Abstract
The tear-producing lacrimal gland is a tubular organ that protects and lubricates the ocular surface. The lacrimal gland possesses many features that make it an excellent model in which to investigate tubulogenesis, but the cell types and lineage relationships that drive lacrimal gland formation are unclear. Using single-cell sequencing and other molecular tools, we reveal novel cell identities and epithelial lineage dynamics that underlie lacrimal gland development. We show that the lacrimal gland from its earliest developmental stages is composed of multiple subpopulations of immune, epithelial and mesenchymal cell lineages. The epithelial lineage exhibits the most substantial cellular changes, transitioning through a series of unique transcriptional states to become terminally differentiated acinar, ductal and myoepithelial cells. Furthermore, lineage tracing in postnatal and adult glands provides the first direct evidence of unipotent KRT5+ epithelial cells in the lacrimal gland. Finally, we show conservation of developmental markers between the developing mouse and human lacrimal gland, supporting the use of mice to understand human development. Together, our data reveal crucial features of lacrimal gland development that have broad implications for understanding epithelial organogenesis.
KEY WORDS: Lacrimal gland, Epithelia, Single cell sequencing, Development, Tubulogenesis
Summary: Single-cell sequencing combined with a battery of molecular tools reveals novel features of cellular dynamics and differentiation during lacrimal gland development in mice.
INTRODUCTION
The lacrimal gland secretes tears to provide the aqueous layer of the tear film, which is essential to ocular surface homeostasis. Tears are synthesized by the secretory end units (acini) of the gland and transported to the ocular surface through an interconnected network of ducts. As for most tubular organs, lacrimal gland development requires a combination of morphogenic cues and interactions between distinct cell types such as those between epithelial and mesenchymal lineages to form a functional organ (Dean et al., 2004; Finley et al., 2014; Voronov et al., 2013). However, owing to our very limited understanding of the cellular composition, progenitor populations, tissue dynamics and lineage relationships within the lacrimal gland, as well as many other tubular organs, the mechanisms driving tubulogenesis remain poorly understood.
In the mouse, the lacrimal gland initiates from the conjunctival epithelium at embryonic day (E)13.5, where a single epithelial bud on an invaginating stalk elongates toward the neural crest-derived periocular mesenchyme (Makarenkova et al., 2000). Upon reaching the mesenchyme at E15, the epithelium undergoes successive rounds of arborization to produce a functional gland by postnatal day 14, a time corresponding to the eyes opening (Makarenkova et al., 2000). The mature lacrimal gland is composed of terminally differentiated acinar cells, ductal cells and myoepithelial cells that cooperate to synthesize and secrete tears in response to neuronal stimulation. However, when these distinct epithelial cells arise during lacrimal gland development and how their transcriptional profiles differ from one another is unknown. Furthermore, the progenitors that maintain these cell types have yet to be discovered. Over the past decade, several studies have attempted to identify lacrimal gland stem cells in order to find strategies to harvest these populations for the regeneration of the adult lacrimal gland (Gromova et al., 2016; Makarenkova and Dartt, 2015; You et al., 2011; Zoukhri et al., 2008). Although these studies suggest the presence of stem cell populations in the lacrimal gland, to date, there is still no direct in vivo evidence of a stem cell population under healthy conditions.
In this study, we have used single cell sequencing and other molecular tools to reveal the cellular composition, cell dynamics and lineage relationships in the developing lacrimal gland. We uncover the presence of cell lineages not previously characterized in the developing gland and a striking diversity within its mesenchymal compartment. We specifically focus on the epithelial lineage, characterizing dynamic features of differentiation and maturation in the acinar, ductal and myoepithelial compartments. Furthermore, by lineage tracing distinct epithelial populations, we uncover new characteristics of epithelial homeostasis and provide the first direct evidence of a lacrimal gland progenitor pool. We also show that the fetal human lacrimal gland displays similar markers of epithelial progenitors and differentiation to its murine counterpart, supporting the use of murine tissue to mimic human tubulogenesis. Together, our results highlight the cellular diversity of the lacrimal gland and provide novel insights into epithelial lineage relationships and dynamics in an exocrine organ.
RESULTS
Single cell sequencing illustrates the cellular diversity in the developing lacrimal gland
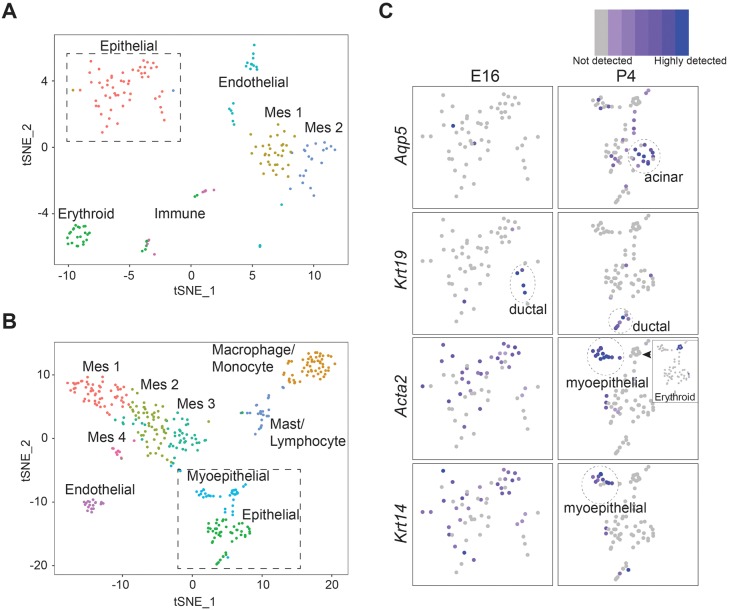
The cellular and molecular composition of the developing lacrimal gland is poorly understood. To define the identity of cell populations during lacrimal gland organogenesis, we performed single cell mRNA sequencing at two time points that span key morphological changes during lacrimal gland development: E16, when lacrimal glands have undergone an initial round of epithelial branching to create future acinar and ductal structures; and P4, when structural features of acini and ducts become recognizable (Finley et al., 2014). Wild-type E16 and P4 lacrimal glands were isolated, dissociated into single cells and subjected to Drop-seq based on the 10× Genomics sequencing platform (Macosko et al., 2015). After filtering was applied, we forwarded 176 cells from E16 lacrimal glands and 359 cells from the P4 timepoint for further analysis. E16 and P4 filtered cells had a mean of 1486 genes and 1388 genes per cell, and 4406 and 3806 unique reads per cell, respectively. Unsupervised graph-based nearest-neighbor clustering uncovered several distinct groups at both time points, as visualized by t-distributed Stochastic Neighbor Embedding (t-SNE; Fig. 1A) (Van Der Maaten et al., 2008). Lineage identities were then assigned to each group based on significant enrichment in canonical cell type marker genes (selected gene list reported in Tables S1-S6). At E16, we identified six distinct clusters belonging to the epithelial (Epcam, Krt8 and Krt18), mesenchymal/neural crest (Lum, Col1a1 and Col3a1) and endothelial (Esam, Cav1, Igfbp7 and Sparcl1) cell lineages, as well as to innate immune (Coro1a, Fcer1g, Apoe, Cd52 and Cd68) and erythroid (Hba-a2, Snca and Alas2) cells (Fig. 1A; Figs S1 and S2; Tables S1 and S2). To validate the presence of these cell lineages at E16 using an orthogonal technique, we also identified epithelial (ECAD, EPCAM), mesenchymal (VIM), endothelial (PECAM1) and immune (F4/80) cell lineages by immunohistological analysis (Fig. S3). In addition, at single-cell resolution, the mesenchymal cell lineage was divided into two distinct but neighboring clusters (MES1 and MES2; Fig. 1A). Although both populations expressed common mesenchymal markers (Vim, Lum and Col1a1), differential expression of several genes (Serpinf1 and Kera) was apparent between the MES1 and the MES2 cluster (Table S2, Fig. S2A). Some genes, such as Gdf10 and Gap43, were restricted to a single mesenchymal cluster (Fig. S2A, Table S2, MES1 or MES2, respectively). These data illustrate a heterogeneity within the mesenchyme early in lacrimal gland development that may predict diversity in mesenchymal function. Thus, even at early stages, the lacrimal gland is composed of multiple distinct cell lineages.
Fig. 1.
Single-cell sequencing reveals the cellular composition, lineage diversity and epithelial lineage relationships in the developing lacrimal gland. (A,B) t-SNE projection of cells isolated from E16 (A) and P4 (B) lacrimal glands. Distinct clusters indicate unique cell populations (see labels). (C) Zoomed images of boxed region of epithelial compartments at E16 (A) and P4 (B), illustrating specific epithelial markers. Inset shows erythroid contamination (Hbba-2) within the myoepithelial compartment.
