Abstract
Mitochondrial dysfunction can cause female infertility. An important unresolved issue is the extent to which incompatibility between mitochondrial and nuclear genomes contributes to female infertility. It has previously been shown that a mitochondrial haplotype from D. simulans (simw501) is incompatible with a nuclear genome from the D. melanogaster strain Oregon-R (OreR), resulting in impaired development, which was enhanced at higher temperature. This mito-nuclear incompatibility is between alleles of the nuclear-encoded mitochondrial tyrosyl-tRNA synthetase (Aatm) and the mitochondrial-encoded tyrosyl-tRNA that it aminoacylates. Here, we show that this mito-nuclear incompatibility causes a severe temperature-sensitive female infertility. The OreR nuclear genome contributed to death of ovarian germline stem cells and reduced egg production, which was further enhanced by the incompatibility with simw501 mitochondria. Mito-nuclear incompatibility also resulted in aberrant egg morphology and a maternal-effect on embryonic chromosome segregation and survival, which was completely dependent on the temperature and mito-nuclear genotype of the mother. Our findings show that maternal mito-nuclear incompatibility during Drosophila oogenesis has severe consequences for egg production and embryonic survival, with important broader relevance to human female infertility and mitochondrial replacement therapy.
KEY WORDS: Mitochondria, Mitochondrial-nuclear incompatibility, Oogenesis, Stem cell, Embryogenesis, Drosophila
Summary: In Drosophila, mito-nuclear incompatibility has severe consequences for oogenesis and embryonic survival, a finding that has broader relevance to human female infertility and mitochondrial replacement therapy.
INTRODUCTION
Mitochondria are essential organelles that produce ATP through oxidative phosphorylation and participate in a number of other cellular processes (Cloonan and Choi, 2013; Tait and Green, 2010; Vyas et al., 2016). Although mitochondria have their own genomes, the vast majority of mitochondrial proteins are encoded in the nucleus (∼1500) (Meisinger et al., 2008). Studies in a number of organisms have shown that incompatibility between mitochondrial and nuclear genomes can have deleterious effects, and can contribute to reproductive isolation between populations (Burton and Barreto, 2012; Gibson et al., 2013; Hoekstra et al., 2013; Lamelza and Ailion, 2017; Ma et al., 2016; Narbonne et al., 2012; Sloan et al., 2017; Spirek et al., 2014). However, in only a few cases have the specific genes responsible for mito-nuclear incompatibility been identified (Chou et al., 2010; Lee et al., 2008; Meiklejohn et al., 2013; Singh and Brown, 1991; Spirek et al., 2014). It is known that mitochondrial dysfunction can severely compromise female fertility, and that maternal inheritance of sub-functional mitochondria can reduce embryonic survival (Bentov et al., 2011; Demain et al., 2016; Ge et al., 2012; Tilly and Sinclair, 2013). An important unresolved issue is how incompatible interactions between specific alleles of mitochondrial and nuclear genes contribute to female reproductive failure.
We and others have previously described a specific mito-nuclear incompatibility between alleles from two closely related Drosophila species (Holmbeck et al., 2015; Meiklejohn et al., 2013; Montooth et al., 2010). A strain with the mitochondrial genome from D. simulans strain w501 (simw501) and with the nuclear genome from the D. melanogaster strain Oregon-R, hereafter denoted as (simw501); OreR, had delayed development, disrupted larval metabolic rate, compromised locomotion, bristle defects and reduced fecundity (Hoekstra et al., 2013; Montooth et al., 2010; Meiklejohn et al., 2013). These phenotypes were most severe at higher temperatures, similar to other strains with compromised mitochondrial function, which is consistent with metabolism being a temperature-sensitive process (Clarke and Fraser, 2004; Ghosh et al., 2013; Hoekstra et al., 2013). In contrast, a strain with the same D. simulans mitochondria but with the D. melanogaster Austria nuclear genome, (simw501); AutW132, was phenotypically normal during larval development. Also phenotypically normal were combinations of the Oregon-R mitochondria (ore) with either D. melanogaster Austria or Oregon-R nuclear genomes, hereafter (ore); AutW132 and (ore); OreR. The molecular basis for the mito-nuclear incompatibility in the (simw501); OreR strain was shown to be an allelic interaction between the mt-tRNAtyr encoded in the simw501 mitochondria and the mt-tRNAtyr synthetase (Aatm) encoded by the D. melanogaster OreR nuclear genome (Meiklejohn et al., 2013). The simw501 mitochondrial mt-tRNAtyr polymorphism changes a G:C to G:U in the stem of the tRNA anticodon arm, and the OreR nuclear polymorphism changes a highly conserved alanine to valine at position 275 next to the synthetase ATP binding pocket AatmA275V. Consistent with the predicted effects of this interaction on mitochondrial protein translation, this incompatibility decreases oxidative phosphorylation activity specifically for only those complexes that require mitochondrial-translated proteins (Meiklejohn et al., 2013).
The incompatible (simw501); OreR strain serves as a model for deciphering how mitochondrial dysfunction contributes to human disease. Mutation of the human ortholog of Aatm, YARS2, as well as other mt-tRNA synthetases cause a spectrum of heritable diseases (Riley et al., 2010). A current challenge is to understand why mutations in different mt-tRNA synthetases result in different clinical presentations (Jiang et al., 2016; Konovalova and Tyynismaa, 2013). The results from the (simw501); OreR strain are consistent with the idea that mito-nuclear incompatibility in specific individuals may contribute to the variability in clinical phenotypes. Moreover, the reduced fecundity of the (simw501); OreR flies suggest the mito-nuclear incompatibility may impair gametogenesis and embryonic survival, but the cellular basis for this reproductive failure has not been investigated.
In this study, we investigate the impact of mito-nuclear incompatibility on Drosophila oogenesis and female fertility. Each Drosophila ovary is composed of ∼16-20 ovarioles, which contain an array of progressively more mature egg chambers (Bilder and Haigo, 2012; Lin and Spradling, 1993) (Fig. 1A,B). Egg chambers are composed of one oocyte and 15 germline sister nurse cells surrounded by an epithelial sheet of somatic follicle cells (Fig. 1B). These cells are descendants of germline stem cells (GSCs) and somatic follicle stem cells (FSCs) that reside in the germarium at the tip of the ovariole (Fig. 1B). Egg chambers are formed and bud off from the germarium as the transit-amplifying FSC daughter cells surround the germline cells. These egg chambers then migrate posteriorly down the ovariole as they mature through 14 morphologically defined stages (Fig. 1B). During early oogenesis, mitochondria greatly increase in number, with some transported into the oocyte, while others remain in nurse cells and are rapidly transferred into the oocyte during later oogenesis (Cox and Spradling, 2003; Hill et al., 2014). In fly strains that are heteroplasmic for different mtDNA haplotypes, the events of early oogenesis are associated with selection and inheritance of functional mitochondria (Ma et al., 2014). Similar mitochondrial proliferation, transport and selection also occur during mouse oogenesis, and defects in these processes can negatively impact oogenesis and embryonic survival in both fly and mouse (Cox and Spradling, 2003; Lei and Spradling, 2016; Mishra and Chan, 2014; Pepling, 2016; Van Blerkom, 2011).
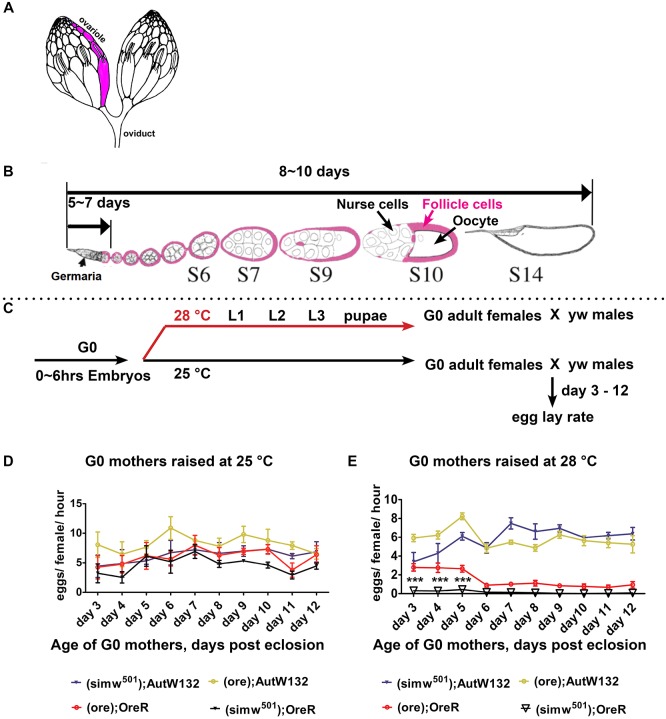
Fig. 1.
The (ore); OreR and (simw501); OreR females have a lower oviposition rate at a higher temperature. (A) An illustration of a pair of Drosophila ovaries with one ovariole indicated in pink. (B) A single ovariole with the developmental timeline of Drosophila oogenesis. Somatic follicle cells (pink) surround the germline nurse cells and oocyte to form an egg chamber. (C) Experimental scheme for the temperature-shift and female egg lay rate assay. (D,E) Oviposition rate of the indicated mito-nuclear females raised at 25°C (D) or 28°C (E) measured over 1 h. Fifty females per genotype, n=six biological replicates; data are mean±s.e.m. ***P<0.001 comparing (simw501); OreR with (ore); OreR using two-way ANOVA with Bonferroni correction. A and B are adapted, with the permission of the Genetics Society of America, from Ables (2015).
Here, we find that mito-nuclear incompatibility during oogenesis has pleiotropic cell and developmental consequences that compromise egg production and embryonic survival. Overall, the results provide a cellular basis for how mito-nuclear incompatibility can reduce organismal fitness and potentially contribute to reproductive barriers. More broadly, our findings are relevant to understanding the impact of mito-nuclear incompatibility on human female infertility, inter-generational inheritance of metabolic defects, and mitochondrial replacement therapy.
