Abstract
Background
Expanding interest in and use of active surveillance for early state prostate cancer has increased need for prognostic biomarkers. Using a multi-institutional tissue microarray resource including over 1000 radical prostatectomy samples, we sought to correlate Ki67 expression captured by an automated image analysis system with clinico-pathologic features and validate its utility as a clinical grade test in predicting cancer-specific outcomes.
Methods
After immunostaining, the Ki67 proliferation index (PI) of tumor areas of each core (3 cancer cores/case) was analyzed using a nuclear quantification algorithm (Aperio). We assessed whether Ki67 PI was associated with clinico-pathologic factors and recurrence free survival including biochemical recurrence, metastasis or PC death (7-year median follow-up).
Results
In 1004 PCs (~4,000 tissue cores) Ki67 PI showed significantly higher inter-tumor (0.68) than intra-tumor variation (0.39). Ki67 PI was associated with stage (p<0.0001), seminal vesicle invasion (SVI, p=0.02), extracapsular extension (ECE, p<0.0001) and Gleason Score (GS, p<0.0001). Ki67 PI as a continuous variable significantly correlated with recurrence free, overall and disease-specific survival by multivariable Cox proportional hazard model (HR=1.04–1.1, p=0.02–0.0008). High Ki67 score (defined as ≥5%) was significantly associated with worse recurrence free survival (HR=1.47, p=0.0007) and worse overall survival (HR=2.03, p=0.03).
Conclusion
In localized PC treated by radical prostatectomy, higher Ki67 PI assessed using a clinical grade automated algorithm is strongly associated with a higher GS, stage, SVI and ECE, and greater probability of recurrence.
Keywords: Ki67, prognostic, biomarker, prostate cancer
INTRODUCTION
Differentiating indolent from aggressive prostate cancer (PC) is a major priority given the high prevalence of PC in the aging population and the current magnitude of its overtreatment (1–3). While clinical and pathological assessments of tumor characteristics provide prognostic information, there is a broad spectrum of outcomes in individual patients. Substantial investments have been made in identifying biomarkers related to PC behavior, though to date no tissue-based markers have been incorporated in routine clinical practice due to conflicting data, lack of independence from other well established clinico-pathological characteristics, and a paucity of appropriately-designed and powered validation and standardization studies.
The Canary Tissue Microarray of Prostate cancer outcomes (CTMAP) was designed with the primary objective of validating promising candidate biomarkers reported to predict PC recurrence at the time of radical prostatectomy (RP) (4). The CTMAP model of a case-cohort study was designed to allow validation of a biomarker with approximately two-fold sensitivity greater than Gleason score (GS) in predicting PC recurrence. Such a marker would likely benefit clinical practice. Oversampling of recurrent low-grade tumors and non-recurrent high-grade tumors in CTMAP decreases the influence of GS, allowing unbiased validation of independent prognostic biomarkers. This large and well-annotated tissue resource of more than 1,300 randomly selected RP specimens with prolonged follow-up from six academic institutions also reflects the heterogeneity of PC and spectrum of patient management in North America.
Comprehensive literature reviews and a recent meta-analysis of prognostic PC biomarkers indicates that immunohistochemical measurement of Ki67 (MKI67/MIB-1) expression is the tissue biomarker with the most consistent association with the clinical outcomes of PC (5–7). Ki67 has provided independent prognostic value in prostate needle biopsies, transurethral prostate resections and prostatectomy specimens (1, 8, 9) including independent associations with biochemical and clinical recurrence regardless of treatment (9). In addition, Ki67 is an attractive biomarker from a technical perspective due to ease of interpretation with moderate to high intra- and inter-observer reproducibility, relatively high tolerance to typical preanalytical variability, universal use and availability across diagnostic laboratories, and the frequent presence of internal positive controls within sampled tissues (3, 7, 10, 11). As a marker of tumor proliferation, Ki67 has been successfully used in routine pathology practice for differential diagnoses, grading, prognostication and assessment of treatment responses for multiple neoplasms including endocrine and neuroendocrine neoplasms (12, 13), breast cancer (10, 14), trophoblastic tumors, lymphomas, soft tissue and brain tumors (15–17).
In view of the need for biomarkers that aid in the clinical management of men with PC, we sought to determine if automated detection of Ki67 immunohistochemical staining, reported as a percentage of cells expressing Ki67 as a measure of cell proliferation, associated with a specific adverse PC outcome: recurrence after primary therapy. We also sought to determine if Ki67 provided information independent of other risk features and if the magnitude of this additional information was sufficient to impact clinical practice.
PATIENTS AND METHODS
The Canary Tissue Microarray Prostate Cancer (CTMAP) Resource
This study utilized tissue microarrays (TMA) comprised of tissue samples from over 1,300 randomly selected participants treated for PC with RP at 6 institutions between 1995 and 2004 (4). The cohort included approximately equal numbers of samples from men with biochemically recurrent and non-recurrent PC after 5 or more years of follow-up. Recurrent PC was defined by (1) a single serum PSA level >0.2 ng/mL more than 8 week after RP and/or (2) receipt of salvage or secondary therapy after RP and/or (3) clinical or radiologic evidence of metastatic disease after RP. Median follow-up was 7 years (range: 1 day – 21.4 years).
The TMA was constructed to assess biomarkers that provide prognostic information independent of clinical and pathological information. As GS is a powerful predictor of outcome, we oversampled recurrent cases of GS 3+3 and 3+4 as well as non-recurrent cases with GS 4+4. This strategy diminishes the prognostic significance of GS and allows for the validation of biomarkers that correlate with PC clinical outcomes, independent of GS. All six participating sites contributed approximately 200 formalin-fixed and paraffin embedded RP specimens each, which were distributed on 33 TMA blocks. Each TMA block with 11 × 16 layouts was fabricated from 42 RP specimens and 8 normal control tissues including tonsil, prostate, kidney, colon and liver. Each PC, sampled with a 1 mm puncher, was represented by 3 cores obtained from the highest grade cancer in the dominant tumor nodule. In addition, 1 core was obtained from histologically benign prostatic tissue of every patient.
