ABSTRACT
Linear chains of five to hundreds of phosphates called polyphosphate are found in organisms ranging from bacteria to humans, but their function is poorly understood. In Dictyostelium discoideum, polyphosphate is used as a secreted signal that inhibits cytokinesis in an autocrine negative feedback loop. To elucidate how cells respond to this unusual signal, we undertook a proteomic analysis of cells treated with physiological levels of polyphosphate and observed that polyphosphate causes cells to decrease levels of actin cytoskeleton proteins, possibly explaining how polyphosphate inhibits cytokinesis. Polyphosphate also causes proteasome protein levels to decrease, and in both Dictyostelium and human leukemia cells, decreases proteasome activity and cell proliferation. Polyphosphate also induces Dictyostelium cells to begin development by increasing expression of the cell–cell adhesion molecule CsA (also known as CsaA) and causing aggregation, and this effect, as well as the inhibition of proteasome activity, is mediated by Ras and Akt proteins. Surprisingly, Ras and Akt do not affect the ability of polyphosphate to inhibit proliferation, suggesting that a branching pathway mediates the effects of polyphosphate, with one branch affecting proliferation, and the other branch affecting development.
KEY WORDS: Development, Dictyostelium, Proteasome, Polyphosphate, Ras, Akt
Summary: Polyphosphate is present in all eukaryotes, but little is known about its function. Here, we describe how Dictyostelium uses polyphosphate as a signal to initiate development.
INTRODUCTION
The transition from growth to differentiation is a fundamental aspect of biology, and both must be regulated in concert for proper development. Dictyostelium discoideum provides an excellent model for the study of the growth to differentiation transition (GDT) in that Dictyostelium live as unicellular amoeba while sufficient nutrients are available, but upon nutrient depletion and starvation these cells stop proliferating, and aggregate together to form a multicellular structure consisting of stalk cells supporting a mass of spore cells (Marin, 1976). In addition to a ‘musical chairs’ mechanism based on the cell cycle phase that a cell happens to be in at the time of starvation causing an initial choice of differentiation into either a stalk or a spore cell (Gomer and Ammann, 1996), secreted autocrine factors also affect differentiation (Clarke and Gomer, 1995; Maeda, 2005). Since the concentration of a constitutively secreted factor will increase as the cell density increases, some of the secreted factors allow cells to sense the local cell density, and induce a pre-starvation response in which cells begin expressing early developmental genes in anticipation of a high density of cells outgrowing the food supply, thus allowing cells to prepare for the starvation-induced GDT (Clarke et al., 1988; Clarke and Gomer, 1995; Maeda, 2005). Three pre-starvation factors have been described, although they have not been identified (Maeda, 2005).
We previously identified inorganic polyphosphate as a molecule secreted continually by growing cells (Suess and Gomer, 2016). At high cell densities, where cells are about to starve, polyphosphate inhibits cytokinesis more than it inhibits cell growth (the accumulation of mass), which then allows the starved cells to have as much stored nutrients as possible (Suess and Gomer, 2016). Polyphosphate is an ancient and highly conserved molecule consisting of a linear chain of orthophosphates bound by high energy phospho-anhydride bonds (Brown and Kornberg, 2004; Rao et al., 2009). Recent work has highlighted the increasing roles of extracellular polyphosphate in a variety of eukaryotic cellular responses, including roles in coagulation, contact pathway activation, inflammation and proliferation (Smith et al., 2006; Gajsiewicz et al., 2017; Morrissey et al., 2012; Wang et al., 2003). Polyphosphate increases matrix metalloproteinase-3 expression and activity in odontoblast-like cells, induces rapid ERK1 and ERK2 (ERK1/2, also known as MAPK3 and MAPK1) phosphorylation in SaOS-2 cells, and inhibits cyclin D1 expression through IKKα and ERK1/2 in endothelial cells; however, in general the intracellular signaling components activated by extracellular polyphosphate remain largely unknown (Ozeki et al., 2015; Lui et al., 2016; Hassanian et al., 2016). Determining the signaling pathways initiated by extracellular polyphosphate in Dictyostelium may provide insight into how this ubiquitous molecule mediates various cellular responses in more complex systems.
Although polyphosphate is unusual as it is not a protein, peptide or organic molecule, it has many of the characteristics of Dictyostelium pre-starvation factors. Polyphosphate is continually secreted during growth and increases as a function of cell density, while it also shows increased extracellular accumulation upon a decrease in available nutrients (Suess and Gomer, 2016). In this report, we show that polyphosphate is a pre-starvation factor that uses a signal transduction pathway involving Ras and Akt proteins to prime cells for development, and that, surprisingly, this pathway is not involved in polyphosphate-induced proliferation inhibition.
RESULTS
Polyphosphate changes the Dictyostelium proteome
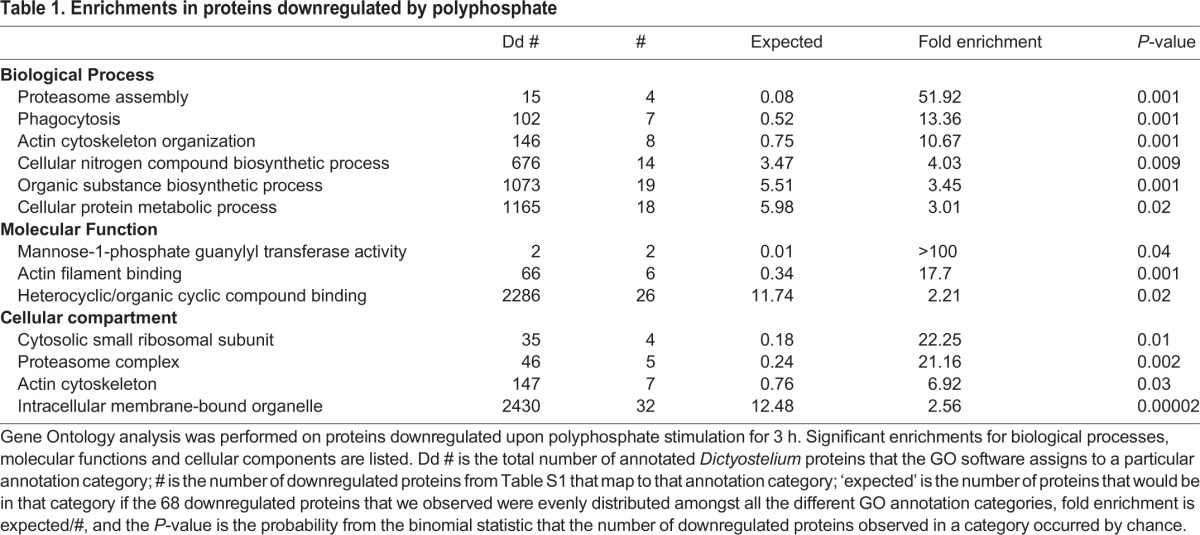
To elucidate the effects of polyphosphate on cells, we undertook a proteomic analysis of cells treated with or without polyphosphate. Polyphosphate downregulated 67 proteins by an average of at least 0.65 relative to control, and upregulated 28 proteins by an average of at least 1.75 across four sample sets (Table S1). Polyphosphate did not significantly affect the amounts of 2459 proteins in the proteomics data (Table S1), and did not discernably change the distribution of protein bands on a Coomassie-stained SDS-polyacrylamide gel of total cell proteins (Fig. 1B), indicating that the effects of polyphosphate are relatively subtle. Gene ontology (GO) analysis indicated that polyphosphate downregulated the proteasome assembly proteins Psmg1, Psmg2, Psmd4 and Psmd8, and the proteasome complex proteins Psmb1, Psmb4-1, Psmb5, Psmd4 and Psmd8, as well as actin cytoskeleton proteins (Table 1; Table S1). Proteins that were upregulated by polyphosphate showed no significant enrichments in any GO categories.