RESULTS
Females from (simw501); OreR and (ore); OreR strains have compromised fertility
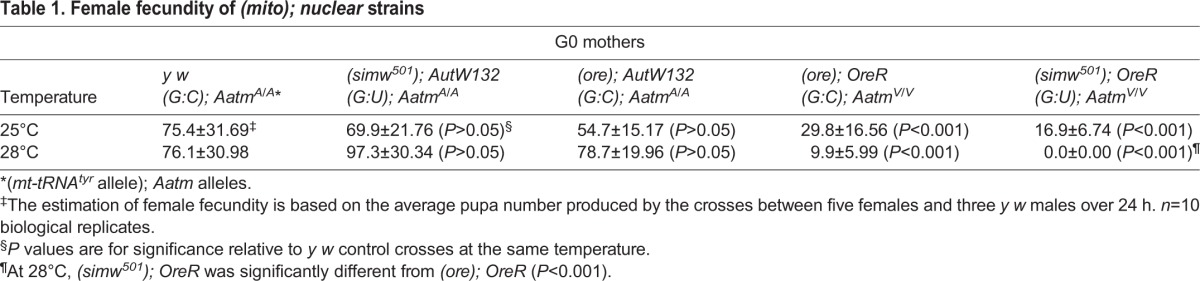
Previous results suggested that the mito-nuclear incompatible (simw501); OreR strain had reduced fecundity relative to the other mito-nuclear combinations, which was more severe at the non-permissive temperature of 28°C (Hoekstra et al., 2013; Meiklejohn et al., 2013). To specifically evaluate the contribution of female infertility to this reduced fecundity, and to eliminate the contribution of male sterility, we outcrossed (simw501); OreR females to males from a yellow (y) white (w) lab strain at 25°C and 28°C. Fecundity was measured by counting the number of pupae in the next generation, and compared with the number of offspring from y w females and females from strains previously shown to have compatible mito-nuclear combinations – (simw501); AutW132, (ore); AutW132 and (ore); OreR, all of which were outcrossed to y w males (Table 1). The number of offspring was not significantly different among the (simw501); AutW132, (ore); AutW132 and y w females at either 25°C or 28°C (Table 1). Whereas the (ore); OreR females had significantly fewer offspring than y w females at 25°C and 28°C, they had significantly more offspring than (simw501); OreR females at 28°C, which were completely infertile at this temperature (Table 1). These results indicate that the OreR nuclear genotype contributes to a reduced female fecundity, which is further enhanced by the simw501 mitochondrial genotype and higher temperature.
Table 1.
Female fecundity of (mito); nuclear strains
The nuclear OreR and mitochondrial simw501 genotypes contribute to a temperature-dependent decline in egg production
We evaluated whether the low fecundity of the (simw501); OreR and (ore); OreR females was because of a reduced egg production. We raised mito-nuclear females at 25°C or 28°C, crossed them to y w males and then measured egg lay rate (oviposition) on different days of adulthood (Fig. 1C). At 25°C, all four types of mito-nuclear females had a similar oviposition rate over 3-12 days of adulthood, suggesting this temperature is permissive for egg production (Fig. 1D). This result differs from those of Meikeljohn et al., who reported that (simw501); OreR and (ore); OreR have lower oviposition rates relative to (simw501); AutW132 and (ore); AutW132 at 25°C, perhaps because we measured oviposition rate over 1 h in the morning, whereas Meikeljohn et al. calculated oviposition over an entire day (Meiklejohn et al., 2013). In contrast, at 28°C the egg production from both (ore); OreR and (simw501); OreR was very low, especially in older females, and was significantly different from both (simw501); AutW132 and (ore); AutW132 (Fig. 1E). The low oviposition rate of both the (ore); OreR and (simw501); OreR females relative to the AutW132 nuclear strains suggests that the OreR nuclear genotype contributes to a temperature-sensitive decline in egg production. Moreover, the significantly lower egg lay rate of (simw501); OreR compared with (ore); OreR in younger females, and the lack of an effect of the simw501 mitochondria in the AutW132 nuclear background, suggests that the mito-nuclear incompatibility of (simw501); OreR further compromises egg production.
The nuclear OreR and mitochondrial simw501 genotypes contribute to temperature-dependent defects in ovary development and egg chamber production
To define the problem with egg production in (ore); OreR and (simw501); OreR, we first examined the gross morphology of the ovaries and ovarioles in the adult females on day three of adulthood (Fig. 1A,B). Ovariole development starts during the early pupal stage when somatic cells form a stem cell niche around germ cells, which are destined to become adult germline stem cells (GSCs) (Gancz et al., 2011). The adult ovaries of (ore); OreR and (simw501); OreR females were smaller than those of (simw501); AutW132 or (ore); AutW132 (Fig. S1A) and were composed of significantly fewer ovarioles, a phenotype that was exacerbated at 28°C and most severe in the (simw501); OreR strain (Fig. S1B). These results suggest that (ore); OreR and (simw501); OreR females have temperature-sensitive defects in gonadogenesis, resulting in a reduced adult ovariole number that contributes to the lower egg production rates.
We next examined stages of oogenesis in adult females by confocal microscopy of ovarioles labeled with the fluorescent DNA dye DAPI. When females were raised at 25°C, on day three of adulthood all four strains had normal germaria and distributions of egg chambers representing different stages of oogenesis (Fig. 2A-D, Fig. S2A). When raised at 28°C, the (simw501); AutW132 and (ore); AutW132 females again had normal germaria and stages of oogenesis (Fig. 2E,F, Figs S2B and S3A-B″). In contrast, in the (ore); OreR and (simw501); OreR females raised at 28°C, ∼70-80% of the ovarioles were missing egg chambers from early and mid-stages of oogenesis (Fig. 2G,H, Figs S2B and S3C-D″). It is known that in response to metabolic and other stresses a vitellogenic checkpoint results in reduced egg chamber production and autophagy of egg chambers during stages 7-9 (Pritchett et al., 2009). Some ovarioles in both (ore); OreR and (simw501); OreR had degenerating egg chambers that were labeled with Lysotracker, indicating that they were undergoing autophagy (data not shown). Unlike the vitellogenic checkpoint, however, these degenerating chambers were also seen during earlier stages of oogenesis, including stage 1. In many ovarioles, stages 1-7 were completely absent and germaria were directly attached to stage 8 or later egg chambers, with no evidence of nascent stage 1 egg chambers (germarium region 3) (Fig. 2G,H, Fig. S3C-D″). This last phenotype suggests that in the (ore); OreR and (simw501); OreR females after an initial period of normal oogenesis egg chamber production from the germarium ceases in many ovarioles. These failures in oogenesis are consistent with the observed temperature-sensitive decline in egg oviposition rate in the (ore); OreR and (simw501); OreR females (Fig. 1E).
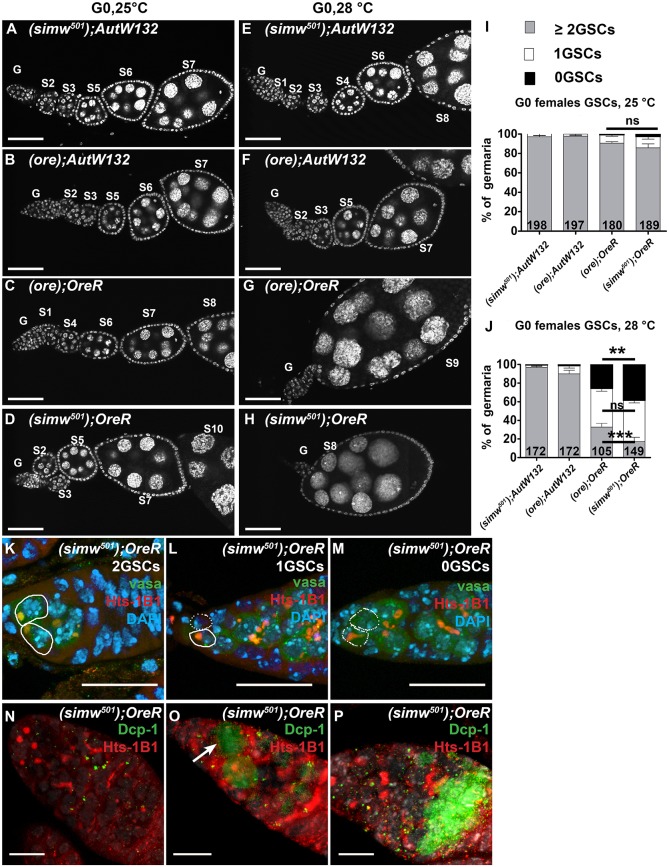
Fig. 2.
The OreR nuclear genotype contributes to a temperature-sensitive ovarian failure that is enhanced by simw501 mitochondria. Ovarioles labeled with DAPI from different mito-nuclear females at day 3 of adulthood raised at either 25°C (A-D) or 28°C (E-H). The germarium (G) and stages (S) of egg chamber maturation are indicated. Scale bars: 50 µm. (I,J) Quantification of the percentage of germaria with different numbers of germline stem cells (GSCs) from the indicated mito-nuclear females raised at either 25°C (I) or 28°C (J). The numbers on the bars represent the total number of germaria analyzed. Comparison of (ore); OreR and (simw501); OreR at 28°C, ***P<0.001 for two GSC, **P<0.01 for zero GSC, ns, non-significant for one GSC. Data are mean±s.e.m. (K-M) Germaria from (simw501); OreR females raised at 28°C with two (K), one (L) or zero (M) GSCs, labeled with antibodies against Hts (red) and Vasa (green) proteins, and DAPI (blue). Solid outlines: GSCs with Hts-labeled spherical spectrosomes. Dotted outlines: niche positions without a GSC. Scale bars: 20 µm. (N-P) Germline cell death in germaria from (simw501); OreR females raised at 28°C, labeled with antibodies against Hts (red) and cleaved caspase Dcp-1 (green). Images show germaria with no Dcp-1 labeling (N), a Dcp-1 labeled GSC (arrow) (O) or a Dcp-1 labeled 16 cell cyst (P). Scale bars: 10 µm.
The (simw501); OreR and (ore); OreR females have a temperature-sensitive defect in germline stem cell maintenance and early germline cell survival
To further define the defect in egg chamber production, we examined the earliest stages of oogenesis in the germarium. To measure germline stem cell (GSC) number, we labeled ovaries with antibodies against the germline protein Vasa and the Hu li tai shao (Hts) protein, the Drosophila adducin ortholog that associates with a spherical ‘spectrosome’ in the cytoplasm of GSCs (Lin et al., 1994). In a normal ovariole, anti-Hts labels spectrosomes in two or more GSCs that reside in the stem cell niche at the anterior tip of the germarium (Losick et al., 2011). The GSC daughter cells, called cystocytes, divide four times with incomplete cytokinesis to form interconnected germline cysts. These cysts contain a branched cytoskeletal body known as the fusome, which also labels with anti-Hts (Lin et al., 1994). Most germaria from all four mito-nuclear strains raised at 25°C had the normal two or more GSCs (Fig. 2I). When raised at 28°C, most germaria from (simw501); AutW132 and (ore); AutW132 females also had two or more GSCs (Fig. 2J, Fig. S3A-B″). In contrast, many germaria from the (simw501); OreR and (ore); OreR at 28°C had only one or zero GSCs and a reduced number of cystocytes, a temperature-dependent reduction of early germline cells that was significantly more severe in the (simw501); OreR strain (Fig. 2J-M, Fig. S3C-D″). Labeling of germaria with an antibody against the cleaved Caspase Dcp-1 indicated that GSCs and their daughter cystocytes were undergoing programmed cell death (PCD) at an elevated rate in the (simw501); OreR and (ore); OreR females specifically at 28°C (Fig. 2N-P, Fig. S2C-D) (McCall and Peterson, 2004). The (simw501); OreR females had the highest frequency of germline PCD, consistent with them having the lowest GSC number at 28°C (Fig. 2J, Figs S2D and S3D-D″). TUNEL labeling confirmed that GSCs and cystocytes were indeed undergoing PCD (data not shown). These results suggest that death of GSCs and cystocytes contributes to the temperature-dependent decline in egg chamber production in the (simw501); OreR and (ore); OreR females. The much lower frequency of this phenotype in the (simw501); AutW132 and (ore); AutW132 strains suggests that the OreR nuclear genotype contributes to early germline cell death, which is further exacerbated by the mito-nuclear incompatibility in the (simw501); OreR strain.
