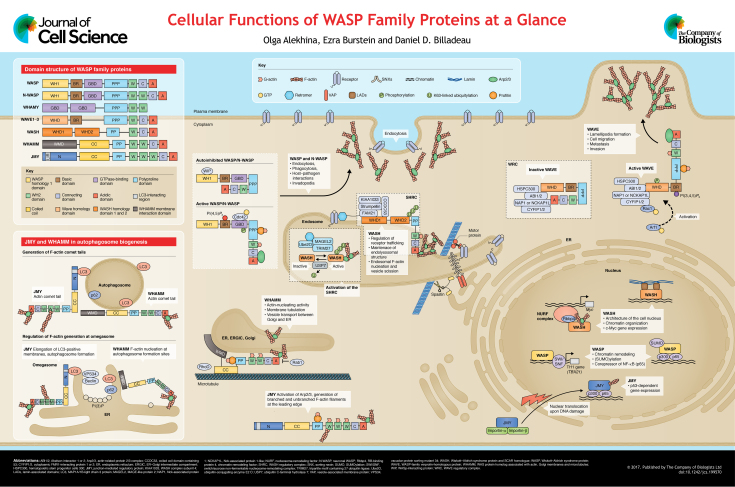
ABSTRACT
Proteins of the Wiskott–Aldrich syndrome protein (WASP) family function as nucleation-promoting factors for the ubiquitously expressed Arp2/3 complex, which drives the generation of branched actin filaments. Arp2/3-generated actin regulates diverse cellular processes, including the formation of lamellipodia and filopodia, endocytosis and/or phagocytosis at the plasma membrane, and the generation of cargo-laden vesicles from organelles including the Golgi, endoplasmic reticulum (ER) and the endo-lysosomal network. Recent studies have also identified roles for WASP family members in promoting actin dynamics at the centrosome, influencing nuclear shape and membrane remodeling events leading to the generation of autophagosomes. Interestingly, several WASP family members have also been observed in the nucleus where they directly influence gene expression by serving as molecular platforms for the assembly of epigenetic and transcriptional machinery. In this Cell Science at a Glance article and accompanying poster, we provide an update on the subcellular roles of WHAMM, JMY and WASH (also known as WASHC1), as well as their mechanisms of regulation and emerging functions within the cell.
KEY WORDS: WASP, N-WASP, WAVE, WHAMM, WASH, JMY, WHAMY, Arp2/3, Actin
Summary: This article provides insight into new functions of WASP family proteins from regulating the biogenesis of autophagosomes to recently identified roles in the nucleus.
Introduction
Actin monomers are dynamically incorporated into filamentous actin (F-actin) to perform crucial cellular functions including cell migration/invasion, cell–cell adhesion, endocytosis/phagocytosis, cytokinesis and intracellular membrane transport, to name a few. Over two decades ago, the gene mutated in the rare X-linked immunodeficiency Wiskott–Aldrich syndrome was identified to encode the hematopoietically expressed Wiskott–Aldrich syndrome protein (WASP; also known as WAS) (Derry et al., 1994). Since then, the list of mammalian WASP family proteins has grown and includes five subfamilies: WASP and neuronal-WASP (N-WASP; also known as WASL), the three WASP family verprolin homolog isoforms (WAVE1–WAVE3; also known as SCAR1–SCAR3 and WASF1–WASF3), WASP homolog associated with actin, membranes and microtubules (WHAMM), WASP and SCAR homolog (WASH; also known as WASHC1), and junction-mediating regulatory protein (JMY) (Campellone and Welch, 2010). Although they all share the conserved WCA (for ‘WH2, connecting and acidic’) domain required for Arp2/3 activation and F-actin nucleation, they also contain unique domains, which regulate their assembly into macromolecular complexes, their subcellular localization and/or their interaction with proteins that regulate their activity. In this way, branched F-actin formation by the Arp2/3 complex can be spatially and temporally regulated on membranes throughout the cell and integrated downstream of a host of intracellular signaling pathways. The regulation and cellular functions of WASP/WAVE family proteins have been the topic of several recent reviews (Burianek and Soderling, 2013; Campellone and Welch, 2010; Rottner et al., 2010) (Box 1). In addition, several additional WASP family proteins have been identified in invertebrates (Box 2). Herein, we will focus on the recently identified roles for WHAMM, JMY and WASH family members at organelles, cytoplasmic membranes and emerging roles in the nucleus.
Box 1. WASP and WAVE regulation and function.
The WASP homology 1 (WH1) domain interacts with WASP-interacting protein (WIP; also known as WIPF1) or other WIP homologs (de la Fuente et al., 2007; Ramesh et al., 1997) and is required to stabilize WASP. Direct binding to the GTPase-binding domain (GBD) of WASP by the GTP-bound form of Cdc42, a member of the Rho GTPase family, relieves an autoinhibitory fold (Kim et al., 2000). Phosphatidyl-inositol (4,5)-bisphosphate binding to the N-WASP basic region (BR) promotes F-actin nucleation synergistically with Cdc42 binding (Rohatgi et al., 2000). Profilin–G-actin binding to the polyproline region provides a pool of monomeric G-actin for F-actin generation. While WASP has been primarily implicated in immune synapse formation in T cells, and phagocytosis and podosome formation in monocytes (Massaad et al., 2013), N-WASP has been linked to endocytosis, host–pathogen interactions and invadopodia (Burianek and Soderling, 2013).
WAVE proteins are intrinsically inactive and exist in a pentameric complex known as the WAVE regulatory complex (WRC), which includes ABI1 or ABI2, NAP1 (also known as NCKAP1) or NCKAP1L, CYFIP1 or CYFIP2 and HSPC300 (Chen et al., 2010; Eden et al., 2002; Ismail et al., 2009; Stradal et al., 2004) (see poster). Rac1 has been shown to activate WAVE proteins through an interaction with the WRC component CYFIP1/2 (Chen et al., 2010). In addition, Arf1 has been found to cooperate with Rac1 in activating the WRC (Koronakis et al., 2011). Another layer of activation involves the binding of phosphatidylinositol (3,4,5)-trisphosphate [PI(3,4,5)P3] to the WAVE BR and phosphorylation of WAVE, which can further increase their activity toward Arp2/3 (Lebensohn and Kirschner, 2009). Through these interactions, the WRC can become activated at membranes where it promotes lamellipodia formation, thereby driving cell migration, metastasis and invasion in multiple tumor models (Burianek and Soderling, 2013). In addition, a WRC-interacting receptor sequence (WIRS) was identified in a number of membrane receptors that directly bind to the WRC components CYFIP1 and ABI1/ABI2 (Chen et al., 2014). Significantly, FAT2 (also known as Kugelei) contains such an element, and through this interaction, the WRC drives collective cell migration in Drosophila (Squarr et al., 2016).