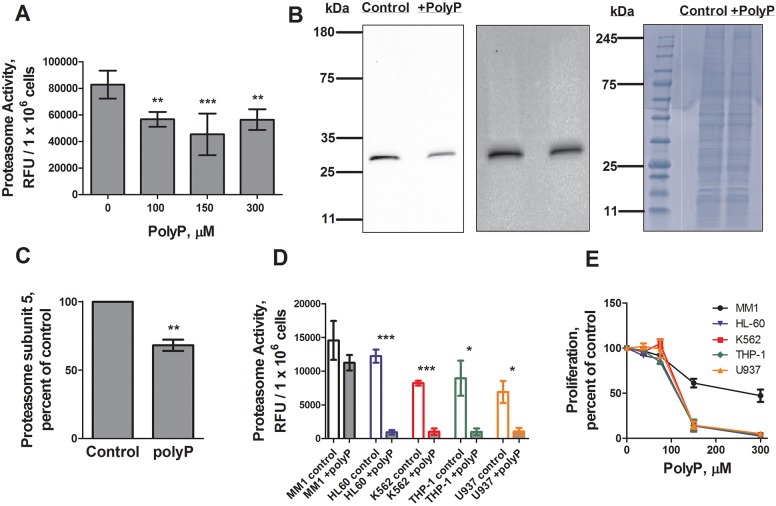
Fig. 1.
Polyphosphate decreases proteasome activity. (A) Cells were cultured with the indicated concentrations of polyphosphate for 4 h and proteasome activity levels were measured and normalized to no-polyphosphate (polyP) controls. (B,C) Cells were cultured with 150 µM polyphosphate for 4 h and proteasome subunit 5 levels were measured by western blotting (a representative image of four blots is shown) and normalized to no-polyphosphate controls. A longer exposure (middle) showed no additional bands. Total protein loading control from an aliquot of the samples used for the proteasome subunit 5 western blot (representative image of four gels) is also shown (right). (D) Human leukemia cell lines were cultured with 0 (control) or 150 µM polyphosphate for 24 h and proteasome activity levels were measured. Differences between the various control values were not statistically significant (one-way ANOVA, Tukey's test). *P<0.05, ***P<0.001 (t-tests). (E) Cells were cultured with the indicated amount of polyphosphate for 24 h and cell counts were normalized to the no-polyphosphate control, so that 100% proliferation is the amount of proliferation with no polyphosphate, and 0% proliferation represents no increase in cell number. 150 and 300 µM polyphosphate significantly decreased cell numbers compared to no polyphosphate for MM1 (P<0.05 for both 150 and 300 µM), HL-60 (P<0.01 and P<0.001, respectively), K562 (P<0.01 and P<0.001, respectively), THP-1 (P<0.01 and P<0.001, respectively) and U937 (P<0.01 and P<0.001, respectively). *P<0.05, **P<0.01, ***P<0.001 (t-tests). All values are mean±s.e.m., n≥3.
Table 1.
Enrichments in proteins downregulated by polyphosphate
Polyphosphate inhibits proteasome activity
To determine whether polyphosphate affects the proteasome activity of Dictyostelium cells, proteasome activity was measured in mid-log Ax2 cells after a 4 h incubation with polyphosphate. Polyphosphate inhibited proteasome activity (Fig. 1A), and, as determined by western blotting, decreased levels of the proteasome subunit Psmb5 after a 4 h incubation with polyphosphate, mirroring the decrease observed by proteomics (Fig. 1B,C; Table S1). Taken together, the data suggest that polyphosphate decreases proteasome activity by decreasing levels of proteasome proteins.
Polyphosphate inhibits proteasome activity and proliferation of leukemia cell lines
Proteasome inhibition has been suggested as a potential therapeutic for some cancers, as many malignant cells exhibit high proteasome activity, and proteasome inhibition often results in apoptosis (Naujokat and Hoffmann, 2002; Frankland-Searby and Bhaumik, 2012; Adams, 2004; Schmidt and Finley, 2014; Bassermann et al., 2014). To determine whether the ability of polyphosphate to inhibit proteasome activity and proliferation is conserved, we tested the effect of polyphosphate on five human leukemia cell lines (Ziegler-Heitbrock et al., 1988; Gallagher et al., 1979; Klein et al., 1976; Tsuchiya et al., 1980; Sundström and Nilsson, 1976). None of the cell lines tested had significant differences in basal proteasome activity (Fig. 1D). Polyphosphate strongly inhibited the proteasome activity of four out of five lines tested (Fig. 1D). At 100 µM, polyphosphate had a modest effect on Dictyostelium proliferation, whereas at 150 µM polyphosphate strongly inhibited proliferation (Suess and Gomer, 2016). Polyphosphate also inhibited the proliferation of the leukemia cell lines, and remarkably these cells showed the same sharp threshold in the response curve (Fig. 1E). These results suggest that the effect of polyphosphate on proteasome activity and proliferation may be conserved from Dictyostelium to humans.
Polyphosphate induces development in the presence of nutrients
Adhesion regulating molecule-1 (Adrm1, also called Rpn13) is a ubiquitin receptor for the proteasome (Husnjak et al., 2008). Phosphoproteome analysis suggested that polyphosphate may have caused Adrm1 phosphorylation at residue T280 in three of the four trials, although polyphosphate did not significantly affect levels of Adrm1 (Table S1). To determine whether cells lacking Adrm1 have altered proteasome activity, we examined the proteasome activity levels of wild-type and adrm1− cells (Cherix et al., 2006). Compared to wild-type cells, adrm1− cells had abnormally low levels of proteasome activity (Fig. 2A). Unlike wild-type cells, Dictyostelium cells lacking Adrm1 transition from vegetative growth to development while still in the presence of nutrients, suggesting a defect in the pre-starvation response and/or the GDT (Cherix et al., 2006). To determine whether the starvation-induced GDT is associated with a decrease in proteasome activity, the proteasome activity levels of starved cells was measured. Starvation decreased proteasome activity by ∼50% after 30 min, similar to the reduction caused by polyphosphate, and a further decrease was observed over the next 6 h (Fig. 2A). These results suggest that a decrease in proteasome activity occurs during the GDT.