Many of the egg chambers produced by (ore); OreR and (simw501); OreR females had more than the normal 15 germline cells (Fig. S3E-G). Given that there were not more, and in fact fewer, cystocytes per cyst in the germarium, the >15 germline cells per egg chamber is not the result of extra cystocyte divisions. Instead, this phenotype is likely the result of a follicle cell epithelium encapsulating two germline cysts during the formation of a single stage 1 egg chamber. This phenotype is similar to other mutants that have a deficit of transit-amplifying follicle cells in the germarium, suggesting that (ore); OreR and (simw501); OreR have problems with early follicle cell proliferation or survival, or both (Cicek et al., 2016; Forbes et al., 1996).
The (simw501); OreR incompatible strain has a unique temperature-dependent maternal effect on embryo hatch rate
The data suggested that (simw501); OreR and (ore); OreR females share similar temperature-sensitive early oogenesis phenotypes. Progeny counts had indicated, however, that whereas (ore); OreR females had reduced number of progeny at 28°C, (simw501); OreR females were completely infertile (Table 1). Therefore, the similar ovary phenotypes in these two strains failed to account for the difference in their female fecundity. Moreover, while progressive GSC loss was somewhat more severe in the (simw501); OreR strain, these females did produce some normal-looking egg chambers, yet no progeny survived to the pupal stage. We therefore examined whether a difference in the survival of offspring from (simw501); OreR and (ore); OreR females accounts for their difference in fecundity.
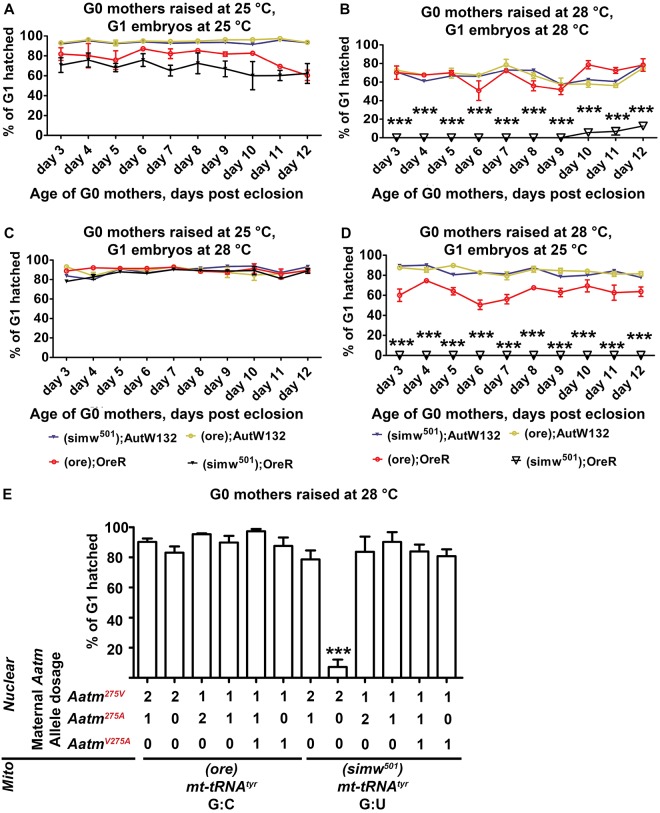
As before, we raised females from the four mito-nuclear strains at either 25°C or 28°C (the G0 generation) and then crossed them to y w males. We collected embryos from these females during different days of adulthood at 25°C or 28°C, allowed these G1 embryos to develop at the same temperature, and counted the fraction that hatched into larvae. When G0 mothers were raised at 25°C, G1 embryos from all four mito-nuclear strains had similar hatch rates (Fig. 3A). When mothers were raised at 28°C, however, the G1 embryos from (simw501); OreR mothers had hatch rates that were significantly lower than those in the other three strains (Fig. 3B). To assess whether this is a temperature-sensitive process in the mother or embryo, or both, we again raised females at 25°C or 28°C, but this time shifted their embryos to the reciprocal temperature after a 1 h egg lay. When mothers were raised at 25°C and embryogenesis was at 28°C, all four strains had similar high hatch rates (Fig. 3C). When mothers were raised at 28°C and embryogenesis was at 25°C, however, the G1 embryos from (simw501); OreR mothers again had hatch rates that was significantly lower than the other three mito-nuclear strains (Fig. 3D). These results suggest that the hatch rate of embryos is dependent on the temperature of the (simw501); OreR mothers.
Fig. 3.
A temperature-sensitive mito-nuclear incompatibility in the mother severely reduces embryonic hatch rate. (A-D) Embryonic hatch rate depends on the temperature of the (simw501); OreR mother. G0 mothers were raised at 25°C or 28°C and their G1 embryos allowed to develop at the same or reciprocal temperature. Embryonic hatch rates were measured for mothers of different ages post eclosion (x axis). Data are mean percentage of G1 hatched eggs±s.e.m. for three biological replicates. (A) Mothers at 25°C and embryos at 25°C. (B) Mothers at 28°C and embryos 28°C. (C) Mothers at 25°C and embryos at 28°C. (D) Mothers at 28°C and embryos 25°C. ***P<0.001 comparing (ore); OreR and (simw501); OreR. n=3. (E) The compatible AatmA allele in the mother rescues embryonic hatch rate. Hatch rates of embryos were measured from mothers with different mitochondria and doses of nuclear Aatm alleles, as indicated below the x axis. Mothers were at 28°C and embryos at 25°C. n=3 biological replicates; data are mean±s.e.m. (***P<0.001). See Fig. S5 for cross scheme.
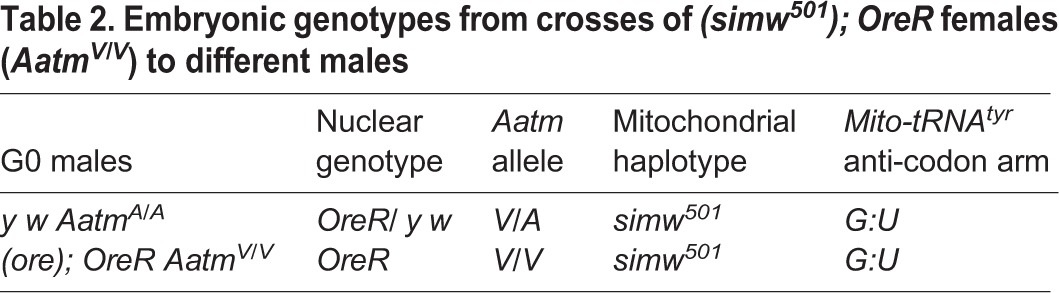
Although the embryos from these crosses inherit the simw501 mitochondria from their mother, they are heterozygous for AatmV/AatmA because their y w fathers are homozygous for the compatible AatmA allele (Table 2). The incompatible AatmV allele has previously been shown to be recessive to the compatible AatmA allele for larval development. Nevertheless, in our experiments inheritance of the AatmA allele from the y w father failed to rescue the maternal effect on embryonic hatch rate. We could not cross to (simw501); OreR males because they had reduced fertility at 28°C. Therefore, to directly assess whether embryos with a (simw501); OreR (AatmV/AatmV) genotype are temperature sensitive, we outcrossed (simw501); OreR females to fertile (ore); OreR males, which have the AatmV allele (Fig. S4A). The hatch rate of homozygous AatmV/AatmV embryos was not affected by the developmental temperature, but was dependent on the developmental temperature of the (simw501); OreR mother (Fig. S4B). These results suggest that the incompatible (simw501); OreR females have no surviving progeny because of a strict maternal effect that is primarily dependent on the genotype and temperature of the mother.
Table 2.
Embryonic genotypes from crosses of (simw501);OreR females (AatmV/V) to different males
Incompatibility between the nuclear AatmV and mitochondrial tRNATyr (G:U) alleles causes the temperature-sensitive maternal effect
We next addressed whether the incompatibility between the nuclear AatmV and mitochondrial tRNATyr (G:U) alleles in (simw501); OreR was the cause of the maternal effect. To do this, we conducted a series of rescue crosses using strains with different Aatm transgenes on the third chromosome that have either the incompatible OreR allele (AatmV), the compatible AutW132 allele (AatmA) or the OreR AatmV allele mutated to the compatible AatmA allele (AatmV275A) (Fig. S5) (Meiklejohn et al., 2013). These rescue strains were heterozygous on the second chromosome for a deletion of the endogenous Aatm gene over the CyO balancer, which has the compatible AatmA allele (Meiklejohn et al., 2013). This resulted in progeny females that had different doses of the AatmA, AatmV or AatmV275A alleles and either ore or simw501 mitochondria. We then tested these females (the G0 mothers) for the maternal effect on embryonic hatch rate (Fig. S5). We could not obtain adult females that only had the AatmV allele with the simw501 mitochondria when raised continuously at 28°C because of insufficient rescue of larval development. Therefore, for all the crosses, we shifted larvae from 25°C to 28°C at second instar to increase survival and obtain adult females. These G0 adult females were crossed to y w males, allowed to lay eggs for 1 h at 28°C, and their embryos shifted to 25°C to specifically assay the maternal effect. The results indicated that only one copy of the AatmA allele or AatmV275A allele was sufficient to rescue the temperature-sensitive maternal effect on embryonic hatch rate (Fig. 3E). Conversely, mothers that inherited only the AatmV allele with simw501 mitochondria displayed a strong temperature-sensitive maternal effect on hatch rate (Fig. 3E). Thus, it appears that, similar to the previous results for larval development, the compatible AatmA is dominant to the incompatible AatmV for the maternal effect on embryogenesis. These rescue results suggest that it is the incompatibility between the nuclear AatmV and mitochondrial tRNATyr (G:U) alleles that is responsible for the temperature-sensitive maternal effect on offspring survival.
The temperature-sensitive period of the (simw501); OreR maternal effect begins during pupal development of the mother
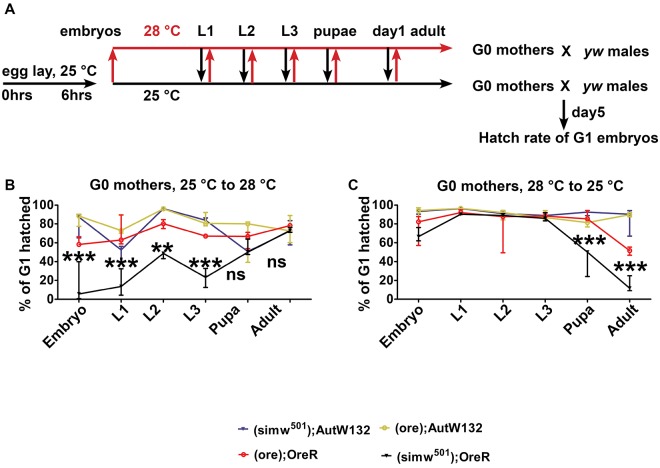
To define the temperature-sensitive period for the (simw501); OreR maternal effect, we performed reciprocal temperature-shift experiments. The G0 females from all four mito-nuclear strains began development at either 25°C or 28°C, but were then shifted to the reciprocal temperature at different times of their larval, pupal or adult life (Fig. 4A). The resulting adult G0 females were crossed to y w males, allowed to lay eggs on day five of adulthood, and the hatch rate of their G1 embryos measured. Embryonic hatch rate from (simw501); OreR mothers was significantly lower than the other strains when these females were shifted from permissive (25°C) to restrictive (28°C) temperature before early pupal development (Fig. 4B). The reciprocal shift from restrictive (28°C) to permissive (25°C) resulted in lower embryonic hatch rates when the (simw501); OreR mothers were shifted after larval development (Fig. 4C). Together, these reciprocal shift experiments suggest that the temperature-sensitive period of the (simw501); OreR maternal effect begins during early pupal development, a period that coincides with ovariole morphogenesis and the onset of oogenesis (Gancz et al., 2011).