This study was conducted with a multi-institutional agreement and approvals from the institutional review boards at University of Washington, Stanford University, University of British Columbia, University of California San Francisco, University of Texas Health Sciences Center San Antonio, Veterans Affairs Puget Sound Health Care System, and Fred Hutchinson Cancer Research Center (FHCRC; Coordinating Center). De-identified demographic, clinical, and pathologic data is maintained in a central data repository at FHCRC managed by the Coordination Center.
Ki67 immunostaining
Unstained 4 μM TMA sections were deparaffinized on an automated immunostainer (Bond III (TM), Leica Biosystems, Germany) using a proprietary Bond Dewax solution. Three applications of the Dewax solution were followed by three applications of 100% ethanol and then three applications of Bond Wash solution. The immunostaining was performed in a CAP certified diagnostic immunohistochemistry laboratory according to a standardized protocol. In brief, antigen retrieval was performed on Bond III (TM) using ER2 buffer (pH 9.0) for 30 minutes. After rinsing and endogenous peroxidase blocking, a post primary IgG linker was applied followed by several rinses with the Bond wash solution and a de-ionized water rinse. The slides were incubated for 15 minutes with a mouse monoclonal IgG1 antibody against Ki67 (clone MIB-1, dilution 1:200, Cat# M7240, DAKO, Carpinteria, CA). Slides were then rinsed multiple times with Bond Wash solution, a polymer anti-mouse Poly-HRP-IgG was applied, and slides were incubated for 8 minutes with polymer detection reagent (Bond Polymer Refine Detection kit, Leica). This was followed by multiple rinses, reacted with 3,3″-Diaminobenzidine tetrahydrochloride chromogen for 10 minutes, and counterstained with hematoxylin for 5 minutes. Negative controls for the immunostaining were prepared by omitting the primary antibody step and substituting it with non-immune mouse serum. Normal tonsil tissue cores present on each TMA section served as internal positive controls for Ki67 staining.
Automated Image Analysis
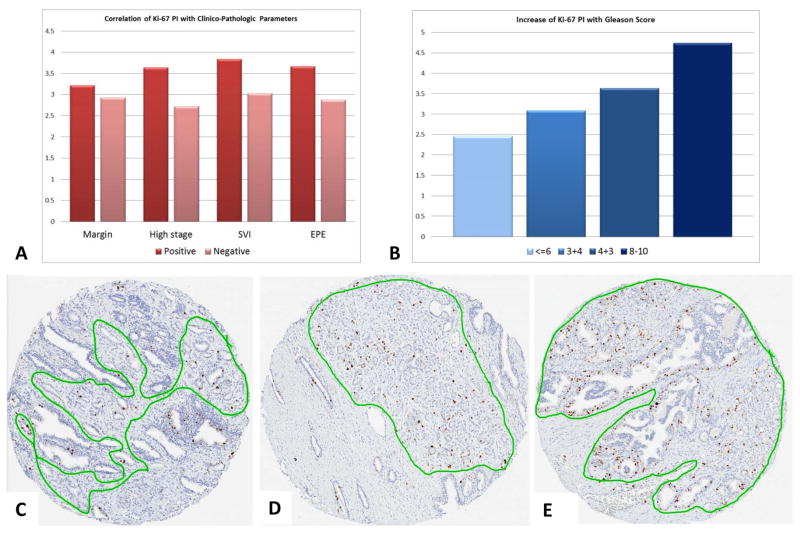
The TMA slides were scanned on Aperio ScanScope AT (Aperio Technologies, Vista, CA, USA) at 200× magnification. High resolution digital image files were created and saved in the web-based Spectrum Plus digital slide manager and segmented using TMALab to further automate the analysis of the individual gridded tissue cores. The tumor areas were manually annotated by an experienced genitourinary pathologist (MT) to maximally exclude non-tumor stroma, benign glands and inflammatory cells [Figure 1]. The Ki67 staining of the cells comprising the tumor areas of each core was determined using a nuclear quantification algorithm which was tuned by an experienced user (MT) to allow reliable identification of all Ki67 positive nuclei and hematoxylin counterstained negative nuclei (18). The threshold for size and shape of tumor cells was manually calibrated to maximally exclude stromal cells and lymphocytes as previously reported (19). Cases from TMA slides with weak or negative internal control cases (tonsil tissue) after repeat staining were excluded from further analysis. The Ki67 proliferation index (PI) was automatically calculated by the software as a ratio (%) of positively stained nuclei to all nuclei. For each core a median of 3,019 tumor nuclei were counted (range 19–12,091). Tumor PIs for each patient were averaged from 3 analyzed cores. The maximum score % Ki67 positivity per tumor was calculated as a surrogate of the ‘hotspot’ reflecting the area of most intense proliferation (20, 21). All cases were coded and analyzed by a pathologist in a blinded fashion, without any knowledge of patient outcome.
Figure 1.
Statistical Analysis
Patient characteristics were collected in the clinical data set and included pre-operative serum PSA level, pathology stage, GS, seminal vesicle invasion (SVI), extracapsular extension (ECE), and surgical margin status (positive or negative). Of 1326 patients with clinical data, 1004 patients had complete high-quality Ki67 tumor data available for analysis with acceptable strong uniform TMA staining of the positive controls.
The summary statistics of patient characteristics are provided in frequencies and percentages (see Table 1). Ki67 and pre-operative PSA data were summarized using mean, SD, and range. The Wilcoxon rank sum test or Kruskal-Wallis test were used to compare Ki67 PI between patient groups. The Pearson correlation test was used to assess association between Ki67 PI and pre-operative PSA. Inter- and intra-tumor variation of Ki67 positive nuclei were estimated using variance component analysis. Ki67 were also dichotomized by either weighted average of tumor Ki67 or maximum percent positive nuclei using 5% cutoff point reported previously for RP/TURP cohorts as an independent prognosticator in multivariable analyses (3, 7, 22–24).