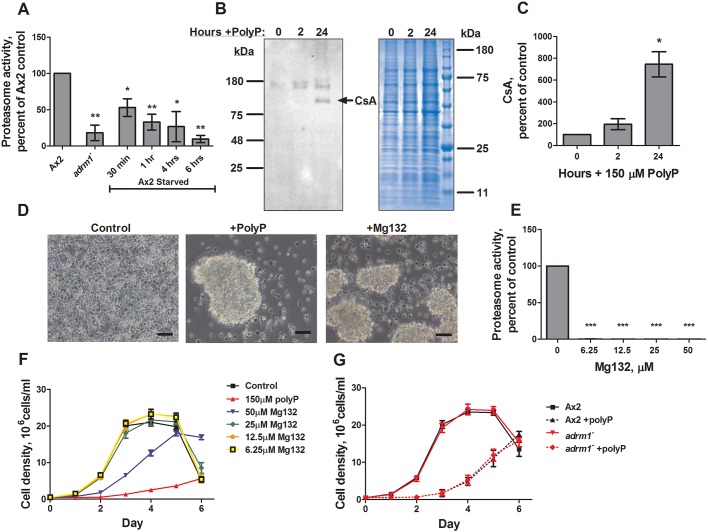
Fig. 2.
Polyphosphate induces cell aggregation. (A) Proteasome activity of mid-log Ax2 and adrm1−, or Ax2 cells starved in PBM for the indicated times, were measured and normalized to mid-log Ax2 controls. (B,C) Cells were cultured with 150 µM polyphosphate (PolyP) for the indicated times and CsA levels were measured by western blotting (a representative image of four blots is shown) and normalized to the no-polyphosphate controls. Total protein loading control from an aliquot of the samples used for the CsA western blot is also shown (representative image of four gels). (D) Cells were cultured in 25% HL5 in the presence or absence of 150 µM polyphosphate or 50 µM Mg132 for 24 h (representative image of 10 experiments). Scale bars: 50 µm. (E) Cells were incubated with the indicated amounts of Mg132 for 4 h in HL5, and proteasome activity levels were measured and normalized to no-Mg132 controls. (F) Cells in HL5 were cultured with the indicated amount of Mg132, and counted every 24 h. (G) Ax2 and adrm1− cells were cultured in HL5 with or without 150 µM polyphosphate and counted every 24 h. All values are mean±s.e.m., n≥3. *P<0.05, **P<0.01, ***P<0.001 (paired t-tests compared to Ax2 for panel A, 0 h for panel C, and 0 Mg132 for panel E).
Unlike wild-type cells, adrm1− cells express early onset development genes and begin to aggregate while in the presence of nutrients (Cherix et al., 2006). To determine whether high levels of extracellular polyphosphate might have effects similar to the loss of Adrm1, we cultured cells with polyphosphate in HL5, a nutrient-rich medium, for 24 h. Using spectrophotometry to measure total protein, we had previously observed that polyphosphate increases protein per cell by 1.57±0.06 fold (Suess and Gomer, 2016), and by scanning Coomassie-stained gels (Fig. 2B), we observed here that polyphosphate increased protein per cell by 1.3±0.2 fold (mean ± s.e.m., n=4). Western blots stained using a monoclonal antibody against CsA (also known as CsaA), an early onset development protein involved in cell–cell adhesion and aggregation (Murray et al., 1981, 1983), showed that polyphosphate increased levels of CsA by 7.4±1.0 fold (Fig. 2B,C), and this increase was significantly greater than the increase in total protein (P<0.02, t-test). The monoclonal anti-CsA antibody identifies an antigenic determinant common to other proteins that are developmentally regulated (Murray et al., 1983), and the polyphosphate-induced bands at ∼150 kDa suggest that polyphosphate may affect other developmentally regulated proteins. Taken together, these results indicate that polyphosphate can increase the expression of at least one developmental gene.
In 25% HL5, control cells did not appear to aggregate after 24 h, while 150 µM polyphosphate caused cells to form aggregates similar to those seen in cells lacking Adrm1 (Fig. 2D). Cells treated with less than 130 µM or more than 170 µM polyphosphate did not form aggregates (Table 2). Equivalent levels of NaPO4 (150–2400 μM) did not cause aggregates, indicating that the induced aggregation was not due to increased phosphate levels (data not shown). Cells cultured in 30% HL5 did not aggregate in response to polyphosphate, suggesting that sufficient nutrient availability inhibits polyphosphate-induced aggregation (Table 2). Polyphosphate also did not cause aggregation when added to cells at low cell density (Table 2). Taken together, these results indicate that at sufficiently high cell densities and sufficiently low levels of nutrients, the accumulation of extracellular polyphosphate can act as a pre-starvation factor, allowing cells to effectively anticipate starvation and begin development.
Table 2.
The effect of polyphosphate on cell aggregation

Proteasome inhibition alone is not sufficient to induce development
To further elucidate the role of proteasome inhibition during the process of GDT, cells were treated with the proteasome inhibitor Mg132 (Rock et al., 1994). High concentrations of Mg132 (25 and 50 μM) consistently induced development in the presence of nutrients (Fig. 2D, Table 2). This effect was only observed at Mg132 concentrations well above those that strongly inhibit proteasome activity (6.25 μM) (Fig. 2E), suggesting either that Mg132 has off-target effects that induce development, such as targeting cysteine or serine proteases (Lee and Goldberg, 1998), or that there are discrepancies between the effect of Mg132 on the in vitro proteasome activity assay and in vivo aggregation assay. To resolve this, we treated cells with lactacystin, a specific inhibitor of the 20S proteasome (Fenteany et al., 1995). Unfortunately lactacystin at normal working levels and in excess had no discernable effect on proteasome activity or aggregation (Fig. S1A,B), confirming previous reports that lactacystin, which is cell permeable in other systems, does not inhibit the proteasome when added to Dictyostelium cells (Mohanty et al., 2001; Langenick et al., 2008). These results suggest that proteasome inhibition alone is not sufficient to induce GDT, although further investigation would be needed to confirm this observation.
The proteasome is essential for cell cycle progression in many eukaryotic cell lines (Naujokat and Hoffmann, 2002; Bassermann et al., 2014). To determine whether polyphosphate inhibits proliferation through its effect on the proteasome, cells were treated with Mg132, and proliferation was measured. Surprisingly, Mg132 concentrations that strongly inhibit proteasome activity and induce aggregation did not significantly affect proliferation, although 50 µM Mg132 did inhibit proliferation (Fig. 2F). In addition, adrm1− cells, which have abnormally low proteasome activity (Fig. 2A), also showed normal proliferation and were sensitive to polyphosphate inhibition (Fig. 2G), suggesting that the inhibitory effect of polyphosphate on proliferation is not due to proteasome inhibition.