Finally, oligomerization of WASP and WAVE proteins has been shown to synergistically increase Arp2/3-dependent F-actin nucleation (Padrick et al., 2008). This is accomplished by two WCA domains interacting with two unique sites on the Arp2/3 complex simultaneously to promote actin nucleation (Padrick et al., 2011).
Box 2. WASP family members WHAMY, WAML and WAWH.
Evolutionary investigation of WASP family members in other organisms has revealed at least three other families of WASP proteins known as WHAMY, WAML (WASP and MIM-like) and WAWH (WASP without WHI domain) (Kollmar et al., 2012; Veltman and Insall, 2010). WHAMY is found in Drosophila, whereas WAML and WAWH have only been found in amoeba, Apusozoa and the anole lizard (Kollmar et al., 2012). At this point, functional studies of WAML and WAWH have not been performed. Interestingly, WAML combines the WCA domain of WASP family proteins with the membrane-deforming functions of IRSp53-Mim-homology domain (IMD)-containing proteins like missing-in-metastasis (MIM) and insulin receptor tyrosine kinase substrate p53 (IRSp53), thus likely coordinating F-actin generation with membrane deformation.
WHAMY was originally suggested to be a WHAMM and JMY family member (Veltman and Insall, 2010). However, it was recently shown that WHAMY arose in Drosophila as a result of an N-WASP gene duplication (Brinkmann et al., 2016). It is of interest that WHAMY possesses two GBDs at its N-terminus, which have specificity for active Rac1 and not Cdc42. WHAMY was found to localize to the leading edge of migrating cells and on cytoplasmic vesicles that were positive for Rab11. Interestingly, WHAMY localization to the leading edge and filopodial tips of spreading cells required the two GBD motifs, and deletion of WHAMY in macrophages showed an important role in generating membrane protrusions and regulating cell migration. Significantly, WHAMY did not stimulate Arp2/3 actin nucleation in vitro, but instead promoted fast filament growth that was stimulated by active Rac1, suggesting that WHAMY may exist in an auto-inhibited conformation. Further studies revealed that WHAMY interacts with N-WASP and could potentiate N-WASP activity toward Arp2/3 in vitro. Taken together, it was proposed that WHAMY works together with N-WASP where WHAMY might generate linear mother filaments near Arp2/3 to promote N-WASP branched F-actin generation. Indeed, genetic studies in Drosophila are suggestive of synergistic interactions between WHAMY and N-WASP in actin nucleation regulation, sensory organ development and myoblast fusion during embryonic muscle formation. Thus, although WHAMY is related to N-WASP and has retained the functions of WASP in development, it has also gained new functions in cell motility.
Regulation and cytoplasmic functions of WHAMM, JMY and WASH
WHAMM
WHAMM possesses an N-terminal WHAMM membrane interaction domain (WMD), a coiled-coil (CC) microtubule (MT)-binding domain, a proline-rich sequence, and a C-terminal WWCA motif involved in regulating actin nucleation (Campellone et al., 2008) (see poster). WHAMM has been found to localize to the endoplasmic reticulum (ER)–Golgi intermediate compartment (ERGIC) where it regulates vesicle transport (Campellone et al., 2008). Rab1 (also known as Rab1a) has been shown to inhibit WHAMM-mediated actin assembly in vitro and regulate WHAMM membrane recruitment through an interaction with the WMD (Russo et al., 2016). More recent studies have shown that RhoD interacts with WHAMM to regulate Golgi structure and the transport of cargo to the plasma membrane (Blom et al., 2015; Gad et al., 2012). Cryoelectron microscopy revealed that WHAMM binds to the outer surface of MT protofilaments via its CC domain and forms helical head-to-tail structures along MTs (Shen et al., 2012). MT binding induces a structural change that exposes the N-terminal WMD for membrane binding and tubulation activity, while masking the C-terminal WWCA motif, thus preventing actin polymerization (Liu et al., 2017). In this way, the coordinated actions of the actin and MT cytoskeletons can regulate membrane deformation and tubulation to generate cargo-laden vesicles for transport.
JMY
JMY was originally identified as a novel p300 (also known as EP300) transcriptional cofactor that augmented p53-dependent (p53 is also known as TP53) transcription in response to DNA damage (Shikama et al., 1999). Subsequently, JMY was found to posses a WCA domain containing three WH2 domains (WWWCA), a proline-rich sequence and a coiled-coil domain at its N-terminus (Coutts et al., 2009; Zuchero et al., 2009) (see poster). JMY has the ability to activate Arp2/3 and produce branched F-actin filaments, but also generates unbranched filaments via its tandem WH2 domains (Coutts et al., 2009; Zuchero et al., 2009). Under non-stressed conditions, JMY can be seen at the leading edge of the cell, but upon DNA damage, JMY accumulates in the nucleus (Zuchero et al., 2009, 2012). The mechanism regulating JMY nuclear import in response to DNA damage was revealed when a bipartite nuclear localization signal (NLS) sequence was found within the tandem WH2 domains (Zuchero et al., 2012). When in the cytosol, G-actin bound to the tandem WH2 domains obscures the NLS and inhibits importin binding, thus preventing JMY nuclear accumulation. Upon DNA damage, there is enhanced cytoplasmic actin polymerization, which sequesters monomeric actin into F-actin. This effectively decreases the binding of G-actin to the WH2 domains, thereby exposing the NLS; this then results in importin binding, JMY nuclear accumulation and p53-dependent gene expression (Zuchero et al., 2012).