Based on genetic markers and unsupervised clustering, six groups were identified in E16 lacrimal glands whereas nine distinct groups were detected at P4, suggesting the emergence of distinct populations during postnatal development (Fig. 1B). Significant enrichment of canonical cell type marker genes confirmed the maintenance of epithelial (Krt8 and Epcam), mesenchymal (Ly6a and Mfap5), endothelial (Esam and Cav1) and immune cells (Coro1a and Lsp1) at P4, which was confirmed by immunohistological analysis (Figs S1B and S2B, Tables S3-S5). At P4, lacrimal gland immune cells clustered into two individual groups with gene signatures indicative of lymphocytes/mast cells (Cd52a and Lat) and macrophage/monocytes (Cd14 and Cd68; Fig. S1B and Table S5). In addition, the mesenchyme expanded from two distinct clusters at E16 to four distinct clusters at P4, exemplifying previous unappreciated features of mesenchymal differentiation (Fig. 1B). Although these clusters retained several canonical mesenchymal markers (Vim, Lum and Kera), several genes were significantly enriched within mesenchymal subsets (MES1, Gap43; MES2, Ndufa4l2; MES3, Sec23a; MES4, Ckap2), unveiling new dynamics of mesenchymal differentiation postnatally (Table S3, Fig. S2B). Together, these results suggest an increasing diversity of cell types between early and postnatal lacrimal gland development. Furthermore, we show previously uncharacterized immune contributions, maturation of the vasculature and the existence of distinct subpopulations of mesenchyme within both the embryonic and postnatal lacrimal glands.
Single-cell sequencing provides novel insight into lacrimal gland epithelial differentiation
Given the evidence for the substantial maturation and differentiation of the mesenchymal, immune and endothelial lineages, we carefully dissected features of epithelial differentiation at both E16 and P4. At E16, we observed a single broad cluster marked by Krt8, Krt18, Fxyd3 and Epcam, indicative of largely undifferentiated epithelial cells (Fig. 1A; Table S4, Fig. S4A). Consistent with this, Aqp5, traditionally associated with differentiation towards a secretory lineage, was not readily detected (Fig. 1C). However, cells expressing Krt19, an established ductal-enriched cytokeratin, clustered away from the general epithelial population, suggesting the commitment of early and possibly immature ductal cells as early as E16 (Fig. 1C). In addition, markers of the myoepithelial lineage, Krt14 and Acta2, demonstrated comparable patterns of expression throughout the broad epithelial cluster, hinting at possible precursors to the myoepithelial lineage as early as E16 (Fig. 1C).
At P4, we detected canonical markers of the three epithelial lineages within the Krt8/Krt18/Fxyd3-positive epithelial cluster (Fig. S5A,B). Intriguingly, acinar markers (Aqp5 and Pip; Fig. 1C and Fig. S5B) (Mirels et al., 1998) were closely associated with the myoepithelial cell cluster (expressing Acta2 and Krt14; Fig. 1B,C), suggesting the possibility of a common lineage between these two epithelial cell types. Myoepithelial cells also expressed genes associated with myoepithelial shape and behavior (Cnn1 and Actg2), an early sign of myoepithelial cell maturation (Fig. S4B). We noted that this cluster exhibited some erythroid cell contamination (marked by Hbba-2); however, importantly, the distinct gene expression signature for erythroid cells did not overlap with Krt14+Acta2+ cells (Fig. 1C). Similar to observations at E16, cells expressing markers of the ductal lineage (Krt19) grouped together. In addition, Sftpd, which is enriched in the ducts of the adult human lacrimal gland (Madsen et al., 2000), was readily detected within this cluster, further validating these as ductal cells (Fig. S5C). Thus, using single-cell analysis, we have defined distinct epithelial populations within the developing lacrimal gland and described dynamic changes in their differentiation status between embryonic and postnatal development.
RNA analysis highlights epithelial dynamics
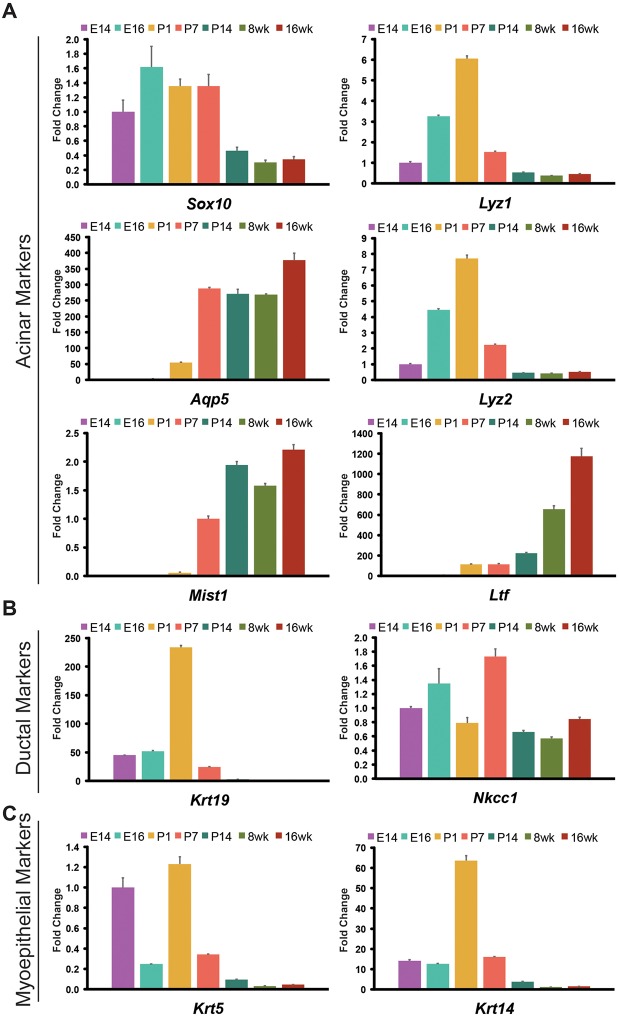
To confirm the gradual differentiation of epithelial cells during lacrimal gland development, we measured transcriptional changes in a panel of genes implicated in glandular epithelial cell differentiation from E14 to adulthood (Fig. 2). We prioritized three genes, Sox10, Aqp5 and Mist1 (Bhlha15 – Mouse Genome Informatics), which were previously associated with acinar cells in the lacrimal gland and other glandular organs, to evaluate acinar cell differentiation. SOX10 is a transcription factor enriched in end buds of developing lacrimal glands where it acts downstream of FGF10 and SOX9 to promote acinar cell formation (Chen et al., 2014). In adult murine lacrimal glands, AQP5, a water channel involved in fluid secretion, and MIST1, a transcriptional regulator of the secretory program, label acinar cells, but their developmental expression patterns are unknown (Ishida et al., 1997; Pin et al., 2000, 2001). As expected, Sox10 was readily detected throughout early embryonic stages of lacrimal gland development, but its expression decreased substantially after P7 (Fig. 2A). Consistent with Aqp5 and Mist1 marking the gradual acquisition of the secretory program, transcripts of both genes, but particularly Mist1, were upregulated after P1 and reached a homeostatic level after P14, coinciding with eyes opening (Fig. 2A). Likewise, levels of Ltf, a secreted protein important for lacrimal gland function, were low at early stages of lacrimal gland development and increased dramatically in the adult lacrimal gland (Janssen and van Bijsterveld, 1983) (Fig. 2A). However, unlike Aqp5 and Mist1, Ltf levels continuously increased as mice aged. Although Lyz1 and Lyz2 are also important secretory proteins for proper ocular homeostasis and have been used as functional markers for the lacrimal gland, their expression peaked at P1, rather than at the later time points assessed (Janssen and van Bijsterveld, 1983; Klaeger et al., 1999) (Fig. 2A). Interestingly, our single-cell analysis at P4 indicated that transcripts of Lyz2 were enriched specifically within the macrophage/monocyte cell lineages (Fig. S1B), suggesting that lysozyme RNA was contributed, at least in part, by non-epithelial cell types in isolated lacrimal glands.
Fig. 2.
Gene expression analysis reveals dynamic expression of epithelial markers during lacrimal gland development. All samples were normalized to E14 lacrimal glands unless otherwise indicated. (A) qPCR analysis of acinar cell markers indicates early expression of Sox10, and gradual acquisition of Aqp5 and Mist1. Levels of Ltf, a marker of mature acini, increased steadily as mice aged. We found Lyz1 and Lyz2, previously considered to be produced by adult acinar cells, were expressed early but levels were reduced over time. (B) Krt19 (normalized to 8-week lacrimal glands) was robustly expressed early in lacrimal gland development but decreased at later stages, whereas Nkcc1 displayed variable and dynamic expression until adult stages. (C) Myoepithelial markers Krt5 and Krt14 (normalized to 16-week lacrimal glands) peaked at P1 and steadily decreased at later stages. All qPCR experiments were completed in biological triplicates (data are mean±s.e.m.).
In contrast to markers of the acinar lineage, ductal markers were abundant during embryonic stages but decreased substantially after birth, a time coinciding with rapid expansion of the end buds (Fig. 2B). Krt19 was readily detected embryonically and peaked by P1 before it declined continuously at later time points (Fig. 2B) (Bouwens et al., 1994). Slc12a2 (also known as Nkcc1), a sodium/potassium chloride co-transporter important for fluid secretion that is enriched in the ducts of the adult lacrimal gland (Evans et al., 2000; Walcott et al., 2005), was dynamically expressed during early stages of lacrimal gland development (Fig. 2B). However, unlike Krt19, levels of Nkcc1 transcripts remained constant at later stages (Fig. 2B). In addition to ductal markers, we also assessed expression of Krt5 and Krt14, which are commonly associated with basal epithelial and myoepithelial cells (Hirayama et al., 2016; Makarenkova and Dartt, 2015). These showed abundant transcript levels during embryonic development, with a dramatic peak in expression at P1 that quickly decreased at later stages of development (Fig. 2C). Together, these results highlight the dynamic nature of epithelial differentiation during the acquisition of a mature functional organ and suggest a transition from an embryonic gland primarily composed of ductal structures to an adult gland predominately composed of acini interconnected through the ductal system.