Akt proteins and RasC mediate the effects of polyphosphate-induced development and proteasome inhibition
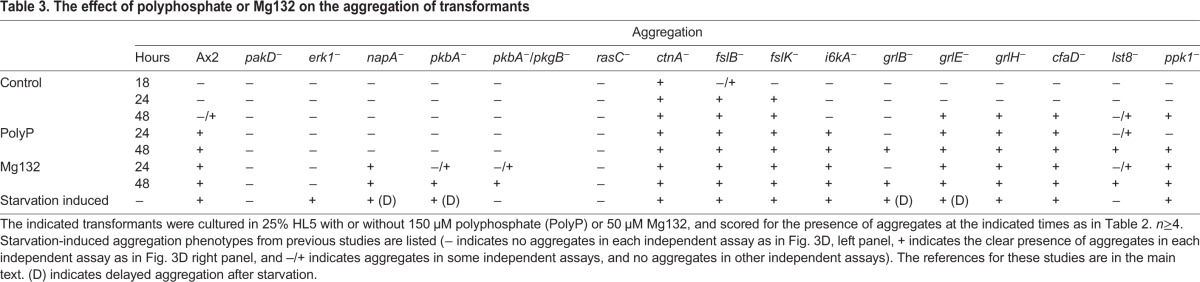
A variety of available transformants were screened in 25% HL5 to identify cells with abnormal responses to polyphosphate. Cells lacking RasC (Daniel et al., 1994), the p21-activated kinase PakD (Garcia et al., 2014), Erk1 (also known as ErkA) (Gaskins et al., 1994), the Dictyostelium Akt homologs Pkba and PkgB (Meili et al., 1999, 2000), and the suppressor of cAMP receptor (SCAR) component NapA (Ibarra et al., 2006) were all nonresponsive to polyphosphate-induced development (Table 3). Cells lacking the secreted factor CtnA (Brock and Gomer, 1999), the frizzled-like receptors FslB and FslK (Prabhu and Eichinger, 2006), the inositol hexakisphosphate kinase I6kA (Luo et al., 2003), the G-protein coupled receptors GrlB, GrlE and GrlH (Prabhu and Eichinger, 2006) the autocrine factor CfaD (Bakthavatsalam et al., 2008), the target of rapamycin complex 2 (TORC2) component Lst8 (Lee et al., 2005), the Dictyostelium polyphosphate kinase Ppk1 (Livermore et al., 2016), and the PTEN homolog CnrN (Tang and Gomer, 2008) did form aggregates in response to polyphosphate (Table 3). Of the transformants that were unresponsive to polyphosphate, cells lacking RasC, PakD and Erk1 were unresponsive to Mg132-induced development while cells lacking Akt (i.e. lacking only Pkba, or both Pkba and PkgB) and NapA were responsive (Table 3). To determine whether polyphosphate might affect signaling pathways used for starvation-induced aggregation, we included previously described aggregation phenotypes for all transformants tested (Table 3) (Brock and Gomer, 1999; Luo et al., 2003; Wu and Janetopoulos, 2013; Bakthavatsalam et al., 2008; Lee et al., 2005; Livermore et al., 2016). Cells lacking both Akt homologs PkbA and PkgB, RasC or PakD have abolished aggregation in response to polyphosphate and starvation (Meili et al., 2000; Lim et al., 2001; Garcia et al., 2014), while cells lacking PkbA or NapA show delayed aggregation upon starvation but do not respond to polyphosphate-induced aggregation (Meili et al., 1999; Ibarra et al., 2006). Cells lacking Erk1 undergo starvation-induced aggregation, but form smaller aggregates than do wild-type cells and become stalled at later stages of development (Hadwiger and Nguyen, 2011), yet are unresponsive to polyphosphate-induced aggregation. These results suggest that some pathway components of polyphosphate and starvation-induced aggregation are shared, while polyphosphate also utilizes unique components. Interestingly, cells lacking NapA show normal aggregation in response to Mg132 despite defects observed in polyphosphate- and starvation-induced aggregation, while cells lacking PkbA alone or both PkbA and PkgB showed delayed aggregation in response to Mg132.
Table 3.
The effect of polyphosphate or Mg132 on the aggregation of transformants
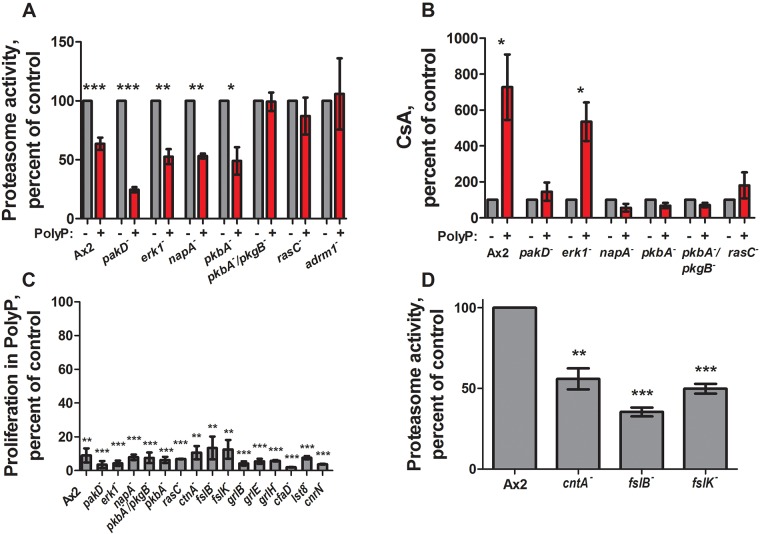
Cells identified to be unresponsive to polyphosphate-induced development were examined for further defects in polyphosphate-induced proteasome inhibition, CsA increase, or proliferation inhibition. Cells lacking RasC and both Dictyostelium Akt proteins were unresponsive to polyphosphate inhibition of proteasome activity (Fig. 3A). Cells lacking Akt proteins, RasC, PakD or NapA did not exhibit polyphosphate-induced CsA expression, while cells lacking Erk1 responded normally (Fig. 3B). All of the cell lines tested exhibited polyphosphate-mediated inhibition of proliferation (Fig. 3C).
Fig. 3.
Some transformants have impaired polyphosphate signaling. (A) Cells were cultured with 150 µM polyphosphate for 4 h and proteasome activity levels were measured and normalized to no-polyphosphate (PolyP) controls. (B) Cells were cultured with 150 µM polyphosphate for 24 h and CsA levels were measured and normalized to no-polyphosphate controls. (C) Cells were cultured starting at 5×105 cells/ml with or without 150 µM polyphosphate for 24 h, and the increase in cell number in the presence of polyphosphate was calculated as a percentage of the increase in cell number in the absence of polyphosphate. (D) Cells were cultured in HL5 and proteasome activity levels were measured and normalized to Ax2 controls. All values are mean±s.e.m., n≥3. *P<0.05, **P<0.01, ***P<0.001 (paired t-tests).
Polyphosphate did not cause a further decrease in proteasome activity in adrm1− cells (Fig. 3A); however, upon starvation a decrease in proteasome activity was observed (Fig. S2A). Polyphosphate did not affect the precocious aggregation of adrm1− cells in 25% HL5, nor did it induce aggregation in higher concentrations of HL5 (Fig. S2B). Cells lacking Adrm1 phenocopy many of the effects initiated by extracellular polyphosphate including expression of CsA and formation of aggregates in the presence of nutrients (Cherix et al., 2006), as well as decreased vegetative proteasome activity. Whether or not polyphosphate affects Adrm1 in addition to decreasing other proteasome subunits to inhibit proteasome activity warrants further investigation.