Fig. 4.
The temperature-sensitive period in the mito-nuclear incompatible mothers begins at the L3-to-pupa transition. (A) Experimental scheme for the reciprocal temperature-shift experiments. The G0 females were shifted from 25°C to 28°C (red arrows) or from 28°C to 25°C (black arrows) during different larval stages (L), white pre-pupae (pupae) or day 1 of adulthood. The resulting G0 adult females were crossed to y w males, and on day five post-eclosion G1 embryos were collected, allowed to develop at 25°C, and hatch rate counted after 36 h of embryogenesis. (B) Hatch rate of G1 embryos when their G0 mothers were shifted from a permissive (25°C) up to a restrictive (28°C) temperature at the indicated developmental times. (C) Hatch rate of G1 embryos when their G0 mothers were shifted from a restrictive (28°C) to a permissive (25°C) temperature at the indicated developmental times. n=3 biological replicates; data are mean±s.e.m. **P<0.01 and ***P<0.001 for comparison between (ore); OreR and (simw501); OreR.
The temperature-sensitive period for the mito-nuclear maternal effect corresponds to early oogenesis
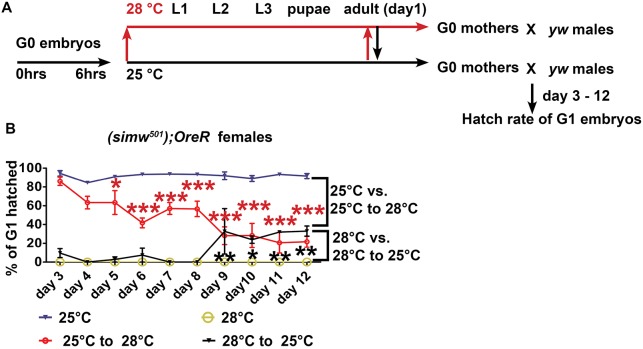
Curiously, shifting the temperature of (simw501); OreR females during adulthood did not alter the maternal effect on hatch rate (Fig. 4B,C). However, this maternal effect was assayed only on day five of adulthood. Therefore, to further investigate the dynamics of the temperature-sensitive maternal effect, we performed reciprocal temperature-shift experiments with females on day one of adulthood, and then measured hatch rate of their embryos from day three to 12 of adulthood (Fig. 5A). As expected, the (simw501); OreR females that were shifted from 25°C to 28°C initially laid embryos with a very high hatch rate (Fig. 5B). This hatch rate then gradually declined to significantly lower levels by day five of adulthood, and continued to decline to very low levels until day 12 (Fig. 5B). Conversely, embryos from mothers shifted from 28°C to 25°C initially had a very low hatch rate. Despite shifting these mothers to permissive temperature on day one of adulthood, this low hatch rate persisted until day eight, with hatch rates rising only by day nine (Fig. 5B).
Fig. 5.
Adult temperature shifts have a delayed impact on the maternal effect. (A) (simw501); OreR females were raised at 25°C or 28°C, kept at the same temperature, or shifted to the reciprocal temperature on day 1 of adulthood, and then crossed to y w males. Embryos from these females were collected on days 3-12 of adulthood, allowed to develop at 25°C and hatch rates were measured at 36 h. (B) Hatch rates of embryos from (simw501); OreR mothers who were treated as described in A. n=3 biological replicates; data are mean±s.e.m.. Red asterisks represent P values for comparison of 25-28°C shift (red line) versus constant 25°C (blue line). Black asterisks represent P values for comparison of 28-25°C shift (black line) versus constant 28°C (gold line). *P<0.05; **P<0.01; ***P<0.001. n=3.
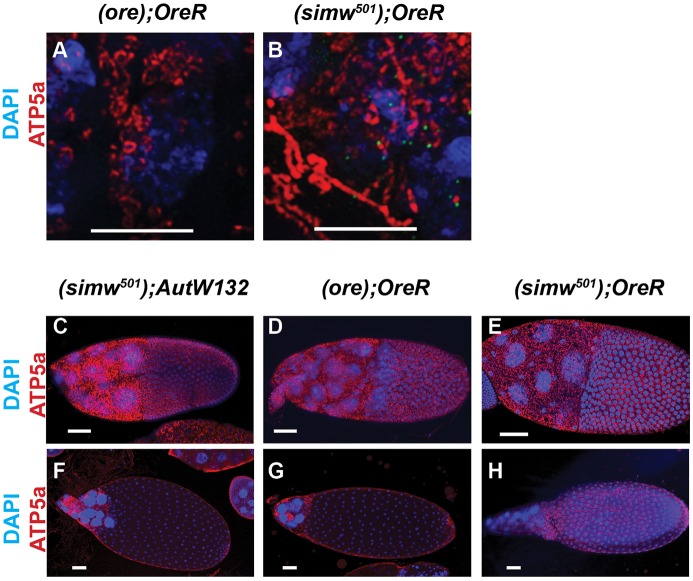
The nine-day lag between the temperature shift and the rise in hatch rate is approximately equal to the time it takes in oogenesis for a GSC daughter cell in the germarium to develop into a mature stage 14 egg (Fig. 1B) (Lin and Spradling, 1993). This similarity in timing suggested that the temperature-sensitive period for the mito-nuclear incompatibility may coincide with events of early oogenesis in the germarium. Indeed, early oogenesis is a crucial time for mitochondrial dynamics when germline mitochondria undergo a period of rapid proliferation and increase greatly in number, with some mitochondria actively transported into the differentiating oocyte to form part of a distinct cluster called the Balbiani body (Cox and Spradling, 2003; Hill et al., 2014; Lei and Spradling, 2016; Pepling, 2016; Pepling et al., 2007). To address whether these processes were affected, we labeled mitochondria with antibodies against ATP5a, a subunit of the F1 ATP synthase complex, and examined them by confocal and super-resolution microscopy (Godbout et al., 1993). This analysis suggested that the (ore); OreR and (simw501); OreR females at 28°C have normal mitochondrial number in the germarium (Fig. 6A,B). In many (simw501); OreR germaria, however, mitochondria formed a more tubular network, although this was difficult to quantify (Fig. 6B). Mitochondrial labeling of (simw501); OreR was similar to all the other mito-nuclear strains in both somatic and germline cells of later stage egg chambers (Fig. 6C-H). Thus, the combined data suggest that mito-nuclear incompatibility does not result in observable reductions in the number of mitochondria in developing egg chambers. It remains possible, however, that early oogenesis is a temperature-sensitive period for mitochondrial quality that later manifests as reduced embryonic hatch rate, consistent with previous evidence that the mito-nuclear incompatibility in (simw501); OreR results in reduced mitochondrial oxidative phosphorylation capacity (Meiklejohn et al., 2013).
Fig. 6.
Mitochondrial abundance and morphology in the ovary. (A,B) Super resolution image of germaria labeled with anti-ATP5a and DAPI from (ore); OreR (A) and (simw501); OreR (B) females raised at 28°C. Scale bars: 5 µm. (C-H) Confocal images of a stage 10 (C-E) and stage 12-13 (F-H) egg chambers labeled with anti-ATP5a and DAPI from (simw501); AutW132 (C,F), (ore); OreR (D,G) and (simw501); OreR (E,H) females that were raised at 28°C. Scale bars: 50 µm.
Maternal mito-nuclear incompatibility in (simw501); OreR has pleiotropic effects on egg morphology, fertilization, and embryonic cell divisions
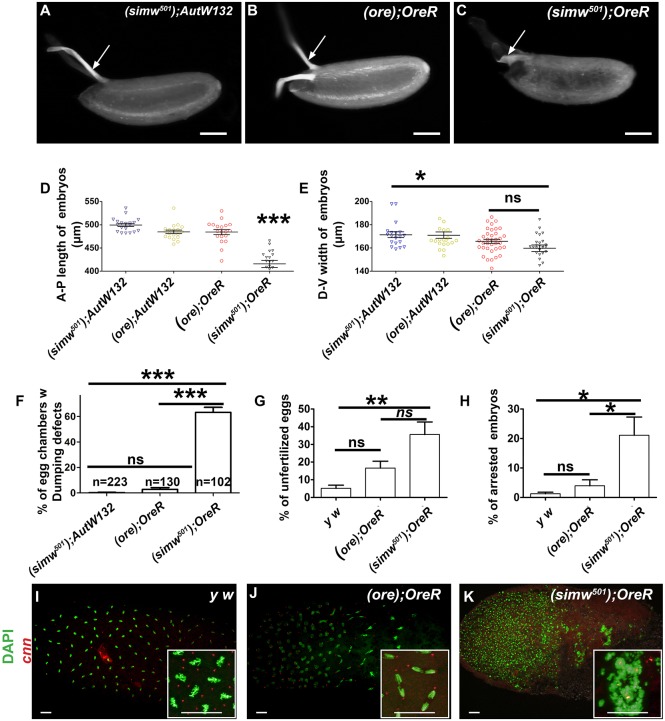
We next examined the eggs laid by (simw501); OreR mothers to determine how maternal mito-nuclear incompatibility inhibits embryonic hatch rate. At 28°C, (simw501); OreR mothers laid eggs that were smaller with shells of unusual morphology (Fig. 7A-C, Fig. S6A-C). The A-P axis length of these eggs was significantly shorter than those from other mito-nuclear mothers by ∼20%, without an increase in egg width, indicating that eggs laid by (simw501); OreR have reduced volume (Fig. 7D,E). This reduction in egg length without a significant change in width is similar to other mutants that have incomplete ‘dumping’ of nurse cell cytoplasm into the oocyte during late oogenesis (Bilder and Haigo, 2012; Cooley et al., 1992). An examination of late stages of oogenesis indicated that the (simw501); OreR females did indeed have a very high fraction of egg chambers with incomplete nurse cell dumping (Fig. 7F). In addition, many of the eggs laid by (simw501); OreR mothers had soft, gelatinous eggshells, a structure that is synthesized by somatic follicle cells. These follicle cells developmentally amplify the copy number of eggshell protein (Chorion) genes late in oogenesis (Calvi, 2006; Calvi et al., 1998; Spradling and Mahowald, 1980). However, labeling of ovaries with the nucleotide analog EdU indicated that developmental amplification of chorion genes was not impaired, suggesting that problems with eggshell synthesis are downstream of developmental gene amplification (Fig. S6D-F) (Calvi and Lilly, 2004; Calvi et al., 1998). Many of these eggs had unusual branching morphology of their eggshell dorsal appendages, with some of them wrapping laterally around the egg. Labeling of egg chambers with antibodies against the dorsal determinant Gurken, however, did not provide evidence for a disruption of D-V patterning (Fig. S6G-I). Together, these phenotypes suggest that some of the (simw501); OreR female infertility may be caused by defects in both germline nurse cells and somatic follicle cells.