Spatiotemporal analysis confirms progressive acinar differentiation
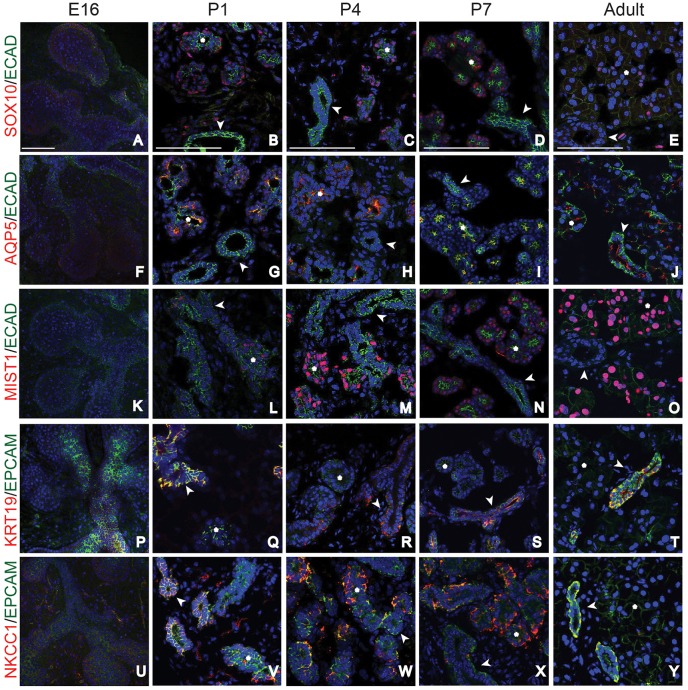
To confirm the spatiotemporal appearance of the epithelial cell types inferred from our gene expression results, we performed immunofluorescent analysis of the lacrimal gland at multiple developmental stages (Fig. 3, Fig. S6). We first assessed the expression of the acinar/secretory cell markers SOX10, AQP5 and MIST1 at each time point. Although SOX10 has been previously reported to be localized to the end buds of early lacrimal glands (Chen et al., 2014), we did not detect robust nuclear expression of SOX10 protein at E16 despite our qPCR analysis, indicating the presence of transcripts (compare Fig. 3A with Fig. 2A). Similarly, at E16, AQP5 was lowly expressed (Fig. 3F) and MIST1 was absent in the epithelia (Fig. 3K). The low expression or absence of these proteins confirmed the limited epithelial differentiation observed in E16 glands by single cell analysis (Fig. 1A). However, by P1 both SOX10 and AQP5 protein were readily detected (Fig. 3B,G) and MIST1+ acinar cells were apparent by P4 (Fig. 3L,M). Interestingly, AQP5 localization changed within the acinar compartment during postnatal development, correlating with epithelial polarization. AQP5 protein was located throughout the acinar cell plasma membrane at P1, but became restricted to the apical membrane by P7 (compare Fig. 3G with I). Based on these findings, we conclude that acinar cell differentiation and the beginning of the secretory program occur at or before P1 and P4, respectively.
Fig. 3.
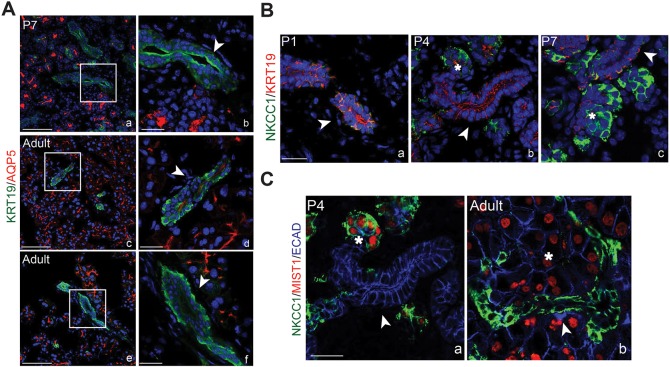
Spatiotemporal analysis reveals dynamic patterns of acinar and duct markers during lacrimal gland development. (A-E) E16 to adult lacrimal glands were immunostained for acinar and ductal markers. SOX10 was readily detected in acinar cells by P1, but slowly declined at later stages of lacrimal gland development. (F-J) AQP5 expression increased at postnatal stages, was prominent in acinar cells before P4, and was apically restricted in acini and small ducts by P7. (K-O) MIST1 was expressed in some acinar cells by P4, and in most acinar cells by P7 and adulthood. (P-T) KRT19 marked ductal cells at all time points evaluated. (U-Y) NKCC1 was broadly expressed at E16, but switched between ductal and acinar compartments between P1 and P7. By adulthood, it was specifically expressed in ducts. Asterisks mark acini and arrowheads mark ducts. Scale bars: 100 μm.
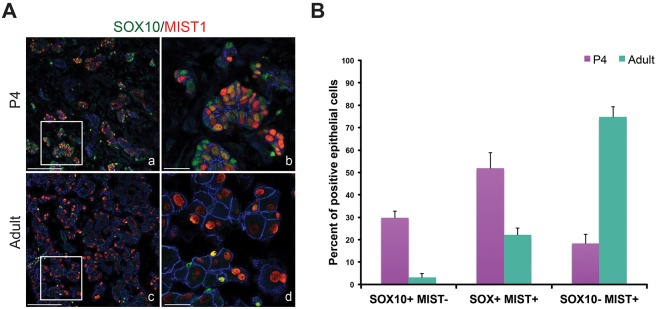
To further investigate the establishment of functional acinar cells, we analyzed expression patterns of SOX10 and MIST1 from P4 to adult stages. We found that SOX10 expression gradually decreased over time, whereas there was a corresponding increase in MIST1+ cells, suggesting that SOX10+ precursors were differentiating. Immunofluorescent analysis of MIST1 and SOX10 at P4 confirmed distinct acinar cell populations that were positive for MIST1 or SOX10, or both (Fig. 4A). Furthermore, we also observed that SOX10+ cells deficient in MIST1 were localized at the border of acini adjacent to the mesenchyme, whereas MIST1-containing cells were located throughout the epithelium (Fig. 4A). These three cell populations were conserved in adult lacrimal glands but they exhibited distinct cell numbers between P4 and adult stages. Whereas 30% of ECAD+ acinar cells were SOX10+MIST1− and 20% were SOX10−MIST1+ at P4 (Fig. 4B), in adult lacrimal glands, 75% were SOX10−MIST1+ and only 3% were SOX10+MIST1−. Thus, these data suggest that SOX10+ cells may undergo a transitory SOX10+MIST1+ state to become mature SOX10−MIST1+ acinar cells. Combined with the gene expression analysis, these data indicate that the acinar lineage is highly dynamic with the rapid conversion of SOX10+ precursors into differentiated MIST1+ secretory cells during postnatal development.
Fig. 4.
SOX10 and MIST1 mark subpopulations of acinar cells in developing and adult acini. (A) SOX10 and MIST1 co-staining revealed the subpopulations of acinar cells at P4 and adult stages. Three types of epithelial cells were readily detected in acini, including SOX10+MIST1−, SOX10+MIST1+ and SOX10−MIST1+ cells. b,d are magnifications of boxed regions in a,c, respectively. Scale bars: 100 μm in a,c; 25 μm in b,d. (B) Quantification of the epithelial cells expressing SOX10 and MIST1 at P4 (n=3) and adult stages (n=3) (data are mean±s.e.m.).
Spatiotemporal analysis reveals unexpected characteristics of ductal differentiation
Ducts are essential for transport of tears to the ocular surface. However, very little is known about their development or maturation. Our single cell analysis confirmed that ductal and acinar cells form unique populations by P4 but features of ductal cell development, including the acquisition of a secretory phenotype, remain unclear (Fig. 1C). Immunostaining confirmed KRT19 is an early and stable marker of the ductal lineage, with expression detected from E16 to adult stages (Fig. 3P-T). To determine the timing of duct cell differentiation, we immunostained for AQP5 and NKCC1. AQP5 has been previously documented in ducts of adult lacrimal glands (Ishida et al., 1997) but the dynamics of its expression have not been reported. We confirmed noticeable expression of AQP5 in many of the ducts of adult lacrimal glands (Fig. 3J) and co-labeling for KRT19 and AQP5 revealed apical AQP5 in most intercalated ductal populations (∼86% of all KRT19+ ducts) (Fig. 5A, Fig. S7). However, at and before P7, AQP5 was largely absent from ducts (27% of all KRT19+ ducts), suggesting that the acquisition of secretory function in duct cells occurs near or at this time point (Fig. 5A, Fig. S7). Consistent with this outcome, NKCC1 expression was robustly detected in ducts after P7 (Fig. 3X,Y). However, it exhibited a unique expression pattern during embryonic and early postnatal development (Fig. 3U-Y). At E16, NKCC1 was detected in both the epithelial and mesenchymal compartments (Fig. 3U), similar to what has been reported in the fetal lung (Brennan et al., 2016). By P1, NKCC1 expression became localized primarily to the KRT19+ ductal compartment of the epithelium, with little expression in the mesenchyme (Fig. 3V, Fig. 5B). Surprisingly, enrichment of NKCC1 switched from the ducts to acinar cells between P1 and P7 (Fig. 3U-X, Fig. 5B). This was further confirmed by co-staining for NKCC1 and MIST1 (Fig. 5C). However, after P7, NKCC1 became more prevalent in the ductal compartment and was absent from acinar cells (Fig. 3Y, Fig. 5C). Thus, NKCC1 is dynamically expressed within mesenchymal and distinct epithelial compartments during development. Together, these data exemplify the diversity of ductal populations in the lacrimal gland as well as the dynamic expression of genes during ductal maturation. In addition, they also suggest that NKCC1 has diverse roles during lacrimal gland development.