We also identified mutants that began the GDT precociously in the presence of nutrients, similar to what is seen cells lacking Adrm1. Cells lacking the autocrine factor CtnA or the frizzled-like receptors FslB or FslK, formed multicellular aggregates by 24 h in 25% HL5 (Table 3) while also exhibiting abnormally low proteasome levels (Fig. 3D), suggesting that CtnA, FslB, and FslK have roles in nutrient sensing or the pre-starvation response.
Polyphosphate induces Akt phosphorylation
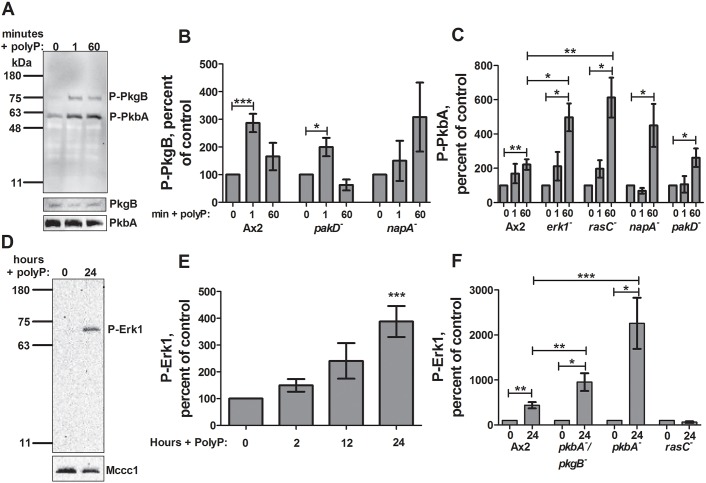
To determine the role of Akt proteins in the polyphosphate response, levels of phosphorylated Akt proteins were measured in Ax2 cells following polyphosphate treatment in 100% HL5 using an antibody that recognizes phosphorylated T278 of PkbA and T309 of PkgB (Kamimura et al., 2008). Polyphosphate induced phosphorylation of the Dictyostelium Akt homolog PkgB within 1 min and the Akt homolog PkbA after 1 h (Fig. 4A, top, B,C). Polyphosphate did not appear to affect levels of total PkgB or PkbA in 60 min (Fig. 4A, bottom). Cells lacking PakD showed normal PkgB phosphorylation upon polyphosphate stimulation, while cells lacking NapA did not show a significant increase in phosphorylation (Fig. 4B). Phosphorylated PkgB was undetectable in cells lacking Erk1 or RasC. All four mutants showed activation of PkbA, suggesting that PkbA activation is upstream of these components of the pathway. PkbA phosphorylation was enhanced in cells lacking RasC or Erk1 (Fig. 4C), suggesting that Erk1 and RasC somewhat inhibit PkbA phosphorylation. Upon cAMP stimulation, cells lacking RasC show delayed phosphorylation of both PkbA and PkgB (Lim et al., 2001; Bolourani et al., 2006; Cai et al., 2010). To determine whether PkgB also has delayed polyphosphate-induced phosphorylation in RasC and Erk1 null backgrounds, phosphorylation was analyzed at 2, 5 and 10 min after polyphosphate stimulation. Phosphorylated PkgB was undetectable at these timepoints in cells lacking Erk1 or RasC, suggesting that cAMP and polyphosphate signaling pathways have unique features (Fig. S3A). These data suggests that Akt activation is an early component of the polyphosphate signal transduction pathway that is necessary for both the transition to development and proteasome inhibition.
Fig. 4.
Polyphosphate-stimulated Akt and Erk1 phosphorylation in transformants. (A) Ax2 cells were cultured with 150 µM polyphosphate (PolyP) for the indicated times, and phosphorylated Akt (P-PkgB and P-PkbA) was detected by western blotting (a representative image of six blots is shown). Staining with anti-PkgB and anti-PkbA antibodies is also shown. (B,C) Cells were cultured in 150 µM polyphosphate for the indicated times and levels of phosphorylated PkgB (B) and PkbA (C) were measured and normalized to no-polyphosphate controls. (D) Cells were cultured with 150 µM polyphosphate for 24 h and phosphorylated Erk1 levels were measured by western blotting (a representative image of 12 blots is shown). Staining for the loading control Mccc1 is also shown. (E) Ax2 cells were cultured with 150 µM polyphosphate for the indicated times and phosphorylated Erk1 levels were measured by western blotting and normalized to no-polyphosphate controls. (F) The indicated cells were cultured with 150 µM polyphosphate for 24 h and phosphorylated Erk1 levels were measured and normalized to no-polyphosphate controls. All values are mean±s.e.m., n≥3. *P<0.05, **P<0.01, ***P<0.001 (paired t-tests).
Polyphosphate induces Erk1 phosphorylation in a delayed response
We next investigated the role of Erk1 activation in polyphosphate-mediated induction of the GDT by using a monoclonal antibody that recognizes phosphorylation of the conserved TxY motif (Schwebs and Hadwiger, 2015). Erk1 is activated in a delayed response to extracellular cAMP and folic acid (Schwebs and Hadwiger, 2015). Polyphosphate induced Erk1 phosphorylation after 24 h in 100% HL5, with no significant induction after 2 and 12 h, suggesting that Erk1 phosphorylation is a secondary or indirect response, perhaps after aggregation competence is obtained and development is initiated (Fig. 4D,E). Polyphosphate did not induce increased Erk1 phosphorylation in rasC− cells; however, overall levels of phosphorylated Erk1 were higher in these cells compared to those in wild type, suggesting that RasC negatively regulates Erk1 phosphorylation (Fig. 4F; Fig. S3B,C). Phosphorylated Erk1 was undetectable under all conditions in cells lacking PakD and NapA. Cells lacking the Akt homologs PkbA and PkgB showed enhanced Erk1 phosphorylation upon polyphosphate treatment. Taken together, these results indicate that PakD and NapA are necessary for polyphosphate-induced Erk1 activation, while Akt somewhat inhibits polyphosphate-induced Erk1 activation, and RasC inhibits basal levels of phosphorylated Erk1.