Table 1.
Summary of Ki67 by patient clinico-pathologic characteristics
| Characteristics | Weighted Average Ki67 Percent Positive Nuclei | ||||||
|---|---|---|---|---|---|---|---|
| N | Mean | SD | Min | Median | Max | P-value | |
| Margin | |||||||
| Missing | 167 | 3.31 | 3.16 | 0.07 | 2.53 | 23 | |
| Negative | 552 | 2.93 | 2.81 | 0 | 2.04 | 26.01 | |
| Positive | 285 | 3.22 | 3.08 | 0 | 2.23 | 25.5 | 0.21 |
| Pathology Stage | |||||||
| Missing | 193 | 3.4 | 3.42 | 0 | 2.38 | 23 | |
| pT2 | 567 | 2.72 | 2.58 | 0 | 1.94 | 26.01 | |
| pT3/pT4 | 244 | 3.64 | 3.25 | 0 | 2.79 | 25.5 | <.0001 |
| Seminal Vesicle Invasion | |||||||
| Missing | 13 | 2.52 | 1.85 | 0.89 | 2.18 | 7.42 | |
| No | 925 | 3.03 | 2.95 | 0 | 2.14 | 26.01 | 0.02 |
| Yes | 66 | 3.84 | 3.11 | 0.21 | 3.41 | 15.62 | |
| Extra-Capsular Extension | |||||||
| Missing | 9 | 2.09 | 1.63 | 0.57 | 1.46 | 5.3 | |
| No | 742 | 2.88 | 2.78 | 0 | 2.02 | 26.01 | <.0001 |
| Yes | 253 | 3.67 | 3.38 | 0 | 2.69 | 25.5 | |
| Gleason Score | |||||||
| Missing | 8 | 3.27 | 4.44 | 0.56 | 1.36 | 13.63 | |
| <=6 | 400 | 2.46 | 2.1 | 0 | 1.93 | 14.68 | <.0001 |
| 3+4 | 370 | 3.09 | 2.59 | 0 | 2.42 | 23 | |
| 4+3 | 125 | 3.64 | 3.2 | 0.09 | 2.4 | 15.62 | |
| 8–10 | 101 | 4.74 | 5.12 | 0.22 | 2.77 | 26.01 | |
| Total | 1004 | 3.07 | 2.95 | 0 | 2.19 | 26.01 | |
P-values by Wilcoxon rank sum test or Kruskal-Wallis test, where appropriate.
The primary endpoints of this analysis were: 1) recurrence-free survival (RFS) post-surgery, where an event was defined as any PC recurrence (biological, clinical/radiological, or treatment with salvage therapy) or metastasis or death due to PC; 2) overall survival (OS), where an event was defined as death of any cause; and 3) disease-specific survival (DSS), where an event was defined as PC metastasis or death due to PC. The baseline was set as the date of RP.
There were four groups of patients defined in this study: 1) non-recurrence (48% cases), 2) recurrence within 5 years of RP (40% cases), 3) lost-to-follow up within 5 years of RP (6% cases), and 4) recurrence more than 5 years after RP (6% cases). The survival of patient groups was estimated using the Kaplan-Meier method and groups were compared using the log rank test. A univariable Cox model was used to assess the effects of Ki67 PI and other patient characteristics on RFS, OS, and DSS. A backwards elimination procedure including PSA, GS, age, margin, ECE, SVI, and Ki67 covariates was used to identify a final multivariable Cox model with all significant factors for each survival endpoint. The summaries of concordance index of different Cox proportional hazard models for RFS, types of recurrence and salvage therapy
All tests were two-sided and p-values of 0.05 or less were considered statistically significant. Statistical analysis was carried out using SAS version 9 (SAS Institute, Cary, NC). Plots were produced using Spotfire S+ 8.2 (TIBCO Inc., Palo Alto, CA).
RESULTS
Correlating Ki67 Proliferation Index with clinico-pathological characteristics
We determined the Ki67 PI in > 4,000 prostate tissue cores from 1004 RPs using the Aperio system for automated detection of staining and quantification of positive nuclei. The Ki67 PI ranged from 0 to 35.6% with weighted average median value of 2.19%. We found significant associations between Ki67 PI and pathologic stage, SVI, ECE, and GS (Table 1). More specifically, Ki67 PI was significantly higher in patients with pathologic stages pT3/pT4 vs. stage pT2 (p<0.0001), as well as cases with SVI and ECE (p=0.02 and p<0.0001, respectively). There was also a statistically significant increase in Ki67 PI from lower to higher GS (Figure 1). Strong positive correlation between higher GS and higher Ki67 PIs were found for weighted average and maximum Ki67 PI by Kruskal-Wallis test (p<0.0001). No significant association was found between Ki67 PI and positive surgical margin (PSM) status or pre-operative PSA by Wilcoxon rank sum test and Pearson’s correlation (p=0.21 and p=0.36, respectively).
The intra-tumor variance of Ki67 expression, estimated from different tumor cores, was 0.39 (95% CI: 0.36–0.41), whereas the inter-tumor variation was 0.68 (95% CI: 0.61–0.76). The inter-tumor variation explained 64% of total variation.