Fig. 5.
AQP5 and NKCC1 are dynamically expressed in the epithelium during lacrimal gland development. (A) Immunolabeling for AQP5 and KRT19 in P7 (a,b) and adult (c-f) lacrimal glands showed that AQP5 was expressed in small ducts after P7. However, AQP5 was absent in large ducts (e). Scale bars: 100 μm in a,c,e; 25 μm in b,d,f. b, d and f are higher magnification images of boxed regions in a, c and e, respectively. (B) NKCC1 expression switched between duct and acinar cells from P1 to adult stages. At P1, NKCC1 expression overlapped with KRT19 (a), but less overlap was apparent at P4 and P7 (b,c). (C) Co-staining with MIST1 confirmed the expression of NKCC1 in acini at P4 (a), but an exclusive switch back to the MIST1− ductal cells in adult lacrimal gland (b). Scale bar: 25 μm. Arrowheads indicate ducts and asterisks indicate acini.
Novel insights into myoepithelial cell differentiation and origin
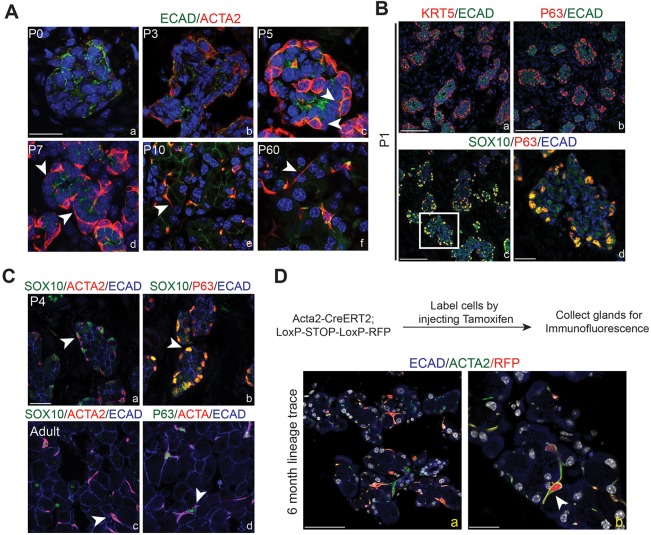
The dynamics of myoepithelial cell development and the origin of the myoepithelial cell lineage in the lacrimal gland remain unclear. Our single cell analysis suggested the presence of a differentiated myoepithelial lineage by P4, as evident by the emergence of a distinct epithelial cluster that was Epcam+, Acta2+ and Krt14+ (Fig. 1B, Fig. S4B). These data were consistent with past reports that described ACTA2+ epithelial cells as early as P3 in lacrimal glands (Wang et al., 1995). To define the precise timing of myoepithelial cell differentiation and maturation, lacrimal glands were imaged using ACTA2 to visualize myoepithelial cell morphology. ACTA2+ epithelial cells were undetected at P1 in lacrimal glands, consistent with low expression of Acta2 before P3 (Fig. 6A). By P3, ACTA2-expressing myoepithelial cells were clearly observed at the borders of the acini of the lacrimal gland. However, these cells appeared morphologically indistinguishable from neighboring epithelial cells, with round nuclei and no processes (Fig. 6A). By P5, myoepithelial cells exhibited membrane extensions as well as more compact nuclei (Fig. 6A), and the number and length of processes increased over time. In P10 lacrimal glands, myoepithelial cells had acquired their classical stellate shape with condensed nuclei and prominent, narrow processes stretching across acini. These features were maintained in the myoepithelial cells of adult lacrimal glands (Fig. 6A). Thus, while markers of myoepithelial identity were apparent at P3, morphological features of myoepithelial cells were gradually acquired and complete by eye opening.
Fig. 6.
Myoepithelial cells are present early in postnatal development but do not contribute to other epithelial lineages in the lacrimal gland. (A) Imaging of myoepithelial cell emergence. ACTA2 protein was not observed at P0 (a), but was detected by P3 (b) in rounded cells within the acini bordering the mesenchyme. Myoepithelial cells began to emerge from the columnar epithelium by P5 (c). By P7 (d), these cells began to radiate processes, assuming their stellate mature appearance by P10 (e), which was maintained in adult glands (f). Arrowheads mark processes. Scale bar: 25 μm. (B) KRT5 (a) and P63 (b) were expressed at P1 in cells of acini immediately adjacent to the mesenchyme. P63+ cells also expressed SOX10 (c). High-magnification image in d (from boxed region in c) confirmed co-expression of these two markers Scale bars: 100 μm in a-c; 25 μm in d. (C) By P4, SOX10+ cells at the border of acini co-expressed ACTA2 (a) and P63 (b). Likewise in adult lacrimal glands, ACTA2+ECAD+ myoepithelial cells remained SOX10+ (c) and P63+ (d). Arrowheads indicate positive cells. (D) Genetic lineage tracing using the Acta2 promoter indicated that myoepithelial cells do not contribute to acinar or ductal lineages. Recombination was induced by tamoxifen injection in adult Acta2CreERT2;Rosa26RFP mice, and cells were traced for 6 months. RFP+ cells co-expressed ACTA2 (a). b is a higher magnification image of the boxed area. Scale bars: 100 μm in a; 25 μm in b. Arrowhead indicates positive cells.
Next, we addressed the origin of the myoepithelial lineage. Although ACTA2 protein was not detected at E16 (data not shown), our single-cell sequencing analysis detected Acta2 mRNA in Krt14+ cells as early E16, suggesting myoepithelial cells might be specified earlier than when protein is detectable. To determine whether the myoepithelial lineage is present prior to ACTA2 protein expression, we took advantage of other notable markers of myoepithelial cells, including KRT5, KRT14 and P63 (Hirayama et al., 2016; Makarenkova and Dartt, 2015). Our qPCR analysis indicated that Krt5 and Krt14 peaked at P1, hinting that myoepithelial cells may be committed earlier than P3 (Fig. 2C). Furthermore, immunostaining confirmed the presence of both KRT5+ECAD+ and P63+ECAD+ cells at the border of P1 acini (Fig. 6B). This localization was reminiscent of SOX10+MIST1− cells previously observed above (Fig. 4A). Indeed, co-staining of P63 and SOX10 uncovered high SOX10 expression in P63+ cells at P1 (Fig. 6B). Likewise, as expected, KRT14+ cells bordering acini robustly expressed SOX10 (∼95% of KRT14+ expressed SOX10) (Fig. S8). SOX10+P63+ cells were also present at P4 and expressed ACTA2, establishing these as true myoepithelial cells (Fig. 6C). Imaging adult lacrimal glands also indicated the maintenance of both SOX10 and P63 in myoepithelial cells (Fig. 6C). Together, these data and our single-cell sequencing data provide strong evidence that the emergence of the myoepithelial lineage occurs before ACTA2 protein expression and that myoepithelial precursors co-express SOX10, P63, KRT14 and KRT5. Furthermore, the co-expression of SOX10 in acinar cells and myoepithelial cells suggest a lineage relationship between these cell types, illuminating previously unknown characteristics of epithelial cell differentiation and commitment.
Finally, we determined whether mature myoepithelial cells become lineage restricted or whether they are capable of producing ductal or acinar cells. Previous studies have implicated myoepithelial cells as epithelial progenitors, but genetic lineage tracing of these cells has not been reported (Makarenkova and Dartt, 2015; Prater et al., 2014; Zoukhri et al., 2008). We genetically labeled myoepithelial cells in adult RFP reporter mice using the Acta2CreERT2 allele (Wendling et al., 2009). After a 6-month chase, all RFP-labeled cells expressed ACTA2 (∼66±14% of all myoepithelial cells), indicating that myoepithelial cells are long living or capable of replenishing themselves (Fig. 6D). In addition, absent RFP labeling in other epithelial lineages suggests that myoepithelial cells are unlikely contributors to the acinar and ductal lineages, and that other progenitor pools may exist for acinar and ductal cells.