WASH
WASH (WASH complex subunit 1; WASHC1) contains an N-terminal WASH homology domain 1 (WHD1), a tubulin-binding WASH homology domain 2 (WHD2), a proline-rich region and C-terminal WCA domain. WASH is intrinsically inactive and exists in a macromolecular pentameric complex named the WASH regulatory complex (SHRC) that also includes FAM21A or FAM21C (WASHC2A and WASHC2C), strumpellin (WASHC5), KIAA1033 (WASHC4) and CCDC53 (WASHC3) (Derivery et al., 2009; Gomez and Billadeau, 2009; Jia et al., 2010) (see poster). Interestingly, the SHRC appears structurally related to the WAVE regulatory complex (WRC), both in terms of complex assembly and the similarity in shape and size as determined by electron microscopy of the two complexes (Jia et al., 2010). While studies in Drosophila melanogaster have revealed a role for Rho1 in activating WASH (Liu et al., 2009), mammalian RhoA did not activate the SHRC in vitro (Jia et al., 2010). WASH activity is regulated by K63-linked polyubiquitylation, which serves to change the conformation of WASH, resulting in exposure of its C-terminal WCA domain (Hao et al., 2013). This is mediated by a TRIM27-containing ubiquitin ligase complex that also contains MAGEL2 and UBE2O, and is regulated by the deubiquitinase USP7 (Hao et al., 2015). Finally, although deletion of WASH in Drosophila was initially found to impair oogenesis and larval development (Linardopoulou et al., 2007; Liu et al., 2009), a recent study using a different WASH mutant showed that homozygous wash mutant flies are viable and fertile (Nagel et al., 2017). This discrepancy was likely caused by a second-site lethal mutation in the Drosophila strain used in the former studies. In contrast, genetic deletion of WASH in mice results in early embryonic lethality (Gomez et al., 2012; Xia et al., 2013). Interestingly, mutations in the genes encoding KIAA1033 and strumpellin have been linked to intellectual disability (ID) syndromes (Elliott et al., 2013; Ropers et al., 2011), and separate mutations in strumpellin have been found in patients with hereditary spastic paraplegia (HSP) (Valdmanis et al., 2007). However, the mechanism by which WASH deregulation contributes to the pathology of either disease is unclear.
WASH localizes to endosomes (Derivery et al., 2009; Gomez and Billadeau, 2009; Harbour et al., 2012; Jia et al., 2012; Monfregola et al., 2010; Monteiro et al., 2013; Ryder et al., 2013), and WASH-generated F-actin not only regulates the architecture of the endolysosomal system, but also spares specific cargo from lysosomal degradation or missorting (Bartuzi et al., 2016; Derivery et al., 2009, 2012; Duleh and Welch, 2010; Gomez and Billadeau, 2009; Gomez et al., 2012; Monteiro et al., 2013; Zech et al., 2011). WASH is recruited to retromer-positive endosomal subdomains via the interaction of the extended C-terminal tail of FAM21 with the retromer subunit VPS35 (Harbour et al., 2012; Jia et al., 2012). Interestingly, a mutation in VPS35 (D620N) (Vilariño-Güell et al., 2011; Zimprich et al., 2011) that is associated with early- onset Parkinson's disease has been shown to diminish the interaction of the SHRC with VPS35 and impair vesicular trafficking from the late endosome (Follett et al., 2013; McGough et al., 2014; Zavodszky et al., 2014). Studies in Drosophila have recently shown important roles for WASH in regulating integrin receptor trafficking and lysosome acidification (Nagel et al., 2017). A similar role for WASH in the recycling of integrins has been seen in mammalian cells (Zech et al., 2011). Moreover, WASH regulates the retrieval of the vacuolar (V)-ATPase from post-lysosomal compartments, and the recycling of plasma membrane components from early macropinosomes and phagosomes in Dictyostelium (Buckley et al., 2016; Carnell et al., 2011). Thus, the ability of WASH to regulate receptor recycling and lysosome neutralization has been highly conserved throughout evolution.
WASH-depleted cells contain cargo-laden tubules (Derivery et al., 2009; Gomez and Billadeau, 2009) and display a collapse of the endolysosomal system (Gomez et al., 2012). These observations have promoted the hypothesis that WASH-generated F-actin regulates tubule fission through the generation of F-actin-mediated forces (Derivery et al., 2009). More recently it was shown that minus-end and plus-end MT motor complexes are recruited to WASH-labeled sorting domains where they coordinate a ‘tug-of-war’, causing endosomal tubulation and ultimately fission of vesicles containing MT1-MMP (also known as MMP14) (Marchesin et al., 2015). In addition, three reports have implicated the ER in regulating vesicular scission at WASH-containing endosomal sorting domains. The first study showed that the ER contacts FAM21-positive endosomes at sites of constricting tubules prior to dynamin-mediated fission (Rowland et al., 2014). The second study demonstrated that the ER-anchored vesicle-associated membrane protein (VAMP)-associated proteins (VAPs) directly interact with retromer-associated SNX2 to regulate phosphatidylinositol 4-phosphate [PI(4)P] levels and WASH activity (Dong et al., 2016). The final study showed that the ER-associated MT-severing protein spastin, which is mutated in HSP, drives endosomal tubule fission at ER–endosome contact sites (Allison et al., 2017). Interestingly, more tubules or collapsed tubules were prevalent in the absence of spastin, as was the acquisition of an abnormal lysosomal morphology. Similar effects on lysosome morphology were observed when strumpellin, which is also mutated in HSP, was depleted (Allison et al., 2017). Thus, vesicle scission at WASH-containing endosomes requires not only F-actin, but also the coordinated involvement of MTs, their associated motor and severing proteins, and the ER.
Centrosomal F-actin was previously observed (Hubert et al., 2011), but it was not until recently that WASH was identified as the nucleation-promoting factor involved in its generation (Farina et al., 2015). Farina et al. found that the accumulation of WASH on centrioles required pericentriolar material protein 1, MTs and dynein. Significantly, depletion of WASH impaired centrosomal F-actin nucleation (Farina et al., 2015). However, the mechanism by which WASH is activated at the centrosome, as well as the contribution of F-actin nucleation to centrosome architecture, positioning and organelle interaction, and the impact of F-actin on primary cilia formation and function remain to be determined.
Role of the WASP family proteins in autophagy
JMY and WHAMM
Autophagy is an evolutionarily conserved catabolic process used by cells to degrade cytoplasmic proteins and organelles in the lysosome (Kroemer et al., 2010). Although the formation of autophagosomes can occur at multiple cellular membranes, ER subdomains enriched in phosphatidylinositol 3-phosphate [PI(3)P], called omegasomes, are a major site for the generation of nascent autophagosomes (Feng et al., 2014). A central question in autophagosome formation is how the omegasome membrane shape is initiated and ultimately elongates and matures into an autophagosome. It is known that the actin cytoskeleton is required during the early events of autophagosome formation (Aguilera et al., 2014), but whether Arp2/3 or any WASP family members are involved in this process was unknown. Recently, two independent groups have identified roles for JMY and WHAMM in autophagosome biogenesis (Coutts and La Thangue, 2015; Kast et al., 2015) (see poster). JMY was found to promote the formation of elongated membranes containing LC3 (MAP1LC3B), a marker of autophagosomes (Coutts and La Thangue, 2015). Significantly, the authors identified an LC3-interacting region (LIR) in the N-terminus of JMY, which was required for the recruitment of JMY to LC3-containing autophagosomes. Mutation of the LIR prevented JMY colocalization with foci of F-actin following autophagy induction, suggesting that JMY was involved in generating F-actin at sites of autophagosome formation. Indeed, JMY harboring a W981A mutation, which abrogates its ability to activate Arp2/3, did not impact its ability to localize with LC3, but did diminish the extent of F-actin nucleation at the autophagosome. Since JMY can nucleate F-actin by Arp2/3-dependent and -independent mechanisms (Coutts et al., 2009; Zuchero et al., 2009), it is likely that the residual F-actin present at the autophagosomes is being generated by the WH2 domains of JMY. Consistent with this, re-expression of a ΔWWWCA JMY mutant in JMY-depleted cells resulted in decreased LC3 cleavage compared to that seen with the W981A mutant and reduced cell viability under conditions of autophagy (Coutts and La Thangue, 2015). Taken together, these data indicate that JMY influences autophagosome formation and cell viability in an actin-mediated manner via Arp2/3-dependent and -independent mechanisms.