Fig. 7.
The (simw501); OreR mito-nuclear incompatibility compromises egg morphology, fertilization and embryonic cell divisions. (A-C) Bright-field images of eggs laid by (simw501); AutW132 (A) (ore); OreR (B) and (simw501); OreR (C) females at 28°C. Eggs from (simw501); OreR females are shorter, with soft eggshells, and unusual dorsal appendage morphology (arrow). Scale bars: 100 µm. (D,E) Measurement of anterior-posterior length (D) and dorsal-ventral width (E) of ∼0-2 h embryos from mothers of the indicated genotypes at 28°C (*P<0.05, ***P<0.001. n=19 per genotype). (F) Percentage of stage 11-13 egg chambers with reduced transfer of nurse cell cytoplasm into oocyte. (G) Percentage of unfertilized embryos from mothers of the indicated genotypes at 28°C determined by anti-Cnn labeling (n=4, **P<0.01). (H) Percentage of fertilized embryos that were arrested by 8 h after egg lay (AEL). (n=4, *P<0.05). (I-K) Confocal images of embryos during nuclear cleavage cycles, 0-2 h AEL. Embryos are from mothers raised at 28°C and of the indicated genotype. Centrosomes are labeled with anti-Cnn antibody (red) and nuclear DNA is labeled with DAPI (green). The insets show higher magnifications of mitotic chromosome segregation. Scale bars for panels and insets: 20 µm.
To gain further insight into the (simw501); OreR maternal effect, we next examined stages of embryogenesis. To focus specifically on the maternal effect, mothers were raised at 28°C, mated to y w males, and the embryos then allowed to develop at 25°C. Many of the eggs from (simw501); OreR mothers appeared unfertilized. To quantify sperm entry into the oocyte, we labeled with antibodies against the paternally-supplied centrosomal protein Centrosomin (Cnn) (Eisman et al., 2015; Megraw et al., 1999). This analysis revealed that at 28°C ∼36% of eggs from (simw501); OreR mothers were unfertilized, a fraction that was significantly higher than the 5% unfertilized fraction produced by control y w mothers (Fig. 7G). Although the fraction of unfertilized eggs from (ore); OreR mothers at 28°C was also elevated (∼17%), this number was not significantly different from y w (Fig. 7G). Sperm enter the egg through a hollow cone called the micropyle, a specialized structure of the vitelline membrane (Nonidez, 1920; Suzanne et al., 2001). For some eggs from (simw501); OreR mothers, the micropyles were occluded by aberrant eggshell structures, at least partially explaining the fertilization defect (Fig. S6J-L).
Although 64% of embryos from (simw501); OreR mothers were fertilized, hatch rate was close to zero. For y w embryos, the duration of embryogenesis was ∼22 h with hatch rates of over 90%, whereas almost all embryos from (simw501); OreR mothers failed to hatch even after 36 h. While embryos arrested at distributed stages, ∼70% of fertilized embryos had defects in syncytial nuclear divisions during the first 2 h of embryogenesis (Fig. 7H,K). These embryos frequently had unevenly spaced nuclei and bridging/lagging mitotic chromosomes, indicative of syncytial nuclear positioning and division defects (Fig. 7K). Taken together, these data suggest that mito-nuclear incompatibility in the (simw501); OreR mothers results in severe pleiotropic effects on egg morphology, fertilization and embryonic cell divisions.
DISCUSSION
In this study, we show that a specific mito-nuclear incompatibility results in a suite of cell and developmental phenotypes that contribute to complete temperature-sensitive infertility of (simw501); OreR females. The data indicate that both (ore); OreR and (simw501); OreR have germline cell death and a decline in egg production with female age, suggesting that the OreR nuclear genotype contributes to reduced female fecundity (Hoekstra et al., 2013; Meiklejohn et al., 2013; Montooth et al., 2010). These phenotypes were more severe for (simw501); OreR than (ore); OreR, and were absent in (simw501); AutW132, suggesting that mito-nuclear incompatibility enhances this ovarian failure. The allelic mito-nuclear incompatibility in (simw501); OreR strain caused a unique temperature-sensitive maternal effect that resulted in complete infertility. Our results provide a cell and developmental framework for understanding how mito-nuclear incompatibility can contribute to reproductive barriers among divergent populations and cause human disease.
The nuclear Oregon-R and mitochondrial simw501 genotypes contribute to ovarian failure
The (ore); OreR and (simw501); OreR females both had a temperature-sensitive reduction in ovariole number and a progressive ovarian failure caused by egg chamber degeneration and programmed cell death of germline cells in the germarium, suggesting that the OreR nuclear genotype contributes to these phenotypes (Fig. 8). It has been previously reported that dividing germline cystocytes undergo a Caspase-dependent cell death in response to metabolic stresses (Drummond-Barbosa and Spradling, 2001; Ikeya et al., 2002; Morrison and Spradling, 2008). Activated Caspase labeling of GSCs was surprising, however, given that it has been reported that GSCs and other stem cells are resistant to apoptosis after irradiation (Xing et al., 2015). It is possible that mitochondrial stress is a more potent inducer of apoptosis in stem cells than the genotoxic stress caused by irradiation. Given that ovarioles form around presumptive GSCs during early pupal stages of development, the reduction in ovariole number suggests that GSCs may also be lost before adulthood (Belles and Piulachs, 2015; Gilboa and Lehmann, 2006; Song et al., 2002).
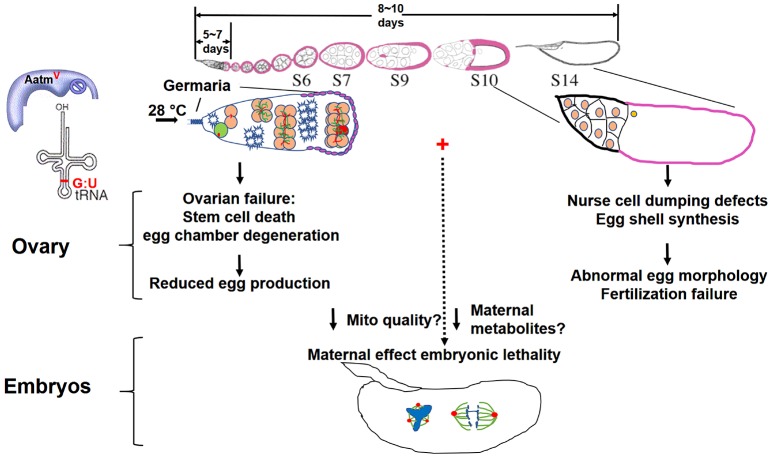
Fig. 8.
Temperature-sensitive mito-nuclear incompatibility has pleiotropic effects on female fertility. At a higher temperature (28°C), the OreR nuclear genotype causes loss of GSCs, reduced egg chamber production and degeneration of egg chambers through apoptosis and autophagy, all of which are enhanced by incompatibility with simw501 mitochondria. These ovarian phenotypes together contribute to greatly decreased egg production. Temperature-sensitive mito-nuclear incompatibility in (simw501); OreR females caused unique nurse cell dumping and eggshell synthesis defects during later oogenesis. Below: temperature-sensitive mito-nuclear incompatibility in (simw501); OreR mothers resulted in mitotic errors in their embryos, which may be caused by insufficient maternal metabolites or maternal inheritance of sub-functional mitochondria.
The similar phenotypes of (simw501); OreR and (ore); OreR in the ovary contrast with their different phenotypes in larvae reported by earlier studies, which showed that (simw501); OreR, but not (ore); OreR, has temperature-sensitive defects during larval development (Hoekstra et al., 2013; Meiklejohn et al., 2013). The main effect of the OreR nuclear genome to disrupt oogenesis, but not larval development, is consistent with the known tissue-specific characteristics of other metabolic dysfunctions in development and disease, and the sensitivity of oogenesis to metabolic stress, including that caused by mitochondrial dysfunction (Benkhalifa et al., 2014; Bentov et al., 2011; Boots et al., 2016; Grindler and Moley, 2013; McCall, 2004; Morrison and Spradling, 2008; Sieber et al., 2016). Although our rescue experiments showed that the allelic incompatibility between the AatmV allele and simw501 mitochondria causes the maternal effect embryonic lethality, preliminary observations suggest that the ovarian failure phenotype is multigenic and not just determined by alleles of Aatm. Changing the OreR nuclear background through rescue crosses ameliorated the ovarian failure phenotype. Females that inherited the simw501 mitochondria and only the incompatible AatmV allele still had an ovarian failure phenotype, but this phenotype was less severe than in the parental (simw501); OreR (AatmV/V) females. The rescue-cross females with only the incompatible AatmV allele, but with ore mitochondria, did not have an ovarian failure phenotype, consistent with the interpretation that mito-nuclear incompatibility enhances ovarian failure. Moreover, whereas (simw501); OreR and (ore); OreR had similar early oogenesis phenotypes, these phenotypes were more severe in (simw501); OreR, and absent in (simw501); AutW132, suggesting that they are enhanced by the mito-nuclear incompatibility. The degree to which alleles of Aatm and other genes contribute to the OreR nuclear main effect on ovarian failure is a complex genetic issue that will be addressed by future mapping studies.
Mito-nuclear incompatibility in (simw501); OreR results in complete female infertility
Importantly, the (simw501); OreR females had the unique temperature-sensitive phenotypes of abnormal egg morphology, fertilization failure and maternal-effect embryonic lethality (Fig. 8). The embryos from these females had aberrant nuclear cleavage division and severe chromosome segregation errors during the first 2 h of embryogenesis. This embryonic lethality was a strict maternal effect dependent on the temperature of the mother, which rescue experiments suggest is caused by incompatibility between the nuclear AatmV and mitochondrial tRNAtyr G:U alleles. The temperature-shift experiments suggested that early oogenesis is a temperature-sensitive period, which is known to be a crucial time for mitochondrial proliferation, transport and selection, and alleles of mitochondrial proteins can even influence early oocyte specification (Cox and Spradling, 2003; Hill et al., 2014; Hurd et al., 2016; Ma et al., 2014; Teixeira et al., 2015). Our analysis, however, did not reveal defects in mitochondrial number, transport or oocyte specification. It is likely, therefore, that this mito-nuclear incompatibility impairs mitochondrial quality, consistent with previous evidence that (simw501); OreR mitochondria have reduced oxidative phosphorylation complex activity and an aberrant morphology in muscle (Holmbeck et al., 2015; Meiklejohn et al., 2013). Thus, temperature-sensitive events during early oogenesis in (simw501); OreR mothers may result in persistent sub-functional mitochondria that cannot support energy-demanding processes during later oogenesis and early embryogenesis (Sieber and Spradling, 2015; Sieber et al., 2016).