Ki67 PI analysis as a continuous variable: univariable and multivariable analysis for RFS
The summary of 5-year RFS by Ki67 PI and clinico-pathological factors are shown in supplementary Table 1. The univariable Cox proportional hazard model determined that the weighted average Ki67 PI was significantly correlated with RFS as a continuous variable (per 1% increase, HR=1.04, p=0.002), as well as maximum % Ki67 positivity (per 1% increase, HR=1.03, p=0.005). Other PC characteristics significantly associated with RFS events were PSM, presence of SVI and ECE, and (log) pre-operative PSA as continuous 1 unit incremental values (p<0.0001). Two multivariable Cox proportional hazard models (including 634 cases with 281 recurrences) for RFS demonstrated that higher weighted Ki67 PI average (model 1) and higher maximum Ki67 % positivity (model 2) were both significantly correlated with worse RFS after adjusting for other clinical factors (PSM, SVI, GS, ECE, and pre-operative PSA) (p=0.02–0.0008; Table 2). The summary of concordance index of different Cox proportional hazard models for RFS is shown in supplemental Table 2.
Table 2.
Ki67 analysis as a continuous variable: Multivariable Cox proportional hazard models for RFS, OS, and DSS.
| Model | Factor | Comparison | Hazard Ratio | 95% LCL | 95% UCL | Overall p-value |
|---|---|---|---|---|---|---|
| RFS Model 1 (N = 634, #Events = 281) | Ki67% weighted average | 1% increase | 1.07 | 1.03 | 1.11 | 0.0008 |
| Margin | Pos vs. Neg | 1.41 | 1.09 | 1.84 | 0.01 | |
| Seminal vesicle invasion | Yes vs. No | 1.85 | 1.23 | 2.80 | 0.003 | |
| Gleason score | 3+4 vs. 6 | 1.22 | 0.90 | 1.66 | 0.005 | |
| 4+3 vs. 6 | 1.88 | 1.33 | 2.66 | |||
| 8–10 vs. 6 | 1.42 | 0.96 | 2.09 | |||
| Pathologic stage | pT3/pT4 vs. pT2 | 1.43 | 1.07 | 1.92 | 0.02 | |
| Log(PSA) | 1 unit increase | 1.62 | 1.35 | 1.96 | <.0001 | |
| RFS Model 2 (N = 634, #Events = 281) | Ki67% maximum | 1% increase | 1.04 | 1.01 | 1.07 | 0.007 |
| Margin | Pos vs. Neg | 1.40 | 1.08 | 1.83 | 0.01 | |
| Seminal vesicle invasion | Yes vs. No | 1.85 | 1.22 | 2.79 | 0.004 | |
| Gleason score | 3+4 vs. 6 | 1.24 | 0.91 | 1.67 | 0.003 | |
| 4+3 vs. 6 | 1.91 | 1.35 | 2.70 | |||
| 8–10 vs. 6 | 1.46 | 0.99 | 2.15 | |||
| Pathologic stage | pT3/pT4 vs. pT2 | 1.44 | 1.07 | 1.93 | 0.02 | |
| Log(PSA) | 1 unit increase | 1.61 | 1.34 | 1.95 | <.0001 | |
| OS (N = 984, #Events = 57) | Ki67% positive | 1% increase | 1.09 | 1.01 | 1.16 | 0.02 |
| Gleason Score | 3+4 vs. <=6 | 0.87 | 0.44 | 1.72 | 0.68 | |
| 4+3 vs. <=6 | 1.14 | 0.46 | 2.84 | 0.78 | ||
| 8–10 vs. <=6 | 3.28 | 1.65 | 6.51 | 0.0007 | ||
| DSS (N = 874, #Events = 44) | Ki67% positive | 1% increase | 1.10 | 1.02 | 1.18 | 0.02 |
| Log(PSA) | 1 unit increase | 1.98 | 1.35 | 2.89 | 0.005 | |
| Gleason Score | 3+4 vs. <=6 | 2.27 | 0.93 | 5.52 | 0.07 | |
| 4+3 vs. <=6 | 2.75 | 0.97 | 7.81 | 0.06 | ||
| 8–10 vs. <=6 | 5.13 | 1.92 | 13.75 | 0.001 |
Recurrence free survival (RFS) event is defined as any recurrence, metastasis, or prostate cancer death. Models 1 and 2 are separate models using Ki67 weighted average % positive (PI=2.19) and maximum % positive (PI=3.11), respectively. Overall survival (OS) event is defined as death of any cause. Disease-specific survival (DSS) event is defined as prostate cancer metastasis or death due to PC. Both OS and DSS have only one final model. Only significant factors were included in the final models.
Ki67 PI analysis as a continuous variable: univariable and multivariable analysis for OS and DSS
The univariable Cox proportional hazard analysis demonstrated that significantly worse OS was associated with increasing Ki67 PI (p=0.003) or pre-operative PSA (p=0.018), stage pT3/pT4 vs. pT2 (p=0.01), presence of SVI (p<0.0001) or ECE (p=0.01), and GS of 8 or higher (vs. 6 or lower) (p=0.008). Worse DSS was also strongly associated with increasing Ki67 PI (p=0.004) and pre-operative PSA (p<0.0001), PSM (p=0.02), stage pT3/pT4 vs. pT2 (p<0.0001), presence of SVI (p=0.01), and GS of 6 vs 4+3 (p<0.0001). The multivariable Cox proportional hazard model for OS comprised a total sample size of 984 with 57 events. After adjusting for pre-operative PSA, PSM, pathologic stage, SVI, and ECE, worse OS was significantly associated with increasing by 1% Ki67 PI (HR=1.09, p=0.02) or GS of 8 or higher (vs. 6 or lower) (HR=3.28, p=0.0007). The same model was run for DSS and comprised a total sample size of 874 with 44 events. There were significant associations with worse DSS and increasing by 1% Ki67 PI (HR=1.1, p=0.02), pre-operative PSA (HR=1.98, p=0.005), and GS of 8 or higher (vs. 6 or lower) (HR=5.13, p=0.001). Multivariable cox regression analysis results for RFS, OS and DSS are shown in Table 2.