A separate study revealed an important role for WHAMM in autophagosome biogenesis (Kast et al., 2015). There, the authors observed that during starvation conditions WHAMM was recruited to ER membranes. Interestingly, WHAMM appeared to propel the movement of autophagosome puncta through the generation of actin comet tails in a manner that required its activity toward Arp2/3. Silencing of WHAMM resulted in far fewer actin comet tails, and impaired the size and number of autophagosomes. They further found that the N-terminal WMD domain of WHAMM was sufficient for its recruitment to the ER, suggesting it harbors an ER-targeting motif. Why autophagosome biogenesis requires both WHAMM and JMY remains to be resolved, but they could either be coordinating F-actin nucleation efforts in regions where autophagosomes are generated, or participate at distinct steps in the process.
WASH
The role of WASH in autophagy is controversial. It has been shown that WASH deficiency in mice results in extensive autophagy, which likely leads to early embryonic lethality (Xia et al., 2013). WASH was found to associate with autophagosomes independently of its nucleation-promoting factor (NPF) activity and other components of the WASH complex, where it prevented the ubiquitylation of Beclin-1 by AMBRA1, an event required for the activation of VPS34 (also known as PIK3C3) (Xia et al., 2013). A follow-up study revealed another layer of WASH regulation; WASH recruited the E3-ligase RNF2, which ubiquitylated AMBRA1 leading to its degradation, thus preventing autophagy induction (Xia et al., 2014a). In contrast, siRNA-mediated silencing of WASH in neuroblastoma cells impaired ATG9A trafficking, an event required for autophagosome biogenesis, resulting in increased cell death of WASH-depleted cells in response to starvation conditions (Zavodszky et al., 2014). These results implicate WASH as a positive regulator of autophagy. It is unclear how to reconcile these two disparate findings, but it was recently shown that genetic deletion of WASH in Drosophila also leads to excessive autophagy (Nagel et al., 2017). Thus, the conservation of WASH as an inhibitor of autophagy in invertebrates and vertebrates is suggestive of an evolutionary conserved function. Clearly more work will be needed to understand autophagy regulation meditated by WASH in different species.
WASP family proteins in the nucleus
WASP
In addition to the role of JMY in the nucleus as a co-factor for p53, a number of recent studies have identified actin-dependent and actin-independent nuclear functions for other WASP family proteins, from regulating nuclear shape to acting as a scaffold in chromatin-remodeling complexes (CRCs), to gene regulation (for a detailed review, please see Verboon et al., 2015b). These observations have revealed unexpected functions for WASP proteins in the nucleus and have provided insight into their roles in disease.
Loss-of-function mutations in WAS can result in X-linked thrombocytopenia, a bleeding disorder associated with platelet dysfunction, or Wiskott–Aldrich syndrome, where in addition to thrombocytopenia, patients also develop immunodeficiency. Paradoxically, despite having immunodeficiency, Wiskott–Aldrich syndrome patients develop autoimmunity (Massaad et al., 2013). Vyas and colleagues revealed the reason for this paradox in a series of elegant studies. In the absence of WASP, T-helper (TH0) cells primarily developed into TH2 cells, which are more prone to promoting autoimmunity. They showed that WASP accumulates in the nucleus of TH0 cells that have been activated through their T-cell receptor and stimulated to polarize toward a TH1 phenotype, which is driven by the expression of the transcription factor T-BET (encoded by the TBX21 gene) (Taylor et al., 2010). WASP was found to associate with the TBX21 promoter, where it recruited the pre-initiation complex, chromatin-remodeling factors and subunits of the SWI/SNF CRC (Sarkar et al., 2014; Taylor et al., 2010). WASP SUMOylation converts WASP from a transcriptional coactivator into a corepressor of nuclear factor (NF)-κB in cells that are differentiating into TH1 cells (Sarkar et al., 2015). Interestingly, mutants within the WH1 domain, which are associated with full-blown Wiskott–Aldrich syndrome, compromised the recruitment of these factors and impaired TH1 polarization. This was not seen with mutants located in the WCA domain, which are associated with X-linked thrombocytopenia (and not Wiskott–Aldrich syndrome), suggesting that the nuclear effects of WASP in driving TH1 development were independent of its activity toward Arp2/3 (Sadhukhan et al., 2014). Taken together, these data have revealed a novel role for WASP in the nucleus and have begun to explain how disease-causing mutations can determine disease manifestations. Clearly, the nuclear roles of WASP in regulating the differentiation and functions of other immune cell types will be an area of interest for future studies.
WASH
It is noteworthy that Drosophila WASH, as well as strumpellin and SWIP were originally defined as components of the core promoter transcription complex containing TRF2 (also known as TBPL2), which associates with CRCs to regulate the expression of DNA replication-related element-containing genes (Hochheimer et al., 2002). WASH has a highly conserved bipartite NLS and nuclear export signal (NES) (Linardopoulou et al., 2007), and the observed subcellular localizations of WASH in Drosophila cells, as well as in mouse hematopoietic stem cells, range from primarily cytosolic to mostly nuclear (Verboon et al., 2015a; Xia et al., 2014b). In addition to WASH, all components of the SHRC, apart from CCDC53, contain predicted NLS motifs. In fact, nuclear FAM21, but not WASH, is involved in the transcription of NF-κB target genes in pancreatic cancer cells (Deng et al., 2015).
In line with the role of Drosophila WASH in the TRF2 complex, mouse WASH was recently found to almost exclusively localize to the nucleus in long-term hematopoietic stem cells (LT-HSCs) where it associates with components of the nucleosome-remodeling factor (NURF) complex to regulate cellular differentiation (Xia et al., 2014b). In the absence of WASH, the number of LT-HSCs significantly increased as a result of their inability to activate the Myc gene, which is required to drive their differentiation. It is worth noting that WASH directly interacts with the Rbbp4 subunit of the NURF complex and this interaction was required for Rbbp4 recruitment to the Myc promoter (see poster). Moreover, wild-type WASH, but not a mutant lacking the WCA domain could rescue Rbbp4 recruitment to the Myc promoter and consequently repopulation of bone marrow HSCs (Xia et al., 2014b). Thus, these data point to an important WCA-dependent role for WASH in regulating Myc gene expression in LT-HSCs.