Progenitor potential of the basal cell layer in the developing and adult lacrimal gland
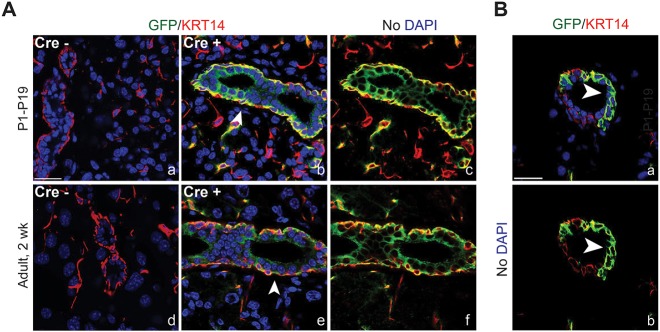
Given the lineage restriction of myoepithelial cells, and the finding that cells expressing Krt5 and Krt14 behave as progenitors in many other epithelial organs (Blanpain and Fuchs, 2009; Papafotiou et al., 2016; Rock et al., 2009), we next determined whether basal cells marked by KRT5 and KRT14 could contribute to all epithelial lineages in the developing and adult lacrimal gland. After confirming that KRT5 (and KRT14) mark a distinct ACTA2-negative basal epithelial population around ducts (Fig. S9), we evaluated the progenitor capacity of these cells using genetic lineage tracing. KRT5+ cells were labeled using mice carrying a Rosa26mTmG reporter and a Krt5CreERT2 allele at either P1 or P60, and traced until P19 or P74, respectively (Indra et al., 1999; Muzumdar et al., 2007). In the absence of Cre, no GFP expression was detected (Fig. 7A). However, in the presence of Cre, KRT5+ basal cells (and myoepithelial cells) were successfully labeled with GFP. At both time points we detected GFP expression in basal and luminal duct cells as well as in myoepithelial cells but not in the acini (Fig. 7A). Furthermore, partial labeling of basal cells indicated the specific contribution of these cells to underlying luminal cells (Fig. 7B), as basal cells that were not labeled did not have underlying GFP+ luminal cells. Furthermore, basal cells maintained GFP expression, highlighting their ability to self-renew. Together, these data establish the first bona fide progenitor pool within the lacrimal gland, and propose a role for unipotent stem cells in lacrimal gland tissue homeostasis.
Fig. 7.
Basal cells are progenitors for ductal, but not acinar, cells. (A) Lineage tracing was performed using Krt5CreERT2;Rosa26mTmG with recombination induced at either P1 (a-c) or adult stages (d-f). In the absence of Cre, no GFP was detected (a,d). However, in young or adult mice, induction of Cre resulted in KRT14+/GFP+ basal cells and GFP+ ductal cells (b,c,e,f). Arrowheads indicate ducts. Scale bar: 25 μm. (B) GFP+ luminal ductal cells were adjacent to GFP+ basal cells but not unlabeled basal cells in partially lineage-traced ducts, suggesting luminal cells are derived from basal cells by asymmetric division. Arrowheads indicate luminal cells derived from asymmetric division. Scale bar: 25 μm.
Molecular profiling of human fetal lacrimal glands
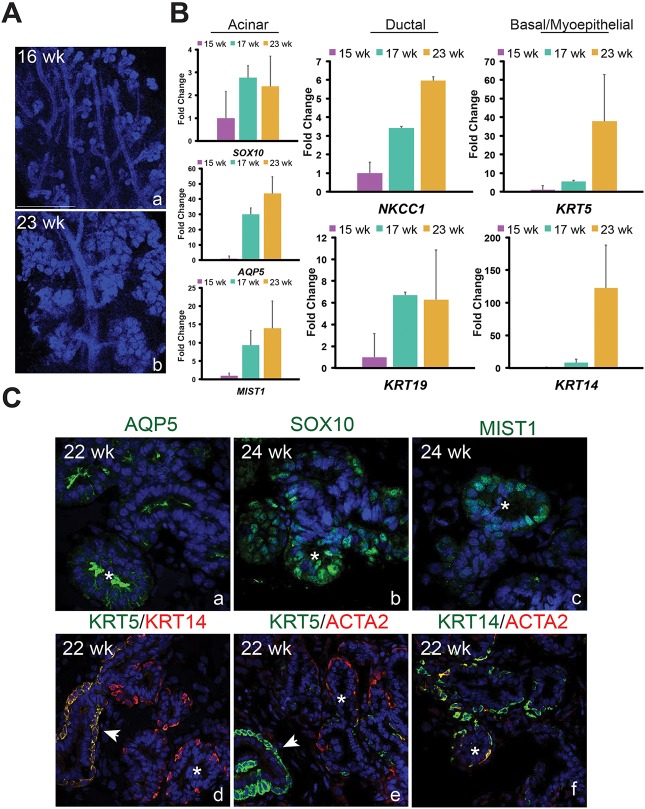
Similar to other organs, whether murine lacrimal gland development recapitulates that of humans remains unclear. Several studies have evaluated adult human lacrimal glands (Paulsen, 2006; Tiwari et al., 2012, 2014). Although the morphological development of the early human lacrimal gland has been described (de la Cuadra-Blanco et al., 2003), to our knowledge no study has investigated the molecular characteristics of the human fetal lacrimal gland or whether its cell identities are similar to the mouse. Such information is crucial to understanding whether epithelial growth and morphogenesis is conserved across species. To further appreciate the scope of the similarities between the two organisms, human lacrimal glands were collected at various stages of development and analyzed for genes we established as differentiation or progenitor markers during mouse lacrimal gland development. Morphologically, the human lacrimal gland is very distinct from its murine counterpart (Fig. 8A). Whereas the mouse lacrimal gland initiates as a single bud and has one primary duct, the human lacrimal gland consists of numerous bud units and multiple large ducts (Fig. 8A). Nonetheless, both undergo extensive branching to form their complex secretory networks (Fig. 8A). Gene expression analysis of human fetal lacrimal glands uncovered comparable trends as observed during mouse development. SOX10 mRNAs were present at 15 weeks whereas AQP5 and MIST1 transcripts were barely detectable (Fig. 8B). However, by 17 weeks, AQP5 and MIST1 were robustly expressed, suggesting that acinar cell differentiation and acquisition of the secretory program occurred at or before this time point. In addition, expression of ductal genes KRT19 and NKCC1 also increased (∼7-fold and ∼3-fold, respectively) from 15 to 17 weeks, as did expression of myoepithelial/progenitor markers KRT5 and KRT14 (5.5- to 8-fold). However, unlike KRT19 and NKCC1, the transcript levels of which stabilized or increased subtly after 17 weeks, there was a dramatic enrichment in KRT5 and KRT14 mRNA by 23 weeks, with an increase of ∼37- and 127-fold, respectively, over 15-week levels (Fig. 8B). A similar leap in Krt5 and Krt14 expression appeared in the mouse tissue from E16 to P1 (Fig. 2C), possibly indicating the expansion of KRT5+ and KRT14+ cell populations that occurs in both species during this period.
Fig. 8.
Phenotypic and molecular features of developing human and murine lacrimal glands are highly conserved. (A) The human fetal lacrimal gland undergoes extensive epithelial branching between 16 weeks (a) and 23 weeks (b). Scale bar: 500 μm. (B) qPCR analysis showed human lacrimal glands express several lineage markers observed in the murine lacrimal gland. All qPCR experiments used three biological replicates (data are mean±s.e.m.). (C) Immunostaining revealed that AQP5, SOX10 and MIST1 localize to the acini of fetal human lacrimal gland. As in mouse tissue, AQP5 was enriched in the apical membrane of acini and small ducts (a). SOX10 and MIST1 were exclusively found in acinar cells (b,c). Unlike in the murine lacrimal gland, KRT5 was barely detectable in myoepithelial cells when compared with basal cells, whereas KRT14 protein levels were similar in both cell types (d-f). Arrowheads indicate ducts and asterisks indicate acini.
Immunofluorescent analysis also revealed conserved features of lacrimal gland development between mice and humans. These included the appearance of ACTA2+ myoepithelial cells exclusively around acini at a time corresponding to the expression of AQP5 (compare Figs 6C and 8C), and the presence of SOX10+ and MIST1+ cells in the developing acini but not the ducts (compare Figs 3 and 8C). In addition, like murine tissue, AQP5 was detected in acini and small ducts (Fig. 8C) but absent in the larger ductal system (data not shown), suggesting human and murine ducts have a similar function. However, we observed distinct expression patterns of KRT5 and KRT14 in humans compared with mice. In both species, KRT5 and KRT14 appeared to be expressed at relatively comparable levels in basal cells (Figs 7A and 8C). However, unlike murine tissue, KRT5 protein in human glands was barely detectable in ACTA2+ myoepithelial cells compared with KRT14 levels (Fig. 8C), suggesting that the requirement of these keratins in myoepithelial cells may differ between species.
DISCUSSION
Our data highlight the large number of cell types and differentiation events that must be coordinated to create complex tubular organs. In this study, we employ a bevy of tools to establish the cell lineages that contribute to embryonic and postnatal lacrimal development. Furthermore, we achieve unprecedented insight into epithelial lineage relationships and the dynamics of differentiation within the developing lacrimal gland. In addition, we show in vivo evidence for a novel progenitor population in the mouse lacrimal gland, and provide the first molecular characterization of the human fetal lacrimal gland. Overall, this body of work represents a major advance in our understanding of lacrimal gland development and lays the foundation for numerous future studies using the lacrimal gland as a model to understand conserved attributes of tubular development.
An increasing number of reports have investigated signaling mechanisms governing branching morphogenesis in the early lacrimal gland (Dean et al., 2004; Makarenkova et al., 2000; Pan et al., 2008, 2010; Qu et al., 2011; Voronov et al., 2013). These studies have revealed multiple mechanisms underlying the initial development of the organ. However, features of late embryonic and postnatal lacrimal gland development, a time frame that encompasses vast morphological changes, and the mechanisms that regulate them, have not been described. Employing single-cell analysis at E16 and P4, we uncovered a surprising diversity of cell lineages present within the developing lacrimal gland, including epithelial, mesenchymal, immune and endothelial lineages. The identification of multiple types of immune cells within the developing gland, as well as the remarkable heterogeneity of the mesenchyme suggests the involvement of as yet undetermined mechanisms regulating developmental programs. Future studies will be necessary to further characterize the diversity and heterogeneity uncovered from our single-cell sequencing data analysis.