Defining clinically useful cutpoints for Ki67 PI
We examined the functional form and possible cutoff points of Ki67 PI and pre-operative PSA using Martingale residual plots (Supplementary Figure 1). The linear form of Ki67 and logarithmic transformation of pre-operative PSA were used for modeling. Martingale residual plots showed no discrete cut-point that could be used to dichotomize samples prognostically using log(PSA) or KI67 PI values. Therefore, we tested three different Ki67 cutpoints to dichotomize cases to evaluate associations with clinical endpoints: weighted average (2.19%), maximum per case (3.11%), and at cut-point of 5% which has been used successfully in several previous studies of localized PC (3, 7, 22–24).
The univariable Cox proportional hazard model showed significant correlation with RFS when dichotomized by median classes only for the weighted average (2.19%, HR=0.72, p=0.01), but not for the maximum % positive Ki67 per tumor (3.11%, HR=0.86, p=0.06). The multivariable Cox proportional hazard model for outcomes dichotomized by Ki67 PI weighted average and maximum Ki67 % positivity were not significant. Moreover, no significant relationship with OS and DSS was detected with either Ki67 cutoff of 2.19% or 3.11%.
The multivariable Cox proportional hazard model showed significant correlation with RFS when Ki67 dichotomized at 5% cutoff for both the weighted average (model 1) and for the maximum % positive (model 2). Tumors with Ki67 ≥5% were associated with worse RFS after adjusting for pre-op PSA, margin status, SV invasion status, and Gleason score: HR=1.47, p=0.0007 (model 1) and HR=1.31, p=0.03 (model 2). The multivariable Cox proportional hazard model for OS comprised a total sample size of 992 with 57 events showed significant relationship with worse OS and Ki67≥5% based on weighted average (HR=2.03, 95% CI 1.09–3.8, p=0.03). The same model was run for DSS and comprised a total sample size of 1352 with 51 events. Analysis of DSS demonstrated its significant relationship with Gleason scores (HR=2.6–8.11, p=0.0004) and (log) pre-operative PSA (HR=2.01, p=0.006), but not with Ki67 dichotomized at 5% cutoff. Multivariable cox regression analysis for RFS stratified by center is shown in Table 3.
Table 3.
Ki67 dichotomized at 5% cutoff: multivariable Cox proportional hazard models stratified by center for RFS.
| Model | Factor | Comparison | Hazard Ratio | 95% LCL | 95% UCL | Overall p-value |
|---|---|---|---|---|---|---|
| RFS Model 1 (N = 706, #Events = 319) | Ki67% weighted average | >=5% vs. <5% | 1.471 | 1.110 | 1.950 | 0.0007 |
| Margin | Pos vs. Neg | 1.598 | 1.248 | 2.047 | 0.0002 | |
| Seminal vesicle invasion | Yes vs. No | 2.072 | 1.430 | 3.002 | 0.0001 | |
| Gleason score | 3+4 vs. 6 | 1.297 | 0.972 | 1.732 | 0.004 | |
| 4+3 vs. 6 | 1.808 | 1.273 | 2.568 | |||
| 8–10 vs. 6 | 1.626 | 1.126 | 2.348 | |||
| Log(PSA) | 1 unit increase | 1.536 | 1.288 | 1.831 | <.0001 | |
| RFS Model 2 (N = 706, #Events = 319) | Ki67% maximum | >=5% vs. <5% | 1.314 | 1.033 | 1.672 | 0.03 |
| Margin | Pos vs. Neg | 1.596 | 1.246 | 2.043 | 0.0002 | |
| Seminal vesicle invasion | Yes vs. No | 2.119 | 1.462 | 3.070 | <.0001 | |
| Gleason score | 3+4 vs. 6 | 1.317 | 0.986 | 1.757 | 0.0002 | |
| 4+3 vs. 6 | 1.876 | 1.324 | 2.658 | |||
| 8–10 vs. 6 | 1.670 | 1.159 | 2.406 | |||
| Log(PSA) | 1 unit increase | 1.505 | 1.264 | 1.792 | <.0001 |
Recurrence free survival (RFS) event is defined as any recurrence, metastasis, or prostate cancer death. Models 1 and 2 are separate models using Ki67 weighted average % positive (PI=2.19) and maximum % positive (PI=3.11), respectively. Only significant factors were included in the final models.
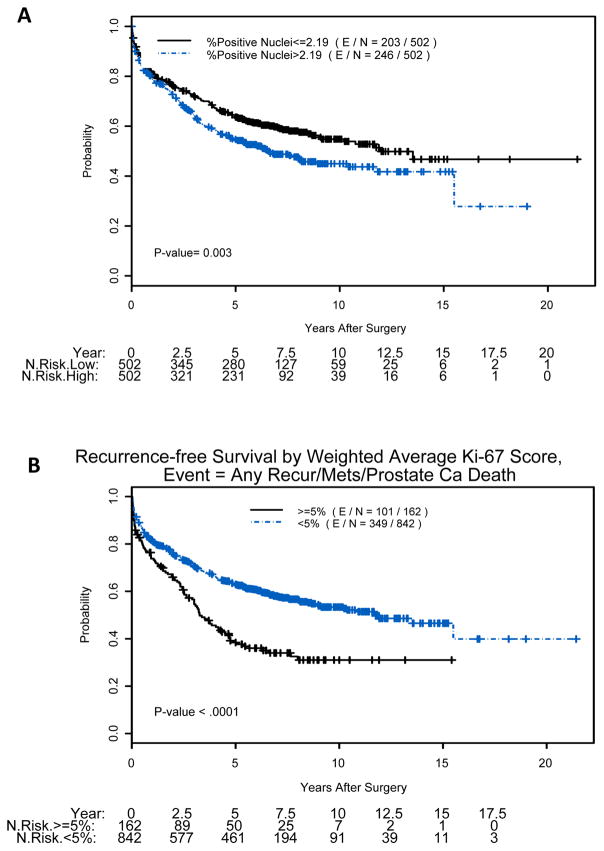
Kaplan-Meier plots for all survival endpoints indicated that patients with tumor Ki67 PI above the median (>2.19%) had a significantly decreased probability of RFS (p=0.003) (Figure 2A), which was even more evident with 5% cut-point (p<0.0001) (Figure 2B). Moreover, Kaplan-Meier survival curves with 5% cutoff point showed a trend toward predicting DSS in tumors with higher Ki67 (p=0.083) and significant difference for OS (p=0.045), but not for cutoffs of 2.19% or 3.11%. All p-values in this paragraph were produced by the log-rank test.