The aberrant eggshell synthesis suggests that some of the processes affected by this incompatibility are in somatic follicle cells. The fertilization failure of these eggs may also be the result of aberrant eggshell and vitelline membrane synthesis by follicle cells (Fig. 8). The incomplete transfer of nurse cell cytoplasm and short egg phenotype resembles that of other ‘dumpless’ mutants, and suggests that mito-nuclear incompatibility also impairs germline functions (Bilder and Haigo, 2012; Frydman and Spradling, 2001; Mahajan-Miklos and Cooley, 1994). Rapid transfer of nurse cell cytoplasm depends on actin polymerization and contraction, and is sensitive to ATP/ADP ratio, perhaps explaining why it is sensitive to mitochondrial dysfunction (Huelsmann et al., 2013). A recent report indicates that compromised function of the muscular ovariole sheath can also affect nurse cell dumping and results in short eggs with reduced ooplasm (Andersen and Horne-Badovinac, 2016). Thus, it is also possible that the reduced nurse cell dumping is a non-autonomous effect of an ovariole sheath myopathy.
The mito-nuclear incompatibility in the (simw501); OreR mothers resulted in an inter-generational lethality of their embryos (Fig. 8). Many embryos had unevenly spaced nuclei and aberrant mitotic chromosome segregation during the first 2 h of embryogenesis. Nuclear spacing and migration is ATP dependent and is enacted by aster microtubules and actin cytoskeleton (Telley et al., 2012). Thus, this energy-demanding process may be sensitive to cellular energy deficits, similar to actin-based nurse cell dumping and muscle function. These aberrant nuclear division and spacing phenotypes may be the result of maternal inheritance of sub-functional mitochondria or insufficient maternally-supplied metabolites. Although we favor the interpretation that the ovarian and embryonic phenotypes are caused by mitochondrial failure in somatic and germline cells of the ovary, it is also possible that they are influenced by non-autonomous effects caused by perturbation of female physiology and neuro-endocrine axis.
The impact of mito-nuclear incompatibility on female fertility
Our data are relevant to how mito-nuclear incompatibility can contribute to reproductive barriers between species (Burton et al., 2006; Chang et al., 2015; Gibson et al., 2013; Ma et al., 2016; Meiklejohn et al., 2013; Narbonne et al., 2012; Paliwal et al., 2014; Spirek et al., 2014). Our results reveal the cellular and developmental bases for the severe effects of this genomic incompatibility on female fertility in a high-temperature environment – a higher-order gene×gene×environment (G×G×E) effect, and suggest that it has the potential to strongly contribute to reproductive barriers among different populations via pleiotropic effects on female fertility (Hoekstra et al., 2013). Although our data show in general how mito-nuclear incompatibilities can generate barriers to reproduction, it is clear that nuclear-nuclear incompatibilities are the major barrier to reproduction between D. simulans and D. melanogaster (Ferree and Barbash, 2009; Phadnis et al., 2015). A recent report indicated that incompatibility between nuclear and mitochondrial genomes can also compromise female fertility in mouse, although the allelic basis for this incompatibility is not known (Ma et al., 2016). Together with our findings, these data suggest that mito-nuclear interactions may contribute to female infertility in humans.
Mutations in the human ortholog of Aatm, YARS2, cause myopathy, anemia and optic neuropathy (Jiang et al., 2016; Jordanova et al., 2006; Riley et al., 2010), and it has recently been reported that incompatibility between alleles of YARS2 and a specific mitochondrial haplotype worsens Leber's hereditary optic neuropathy (Jiang et al., 2016; Konovalova and Tyynismaa, 2013). Mutation of the human mitochondrial histidine tRNA synthetase HARS2 is known to cause ovarian dysgenesis, but an effect of YARS2 on female fertility has not been reported. Our results suggest that polymorphisms in YARS2 may have undocumented effects on human female fertility that may depend on mitochondrial haplotype, and emphasize that mito- nuclear interactions should be considered when interpreting mechanisms of mitochondrial disease (Lu et al., 2015; Storkebaum et al., 2009). A careful consideration of possible mito-nuclear interactions is also important for informing best practices for mitochondrial replacement therapies that introduce nuclei from two parents into the oocyte cytoplasm of a donor female (Craven et al., 2010; Kang et al., 2016).
MATERIALS AND METHODS
Drosophila genetics
The Drosophila mito-nuclear strains and rescue strains used in this study were previously described and are available in Table S1. The control strain, y w67c23, was obtained from the Bloomington Stock Center and sequenced to confirm that it is homozygous for the AatmA allele.
Temperature-shift experiments and phenotypic assays
Female fecundity in Table 1 was assayed by crossing five virgin females to three y w males over 24 h at either 25°C or 28°C. After removing the adults, the vials were kept at the same temperature and the number of pupae were counted on day 7. Ten biological replicates were conducted for each genotype and temperature.
For oviposition and ovary analyses of Figs 1, 2 and Figs S1, S2, flies were allowed to lay eggs in vials for 6 h at 25°C. After the adults were removed, the vials were either kept at 25°C or shifted to 28°C. The offspring in these vials develop into the G0 mothers that were subsequently assayed. Adult G0 mothers were kept at either 25°C or 28°C, mated to y w males, conditioned on wet yeast, and their oviposition rate and ovaries analyzed beginning on day 3 of adulthood. The ovarian failure phenotypes in Figs 2, 6 and 7, Figs S1, S2, S3 and S6 were analyzed on day 3 of adulthood. For oviposition rate, 50 newly emerged females were mated to 25 y w or (ore); OreR males, conditioned for 2 days on grape plates with wet yeast. Oviposition rate was measured over 1 h in the morning and flies were transferred to fresh yeast daily.
The G0 mothers were crossed to y w males and the hatch rate of the G1 embryos was analyzed at 36 h after egg lay (Fig. 3). To assay maternal-embryonic temperature sensitivity, G0 mothers were allowed to lay eggs for one hour, and then those G1 offspring were kept at the same temperature or shifted to the reciprocal temperature.
In the reciprocal temperature-shift experiments in Fig. 4, the G0 females were allowed to develop at 25°C or 28°C until they reached the indicated developmental stages and were then shifted to the reciprocal temperature, followed by measurement of hatch rate of their embryos as described. For rescue crosses, (ore); OreR and (simw501); OreR virgin females were crossed to the males from thee different rescue strains. The G0 females were raised at 25°C until second-instar larvae and were then shifted to 28°C for further development.
For Fig. 5, the G0 adult females were mated to y w males and the temperature was shifted on day one of adulthood, followed by measurement of G1 hatch rate beginning on day 3.
For embryo size measurements, embryos at 0∼2 h of development were dechorionated and measured along their A-P and D-V axis using Openlab imaging software (Fig. 7). Nurse cell dumping was quantified by estimating the A-P axis length ratio of nurse cell compartment to oocyte in stage 12-14 egg chambers.
Immunofluorescence microscopy
Ovaries and embryos were fixed and labeled as previously described (Calvi and Lilly, 2004). The following antibodies and concentrations were used: mouse anti-Hts [1:20 dilution, Developmental Studies Hybridoma Bank (DHSB)]; rabbit anti-cleaved Dcp-1 (1:100 dilution, Cell Signaling Technology, 9578); mouse anti-ATP5a (1:100 dilution, Abcam, ab14748); rat anti-Vasa (1:100 dilution, DHSB); and mouse anti-Gurken (2 µg/ml, Hybridoma bank). EdU labeling of amplicon foci was carried out as previously described (Calvi and Lilly, 2004; Paranjape and Calvi, 2016).
Secondary antibodies were Alexa Fluor 488-conjugated goat anti-mouse IgG (4 μg/ml, Invitrogen, 169425); Alexa Fluor 488-conjugated goat anti-rabbit IgG (4 μg/ml, Invitrogen, 1705869); Alexa Fluor 568-conjugated goat anti-mouse IgG (4 μg/ml, Invitrogen, 1698376); Alexa Fluor 568- conjugated donkey anti-rabbit IgG (4 μg/ml, Life Technologies, 1668655); and FITC-conjugated goat anti-rat IgG (2 μg/ml, Life Technologies, A18866). DNA was labeled with 1 μg/ml DAPI (Invitrogen).
For embryo labeling, 0∼2 h embryos were collected on grape plates and rinsed with 0.01% Triton-X 100, dechorionated with 1:1 mixture of bleach and 0.2% NaCl/ 0.02% Triton-X 100, and then slow fixed using a modification of previous protocols (Sullivan et al., 2000). Briefly, the dechorionated embryos were brushed into a glass vial and then 1 ml heptane was added followed by 1 ml of 3.7% formaldehyde in PEM buffer. After shaking the vial for 15 s, the embryos were allowed to fix at room temperature for 15 min. The bottom layer was removed and 1 ml of methanol was added. After vortexing for 10 min, the embryos that did or did not sink were collected separately and hand devitellinized. We did this because alterations to vitelline membrane synthesis in the incompatible strain made even fertilized embryos partially resistant to methanol devitellinization, and to quantify the fertilization rate independent of devitellinization efficiency. All the embryos were rehydrated with PBTA solution and labeled with rabbit anti-Cnn antibody (1:100, a generous gift from T. Kaufman, Indiana University, Bloomington, USA). The unfertilized embryos were quantified based on lack of Cnn labeled centrosomes.
Ovaries and embryos were mounted in Vectashield and imaged with Leica-SP5 scanning confocal microscope. For live embryo imaging, embryos were rinsed with PBS and mounted on supported slides in PBS and micropyles were imaged by DIC with Leica DMRA2 fluorescence microscope. The germaria mitochondrial morphology images were captured on an OMX super resolution microscope (Applied Precision).
Statistics
One-way ANOVA was used for comparison of GSC number or GSC death among all four strains on day three of adulthood. One-way ANOVA was also used to compare the four strains for developmental temperature-shift experiments in Fig. 4, and for rescue experiments in Fig. 3E. For Fig. 5, significance was assessed by two-way ANOVA of temperature and age. For all other analyses, two-way ANOVA of female age and genotype was used to compare the four strains. ANOVA was followed by Tukey's or Bonferroni posttests to evaluate additivity of variables (e.g. female age and genotype) and correct for false discovery rate (FDR), respectively. All statistical analysis was carried out using GraphPad Prism5 statistical packages.
Acknowledgements
We thank R. Eisman and T. Kaufman for advice and the Cnn antibody, and the Bloomington Drosophila Stock Center (BDSC) and Flybase for flies and crucial information. We also thank J. Powers of the IU Light Microscopy Imaging Center (LMIC) for help and advice, and J. Zhu, M Ailion, P Lamelza and members of the Calvi lab for comments on the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: C.Z., K.L.M., B.R.C.; Methodology: C.Z., K.L.M., B.R.C.; Software: C.Z., B.R.C.; Validation: C.Z., K.L.M., B.R.C.; Formal analysis: C.Z., K.L.M., B.R.C.; Investigation: C.Z., B.R.C.; Resources: B.R.C.; Writing - original draft: C.Z., B.R.C.; Writing - review & editing: C.Z., K.L.M., B.R.C.; Visualization: C.Z., B.R.C.; Supervision: K.L.M., B.R.C.; Project administration: B.R.C.; Funding acquisition: B.R.C.