It was recently found that Drosophila WASH has an important role in maintaining the global architecture of the cell nucleus, as WASH-depleted cells or those expressing mutant WASH exhibited defects, including the mislocalization of proteins that mark nuclear compartments, such as the nucleolus, cajal bodies and heterochromatin territories (Verboon et al., 2015a). Interestingly, the link to heterochromatin territories involves a newly described interaction of WASH with the nuclear protein lamin Dm0 (CG6944; a Drosophila B-type lamin). In the absence of either WASH or lamin Dm0, the accessibility of DNA to heterochromatic regions is increased, without any effect on transcription start sites. These observations, together with the role of WASH in mouse LT-HSCs described above, suggest that WASH exerts multi-faceted functions in the nucleus, not only impacting on nuclear shape and chromatin organization, but also regulating gene expression. The contribution of the other SHRC components to this regulation, as well as the impact on other transcriptional programs in other cell types will be an area of interest going forward.
Conclusions
Since the discovery of the WAS gene and the function of WASP as an actin-nucleating promoting factor two decades ago, the WASP family has expanded to eleven members and it has become clear that this family of proteins has evolved to regulate Arp2/3-generated F-actin dynamics and perform other cellular tasks at virtually every organelle in the cell. Significantly, while the vast number of studies on WASP family proteins has focused on their regulation of Arp2/3, emerging findings indicate that WASP proteins and their associated partners have substantial Arp2/3-independent functions. Clearly there remain several unknowns regarding WASP family proteins and future research into the biology of these proteins and complexes is expected to reveal not only new functions throughout the cell, but also mechanisms by which deregulation contributes to human disease.
Acknowledgements
We apologize to colleagues whose work we could not include due to space considerations.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
The work of our laboratories was supported by the National Institutes of Health [AI065474 and AI20959 to D.D.B., DK107733 to E.B. and D.D.B., and DK073639 to E.B.] and the Mayo Foundation for Medical Education and Research. Deposited in PMC for release after 12 months.
Cell science at a glance
A high-resolution version of the poster and individual poster panels are available for downloading at http://jcs.biologists.org/lookup/doi/10.1242/jcs.199570.supplemental
References
- Aguilera M. O., Berón W. and Colombo M. I. (2014). The actin cytoskeleton participates in the early events of autophagosome formation upon starvation induced autophagy. Autophagy 8, 1590-1603. 10.4161/auto.21459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison R., Edgar J. R., Pearson G., Rizo T., Newton T., Günther S., Berner F., Hague J., Connell J. W., Winkler J. et al. (2017). Defects in ER–endosome contacts impact lysosome function in hereditary spastic paraplegia. J. Cell Biol. 216, 1337-1355. 10.1083/jcb.201609033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartuzi P., Billadeau D. D., Favier R., Rong S., Dekker D., Fedoseienko A., Fieten H., Wijers M., Levels J. H., Huijkman N. et al. (2016). CCC- and WASH-mediated endosomal sorting of LDLR is required for normal clearance of circulating LDL. Nat. Commun. 7, 10961 10.1038/ncomms10961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom M., Reis K., Nehru V., Blom H., Gad A. K. B. and Aspenström P. (2015). RhoD is a Golgi component with a role in anterograde protein transport from the ER to the plasma membrane. Exp. Cell Res. 333, 208-219. 10.1016/j.yexcr.2015.02.023 [DOI] [PubMed] [Google Scholar]
- Brinkmann K., Winterhoff M., Onel S.-F., Schultz J., Faix J. and Bogdan S. (2016). WHAMY is a novel actin polymerase promoting myoblast fusion, macrophage cell motility and sensory organ development in Drosophila. J. Cell Sci. 129, 604-620. 10.1242/jcs.179325 [DOI] [PubMed] [Google Scholar]
- Buckley C. M., Gopaldass N., Bosmani C., Johnston S. A., Soldati T., Insall R. H. and King J. S. (2016). WASH drives early recycling from macropinosomes and phagosomes to maintain surface phagocytic receptors. Proc. Natl Acad. Sci. USA 113, E5906-E5915. 10.1073/pnas.1524532113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burianek L. E. and Soderling S. H. (2013). Under lock and key: spatiotemporal regulation of WASP family proteins coordinates separate dynamic cellular processes. Semin. Cell Dev. Biol. 24, 258-266. 10.1016/j.semcdb.2012.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone K. G. and Welch M. D. (2010). A nucleator arms race: cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 11, 237-251. 10.1038/nrm2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone K. G., Webb N. J., Znameroski E. A. and Welch M. D. (2008). WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell 134, 148-161. 10.1016/j.cell.2008.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnell M., Zech T., Calaminus S. D., Ura S., Hagedorn M., Johnston S. A., May R. C., Soldati T., Machesky L. M. and Insall R. H. (2011). Actin polymerization driven by WASH causes V-ATPase retrieval and vesicle neutralization before exocytosis. J. Cell Biol. 193, 831-839. 10.1083/jcb.201009119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Borek D., Padrick S. B., Gomez T. S., Metlagel Z., Ismail A. M., Umetani J., Billadeau D. D., Otwinowski Z. and Rosen M. K. (2010). Structure and control of the actin regulatory WAVE complex. Nature 468, 533-538. 10.1038/nature09623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Brinkmann K., Chen Z., Pak C. W., Liao Y., Shi S., Henry L., Grishin N. V., Bogdan S. and Rosen M. K. (2014). The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell 156, 195-207. 10.1016/j.cell.2013.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutts A. S. and La Thangue N. B. (2015). Actin nucleation by WH2 domains at the autophagosome. Nat. Commun. 