We also demonstrate the highly dynamic differentiation of epithelial cells during the late embryonic/early postnatal period. Our data indicate that the commitment of the epithelium to the ductal lineage occurs by E16 but that acquisition of the secretory status of small ducts does not occur until at, or after, P7, with the expression of AQP5 and NKCC1. This time point is consistent with a previous study demonstrating the presence of secretory granules at P7, a feature indicative of secretory function (Wang et al., 1995). However, before this time point we observed dramatic changes in the expression patterns of AQP5 and particularly NKCC1 that have not been revealed before in any organ system and strongly suggest that NKCC1 and AQP5 have developmental roles in addition to their function in secretion.
Acinar cell differentiation has not been described previously in the lacrimal gland. Our data indicate that acinar cells gain a secretory phenotype by P4, as illustrated by the acquisition of MIST1 and apical AQP5. Differentiation of acini may also entail a transition from a SOX10+ progenitor state to a differentiated MIST1+ state. However, it remains possible that MIST1+ acinar cells arise independently of a SOX10+MIST1+ intermediate state, and that SOX10−MIST1+ acinar cells outpopulate its SOX10+ counterparts to contribute to the acinar lineage. Future experiments will be necessary to tease apart these possibilities. Our identification of a small subset of SOX10+MIST+ acinar cells in adult tissue indicates the maintenance of a primitive cell type, which may possibly serve as a progenitor for the acinar lineage. Although MIST1+ cells have been reported to populate the acini of the salivary gland by self-duplication, progenitor characteristics of these cells were not determined (Aure et al., 2015). Thus, whether these SOX10+MIST1+ cells display a distinct progenitor capacity in the lacrimal gland will require future study.
The origin of myoepithelial cells in the lacrimal gland has not been reported. Our single cell and immunofluorescent analysis suggest a lineage relationship between acinar and myoepithelial cells, where SOX10+ cells may give rise to the acinar or myoepithelial lineage, or to both. In support of this differentiation capacity, deletion of Sox10 in the lacrimal gland not only impaired acinar cell development but also resulted in absent myoepithelial cells (Chen et al., 2014). Thus, SOX10 is required for the development of myoepithelial cells in addition to acinar cells. Whether SOX10 marks a common progenitor for both lineages or whether it is expressed by progenitor cells that independently give rise to acinar and myoepithelial cells requires further investigation. Nonetheless, our data strongly suggest a novel connection between these two lineages during lacrimal gland development.
Although several studies have sought to identify progenitor pools in the lacrimal gland, this is the first study to directly identify progenitors in healthy uninjured tissues (Gromova et al., 2016; You et al., 2011; Zoukhri et al., 2008). Contrary to predictions from the literature and the progenitor potential of myoepithelial cells in other organs, we found no evidence for the capacity of the myoepithelial lineage to contribute to other epithelial lineages in adult lacrimal glands (Makarenkova and Dartt, 2015; Prater et al., 2014; Zoukhri et al., 2007, 2008). Rather, the maintenance of labeled cells after extensive chasing suggested that myoepithelial cells replenish themselves. We found that the basal cell layer marked by KRT5 behaves as a unipotent progenitor that contributes to the adult ducts. Our lineage-tracing analysis indicates that these cells divide asymmetrically to produce a basal cell and luminal ductal cell. How these cells behave during injury repair will be interesting to discover. Likewise, whether acinar cells, like ductal cells, have a distinct unipotent progenitor requires future investigation. Overall, these data highlight a major advance in our understanding of lacrimal gland epithelial homeostasis.
Despite distinctions in their structure and the timing of epithelial differentiation, our analyses revealed comparable molecular profiles between developing mouse and human lacrimal glands. This outcome is consistent with the similar morphological changes described for developing human and murine lacrimal gland (de la Cuadra-Blanco et al., 2003). Indeed, our gene expression and immunofluorescent analyses confirm that murine markers can be used to define similar populations in human lacrimal glands, although the majority of differentiation occurs embryonically in humans and postnatally in mice. Thus, the murine lacrimal gland is an excellent model for understanding human lacrimal gland development but further interrogation of the two species is required to elucidate potential differences.
Altogether, this report unveils interesting and unappreciated features of epithelial differentiation and turnover, and reveals novel relationships between epithelial lineages within the lacrimal gland. Given the similarity between exocrine gland types, with the expression of many markers being conserved, e.g. NKCC1, KRT5, KRT14 and MIST1, our results are likely applicable to these other organ systems. We also introduce the use of human embryonic lacrimal glands to test lineage relationships and tissue dynamics of human tubular development. As such, this study provides an excellent framework for the application of lacrimal glands as a tractable model system to investigate fundamental processes of tubular development.
MATERIALS AND METHODS
Mouse lines
All animals procedures were approved by the UCSF Institutional Animal Care and Use Committee (IACUC). Timed CD1 females were purchased from Envigo. Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo (Rosa26mTmG), Tg(Krt5-cre/ERT2)1Blh (Krt5CreERT2), Tg(Acta2-cre/ERT2)12Pcn (Acta2CreERT2) and Gt(ROSA)26Sortm1Hjf (Rosa26Rfp) mice were acquired from The Jackson Laboratory.
Lineage-tracing experiments
For lineage-tracing experiments, adult Acta2CreERT2; Rosa26RFP mice were injected with 5 mg of tamoxifen in corn oil by intraperitoneal injection and euthanized after 6 months. P60 Krt5CreERT2; Rosa26mTmG mice were injected with 2.5 mg/20 g of tamoxifen in corn oil by intraperitoneal injection and euthanized 2 weeks later. P1 Krt5CreERT2; Rosa26mTmG mice were injected in the scruff of the neck with 0.3 mg of tamoxifen in 25 µl of corn oil and euthanized at P19.
Human fetal lacrimal gland tissue isolation
Human fetal lacrimal glands were harvested from post-mortem fetuses between 15 and 24 weeks of gestation with patient consent and permission from the ethical committee of the University of California San Francisco. Harvested lacrimal glands were identified by location and glandular appearance and placed in 4% PFA for immunostaining or RNAlater (Qiagen) for gene expression analysis.
Human fetal lacrimal gland processing
Fixed lacrimal glands were incubated in increasing concentrations of sucrose (25-75%), embedded in OCT and sectioned at 12 µm on a cryostat.
Immunofluorescence
Fresh frozen mouse lacrimal glands were sectioned at 10 µm on a cryostat. Sections were fixed with either 4% paraformaldehyde (PFA) for 10 min at room temperature or with ice-cold acetone/methanol (1:1) for 1 min. PFA-fixed tissues were permeabilized with 0.1% Triton in PBS for 10 min. Slides were blocked for 1 h in 10% chicken serum (Vector Labs) in 0.05% PBS-Tween-20 (PBST) and incubated overnight in primary antibody. Antibodies included: rat anti-E-cadherin (1:300, Life Technologies, 13-1900); rabbit anti-KRT5 (1:1000, Covance, PRB-160P); rat anti-KRT19 (1:300, troma III, DSHB); rabbit anti-KRT14 (1:1000, Covance, PRB-155P); rabbit anti-AQP5 (1:100, Millipore, AB3559); goat anti-SOX10 (1:100, Santa Cruz, sc-17342); goat anti-NKCC1 (1:200, Santa Cruz, sc-21545); rat anti-F4/80 (1:50, AbD Serotec, MCA497R); chicken anti-GFP (1:1000, Abcam, AB13970); rabbit anti-P63 (1:200, Santa Cruz, sc-8343); mouse anti-actin, alpha-smooth muscle (1:400, Sigma, C6198); rabbit anti-vimentin (1:300, Abcam, AB92547); rat anti-PECAM1 (clone MEC13.3, 1:300, BD Pharmigen, 550274); rat anti-EpCAM (1:1000, Biolegend, 118210) and rabbit anti-MIST1 (1:500, a gift from Stephen Konieczny, Purdue University, IN, USA). Sections were then incubated with secondary antibodies from Jackson Laboratories or Life Technologies at 1:300 for 1 h at room temperature and stained with Hoescht 33342 (1:1000) or DAPI (1:4000). Sections were imaged using a Leica SP5 confocal microscope and processed using NIH ImageJ software (Abràmoff et al., 2004). SOX10+ and MIST+ were quantified using the ImageJ software by manually counting ECAD+ cells that were positive for either or both SOX10 and MIST1 using the multi-point tool. Four independent images were quantified and averaged.
Whole-mount lacrimal gland immunofluorescence
Lacrimal glands were stained as previously described (Knox et al., 2010). Briefly, E16 lacrimal glands were fixed with either 4% PFA for 20 min or with ice-cold acetone/methanol (1:1) for 1 min. PFA fixed glands were permeabilized with 0.1% Triton-X for 15 min. Lacrimal glands were then blocked in 10% chicken serum (Vector Labs) and 1% BSA (Sigma Aldrich) in PBST for 3 h at room temperature or overnight at 4°C. Lacrimal glands were incubated with primary antibodies for 2 h at room temperature followed by an overnight incubation at 4°C. Glands were then incubated at room temperature with secondary antibodies from Jackson Laboratories or Life Technologies at 1:300 for 2 h and stained with Hoescht 33342 (1:1000) or DAPI (1:4000). Glands were imaged using a Leica SP5 confocal microscope and processed using NIH ImageJ software (Abràmoff et al., 2004). Higher magnification images in Fig. 3 were processed using ImageJ from images represented in Fig. S6.