Funding
This work was supported by funding to B.R.C from the Indiana Clinical and Translational Sciences Institute (National Institutes of Health) (UL1 TR001108) and by an IOS CAREER award (National Science Foundation) (1505247) to K.L.M. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.151951.supplemental
References
- Ables E. T. (2015). Drosophila oocytes as a model for understanding meiosis: an educational primer to accompany “corolla is a novel protein that contributes to the architecture of the synaptonemal complex of Drosophila”. Genetics 199, 17-23. 10.1534/genetics.114.167940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen D. and Horne-Badovinac S. (2016). Influence of ovarian muscle contraction and oocyte growth on egg chamber elongation in Drosophila. Development 143, 1375-1387. 10.1242/dev.131276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belles X. and Piulachs M.-D. (2015). Ecdysone signalling and ovarian development in insects: from stem cells to ovarian follicle formation. Biochim. Biophys. Acta 1849, 181-186. 10.1016/j.bbagrm.2014.05.025 [DOI] [PubMed] [Google Scholar]
- Benkhalifa M., Ferreira Y. J., Chahine H., Louanjli N., Miron P., Merviel P. and Copin H. (2014). Mitochondria: participation to infertility as source of energy and cause of senescence. Int. J. Biochem. Cell Biol. 55, 60-64. 10.1016/j.biocel.2014.08.011 [DOI] [PubMed] [Google Scholar]
- Bentov Y., Yavorska T., Esfandiari N., Jurisicova A. and Casper R. F. (2011). The contribution of mitochondrial function to reproductive aging. J. Assist. Reprod. Genet. 28, 773-783. 10.1007/s10815-011-9588-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilder D. and Haigo S. L. (2012). Expanding the morphogenetic repertoire: perspectives from the Drosophila egg. Dev. Cell 22, 12-23. 10.1016/j.devcel.2011.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boots C. E., Boudoures A., Zhang W., Drury A. and Moley K. H. (2016). Obesity-induced oocyte mitochondrial defects are partially prevented and rescued by supplementation with co-enzyme Q10 in a mouse model. Hum. Reprod. 31, 2090-2097. 10.1093/humrep/dew181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton R. S. and Barreto F. S. (2012). A disproportionate role for mtDNA in Dobzhansky-Muller incompatibilities? Mol. Ecol. 21, 4942-4957. 10.1111/mec.12006 [DOI] [PubMed] [Google Scholar]
- Burton R. S., Ellison C. K. and Harrison J. S. (2006). The sorry state of F2 hybrids: consequences of rapid mitochondrial DNA evolution in allopatric populations. Am. Nat. 168, S14-S24. 10.1086/509046 [DOI] [PubMed] [Google Scholar]
- Calvi B. R. (2006). Developmental DNA amplification. In DNA replication and human disease (ed. DePamphilis M. L.), pp. 233-255. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Calvi B. R. and Lilly M. A. (2004). Fluorescent BrdU labeling and nuclear flow sorting of the Drosophila ovary. Methods Mol. Biol. 247, 203-213. [DOI] [PubMed] [Google Scholar]
- Calvi B. R., Lilly M. A. and Spradling A. C. (1998). Cell cycle control of chorion gene amplification. Genes Dev. 12, 734-744. 10.1101/gad.12.5.734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-C., Rodriguez J. and Ross J. (2015). Mitochondrial-nuclear epistasis impacts fitness and mitochondrial physiology of interpopulation caenorhabditis briggsae hybrids. G3 (Bethesda) 6, 209-219. 10.1534/g3.115.022970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J.-Y., Hung Y.-S., Lin K.-H., Lee H.-Y. and Leu J.-Y. (2010). Multiple molecular mechanisms cause reproductive isolation between three yeast species. PLoS Biol. 8, e1000432 10.1371/journal.pbio.1000432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicek I. O., Karaca S., Brankatschk M., Eaton S., Urlaub H. and Shcherbata H. R. (2016). Hedgehog signaling strength is orchestrated by the mir-310 cluster of microRNAs in response to diet. Genetics 202, 1167-1183. 10.1534/genetics.115.185371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke A. and Fraser K. P. P. (2004). Why does metabolism scale with temperature? Funct. Ecol. 18, 243-251. 10.1111/j.0269-8463.2004.00841.x [DOI] [Google Scholar]
- Cloonan S. M. and Choi A. M. K. (2013). Mitochondria: sensors and mediators of innate immune receptor signaling. Curr. Opin. Microbiol. 16, 327-338. 10.1016/j.mib.2013.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley L., Verheyen E. and Ayers K. (1992). chickadee encodes a profilin required for intercellular cytoplasm transport during Drosophila oogenesis. Cell 69, 173-184. 10.1016/0092-8674(92)90128-Y [DOI] [PubMed] [Google Scholar]
- Cox R. T. and Spradling A. C. (2003). A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development 130, 1579-1590. 10.1242/dev.00365 [DOI] [PubMed] [Google Scholar]
- Craven L., Tuppen H. A., Greggains G. D., Harbottle S. J., Murphy J. L., Cree L. M., Murdoch A. P., Chinnery P. F., Taylor R. W., Lightowlers R. N. et al. (2010). Pronuclear transfer in human embryos to prevent transmission of mitochondrial DNA disease. Nature 465, 82-85. 10.1038/nature08958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demain L. A., Conway G. S. and Newman W. G. (2016). Genetics of mitochondrial dysfunction and infertility. Clin. Genet. 91, 199-207. 10.1111/cge.12896 [DOI] [PubMed] [Google Scholar]
- Drummond-Barbosa D. and Spradling A. C. (2001). Stem cells and their progeny respond to nutritional changes during Drosophila oogenesis. Dev. Biol. 231, 265-278. 10.1006/dbio.2000.0135 [DOI] [PubMed] [Google Scholar]
- Eisman R. C., Phelps M. A. S. and Kaufman T. (2015). An amino-terminal polo kinase interaction motif acts in the regulation of centrosome formation and reveals a novel function for centrosomin (cnn) in Drosophila. Genetics 201, 685-706. 10.1534/genetics.115.181842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferree P. M. and Barbash D. A. (2009). Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol. 7, e1000234 10.1371/journal.pbio.1000234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes A. J., Lin H., Ingham P. W. and Spradling A. C. (1996). hedgehog is required for the proliferation and specification of ovarian somatic cells prior to egg chamber formation in Drosophila. Development 122, 1125-1135. [DOI] [PubMed] [Google Scholar]
- Frydman H. M. and Spradling A. C. (2001). The receptor-like tyrosine phosphatase lar is required for epithelial planar polarity and for axis determination within drosophila ovarian follicles. Development 128, 3209-3220. [DOI] [PubMed] [Google Scholar]
- Gancz D., Lengil T. and Gilboa L. (2011). Coordinated regulation of niche and stem cell precursors by hormonal signaling. PLoS Biol. 9, e1001202 10.1371/journal.pbio.1001202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H., Tollner T. L., Hu Z., Da M., Li X., Guan H. Q., Shan D., Lu J., Huang C. and Dong Q. (2012). Impaired mitochondrial function in murine oocytes is associated with controlled ovarian hyperstimulation and in vitro maturation. Reprod. Fertil. Dev. 24, 945-952. 10.1071/RD11212 [DOI] [PubMed] [Google Scholar]
- Ghosh S. M., Testa N. D. and Shingleton A. W. (2013). Temperature-size rule is mediated by thermal plasticity of critical size in Drosophila melanogaster. Proc. R. Soc. B Biol. Sci. 280, 20130174 10.1098/rspb.2013.0174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson J. D., Niehuis O., Peirson B. R., Cash E. I. and Gadau J. (2013). Genetic and developmental basis of F2 hybrid breakdown in Nasonia parasitoid wasps. Evolution 67, 2124-2132. 10.1111/evo.12080 [DOI] [PubMed] [Google Scholar]
- Gilboa L. and Lehmann R. (2006). Soma-germline interactions coordinate homeostasis and growth in the Drosophila gonad. Nature 443, 97-100. 10.1038/nature05068 [DOI] [PubMed] [Google Scholar]
- Godbout R., Bisgrove D. A., Honoré L. H. and Day R. S., III (1993). Amplification of the gene encoding the alpha-subunit of the mitochondrial ATP synthase complex in a human retinoblastoma cell line. Gene 123, 195-201. 10.1016/0378-1119(93)90124-L [DOI] [PubMed] [Google Scholar]
- Grindler N. M. and Moley K. H. (2013). Maternal obesity, infertility and mitochondrial dysfunction: potential mechanisms emerging from mouse model systems. Mol. Hum. Reprod. 19, 486-494. 10.1093/molehr/gat026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J. H., Chen Z. and Xu H. (2014). Selective propagation of functional mitochondrial DNA during oogenesis restricts the transmission of a deleterious mitochondrial variant. Nat. Genet. 46, 389-392. 10.1038/ng.2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra L. A., Siddiq M. A. and Montooth K. L. (2013). Pleiotropic effects of a mitochondrial-nuclear incompatibility depend upon the accelerating effect of temperature in Drosophila. Genetics 195, 1129-1139. 10.1534/genetics.113.154914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmbeck M. A., Donner J. R., Villa-Cuesta E. and Rand D. M. (2015). A Drosophila model for mito-nuclear diseases generated by an incompatible interaction between tRNA and tRNA synthetase. Dis. Model. Mech. 8, 843-854. 10.1242/dmm.019323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsmann S., Ylänne J. and Brown N. H. (2013). Filopodia-like actin cables position nuclei in association with perinuclear actin in Drosophila nurse cells. Dev. Cell 26, 604-615. 10.1016/j.devcel.2013.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd T. R., Herrmann B., Sauerwald J., Sanny J., Grosch M. and Lehmann R. (2016). Long Oskar Controls Mitochondrial Inheritance in Drosophila melanogaster. Dev. Cell 39, 560-571. 10.1016/j.devcel.2016.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeya T., Galic M., Belawat P., Nairz K. and Hafen E. (2002). Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr. Biol. 12, 1293-1300. 10.1016/S0960-9822(02)01043-6 [DOI] [PubMed] [Google Scholar]
- Jiang P., Jin X., Peng Y., Wang M., Liu H., Liu X., Zhang Z., Ji Y., Zhang J., Liang M. et al. (2016). The exome sequencing identified the mutation in YARS2 encoding the mitochondrial tyrosyl-tRNA synthetase as a nuclear modifier for the phenotypic manifestation of Leber's hereditary optic neuropathy-associated mitochondrial DNA mutation. Hum. Mol. Genet. 25, 584-596. 10.1093/hmg/ddv498 [DOI] [PubMed] [Google Scholar]
- Jordanova A., Irobi J., Thomas F. P., Van Dijck P., Meerschaert K., Dewil M., Dierick I., Jacobs A., De Vriendt E., Guergueltcheva V. et al. (2006). Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat. Genet. 38, 197-202. 10.1038/ng1727 [DOI] [PubMed] [Google Scholar]
