickinson M. E. et al. (2016). Yap and Taz play a crucial role in neural crest-derived craniofacial development. Development 143, 504-515. 10.1242/dev.126920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk B., Hagman J. and Gutierrez-Hartmann A. (1998). Ets transcription factors: nuclear effectors of the Ras-MAP-kinase signaling pathway. Trends Biochem. Sci. 23, 213-216. 10.1016/S0968-0004(98)01211-0 [DOI] [PubMed] [Google Scholar]
- Westerfield M. (2007). The Zebrafish Book. A guide for the Laboratory Use of Zebrafish (Danio rerio), 5th edn. Eugene: University of Oregon Press. [Google Scholar]
- Wythe J. D., Jurynec M. J., Urness L. D., Jones C. A., Sabeh M. K., Werdich A. A., Sato M., Yost H. J., Grunwald D. J., MacRae C. A. et al. (2011). Hadp1, a newly identified pleckstrin homology domain protein, is required for cardiac contractility in zebrafish. Dis. Model. Mech. 4, 607-621. 10.1242/dmm.002204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wythe J. D., Dang L. T. H., Devine W. P., Boudreau E., Artap S. T., He D., Schachterle W., Stainier D. Y. R., Oettgen P., Black B. L. et al. (2013). ETS factors regulate Vegf-dependent arterial specification. Dev. Cell 26, 45-58. 10.1016/j.devcel.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yordy J. S. and Muise-Helmericks R. C. (2000). Signal transduction and the Ets family of transcription factors. Oncogene 19, 6503-6513. 10.1038/sj.onc.1204036 [DOI] [PubMed] [Google Scholar]
- Yuan L., Sacharidou A., Stratman A. N., Le Bras A., Zwiers P. J., Spokes K., Bhasin M., Shih S.-C., Nagy J. A., Molema G. et al. (2011). RhoJ is an endothelial cell-restricted Rho GTPase that mediates vascular morphogenesis and is regulated by the transcription factor ERG. Blood 118, 1145-1153. 10.1182/blood-2010-10-315275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L., Le Bras A., Sacharidou A., Itagaki K., Zhan Y., Kondo M., Carman C. V., Davis G. E., Aird W. C. and Oettgen P. (2012). ETS-related gene (ERG) controls endothelial cell permeability via transcriptional regulation of the claudin 5 (CLDN5) gene. J. Biol. Chem. 287, 6582-6591. 10.1074/jbc.M111.300236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Day D. S., Ho J. W., Song L., Cao J., Christodoulou D., Seidman J. G., Crawford G. E., Park P. J. and Pu W. T. (2013). A dynamic H3K27ac signature identifies VEGFA-stimulated endothelial enhancers and requires EP300 activity. Genome Res. 23, 917-927. 10.1101/gr.149674.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N., Zhao W.-D., Liu D.-X., Liang Y., Fang W.-G., Li B. and Chen Y.-H. (2011). Inactivation of EphA2 promotes tight junction formation and impairs angiogenesis in brain endothelial cells. Microvasc. Res. 82, 113-121. 10.1016/j.mvr.2011.06.005 [DOI] [PubMed] [Google Scholar]