Quantitative PCR analysis
RNA was isolated from lacrimal glands using the RNAqueous Micro Kit (Ambion). RNA samples were then DNase treated (Ambion) and used for cDNA synthesis using the SuperScript III First-Strand Synthesis System (Invitrogen). qPCR was performed using 5 ng of mouse cDNA or 3-5 ng of human cDNA and SYBR Green reagents. Primer sequences are listed in Tables S7 and S8. Melt-curves and primer efficiencies were estimated as previously described (Hoffman et al., 2002). Gene expression was normalized to the Rps18 for mouse and GAPDH for human samples. Reactions were run in triplicate.
Single-cell dissociation of lacrimal glands
Murine LGs from timed-pregnant female mice (CD1; E16 and P4) were dissected and pooled together in PBS. A single-cell suspension was created by incubating lacrimal glands in 1 Wünsch unit of Liberase (Roche)/10 ml PBS (Roche) and DNaseI (Roche) at 37°C for 20-40 min. The enzyme reaction was quenched by the addition of fetal calf serum and the solution was filtered through a 40 µm strainer (BD Falcon) and centrifuged at 400 g for 5 min. Cell pellets were washed in PBS, centrifuged and resuspended in 0.04% BSA (ThermoFisher) at a cell density of 1000 cells/µl.
Single-cell sequencing using a drop-seq platform
Cell suspensions at a density of 1000 cells/µl in PBS+0.04% BSA were prepared for single-cell sequencing using the Chromium Single Cell 3′ Reagent Version 1 Kit and Chromium Controller (10× Genomics) as previously described (Zheng et al., 2017). Briefly, 20,000 cells per reaction were loaded for GEM (gel bead-in-emulsion) generation and barcoding. GEM-RT was performed using a Thermocycler (BioRad; 55°C for 2 h, 85°C for 5 min, held at 4°C). Post GEM-RT cleanup and cDNA amplification was performed to isolate and amplify cDNA for library construction. Libraries were constructed using the Chromium Single Cell 3′ Reagent Kit (10× Genomics) and samples were each sequenced in a single lane using the Illumina HiSeq2500 in Rapid Run Mode using a paired-end flow cell: Read1, 98 cycles; Index1, 14 cycles; Index2, 8 cycles; and Read2, 10 cycles. We used the CellRanger v1.1 and v1.2 software with the default settings to process the raw FASTQ files, aligned the sequencing reads to the ensembl release 84 from the GRCm38 genome build using STAR [(Dobin et al., 2013) ftp://ftp.ensembl.org/pub/release-84/gtf/mus_musculus/Mus_musculus.GRCm38.84.gtf.gz] and generated a filtered unique molecular identifier (UMI) expression profile for each cell, as performed previously (Zheng et al., 2017). For the E16 time point, there were on average 171,266 mean reads per cell and 755 genes per cell with a PCR duplication rate of 94.6%. For the P4 time point, we had 133,420 mean reads per cell and 758 mean genes per cell with a PCR duplication rate of 94.6%. PCR duplicates are identified by their unique random barcode in addition to their cell specific barcode and collapsed down to a single count, referred to as a unique molecular identifier (UMI). Only confidently mapped, non-PCR duplicates with valid barcodes and UMIs were used to generate a gene-barcode matrix for further analysis. A list of numbers of genes detected per cell and number, histograms of genes and UMIs per cell, and summary statistics are provided in Tables S9-S12 and Fig. S10-S13.
Data were analyzed using R and the R package Seurat for single cell analysis (R Development Core Team, 2016; Satija et al., 2015). Cells were processed via the Seurat workflow to remove doublets and unwanted sources of variation. The basic filtering removed genes expressed in fewer than three cells and removed cells with less than 200 genes expressed. Cells with greater than 2500 genes expressed were filtered out in order to minimize the incorporation of doublets in the further rounds of analysis. Complete lists of differentially expressed genes are provided in Tables S13 and S14. Principal component analysis was performed to reduce dimensionality on the scaled and log normalized data matrix. The first 10 PCA components were used to cluster the cells using Seurat's innate algorithm, which optimizes a KNN graph based on Euclidean distance in the PCA space and processes the edge weights of pairs of cells based on their local neighbors. tSNE plots were generated to aid in the 2D representation of multidimensional data independent of the clustering algorithm. The ‘bimod’, likelihood-ratio test for single cell gene expression was used for differential gene analysis (McDavid et al., 2013).
Acknowledgements
The authors acknowledge Mr Noel Cruz Pacheco for his laboratory assistance.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: D.T.F., S.M.K.; Methodology: D.T.F., S.N., J.K.F., K.S.Y., E.E., L.E.B., J.B.S., A.D.T., S.M.K.; Software: K.S.Y., J.B.S., A.D.T., S.M.K.; Validation: D.T.F., S.N., S.M.K.; Formal analysis: D.T.F., S.N., J.K.F., K.S.Y., E.E., A.D.T., S.M.K.; Investigation: D.T.F., S.N., S.M.K.; Resources: S.M.K.; Data curation: D.T.F., S.N., J.K.F., K.S.Y., E.E., A.D.T., S.M.K.; Writing - original draft: D.T.F., S.N., M.T.M., S.M.K.; Writing - review & editing: K.S.Y., E.E., L.E.B., J.B.S., A.D.T., S.M.K.; Visualization: D.T.F., S.N., J.K.F., K.S.Y., E.E., A.D.T., S.M.K.; Supervision: M.T.M., A.D.T., S.M.K.; Project administration: S.M.K.; Funding acquisition: S.M.K.
Funding
This work was supported by the National Eye Institute (1R01EY026492-01 to S.M.K.); the National Cancer Institute (1U19CA179513 to D.T.F.); the National Eye Institute (5F32EY025139-02 to J.K.F.); and a graduate research fellowship from the National Science Foundation (to D.T.F.). Deposited in PMC for release after 12 months.
Data availability
Count matrices for 10x Lacrimal Gland scRNA seq data are available in GEO under accession numbers GSE100106, GSM2671415 and GSM2671416.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.150789.supplemental
References
- Abràmoff M. D., Magalhães P. J. and Ram S. J. (2004). Image processing with imageJ. Biophotonics Int. 11, 36-41. [Google Scholar]
- Aure M. H., Konieczny S. F. and Ovitt C. E. (2015). Salivary gland homeostasis is maintained through acinar cell self-duplication. Dev. Cell 33, 231-237. 10.1016/j.devcel.2015.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C. and Fuchs E. (2009). Epidermal homeostasis: a balancing act of stem cells in the skin. Nat. Rev. Mol. Cell Biol. 10, 207-217. 10.1038/nrm2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwens L., Wang R.-N., De Blay E., Pipeleers D. G. and Kloppel G. (1994). Cytokeratins as markers of ductal cell differentiation and islet neogenesis in the neonatal rat pancreas. Diabetes 43, 1279-1283. 10.2337/diab.43.11.1279 [DOI] [PubMed] [Google Scholar]
- Brennan S. C., Wilkinson W. J., Tseng H.-E., Finney B., Monk B., Dibble H., Quilliam S., Warburton D., Galietta L. J., Kemp P. J. et al. (2016). The extracellular calcium-sensing receptor regulates human fetal lung development via CFTR. Sci. Rep. 6, 21975 10.1038/srep21975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Huang J., Liu Y., Dattilo L. K., Huh S.-H., Ornitz D. and Beebe D. C. (2014). FGF signaling activates a Sox9-Sox10 pathway for the formation and branching morphogenesis of mouse ocular glands. Development 141, 2691-2701. 10.1242/dev.108944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cuadra-Blanco C., Peces-Peña M. D. and Mérida-Velasco J. R. (2003). Morphogenesis of the human lacrimal gland. J. Anat. 203, 531-536. 10.1046/j.1469-7580.2003.00233.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean C., Ito M., Makarenkova H. P., Faber S. C. and Lang R. A. (2004). Bmp7 regulates branching morphogenesis of the lacrimal gland by promoting mesenchymal proliferation and condensation. Development 131, 4155-4165. 10.1242/dev.01285 [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M. and Gingeras T. R. (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15-21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R. L., Park K., Turner R. J., Watson G. E., Van Nguyen H., Dennett M. R., Hand A. R., Flagella M., Shull G. E. et al. (2000). Severe impairment of salivation in Na+/K+/2Cl- cotransporter (NKCC1)-deficient mice. J. Biol. Chem. 275, 26720-26726. [DOI] [PubMed] [Google Scholar]
- Finley J. K., Farmer D., Emmerson E., Cruz Pacheco N. and Knox S. M. (2014). Manipulating the murine lacrimal gland. J. Vis. Exp. e51970. 10.3791/51970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromova A., Voronov D. A., Yoshida M., Thotakura S., Meech R., Dartt D. A. and Makarenkova H. P. (2016). Lacrimal gland repair using progenitor cells. Stem Cells Transl. Med. 10.5966/sctm.2016-0191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama M., Liu Y., Kawakita T., Shimmura S. and Tsubota K. (2016). Cytokeratin expression in mouse lacrimal gland germ epithelium. Exp. Eye Res. 146, 54-59. 10.1016/j.exer.2015.11.020 [DOI] [PubMed] [Google Scholar]