6, 7888 10.1038/ncomms8888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutts A. S., Weston L. and La Thangue N. B. (2009). A transcription co-factor integrates cell adhesion and motility with the p53 response. Proc. Natl. Acad. Sci. USA 106, 19872-19877. 10.1073/pnas.0906785106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente M. A., Sasahara Y., Calamito M., Antón I. M., Elkhal A., Gallego M. D., Suresh K., Siminovitch K. A., Ochs H. D., Anderson K. C. et al. (2007). WIP is a chaperone for Wiskott-Aldrich syndrome protein (WASP). Proc. Natl. Acad. Sci. USA 104, 926-931. 10.1073/pnas.0610275104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z.-H., Gomez T. S., Osborne D. G., Phillips-Krawczak C. A., Zhang J.-S. and Billadeau D. D. (2015). Nuclear FAM21 participates in NF- B-dependent gene regulation in pancreatic cancer cells. J. Cell Sci. 128, 373-384. 10.1242/jcs.161513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derivery E., Sousa C., Gautier J. J., Lombard B., Loew D. and Gautreau A. (2009). The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev. Cell 17, 712-723. 10.1016/j.devcel.2009.09.010 [DOI] [PubMed] [Google Scholar]
- Derivery E., Helfer E., Henriot V. and Gautreau A. (2012). Actin polymerization controls the organization of WASH domains at the surface of endosomes. PLoS ONE 7, e39774-16 10.1371/journal.pone.0039774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derry J. M. J., Ochs H. D. and Francke U. (1994). Isolation of a novel gene mutated in Wiskott-Aldrich syndrome. Cell 78, 635-644. 10.1016/0092-8674(94)90528-2 [DOI] [PubMed] [Google Scholar]
- Dong R., Saheki Y., Swarup S., Lucast L., Harper J. W. and De Camilli P. (2016). Endosome-ER contacts control actin nucleation and retromer function through VAP-dependent regulation of PI4P. Cell 166, 408-423. 10.1016/j.cell.2016.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duleh S. N. and Welch M. D. (2010). WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton 67, 193-206. 10.1002/cm.20437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden S., Rohatgi R., Podtelejnikov A. V., Mann M. and Kirschner M. W. (2002). Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 418, 790-793. 10.1038/nature00859 [DOI] [PubMed] [Google Scholar]
- Elliott A. M., Simard L. R., Coghlan G., Chudley A. E., Chodirker B. N., Greenberg C. R., Burch T., Ly V., Hatch G. M. and Zelinski T. (2013). A novel mutation in KIAA0196: identification of a gene involved in Ritscher-Schinzel/3C syndrome in a First Nations cohort. J. Med. Genet. 50, 819-822. 10.1136/jmedgenet-2013-101715 [DOI] [PubMed] [Google Scholar]
- Farina F., Gaillard J., Guérin C., Couté Y., Sillibourne J., Blanchoin L. and Théry M. (2015). The centrosome is an actin-organizing centre. Nat. Cell Biol. 18, 65-75. 10.1038/ncb3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., He D., Yao Z. and Klionsky D. J. (2014). The machinery of macroautophagy. Cell Res. 24, 24-41. 10.1038/cr.2013.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follett J., Norwood S. J., Hamilton N. A., Mohan M., Kovtun O., Tay S., Zhe Y., Wood S. A., Mellick G. D., Silburn P. A. et al. (2013). The Vps35 D620N mutation linked to Parkinson's disease disrupts the cargo sorting function of retromer. Traffic 15, 230-244. 10.1111/tra.12136 [DOI] [PubMed] [Google Scholar]
- Gad A. K. B., Nehru V., Ruusala A. and Aspenström P. (2012). RhoD regulates cytoskeletal dynamics via the actin nucleation–promoting factor WASp homologue associated with actin Golgi membranes and microtubules. Mol. Biol. Cell 23, 4807-4819. 10.1091/mbc.E12-07-0555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez T. S. and Billadeau D. D. (2009). A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev. Cell 17, 699-711. 10.1016/j.devcel.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez T. S., Gorman J. A., de Narvajas A. A.-M., Koenig A. O. and Billadeau D. D. (2012). Trafficking defects in WASH-knockout fibroblasts originate from collapsed endosomal and lysosomal networks. Mol. Biol. Cell 23, 3215-3228. 10.1091/mbc.E12-02-0101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y.-H., Doyle J. M., Ramanathan S., Gomez T. S., Jia D., Xu M., Chen Z. J., Billadeau D. D., Rosen M. K. and Potts P. R. (2013). Regulation of WASH-dependent actin polymerization and protein trafficking by ubiquitination. Cell 152, 1051-1064. 10.1016/j.cell.2013.01.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y.-H., Fountain M. D. Jr, Tacer K. F., Xia F., Bi W., Kang S.-H. L., Patel A., Rosenfeld J. A., Le Caignec C., Isidor B. et al. (2015). USP7 acts as a molecular rheostat to promote WASH-dependent endosomal protein recycling and is mutated in a human neurodevelopmental disorder. Mol. Cell 59, 956-969. 10.1016/j.molcel.2015.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour M. E., Breusegem S. Y. and Seaman M. N. J. (2012). Recruitment of the endosomal WASH complex is mediated by the extended “tail” of Fam21 binding to the retromer protein Vps35. Biochem. J. 442, 209-220. 10.1042/BJ20111761 [DOI] [PubMed] [Google Scholar]
- Hochheimer A., Zhou S., Zheng S., Holmes M. C. and Tjian R. (2002). TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature 420, 439-445. 10.1038/nature01167 [DOI] [PubMed] [Google Scholar]
- Hubert T., Vandekerckhove J. and Gettemans J. (2011). Actin and Arp2/3 localize at the centrosome of interphase cells. Biochem. Biophys. Res. Commun. 404, 153-158. 10.1016/j.bbrc.2010.11.084 [DOI] [PubMed] [Google Scholar]
- Ismail A. M., Padrick S. B., Chen B., Umetani J. and Rosen M. K. (2009). The WAVE regulatory complex is inhibited. Nat. Struct. Mol. Biol. 16, 561-563. 10.1038/nsmb.1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D., Gomez T. S., Metlagel Z., Umetani J., Otwinowski Z., Rosen M. K. and Billadeau D. D. (2010). WASH and WAVE actin regulators of the Wiskott-Aldrich syndrome protein (WASP) family are controlled by analogous structurally related complexes. Proc. Natl. Acad. Sci. USA 107, 10442-10447. 10.1073/pnas.0913293107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D., Gomez T. S., Billadeau D. D. and Rosen M. K. (2012). Multiple repeat elements within the FAM21 tail link the WASH actin regulatory complex to the retromer. Mol. Biol. Cell 23, 2352-2361. 10.1091/mbc.E11-12-1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast D. J., Zajac A. L., Holzbaur E. L. F., Ostap E. M. and Dominguez R. (2015). WHAMM directs the Arp2/3 complex to the ER for autophagosome biogenesis through an actin comet tail mechanism. Curr. Biol. 25, 1791-1797. 10.1016/j.cub.2015.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A. S., Kakalis L. T., Abdul-Manan N., Liu G. A. and Rosen M. K. (2000). Autoinhibition and activation mechanisms of the Wiskott–Aldrich syndrome protein. Nature 404, 151-158. 10.1038/35004513 [DOI] [PubMed] [Google Scholar]