DISCUSSION
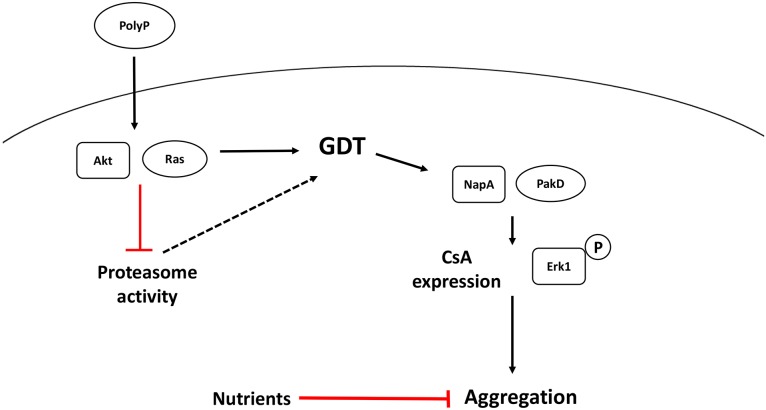
Here, we show that polyphosphate acts as a pre-starvation factor by accumulating extracellularly upon nutrient depletion and/or high cell densities, initiating a signaling pathway that prepares cells for the transition from vegetative growth to development before the inevitable onset of starvation (Fig. 5). Even in the presence of abundant nutrients, polyphosphate inhibits the proteasome, induces Akt and Erk1 activation, and increases expression of the early onset development gene csA. If the nutrients are low and cell density is high, polyphosphate also induces aggregation. This suggests that polyphosphate is able to prime cells for the eventual transition to development, but Dictyostelium cells are able to sense the presence of nutrients as well as a low local cell density, both of which can inhibit aggregation. Ras and Akt proteins are intimately involved with proliferation in other systems (Mitin et al., 2005; Brazil et al., 2004; Brazil and Hemmings, 2001), but remarkably these and other signal transduction pathway components do not affect polyphosphate-mediated inhibition of proliferation.
Fig. 5.
Summary of polyphosphate signal transduction. Levels of extracellular polyphosphate (PolyP) increase as the local cell density increases. Polyphosphate requires Ras and Akt proteins to inhibit the proteasome, and requires Ras, Akt, NapA and PakD proteins to induce CsA expression. Polyphosphate also activates Erk1 in a pathway that requires Ras, NapA, and PakD. A pathway that does not involve any gene product listed in Table 3 inhibits cytokinesis. Although proteasome inhibition correlates with the growth to development transition (GDT), its direct role in the GDT is unclear (dashed line). When nutrient levels are sufficiently low, polyphosphate also induces aggregation.
Polyphosphate was able to inhibit the proteasome of both Dictyostelium cells and human leukemia cell lines, suggesting that this is a conserved function from Dictyostelium to humans. Proteasome inhibition appears to be mediated by Akt proteins and RasC in Dictyostelium. The single Akt knockout pkbA− was sensitive to polyphosphate proteasome inhibition, while cells lacking both homologs were not responsive, perhaps suggesting redundant roles, as has been previously reported (Meili et al., 2000). An association between proteasome activity inhibition and the GDT was observed in all cases in this study; however, only excessive concentrations of Mg132 induced aggregation, suggesting that proteasome inhibition is associated with, but is not sufficient, to induce aggregation. However, this assumption is based on the comparison between an in vitro activity assay after incubation with inhibitors and an in vivo activity output in aggregation. It is possible that discrepancies exist between these two assays, such as the use of alternative substrates, making the interpretation of the direct role of proteasome inhibition difficult.
While Adrm1 null cells phenocopy many of the effects observed upon polyphosphate treatment, whether or not Adrm1 is a target of the polyphosphate signaling pathway is still unclear. A more extensive study would need to be carried out to determine whether polyphosphate does induce a phosphorylation of Adrm1, and if so whether or not this phosphorylation is needed for any of the processes initiated by polyphosphate. Mammalian proteasomes undergo autophagic destruction upon amino acid starvation (Cohen-Kaplan et al., 2017). Whether or not Dictyostelium undergoes a similar process during starvation warrants further investigation.
Polyphosphate-induced CsA expression is dependent on Akt proteins, RasC, NapA and PakD. Erk1 is not necessary for polyphosphate proteasome inhibition or CsA expression; however, it is essential for polyphosphate-induced aggregation. Polyphosphate can activate Erk1 although this response is delayed (i.e. occurs over 24 h). Previous reports have described similar results for Erk1 activation upon cAMP stimulation, and its activation is dependent on cell–cell interactions (Schwebs and Hadwiger, 2015). Perhaps as cells become aggregation competent, in part due to polyphosphate-induced CsA expression, Erk1 activation becomes stronger because of the increase in cell–cell interactions, as Erk1 activation aligns with peak CsA levels. Akt proteins were not necessary for Erk1 activation, while NapA and PakD were both essential. Cells lacking RasC did not show an increase in phosphorylated Erk1 upon polyphosphate treatment; however, the endogenous levels of phosphorylated Erk1 were consistently higher in this background, suggesting that RasC negatively regulates Erk1 activation during vegetative growth. The observations that Erk1 phosphorylation requires NapA and PakD, and is inhibited by RasC, PkbA and PkgB, that PkbA phosphorylation is inhibited by RasC and Erk1, and that PkgB phosphorylation requires Erk1, RasC and NapA, indicate a considerable degree of cross-talk in the polyphosphate signal transduction pathway.
Ras, Erk, Akt and NapA proteins are parts of the signal transduction pathway used by polyphosphate. These components are also involved in Dictyostelium chemotaxis response to cAMP (Veltman et al., 2008; Sasaki and Firtel, 2006), which is mediated by signaling pathways that interact with each other in complex ways, similar to what we have observed. Other studies have identified other regulators of the transition of growth to development, such as YakA (Souza et al., 1998), Gdt1 (Zeng et al., 2000) and Gdt2 (Chibalina et al., 2004), which may interact with the polyphosphate signal transduction pathway. A reasonable assumption would be that polyphosphate induces a unique signaling pathway, leading to the activation of pathways used for the GDT and starvation-induced aggregation. Since polyphosphate appears to have similar effects on Dictyostelium and human cells, additional studies on components of the Dictyostelium GDT and cAMP signal transduction pathways may help to further elucidate the signal transduction pathways used by the ubiquitous and unusual signal polyphosphate.
MATERIALS AND METHODS
Reagents and materials
HL5 medium was from Formedium (Norfolk, UK). Polyphosphate was from Spectrum (New Brunswick, NJ). Blasticidin, Mg132 and low-fat milk powder were from CalBioChem (Boston, MA). Dictyostelium cell lines were obtained from dictyBase (Northwestern University, Chicago, IL). Nylon 17 mm and 5 µm pore size filters were from Sterlitech (Kent, WA). RIPA buffer, RPMI, tissue culture grade six-well plates, and Super Signal West Pico chemiluminescent substrate were from Thermo Fisher Scientific (Waltham, MA). Donkey anti-mouse-IgG and anti-rabbit-IgG horseradish peroxidase (HRP)-conjugated secondary antibodies were from Jackson ImmunoResearch (West Grove, PA). Laemmli sample buffer was from BioRad (Hercules, CA). Proteasome activity assay kits were from Sigma (St Louis, MO). Calf serum and biochemistry grade bovine serum albumin (BSA) powder were from VWR (Radnor, PA). 4–20% Tris-glycine polyacrylamide gels were from Lonza (Walkersville, MD). Anti-CsA (20-121-1) and anti-proteasome subunit 5 (171-337) antibodies were from the Developmental Studies Hybridoma Bank (Iowa City, IA). Anti-phosphorylated Akt (2060S) and anti-phosphorylated Erk1 (9101S) antibodies and protease and phosphatase inhibitor cocktail were from Cell Signaling Technology (Danvers, MA). Lactacystin was from Enzo Life Sciences (Farmingdale, NY).