Figure 2.
DISCUSSION
Our Ki67 validation study was conducted in over 1,000 radical prostatectomies in compliance with MISHFISHIE (Minimum Information specification for in situ hybridization and immunohistochemistry experiments) (25) and REMARK (Reporting recommendations for tumor marker prognostic studies) (26, 27). Our objective was to evaluate whether Ki67 testing provides meaningful prognostication and whether an image-based automated scoring system could meet criteria for use in clinical practice. To our knowledge, this is the largest study to date, with one of the longest periods of follow-up (median: 7 years) (5, 9, 28), and represents a wide spectrum of patients with PC managed at six major centers across North America. We found that increased Ki67 as a continuous variable was significantly associated with stage pT3/pT4, presence of SVI or ECE, and higher GS, but not with PSM and/or pre-operative PSA levels. After adjusting for other factors on multivariable analysis, Ki67 as a continuous variable remained a significant independent predictor for recurrence-free, overall, and disease-specific survival using logistic regression models and Cox proportional hazard models. These findings substantiate Ki67 staining as an independent predictive biomarker for PC outcomes.
Our tested Ki67% cutoffs of weighted average and maximum % positivity per case (hotspot equivalent) were 2.19% and 3.11%, respectively. These values were comparable to several other studies (1, 29, 30), but at the lower end of broadly ranging cutoffs from 2.4% to 26%. The range of Ki67 PI values in these studies could reflect differences in the risk profiles of the patient cohorts, tumor heterogeneity, pre- and post - analytical variables, manual vs automated counting, and different statistical methodologies in determining “suitable cutpoints” as mean, median, maximal, or quartile-based (1, 3, 8, 14, 28, 30, 31). Our relatively low weighted average PI could also be partially explained by the study population that was selected to enrich for recurrent low-grade and non-recurrent high-grade cases, possibly leading to oversampling of cases with lower proliferative indices. However, our patient population reflects contemporary risk groups of prostate cancer diagnosed in North America, particularly patients who are candidates for active surveillance in whom prognostic biomarkers are most needed.
A recent study of PC patients treated on Radiation Therapy Oncology Group (RTOG 94–08) using automated ACIS scoring showed median Ki67 PI similar to what we observed (2.65%), although the automated scoring was slightly lower than the median value obtained by manual scoring in the same cohort (3.85%) (30). Manual scoring likely produces higher PIs because there typically are smaller numbers of nuclei counted and regions with more stained nuclei (hotspots) are likely to be oversampled. In agreement with these observations, median Ki67 PI in our study was nearly identical to RTOG 94–08 (PI=2.19%) while the maximum Ki67 PI % positivity per case in our study (PI=3.11%) was remarkably similar to manually scored hotspots.
Implementation of automated scoring in our high-throughput study was justified since 1) it provides more accurate determination of Ki67 PI due to higher numbers of counted cells (a median of 3,019 tumor nuclei), 2) it reduces human error and fatigue during quantitating of ~4,000 cores, and 3) it eliminates scoring variations by analysis of uniformly batch stained TMA slides with a standard nuclear counting algorithm. Furthermore, the accuracy of automated scoring has been separately validated in studies in which adequate sample sizes for manually counted nuclei were obtained. A comparative study between digital image analysis using the same Aperio XT nuclear algorithm and manual counting of adequate numbers of cells (>2,000 nuclei) showed excellent concordance with interclass correlation of 0.98 (19).
Our data suggest 2 alternate approaches for incorporating Ki67 PI into clinical practice. In multivariable analysis, both median and maximum KI67 PI provided independent prediction of RFS when coupled with other clinical and pathological parameters. In this case, KI67 PI could be incorporated into clinical risk models, such as CAPRA or other nomograms, although appropriate weighting of the models might need to be adjusted for a non-selected population. Alternatively, biomarkers with a purely dichotomous output (positive or negative) that sort patients into high and low risk groups are easier to incorporate into risk models for clinical use (14, 31). Despite our inability to define a cut-point of Ki67 PI in our dataset using the Lowess smoothed plot of Martingate residuals, we did validate that a cut-point of 5%, which had been derived empirically in previous studies (3, 7, 22–24), did provide independent prediction of RFS with a hazard ratio (HR=1.47) comparable to Gleason score (HR=1.29–1.81), margin status (HR=1.59) and log(PSA) increase by 1 unit (HR=1.54), although slightly lower than SVI (HR=2.07). Additionally, Kaplan-Meier plots showed robust curve separation between low and high Ki67 groups at 5% that outperformed median or maximum Ki67 PI per case. Whether 5% is the ideal cut-point is unclear, given the results of our cut-point analysis. However, Kaplan-Meier analysis also showed that tumors with very low Ki67 PI (≤1%) were indistinguishable from those that were 1–5%, implying that the cut-point is likely to be at 5% or greater.