- Kollmar M., Lbik D. and Enge S. (2012). Evolution of the eukaryotic ARP2/3 activators of the WASP family: WASP, WAVE, WASH, and WHAMM, and the proposed new family members WAWH and WAML. BMC Research Notes 5, 88 10.1186/1756-0500-5-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koronakis V., Hume P. J., Humphreys D., Liu T., Horning O., Jensen O. N. and McGhie E. J. (2011). WAVE regulatory complex activation by cooperating GTPases Arf and Rac1. Proc. Natl. Acad. Sci. USA 108, 14449-14454. 10.1073/pnas.1107666108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G., Mariño G. and Levine B. (2010). Autophagy and the integrated stress response. Mol. Cell 40, 280-293. 10.1016/j.molcel.2010.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebensohn A. M. and Kirschner M. W. (2009). Activation of the WAVE complex by coincident signals controls actin assembly. Mol. Cell 36, 512-524. 10.1016/j.molcel.2009.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardopoulou E. V., Parghi S. S., Friedman C., Osborn G. E., Parkhurst S. M. and Trask B. J. (2007). Human subtelomeric WASH genes encode a new subclass of the WASP family. PLoS Genet. 3, e237-e239. 10.1371/journal.pgen.0030237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Abreu-Blanco M. T., Barry K. C., Linardopoulou E. V., Osborn G. E. and Parkhurst S. M. (2009). Wash functions downstream of Rho and links linear and branched actin nucleation factors. Development 136, 2849-2860. 10.1242/dev.035246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Dai A., Cao Y., Zhang R., Dong M.-Q. and Wang H.-W. (2017). Structural Insights of WHAMM's Interaction with Microtubules by Cryo-EM. J. Mol. Biol. 216, 1337-1355. 10.1016/j.jmb.2017.03.022 [DOI] [PubMed] [Google Scholar]
- Marchesin V., Castro-Castro A., Lodillinsky C., Castagnino A., Cyrta J., Bonsang-Kitzis H., Fuhrmann L., Irondelle M., Infante E., Montagnac G. et al. (2015). ARF6–JIP3/4 regulate endosomal tubules for MT1- MMP exocytosis in cancer invasion. J. Cell Biol. 211, 339-358. 10.1083/jcb.201506002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massaad M. J., Ramesh N. and Geha R. S. (2013). Wiskott-Aldrich syndrome: a comprehensive review. Ann. N. Y. Acad. Sci. 1285, 26-43. 10.1111/nyas.12049 [DOI] [PubMed] [Google Scholar]
- McGough I. J., Steinberg F., Da Jia, Barbuti P. A., McMillan K. J., Heesom K. J., Caldwell M. A., Billadeau D. D., Rosen M. K. and Cullen P. J. (2014). Retromer binding to FAM21 and the WASH complex is perturbed by the Parkinson disease-linked VPS35(D620N) mutation. Curr. Biol. 24, 1670-1676. 10.1016/j.cub.2014.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monfregola J., Napolitano G., D'Urso M., Lappalainen P. and Ursini M. V. (2010). Functional characterization of Wiskott-Aldrich syndrome protein and scar homolog (WASH), a bi-modular nucleation-promoting factor able to interact with biogenesis of lysosome-related organelle subunit 2 (BLOS2) and gamma-tubulin. J. Biol. Chem. 285, 16951-16957. 10.1074/jbc.M109.078501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro P., Rossé C., Castro-Castro A., Irondelle M., Lagoutte E., Paul-Gilloteaux P., Desnos C., Formstecher E., Darchen F., Perrais D. et al. (2013). Endosomal WASH and exocyst complexes control exocytosis of MT1-MMP at invadopodia. J. Cell Biol. 203, 1063-1079. 10.1083/jcb.201306162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel B. M., Bechtold M., Rodriguez L. G. and Bogdan S. (2017). Drosophila WASH is required for integrin-mediated cell adhesion, cell motility and lysosomal neutralization. J. Cell Sci. 130, 344-359. 10.1242/jcs.193086 [DOI] [PubMed] [Google Scholar]
- Padrick S. B., Cheng H.-C., Ismail A. M., Panchal S. C., Doolittle L. K., Kim S., Skehan B. M., Umetani J., Brautigam C. A., Leong J. M. et al. (2008). Hierarchical regulation of WASP/WAVE proteins. Mol. Cell 32, 426-438. 10.1016/j.molcel.2008.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick S. B., Doolittle L. K., Brautigam C. A., King D. S. and Rosen M. K. (2011). Arp2/3 complex is bound and activated by two WASP proteins. Proc. Natl. Acad. Sci. USA 108, E472-E479. 10.1073/pnas.1100236108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh N., Anton I. M., Hartwig J. H. and Geha R. S. (1997). WIP, a protein associated with Wiskott–Aldrich syndrome protein, induces actin polymerization and redistribution in lymphoid cells. Proc. Natl. Acad. Sci. USA 94, 14671-14676. 10.1073/pnas.94.26.14671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R., Ho H.-Y. H. and Kirschner M. W. (2000). Mechanism of N-Wasp activation by Cdc42 and phosphatidylinositol 4,5-bisphosphate. J. Cell Biol. 150, 1299-1310. 10.1083/jcb.150.6.1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropers F., Derivery E., Hu H., Garshasbi M., Karbasiyan M., Herold M., Nurnberg G., Ullmann R., Gautreau A., Sperling K. et al. (2011). Identification of a novel candidate gene for non-syndromic autosomal recessive intellectual disability: the WASH complex member SWIP. Hum. Mol. Genet. 20, 2585-2590. 10.1093/hmg/ddr158 [DOI] [PubMed] [Google Scholar]
- Rottner K., Hänisch J. and Campellone K. G. (2010). WASH, WHAMM and JMY: regulation of Arp2/3 complex and beyond. Trends Cell Biol.. 20, 650-661. 10.1016/j.tcb.2010.08.014 [DOI] [PubMed] [Google Scholar]