Dictyostelium and human leukemia cell culture
The cells used were Dictyostelium wild-type Ax2, pakD− (DBS0350281) (Garcia et al., 2014), erk1− (DBS0350622) (Gaskins et al., 1994), pkbA− (DBS0349876) (Meili et al., 1999), pkbA−/pkgB− (DBS0236785) (Meili et al., 2000), rasC− (DBS0236853) (Daniel et al., 1994), napA− (DBS0236596) (Ibarra et al., 2006), adrm1− (DBS0238203) (Cherix et al., 2006), fslB− (DBS0350230) (Prabhu and Eichinger, 2006), fslK− (DBS0350229) (Prabhu and Eichinger, 2006), ctnA− (DBS0235624) (Brock and Gomer, 1999), i6kA− (DBS0236426) (Luo et al., 2003), grlB− (DBS0350074) (Prabhu and Eichinger, 2006), grlE− (DBS0350075) (Prabhu and Eichinger, 2006), grlH− (DBS0350226) (Prabhu and Eichinger, 2006), cfaD− (DBS0302444) (Bakthavatsalam et al., 2008), lst8− (DBS0236517) (Lee et al., 2005), ppk1− (DBS0350685) (Livermore et al., 2016) and cnrN− (DBS0302655) (Tang and Gomer, 2008). Mutant genotypes were verified by PCR, and cells were kept under constant selection. Cells were grown in shaking culture in HL5, and were never allowed to reach cell densities above 4×106 cells/ml. Proliferation curves were made and starvation was performed as previously described (Suess and Gomer, 2016). HL-60, K562, THP-1 and U937 cell lines were from the ATCC (Manassas, VA). MonoMac1 (MM1) cells were from DSMZ-German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany) (Ziegler-Heitbrock et al., 1988; Tsuchiya et al., 1980; Sundström and Nilsson, 1976; Klein et al., 1976; Gallagher et al., 1979). Cells were grown in a BL2 facility in RPMI 1640 with 10% bovine calf serum (BCS) containing 2 mM glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin (all from Lonza, Walkersville, MD) and tested for contamination. For proliferation assays, leukemia cells were incubated with or without polyphosphate in RPMI medium with 2.5% serum at a density of 1×106 cells/ml. After 24 h, cells were counted using a hemocytometer.
Proteomics
Mid-log Ax2 cells were treated with 150 µM polyphosphate for 3 h. 106 cells were collected by centrifugation at 200 g for 3 min, resuspended in 40 µl RIPA buffer containing protease and phosphatase inhibitors, and incubated on ice for 20 min. Lysates were then sonicated for 1 min (Chicago Electric #95563, Camarillo CA), and clarified by centrifugation at 18,000 g at 4°C for 10 min. 30 µl of supernatant was transferred to a tube containing 10 µl of 4× sample buffer, and heated at 95°C for 5 min. 10 µl of sample was run for 5–10 mm into a polyacrylamide gel. The gel was stained with Coomassie Blue for 1 h, and destained overnight in sterile water. The combined protein bands were diced into small pieces, and sent to the University of Texas Southwest proteomics core (http://www.utsouthwestern.edu/research/core-facilities/proteomics-core.html) for trypsin digestion and tandem mas spectrometry (MS/MS) analysis, using the normalized spectral index method for quantification. Gene ontology analysis was performed using the Gene Ontology database (http://geneontology.org/) and the PANTHER classification system.
Proteasome activity assay
1.5×106 cells from mid-log cultures (2×106–4×106 cells/ml) were diluted to 1 ml using HL5 containing the appropriate final concentration of polyphosphate Mg132 or Lactacystin. After 4 or 24 h, cells were collected by centrifugation at 200 g for 3 min and resuspended in PBS. Cell lysates were generated by passing cells through 5 μm syringe filters. 100 µl of cell lysates were incubated with 100 µl of proteasome activity kit assay buffer, then incubated at room temperature overnight for Dictyostelium or at 37°C overnight for leukemia cell lines, and fluorescence was measured with an excitation of 490 nm and emission of 525 nm using a Synergy Mx plate reader (BioTek, Winooski VT).
Western blots
Cells were collected at 200 g for 3 min, followed by collection and resuspension 2× in PBM (20 mM KH2PO4, 1 mM MgCl2, 0.01 mM CaCl2, pH 6.5). Cells were then collected at 200 g for 3 min. The cell pellets were solubilized in Laemmli sample buffer containing protease and phosphatase inhibitors, and heated to 95°C for 5 min. Gel electrophoresis and blotting were performed as described previously (Bakthavatsalam et al., 2008). Blots were blocked in TBS with 4% milk and 2% BSA. Blots were incubated with primary antibody (1:1000 for anti-phosphorylated Akt and anti-phosphorylated Erk1 antibody, 0.2 µg/ml for anti-CsA and anti-proteasome subunit 5) in TBS containing 5% BSA and 0.1% Tween 20 at room temperature for 1 h or overnight at 4°C. After washing 3× in TBST for 5 min, blots were incubated with 1:5000 secondary antibody in TBS with 2% BSA for 1 h at room temperature. After washing 3× in TBST, blots were then incubated with Super Signal West Pico chemiluminescent substrate for 5 min. Signals were visualized with a ChemiDoc XRS+ (Bio-Rad, Hercules, CA) and quantification of relative protein levels was performed with Image Lab (Bio-Rad). For blots with total protein loading controls, identical aliquots of the protein samples were electrophoresed on gels, and proteins were visualized via Coomassie Blue stain. Mccc1 loading control staining for phospho-Erk1 blots was performed as previously described (Davidson et al., 2013; Schwebs and Hadwiger, 2015). Anti-PkbA and anti-PkgB were kind gifts from Pascale Charest, and were each incubated in TBS containing 5% BSA for 1 h at 1:2000.
Aggregation assay
Mid-log cells were collected by centrifugation at 200 g for 3 min, resuspended in HL5 and the centrifugation and resuspension was repeated twice. Cells were resuspended to 1.5×106 cells/ml in HL5 diluted with PBM, and 2 ml was placed in the well of a six-well plate. Plates were incubated at room temperature in humid boxes, then analyzed by microscopy with a 10× phase-contrast objective on a Diaphot inverted microscope (Nikon, Garden City, NY) for aggregation and development. Images of a calibration slide (SWIFT #MA663, Carlsbad, CA) were used for scale bars.
Acknowledgements
We thank Dr Pierre Cosson for providing adrm1− cells. We thank Dr Pascale Charest for providing anti-PkbA and anti-PkgB antibodies.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: P.M.S., R.H.G.; Methodology: P.M.S., R.H.G.; Validation: P.M.S., J.W.; Formal analysis: P.M.S., W.C., R.H.G.; Investigation: P.M.S., J.W., W.C., R.H.G.; Resources: R.H.G.; Writing - original draft: P.M.S.; Writing - review & editing: R.H.G.; Supervision: R.H.G.; Project administration: R.H.G.; Funding acquisition: R.H.G.