- Kang E., Wu J., Gutierrez N. M., Koski A., Tippner-Hedges R., Agaronyan K., Platero-Luengo A., Martinez-Redondo P., Ma H., Lee Y. et al. (2016). Mitochondrial replacement in human oocytes carrying pathogenic mitochondrial DNA mutations. Nature 540, 270-275. 10.1038/nature20592 [DOI] [PubMed] [Google Scholar]
- Konovalova S. and Tyynismaa H. (2013). Mitochondrial aminoacyl-tRNA synthetases in human disease. Mol. Genet. Metab. 108, 206-211. 10.1016/j.ymgme.2013.01.010 [DOI] [PubMed] [Google Scholar]
- Lamelza P. and Ailion M. (2017). Cytoplasmic-nuclear incompatibility between wild isolates of caenorhabditis nouraguensis. G3 (Bethesda) 7, 823-834. 10.1534/g3.116.037101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-Y., Chou J.-Y., Cheong L., Chang N.-H., Yang S.-Y. and Leu J.-Y. (2008). Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell 135, 1065-1073. 10.1016/j.cell.2008.10.047 [DOI] [PubMed] [Google Scholar]
- Lei L. and Spradling A. C. (2016). Mouse oocytes differentiate through organelle enrichment from sister cyst germ cells. Science 352, 95-99. 10.1126/science.aad2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. and Spradling A. C. (1993). Germline stem cell division and egg chamber development in transplanted Drosophila germaria. Dev. Biol. 159, 140-152. 10.1006/dbio.1993.1228 [DOI] [PubMed] [Google Scholar]
- Lin H., Yue L. and Spradling A. C. (1994). The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development 120, 947-956. [DOI] [PubMed] [Google Scholar]
- Losick V. P., Morris L. X., Fox D. T. and Spradling A. (2011). Drosophila stem cell niches: a decade of discovery suggests a unified view of stem cell regulation. Dev. Cell 21, 159-171. 10.1016/j.devcel.2011.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Marygold S. J., Gharib W. H. and Suter B. (2015). The aminoacyl-tRNA synthetases of Drosophila melanogaster. Fly 9, 53-61. 10.1080/19336934.2015.1101196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Xu H. and O'Farrell P. H. (2014). Transmission of mitochondrial mutations and action of purifying selection in Drosophila melanogaster. Nat. Genet. 46, 393-397. 10.1038/ng.2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Marti Gutierrez N., Morey R., Van Dyken C., Kang E., Hayama T., Lee Y., Li Y., Tippner-Hedges R., Wolf D. P. et al. (2016). Incompatibility between nuclear and mitochondrial genomes contributes to an interspecies reproductive barrier. Cell Metab. 24, 283-294. 10.1016/j.cmet.2016.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan-Miklos S. and Cooley L. (1994). Intercellular cytoplasm transport during Drosophila oogenesis. Dev. Biol. 165, 336-351. 10.1006/dbio.1994.1257 [DOI] [PubMed] [Google Scholar]
- McCall K. (2004). Eggs over easy: cell death in the Drosophila ovary. Dev. Biol. 274, 3-14. 10.1016/j.ydbio.2004.07.017 [DOI] [PubMed] [Google Scholar]
- McCall K. and Peterson J. S. (2004). Detection of apoptosis in Drosophila. Methods Mol. Biol. 282, 191-205. 10.1385/1-59259-812-9:191 [DOI] [PubMed] [Google Scholar]
- Megraw T. L., Li K., Kao L. R. and Kaufman T. C. (1999). The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development 126, 2829-2839. [DOI] [PubMed] [Google Scholar]
- Meiklejohn C. D., Holmbeck M. A., Siddiq M. A., Abt D. N., Rand D. M. and Montooth K. L. (2013). An Incompatibility between a mitochondrial tRNA and its nuclear-encoded tRNA synthetase compromises development and fitness in Drosophila. PLoS Genet. 9, e1003238 10.1371/journal.pgen.1003238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger C., Sickmann A. and Pfanner N. (2008). The mitochondrial proteome: from inventory to function. Cell 134, 22-24. 10.1016/j.cell.2008.06.043 [DOI] [PubMed] [Google Scholar]
- Mishra P. and Chan D. C. (2014). Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol. 15, 634-646. 10.1038/nrm3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montooth K. L., Meiklejohn C. D., Abt D. N. and Rand D. M. (2010). Mitochondrial-nuclear epistasis affects fitness within species but does not contribute to fixed incompatibilities between species of Drosophila. Evolution 64, 3364-3379. 10.1111/j.1558-5646.2010.01077.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison S. J. and Spradling A. C. (2008). Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell 132, 598-611. 10.1016/j.cell.2008.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narbonne P., Halley-Stott R. P. and Gurdon J. B. (2012). On the cellular and developmental lethality of a Xenopus nucleocytoplasmic hybrid. Commun. Integr. Biol. 5, 329-333. 10.4161/cib.20334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonidez J. F. (1920). The internal phenomenon of reproduction in Drosophila. Biol. Bull. 39, 207-230. 10.2307/1536488 [DOI] [Google Scholar]
- Paliwal S., Fiumera A. C. and Fiumera H. L. (2014). Mitochondrial-nuclear epistasis contributes to phenotypic variation and coadaptation in natural isolates of Saccharomyces cerevisiae. Genetics 198, 1251-1265. 10.1534/genetics.114.168575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjape N. P. and Calvi B. R. (2016). The histone variant H3.3 is enriched at Drosophila amplicon origins but does not mark them for activation. G3 (Bethesda) 6, 1661-1671. 10.1534/g3.116.028068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepling M. E. (2016). Development. Nursing the oocyte . Science 352, 35-36. 10.1126/science.aaf4943 [DOI] [PubMed] [Google Scholar]
- Pepling M. E., Wilhelm J. E., O'Hara A. L., Gephardt G. W. and Spradling A. C. (2007). Mouse oocytes within germ cell cysts and primordial follicles contain a Balbiani body. Proc. Natl. Acad. Sci. USA 104, 187-192. 10.1073/pnas.0609923104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis N., Baker E. P., Cooper J. C., Frizzell K. A., Hsieh E., de la Cruz A. F., Shendure J., Kitzman J. O. and Malik H. S. (2015). An essential cell cycle regulation gene causes hybrid inviability in Drosophila. Science 350, 1552-1555. 10.1126/science.aac7504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchett T. L., Tanner E. A. and McCall K. (2009). Cracking open cell death in the Drosophila ovary. Apoptosis 14, 969-979. 10.1007/s10495-009-0369-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley L. G., Cooper S., Hickey P., Rudinger-Thirion J., McKenzie M., Compton A., Lim S. C., Thorburn D., Ryan M. T., Giegé R. et al. (2010). Mutation of the mitochondrial tyrosyl-tRNA synthetase gene, YARS2, causes myopathy, lactic acidosis, and sideroblastic anemia--MLASA syndrome. Am. J. Hum. Genet. 87, 52-59. 10.1016/j.ajhg.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber M. H. and Spradling A. C. (2015). Steroid signaling establishes a female metabolic state and regulates SREBP to control oocyte lipid accumulation. Curr. Biol. 25, 993-1004. 10.1016/j.cub.2015.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber M. H., Thomsen M. B. and Spradling A. C. (2016). Electron transport chain remodeling by GSK3 during oogenesis connects nutrient state to reproduction. Cell 164, 420-432. 10.1016/j.cell.2015.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M. and Brown G. G. (1991). Suppression of cytoplasmic male sterility by nuclear genes alters expression of a novel mitochondrial gene region. Plant Cell 3, 1349-1362. 10.1105/tpc.3.12.1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan D. B., Havird J. C. and Sharbrough J. (2017). The on-again, off-again relationship between mitochondrial genomes and species boundaries. Mol. Ecol. 26, 2212-2236. 10.1111/mec.13959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Zhu C. H., Doan C. and Xie T. (2002). Germline stem cells anchored by adherens junctions in the Drosophila ovary niches. Science 296, 1855-1857. 10.1126/science.1069871 [DOI] [PubMed] [Google Scholar]
- Spirek M., Polakova S., Jatzova K. and Sulo P. (2014). Post-zygotic sterility and cytonuclear compatibility limits in S. cerevisiae xenomitochondrial cybrids. Front Genet 5, 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A. C. and Mahowald A. P. (1980). Amplification of genes for chorion proteins during oogenesis in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 77, 1096-1100. 10.1073/pnas.77.2.1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storkebaum E., Leitão-Gonçalves R., Godenschwege T., Nangle L., Mejia M., Bosmans I., Ooms T., Jacobs A., Van Dijck P., Yang X.-L. et al. (2009). Dominant mutations in the tyrosyl-tRNA synthetase gene recapitulate in Drosophila features of human Charcot-Marie-Tooth neuropathy. Proc. Natl. Acad. Sci. USA 106, 11782-11787. 10.1073/pnas.0905339106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan W., Ashburner M. and Hawley R. S. (2000). Drosophila Protocols. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Suzanne M., Perrimon N. and Noselli S. (2001). The Drosophila JNK pathway controls the morphogenesis of the egg dorsal appendages and micropyle. Dev. Biol. 237, 282-294. 10.1006/dbio.2001.0384 [DOI] [PubMed] [Google Scholar]
- Tait S. W. G. and Green D. R. (2010). Mitochondria and cell death: outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 11, 621-632. 10.1038/nrm2952 [DOI] [PubMed] [Google Scholar]
- Teixeira F. K., Sanchez C. G., Hurd T. R., Seifert J. R. K., Czech B., Preall J. B., Hannon G. J. and Lehmann R. (2015). ATP synthase promotes germ cell differentiation independent of oxidative phosphorylation. Nat. Cell Biol. 17, 689-696. 10.1038/ncb3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telley I. A., Gáspár I., Ephrussi A. and Surrey T. (2012). Aster migration determines the length scale of nuclear separation in the Drosophila syncytial embryo. J. Cell Biol. 197, 887-895. 10.1083/jcb.201204019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly J. L. and Sinclair D. A. (2013). Germline energetics, aging, and female infertility. Cell Metab. 17, 838-850. 10.1016/j.cmet.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Blerkom J. (2011). Mitochondrial function in the human oocyte and embryo and their role in developmental competence. Mitochondrion 11, 797-813. 10.1016/j.mito.2010.09.012 [DOI] [PubMed] [Google Scholar]
- Vyas S., Zaganjor E. and Haigis M. C. (2016). Mitochondria and cancer. Cell 166, 555-566. 10.1016/j.cell.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Su T. T. and Ruohola-Baker H. (2015). Tie-mediated signal from apoptotic cells protects stem cells in Drosophila melanogaster. Nat. Commun. 6, 7058 10.1038/ncomms8058 [DOI] [PMC free article] [PubMed] [Google Scholar]