- Rowland A. A., Chitwood P. J., Phillips M. J. and Voeltz G. K. (2014). ER contact sites define the position and timing of endosome fission. Cell 159, 1027-1041. 10.1016/j.cell.2014.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A. J., Mathiowetz A. J., Hong S., Welch M. D. and Campellone K. G. (2016). Rab1 recruits WHAMM during membrane remodeling but limits actin nucleation. Mol. Biol. Cell 27, 967-978. 10.1091/mbc.E15-07-0508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder P. V., Vistein R., Gokhale A., Seaman M. N., Puthenveedu M. A. and Faundez V. (2013). The WASH complex, an endosomal Arp2/3 activator, interacts with the Hermansky-Pudlak syndrome complex BLOC-1 and its cargo phosphatidylinositol-4-kinase type II. Mol. Biol. Cell 24, 2269-2284. 10.1091/mbc.E13-02-0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadhukhan S., Sarkar K., Taylor M., Candotti F. and Vyas Y. M. (2014). Nuclear role of WASp in gene transcription is uncoupled from its ARP2/3-dependent cytoplasmic role in actin polymerization. J. Immunol. 193, 150-160. 10.4049/jimmunol.1302923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar K., Sadhukhan S., Han S.-S. and Vyas Y. M. (2014). Disruption of hSWI/SNF complexes in T cells by WAS mutations distinguishes X-linked thrombocytopenia from Wiskott-Aldrich syndrome. Blood 124, 3409-3419. 10.1182/blood-2014-07-587642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar K., Sadhukhan S., Han S.-S. and Vyas Y. M. (2015). SUMOylation-disrupting WAS mutation converts WASp from a transcriptional activator to a repressor of NF-kB response genes in T cells. Blood 126, 1670-1682. 10.1182/blood-2015-05-646182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q.-T., Hsiue P. P., Sindelar C. V., Welch M. D., Campellone K. G. and Wang H.-W. (2012). Structural insights into WHAMM-mediated cytoskeletal coordination during membrane remodeling. J. Cell Biol. 199, 111-124. 10.1083/jcb.201204010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikama N., Lee C.-W., France S., Delavaine L., Lyon J., Krstic-Demonacos M. and La Thangue N. B. (1999). A novel cofactor for p300 that regulates the p53 response. Mol. Cell 4, 365-376. 10.1016/S1097-2765(00)80338-X [DOI] [PubMed] [Google Scholar]
- Squarr A. J., Brinkmann K., Chen B., Steinbacher T., Ebnet K., Rosen M. K. and Bogdan S. (2016). Fat2 acts through the WAVE regulatory complex to drive collective cell migration during tissue rotation. J. Cell Biol. 212, 591-603. 10.1083/jcb.201508081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stradal T. E. B., Rottner K., Disanza A., Confalonieri S., Innocenti M. and Scita G. (2004). Regulation of actin dynamics by WASP and WAVE family proteins. Trends Cell Biol.. 14, 303-311. 10.1016/j.tcb.2004.04.007 [DOI] [PubMed] [Google Scholar]
- Taylor M. D., Sadhukhan S., Kottangada P., Ramgopal A., Sarkar K., D'Silva S., Selvakumar A., Candotti F. and Vyas Y. M. (2010). Nuclear role of WASp in the pathogenesis of dysregulated TH1 immunity in human Wiskott-Aldrich Syndrome. Sci. Transl. Med. 2, 37ra44 10.1126/scitranslmed.3000813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdmanis P. N., Meijer I. A., Reynolds A., Lei A., MacLeod P., Schlesinger D., Zatz M., Reid E., Dion P. A., Drapeau P. et al. (2007). Mutations in the KIAA0196 gene at the SPG8 locus cause hereditary spastic paraplegia. Am. J. Hum. Genet. 80, 152-161. 10.1086/510782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman D. M. and Insall R. H. (2010). WASP family proteins: their evolution and its physiological implications. Mol. Biol. Cell 21, 2880-2893. 10.1091/mbc.E10-04-0372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verboon J. M., Rincon-Arano H., Werwie T. R., Delrow J. J., Scalzo D., Nandakumar V., Groudine M. and Parkhurst S. M. (2015a). Wash interacts with lamin and affects global nuclear organization. Curr. Biol. 25, 804-810. 10.1016/j.cub.2015.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verboon J. M., Sugumar B. and Parkhurst S. M. (2015b). Wiskott-Aldrich syndrome proteins in the nucleus: aWASH with possibilities. Nucleus 6, 349-359. 10.1080/19491034.2015.1086051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilariño-Güell C., Wider C., Ross O. A., Dachsel J. C., Kachergus J. M., Lincoln S. J., Soto-Ortolaza A. I., Cobb S. A., Wilhoite G. J., Bacon J. A. et al. (2011). VPS35 mutations in Parkinson disease. Am. J. Hum. Genet. 89, 162-167. 10.1016/j.ajhg.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P., Wang S., Du Y., Zhao Z., Shi L., Sun L., Huang G., Ye B., Li C., Dai Z. et al. (2013). WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J. 32, 2685-2696. 10.1038/emboj.2013.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P., Wang S., Huang G., Du Y., Zhu P., Li M. and Fan Z. (2014a). RNF2 is recruited by WASH to ubiquitinate AMBRA1 leading to downregulation of autophagy. Cell Res.. 24, 943-958. 10.1038/cr.2014.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P., Wang S., Huang G., Zhu P., Li M., Ye B., Du Y. and Fan Z. (2014b). WASH is required for the differentiation commitment of hematopoietic stem cells in a c-Myc–dependent manner. J. Exp. Med. 211, 2119-2134. 10.1084/jem.20140169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavodszky E., Seaman M. N. J., Moreau K., Jimenez-Sanchez M., Breusegem S. Y., Harbour M. E. and Rubinsztein D. C. (2014). Mutation in VPS35 associated with Parkinson's disease impairs WASH complex association and inhibits autophagy. Nat. Commun. 5, 1-16. 10.1038/ncomms4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zech T., Calaminus S. D. J., Caswell P., Spence H. J., Carnell M., Insall R. H., Norman J. and Machesky L. M. (2011). The Arp2/3 activator WASH regulates 5 1-integrin-mediated invasive migration. J. Cell Sci. 124, 3753-3759. 10.1242/jcs.080986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A., Benet-Pagès A., Struhal W., Graf E., Eck S. H., Offman M. N., Haubenberger D., Spielberger S., Schulte E. C., Lichtner P. et al. (2011). A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am. J. Hum. Genet. 89, 168-175. 10.1016/j.ajhg.2011.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchero J. B., Coutts A. S., Quinlan M. E., Thangue N. B. L. and Mullins R. D. (2009). p53-cofactor JMY is a multifunctional actin nucleation factor. Nat. Cell Biol. 11, 451-459. 10.1038/ncb1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchero J. B., Belin B. and Mullins R. D. (2012). Actin binding to WH2 domains regulates nuclear import of the multifunctional actin regulator JMY. Mol. Biol. Cell 23, 853-863. 10.1091/mbc.E11-12-0992 [DOI] [PMC free article] [PubMed] [Google Scholar]