Our study has some limitations since it is retrospective and based on radical prostatectomy samples and was designed with specific parameters to help in identifying prognostic biomarkers that are independent of clinical predictors. For example, the selection of balanced numbers of recurrent and non-recurrent Gleason score 3+4 and 4+3 cases and oversampling of recurrent 3+3 and non-recurrent 4+4 cases improved our ability to validate markers that predict outcomes after surgery that are independent of known clinical predictors such as Gleason score. Because of this, the relative weight of the biomarker in predicting outcome cannot be incorporated into existing algorithms, such as CAPRA, that have been developed and tested in non-selected surgical populations. In addition, selection of the region of highest grade in construction of the TMA could limit application of the findings to non-selected random biopsies and further confounded by intratumoral heterogeneity. In addition, there is considerable overlap in Ki67 PI in univariable analysis between favorable and high risk clinical and pathological outcomes, making clinical translation challenging. Very likely, the greatest utility will be derived from combining Ki67 PI with independent predictors, or by focusing more on extreme values, such as the >5% cut-point. Translation to clinically relevant scenarios, such as selection of patients for active surveillance in the low risk population or for adjuvant therapies in high risk localized disease will require testing in prospective cohorts. While our study is based on small samples of cancer, in many ways comparable to standard prostate needle biopsies, it will need to be validated on biopsy samples because of potential issues with sampling error in prostate needle biopsies. It is possible that the advent of image-directed biopsies using multiparametric MRI could significantly improve the performance of prognostic biomarkers because of improved sampling the largest incident lesion. However, this hypothesis needs to be tested.
CONCLUSIONS
In localized PC treated by radical prostatectomy, Ki67 PI provides independent prognostic value for RFS, DSS and OS beyond Gleason score, pathological stage and PSA levels. In our large, multi-institutional cohort, Ki67 PI performed best as a continuous variable and could be incorporated into existing or new predictive nomograms. Additionally, our study suggests that risk stratification for localized PC could be achieved with Ki67 as a dichotomous variable at 5% cutoff. These findings strongly suggest that Ki67 PI should be further tested as a prognostic biomarker in other clinically relevant cohorts such as patients on active surveillance (32), and possibly in patients undergoing image guided biopsies.
Supplementary Material
Acknowledgments
Supported by: The Canary Foundation; the Department of Defense W81XWH-11-1-0380; the NCI Early Detection Research Network U01 CA086402, CA152737, CA08636815; P30 CA054174; and the Pacific Northwest Prostate Cancer SPORE P50 CA097186
We cordially thank a large and dedicated Canary Foundation team of coordinating center staff, local coordinators, lab staff, and physicians who have made this study possible.
Footnotes
CONFLICT OF INTEREST
There are no conflicts of interest for any authors.
References
- 1.Fisher G, Yang ZH, Kudahetti S, Moller H, Scardino P, Cuzick J, et al. Prognostic value of ki-67 for prostate cancer death in a conservatively managed cohort. Br J Cancer. 2013;108(2):271–7. doi: 10.1038/bjc.2012.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Etzioni R, Penson DF, Legler JM, di Tommaso D, Boer R, Gann PH, et al. Overdiagnosis due to prostate-specific antigen screening: Lessons from U.S. prostate cancer incidence trends. J Natl Cancer Inst. 2002;94(13):981–90. doi: 10.1093/jnci/94.13.981. [DOI] [PubMed] [Google Scholar]
- 3.Berney DM, Gopalan A, Kudahetti S, Fisher G, Ambroisine L, Foster CS, et al. Ki-67 and outcome in clinically localised prostate cancer: Analysis of conservatively treated prostate cancer patients from the trans-atlantic prostate group study. Br J Cancer. 2009;100(6):888–93. doi: 10.1038/sj.bjc.6604951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hawley S, Fazli L, McKenney JK, Simko J, Troyer D, Nicolas M, et al. A model for the design and construction of a resource for the validation of prognostic prostate cancer biomarkers: The canary prostate cancer tissue microarray. Adv Anat Pathol. 2013;20(1):39–44. doi: 10.1097/PAP.0b013e31827b665b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kachroo N, Gnanapragasam VJ. The role of treatment modality on the utility of predictive tissue biomarkers in clinical prostate cancer: A systematic review. J Cancer Res Clin Oncol. 2013;139(1):1–24. doi: 10.1007/s00432-012-1351-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sebo TJ, Cheville JC, Riehle DL, Lohse CM, Pankratz VS, Myers RP, et al. Perineural invasion and MIB-1 positivity in addition to gleason score are significant preoperative predictors of progression after radical retropubic prostatectomy for prostate cancer. Am J Surg Pathol. 2002;26(4):431–9. doi: 10.1097/00000478-200204000-00004. [DOI] [PubMed] [Google Scholar]
- 7.Gunia S, Albrecht K, Koch S, Herrmann T, Ecke T, Loy V, et al. Ki67 staining index and neuroendocrine differentiation aggravate adverse prognostic parameters in prostate cancer and are characterized by negligible inter-observer variability. World J Urol. 2008;26(3):243–50. doi: 10.1007/s00345-008-0257-0. [DOI] [PubMed] [Google Scholar]
- 8.Van der Kwast TH. Prognostic prostate tissue biomarkers of potential clinical use. Virchows Arch. 2014;464(3):293–300. doi: 10.1007/s00428-014-1540-7. [DOI] [PubMed] [Google Scholar]
- 9.Zhao L, Yu N, Guo T, Hou Y, Zeng Z, Yang X, et al. Tissue biomarkers for prognosis of prostate cancer: A systematic review and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2014;23(6):1047–54. doi: 10.1158/1055-9965.EPI-13-0696. [DOI] [PubMed] [Google Scholar]