- Hoffman M. P. (2002). Gene expression profiles of mouse submandibular gland development: FGFR1 regulates branching morphogenesis in vitro through BMP- and FGF-dependent mechanisms. Development 129, 5767-5778. 10.1242/dev.00172 [DOI] [PubMed] [Google Scholar]
- Indra A. K., Warot X., Brocard J., Bornert J.-M., Xiao J. H., Chambon P. and Metzger D. (1999). Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 27, 4324-4327. 10.1093/nar/27.22.4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida N., Hirai S.-I. and Mita S. (1997). Immunolocalization of aquaporin homologs in mouse lacrimal glands. Biochem. Biophys. Res. Commun. 238, 891-895. 10.1006/bbrc.1997.7396 [DOI] [PubMed] [Google Scholar]
- Janssen P. T. and van Bijsterveld O. P. (1983). Origin and biosynthesis of human tear fluid proteins. Invest. Ophthalmol. Vis. Sci. 24, 623-630. [PubMed] [Google Scholar]
- Klaeger A. J., Cevallos V., Sherman M. D., Whitcher J. P. and Stephens R. S. (1999). Clinical application of a homogeneous colorimetric assay for tear lysozyme. Ocul. Immunol. Inflamm. 7, 7-15. 10.1076/ocii.7.1.7.8108 [DOI] [PubMed] [Google Scholar]
- Knox S. M., Lombaert I. M. A., Reed X., Vitale-Cross L., Gutkind J. S. and Hoffman M. P. (2010). Parasympathetic innervation maintains epithelial progenitor cells during salivary organogenesis. Science 329, 1645-1647. 10.1126/science.1192046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko E. Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A. R., Kamitaki N., Martersteck E. M. et al. (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202-1214. 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen J., Kliem A., Tornoe I., Skjodt K., Koch C. and Holmskov U. (2000). Localization of lung surfactant protein D on mucosal surfaces in human tissues. J. Immunol. 164, 5866-5870. 10.4049/jimmunol.164.11.5866 [DOI] [PubMed] [Google Scholar]
- Makarenkova H. P. and Dartt D. A. (2015). Myoepithelial cells: their origin and function in lacrimal gland morphogenesis, homeostasis, and repair. Curr. Mol. Biol. Reports 1, 115-123. 10.1007/s40610-015-0020-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarenkova H. P., Ito M., Govindarajan V., Faber S. C., Sun L., McMahon G., Overbeek P. A. and Lang R. A. (2000). FGF10 is an inducer and Pax6 a competence factor for lacrimal gland development. Development 127, 2563-2572. [DOI] [PubMed] [Google Scholar]
- McDavid A., Finak G., Chattopadyay P. K., Dominguez M., Lamoreaux L., Ma S. S., Roederer M. and Gottardo R. (2013). Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics 29, 461-467. 10.1093/bioinformatics/bts714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirels L., Hand A. R. and Branin H. J. (1998). Expression of gross cystic disease fluid protein-15/Prolactin-inducible protein in rat salivary glands. J. Histochem. Cytochem. 46, 1061-1071. 10.1177/002215549804600910 [DOI] [PubMed] [Google Scholar]
- Muzumdar M. D., Tasic B., Miyamichi K., Li N. and Luo L. (2007). A global double-fluorescent cre reporter mouse. Genesis 45, 593-605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Pan Y., Carbe C., Powers A., Zhang E. E., Esko J. D., Grobe K., Feng G.-S. and Zhang X. (2008). Bud specific N-sulfation of heparan sulfate regulates Shp2-dependent FGF signaling during lacrimal gland induction. Development 135, 301-310. 10.1242/dev.014829 [DOI] [PubMed] [Google Scholar]
- Pan Y., Carbe C., Powers A., Feng G.-S. and Zhang X. (2010). Sprouty2-modulated Kras signaling rescues Shp2 deficiency during lens and lacrimal gland development. Development 137, 1085-1093. 10.1242/dev.042820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papafotiou G., Paraskevopoulou V., Vasilaki E., Kanaki Z., Paschalidis N. and Klinakis A. (2016). ARTICLE KRT14 marks a subpopulation of bladder basal cells with pivotal role in regeneration and tumorigenesis. Nat. Commun. 7, 1-11. 10.1038/ncomms11914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen F. (2006). Cell and molecular biology of human lacrimal gland and nasolacrimal duct mucins. Int. Rev. Cytol. 249, 229-279. 10.1016/S0074-7696(06)49005-7 [DOI] [PubMed] [Google Scholar]
- Pin C. L., Bonvissuto A. C. and Konieczny S. F. (2000). Mist1 expression is a common link among serous exocrine cells exhibiting regulated exocytosis. Anat. Rec. 259, 157-167. [DOI] [PubMed] [Google Scholar]
- Pin C. L., Rukstalis J. M., Johnson C. and Konieczny S. F. (2001). The bHLH transcription factor Mist1 is required to maintain exocrine pancreas cell organization and acinar cell identity. J. Cell Biol. 155, 519-530. 10.1083/jcb.200105060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prater M. D., Petit V., Alasdair Russell I., Giraddi R. R., Shehata M., Menon S., Schulte R., Kalajzic I., Rath N., Olson M. F. et al. (2014). Mammary stem cells have myoepithelial cell properties. Nat. Cell Biol. 16, 942–950, 1-7 10.1038/ncb3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X., Carbe C., Tao C., Powers A., Lawrence R., van Kuppevelt T. H., Cardoso W. V., Grobe K., Esko J. D. and Zhang X. (2011). Lacrimal gland development and Fgf10-Fgfr2b signaling are controlled by 2-O- and 6-O-sulfated heparan sulfate. J. Biol. Chem. 286, 14435-14444. 10.1074/jbc.M111.225003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2016). R: A Language and Environment for Statistical Computing R Found. Stat. Comput; Vienna: Austria: 0, {ISBN} 3-900051-07-0. [Google Scholar]
- Rock J. R., Onaitis M. W., Rawlins E. L., Lu Y., Clark C. P., Xue Y., Randell S. H. and Hogan B. L. (2009). Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. USA 106, 12771-12775. 10.1073/pnas.0906850106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R., Farrell J. A., Gennert D., Schier A. F. and Regev A. (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495-502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S., Ali M. J., Balla M. M. S., Naik M. N., Honavar S. G., Reddy V. A. P. and Vemuganti G. K. (2012). Establishing human lacrimal gland cultures with secretory function. PLoS ONE 7, e29458 10.1371/journal.pone.0029458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S., Ali M. J. and Vemuganti G. K. (2014). Human lacrimal gland regeneration: Perspectives and review of literature. Saudi J. Ophthalmol. Off. J. Saudi Ophthalmol. Soc. 28, 12-18. 10.1016/j.sjopt.2013.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Maaten L., Hinton G. and van der Maaten G. H. (2008). Visualizing Data using t-SNE. J. Mach. Learn. Res. 9, 2579-2605. [Google Scholar]
- Voronov D., Gromova A., Liu D., Zoukhri D., Medvinsky A., Meech R. and Makarenkova H. P. (2013). Transcription factors Runx1 to 3 are expressed in the lacrimal gland epithelium and are involved in regulation of gland morphogenesis and regeneration. Invest. Ophthalmol. Vis. Sci. 54, 3115-3125. 10.1167/iovs.13-11791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walcott B., Birzgalis A., Moore L. C. and Brink P. R. (2005). Fluid secretion and the Na+-K+-2Cl- cotransporter in mouse exorbital lacrimal gland. Am. J. Physiol. Cell Physiol. 289, C860-C867. 10.1152/ajpcell.00526.2004 [DOI] [PubMed] [Google Scholar]
- Wang Y. L., Tan Y., Satoh Y. and Ono K. (1995). Morphological changes of myoepithelial cells of mouse lacrimal glands during postnatal development. Histol. Histopathol. 10, 821-827. [PubMed] [Google Scholar]
- Wendling O., Bornert J.-M., Chambon P. and Metzger D. (2009). Efficient temporally-controlled targeted mutagenesis in smooth muscle cells of the adult mouse. Genesis 47, 14-18. 10.1002/dvg.20448 [DOI] [PubMed] [Google Scholar]
- You S., Tariq A., Kublin C. L. and Zoukhri D. (2011). Detection of BrdU-label retaining cells in the lacrimal gland: Implications for tissue repair. Cell Tissue Res. 346, 317-326. 10.1007/s00441-011-1271-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G. X. Y., Terry J. M., Belgrader P., Ryvkin P., Bent Z. W., Wilson R., Ziraldo S. B., Wheeler T. D., McDermott G. P., Zhu J. et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat. Commun.. 16, 14049 10.1038/ncomms14049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoukhri D., Macari E. and Kublin C. L. (2007). A single injection of interleukin-1 induces reversible aqueous-tear deficiency, lacrimal gland inflammation, and acinar and ductal cell proliferation. Exp. Eye Res. 84, 894-904. 10.1016/j.exer.2007.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoukhri D., Fix A., Alroy J. and Kublin C. L. (2008). Mechanisms of murine lacrimal gland repair after experimentally induced inflammation. Investig. Ophthalmol. Vis. Sci. 49, 4399-4406. 10.1167/iovs.08-1730 [DOI] [PMC free article] [PubMed] [Google Scholar]