- 10.Dowsett M, Nielsen TO, A’Hern R, Bartlett J, Coombes RC, Cuzick J, et al. Assessment of Ki67 in breast cancer: Recommendations from the international Ki67 in breast cancer working group. J Natl Cancer Inst. 2011;103(22):1656–64. doi: 10.1093/jnci/djr393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voros A, Csorgo E, Nyari T, Cserni G. An intra- and interobserver reproducibility analysis of the ki-67 proliferation marker assessment on core biopsies of breast cancer patients and its potential clinical implications. Pathobiology. 2013;80(3):111–8. doi: 10.1159/000343795. [DOI] [PubMed] [Google Scholar]
- 12.Bosman FT, Carneiro F, Hruban RH, Theise ND, editors. WHO classification of tumours of the digestive system. 4. Geneva, Switzerland: WHO Press; 2010. [Google Scholar]
- 13.Edge S, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A, editors. AJCC cancer staging manual. 7. New York, NY: Springer; 2009. [Google Scholar]
- 14.Luporsi E, Andre F, Spyratos F, Martin PM, Jacquemier J, Penault-Llorca F, et al. Ki-67: Level of evidence and methodological considerations for its role in the clinical management of breast cancer: Analytical and critical review. Breast Cancer Res Treat. 2012;132(3):895–915. doi: 10.1007/s10549-011-1837-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L, Yang B. Immunohistochemical analysis of p16, p53, and ki-67 expression in uterine smooth muscle tumors. Int J Gynecol Pathol. 2008;27(3):326–32. doi: 10.1097/PGP.0b013e31815ea7f5. [DOI] [PubMed] [Google Scholar]
- 16.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al., editors. HO classification of tumours of haematopoietic and lymphoid tissues. 4. Geneva, Switzeland: WHO Press; 2008. [Google Scholar]
- 17.Rekhtman NBJA. Quick reference handbook for surgical pathologists. 1. Berlin, Heidelberg: Springer; 2011. [Google Scholar]
- 18.Tretiakova M, Turkyilmaz M, Grushko T, Kocherginsky M, Rubin C, Teh B, et al. Topoisomerase IIalpha in wilms’ tumour: Gene alterations and immunoexpression. J Clin Pathol. 2006;59(12):1272–7. doi: 10.1136/jcp.2005.031963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang LH, Gonen M, Hedvat C, Modlin IM, Klimstra DS. Objective quantification of the Ki67 proliferative index in neuroendocrine tumors of the gastroenteropancreatic system: A comparison of digital image analysis with manual methods. Am J Surg Pathol. 2012;36(12):1761–70. doi: 10.1097/PAS.0b013e318263207c. [DOI] [PubMed] [Google Scholar]
- 20.Halvorsen OJ, Haukaas S, Hoisaeter PA, Akslen LA. Maximum ki-67 staining in prostate cancer provides independent prognostic information after radical prostatectomy. Anticancer Res. 2001;21(6A):4071–6. [PubMed] [Google Scholar]
- 21.Gravdal K, Halvorsen OJ, Haukaas SA, Akslen LA. Proliferation of immature tumor vessels is a novel marker of clinical progression in prostate cancer. Cancer Res. 2009;69(11):4708–15. doi: 10.1158/0008-5472.CAN-08-4417. [DOI] [PubMed] [Google Scholar]
- 22.May M, Siegsmund M, Hammermann F, Loy V, Gunia S. Prognostic significance of proliferation activity and neuroendocrine differentiation to predict treatment failure after radical prostatectomy. Scand J Urol Nephrol. 2007;41(5):375–81. doi: 10.1080/00365590701224445. [DOI] [PubMed] [Google Scholar]
- 23.Rubio J, Ramos D, Lopez-Guerrero JA, Iborra I, Collado A, Solsona E, et al. Immunohistochemical expression of ki-67 antigen, cox-2 and Bax/Bcl-2 in prostate cancer; prognostic value in biopsies and radical prostatectomy specimens. Eur Urol. 2005;48(5):745–51. doi: 10.1016/j.eururo.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 24.Miyake H, Muramaki M, Kurahashi T, Takenaka A, Fujisawa M. Expression of potential molecular markers in prostate cancer: Correlation with clinicopathological outcomes in patients undergoing radical prostatectomy. Urol Oncol. 2010;28(2):145–51. doi: 10.1016/j.urolonc.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 25.Deutsch EW, Ball CA, Berman JJ, Bova GS, Brazma A, Bumgarner RE, et al. Minimum information specification for in situ hybridization and immunohistochemistry experiments (MISFISHIE) Nat Biotechnol. 2008;26(3):305–12. doi: 10.1038/nbt1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altman DG, McShane LM, Sauerbrei W, Taube SE. Reporting recommendations for tumor marker prognostic studies (REMARK): Explanation and elaboration. PLoS Med. 2012;9(5):e1001216. doi: 10.1371/journal.pmed.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, et al. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97(16):1180–4. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 28.Kristiansen G. Diagnostic and prognostic molecular biomarkers for prostate cancer. Histopathology. 2012;60(1):125–41. doi: 10.1111/j.1365-2559.2011.04083.x. [DOI] [PubMed] [Google Scholar]
- 29.Shimizu Y, Segawa T, Inoue T, Shiraishi T, Yoshida T, Toda Y, et al. Increased akt and phosphorylated akt expression are associated with malignant biological features of prostate cancer in japanese men. BJU Int. 2007;100(3):685–90. doi: 10.1111/j.1464-410X.2007.07014.x. [DOI] [PubMed] [Google Scholar]
- 30.Verhoven B, Yan Y, Ritter M, Khor LY, Hammond E, Jones C, et al. Ki-67 is an independent predictor of metastasis and cause-specific mortality for prostate cancer patients treated on radiation therapy oncology group (RTOG) 94–08. Int J Radiat Oncol Biol Phys. 2013;86(2):317–23. doi: 10.1016/j.ijrobp.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Royston P, Altman DG, Sauerbrei W. Dichotomizing continuous predictors in multiple regression: A bad idea. Stat Med. 2006;25(1):127–41. doi: 10.1002/sim.2331. [DOI] [PubMed] [Google Scholar]
- 32.Newcomb LF, Brooks JD, Carroll PR, Feng Z, Gleave ME, Nelson PS, et al. Canary prostate active surveillance study: Design of a multi-institutional active surveillance cohort and biorepository. Urology. 2010;75(2):407–13. doi: 10.1016/j.urology.2009.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.