Funding
This work was supported by the National Institutes of Health (grant R01 GM102280). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.203372.supplemental
References
- Adams J. (2004). The development of proteasome inhibitors as anticancer drugs. Cancer Cell 5, 417-421. 10.1016/S1535-6108(04)00120-5 [DOI] [PubMed] [Google Scholar]
- Bakthavatsalam D., Brock D. A., Nikravan N. N., Houston K. D., Hatton R. D. and Gomer R. H. (2008). The secreted Dictyostelium protein CfaD is a chalone. J. Cell Sci. 121, 2473-2480. 10.1242/jcs.026682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassermann F., Eichner R. and Pagano M. (2014). The ubiquitin proteasome system - implications for cell cycle control and the targeted treatment of cancer. Biochim. Biophys. Acta 1843, 150-162. 10.1016/j.bbamcr.2013.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolourani P., Spiegelman G. B. and Weeks G. (2006). Delineation of the roles played by RasG and RasC in cAMP-dependent signal transduction during the early development of Dictyostelium discoideum. Mol. Biol. Cell 17, 4543-4550. 10.1091/mbc.E05-11-1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazil D. P. and Hemmings B. A. (2001). Ten years of protein kinase B signalling: a hard Akt to follow. Trends Biochem. Sci. 26, 657-664. 10.1016/S0968-0004(01)01958-2 [DOI] [PubMed] [Google Scholar]
- Brazil D. P., Yang Z.-Z. and Hemmings B. A. (2004). Advances in protein kinase B signalling: AKTion on multiple fronts. Trends Biochem. Sci. 29, 233-242. 10.1016/j.tibs.2004.03.006 [DOI] [PubMed] [Google Scholar]
- Brock D. A. and Gomer R. H. (1999). A cell-counting factor regulating structure size in Dictyostelium. Genes Dev. 13, 1960-1969. 10.1101/gad.13.15.1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. R. W. and Kornberg A. (2004). Inorganic polyphosphate in the origin and survival of species. Proc. Natl. Acad. Sci. USA 101, 16085-16087. 10.1073/pnas.0406909101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., Das S., Kamimura Y., Long Y., Parent C. A. and Devreotes P. N. (2010). Ras-mediated activation of the TORC2-PKB pathway is critical for chemotaxis. J. Cell Biol. 190, 233-245. 10.1083/jcb.201001129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherix N., Froquet R., Charette S. J., Blanc C., Letourneur F. and Cosson P. (2006). A Phg2-Adrm1 pathway participates in the nutrient-controlled developmental response in Dictyostelium. Mol. Biol. Cell 17, 4982-4987. 10.1091/mbc.E06-07-0619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibalina M. V., Anjard C. and Insall R. H. (2004). Gdt2 regulates the transition of Dictyostelium cells from growth to differentiation. BMC Dev. Biol. 4, 8 10.1186/1471-213X-4-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M. and Gomer R. H. (1995). PSF and Cmf, autocrine factors that regulate gene expression during growth and early development of Dictyostelium. Experientia 51, 1124-1134. 10.1007/BF01944730 [DOI] [PubMed] [Google Scholar]
- Clarke M., Yang J. and Kayman S. C. (1988). Analysis of the prestarvation response in growing cells of Dictyostelium discoideum. Dev. Genet. 9, 315-326. 10.1002/dvg.1020090413 [DOI] [PubMed] [Google Scholar]
- Cohen-Kaplan V., Ciechanover A. and Livneh I. (2017). Stress-induced polyubiquitination of proteasomal ubiquitin receptors targets the proteolytic complex for autophagic degradation. Autophagy. 13, 759-760. 10.1080/15548627.2016.1278327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J., Bush J., Cardelli J., Spiegelman G. B. and Weeks G. (1994). Isolation of two novel ras genes in Dictyostelium discoideum; evidence for a complex, developmentally regulated ras gene subfamily. Oncogene 9, 501-508. [PubMed] [Google Scholar]
- Davidson A. J., King J. S. and Insall R. H. (2013). The use of streptavidin conjugates as immunoblot loading controls and mitochondrial markers for use with Dictyostelium discoideum. BioTechniques 55, 39-41. 10.2144/000114054 [DOI] [PubMed] [Google Scholar]
- Fenteany G., Standaert R. F., Lane W. S., Choi S., Corey E. J. and Schreiber S. L. (1995). Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science 268, 726-731. 10.1126/science.7732382 [DOI] [PubMed] [Google Scholar]
- Frankland-Searby S. and Bhaumik S. R. (2012). The 26S proteasome complex: an attractive target for cancer therapy. Biochim. Biophys. Acta 1825, 64-76. 10.1016/j.bbcan.2011.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajsiewicz J. M., Smith S. A. and Morrissey J. H. (2017). Polyphosphate and RNA differentially modulate the contact pathway of blood clotting. J. Biol. Chem. 292, 1808-1814. 10.1074/jbc.M116.754325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher R., Collins S., Trujillo J., Mccredie K., Ahearn M., Tsai S., Metzgar R., Aulakh G., Ting R., Ruscetti F. et al. (1979). Characterization of the continuous, differentiating myeloid cell line (HL-60) from a patient with acute promyelocytic leukemia. Blood 54, 713-733. [PubMed] [Google Scholar]
- Garcia M., Ray S., Brown I., Irom J. and Brazill D. (2014). Pakd, a putative p21-activated protein kinase in Dictyostelium discoideum, regulates actin. Eukaryot. Cell 13, 119-126. 10.1128/EC.00216-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskins C., Maeda M. and Firtel R. A. (1994). Identification and functional analysis of a developmentally regulated extracellular signal-regulated kinase gene in Dictyostelium discoideum. Mol. Cell. Biol. 14, 6996-7012. 10.1128/MCB.14.10.6996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomer R. H. and Ammann R. R. (1996). A cell-cycle phase-associated cell-type choice mechanism monitors the cell cycle rather than using an independent timer. Dev. Biol. 174, 82-91. 10.1006/dbio.1996.0053 [DOI] [PubMed] [Google Scholar]
- Hadwiger J. A. and Nguyen H.-N. (2011). MAPKs in development: insights from Dictyostelium signaling pathways. Biomol. Concepts 2, 39-46. 10.1515/bmc.2011.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanian S. M., Ardeshirylajimi A., Dinarvand P. and Rezaie A. R. (2016). Inorganic polyphosphate promotes cyclin D1 synthesis through activation of mTOR/Wnt/beta-catenin signaling in endothelial cells. J. Thromb. Haemost. 14, 2261-2273. 10.1111/jth.13477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnjak K., Elsasser S., Zhang N., Chen X., Randles L., Shi Y., Hofmann K., Walters K. J., Finley D. and Dikic I. (2008). Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453, 481-488. 10.1038/nature06926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra N., Blagg S. L., Vazquez F. and Insall R. H. (2006). Nap1 regulates Dictyostelium cell motility and adhesion through SCAR-dependent and -independent pathways. Curr. Biol. 16, 717-722. 10.1016/j.cub.2006.02.068 [DOI] [PubMed] [Google Scholar]
- Kamimura Y., Xiong Y., Iglesias P. A., Hoeller O., Bolourani P. and Devreotes P. N. (2008). PIP3-independent activation of TorC2 and PKB at the cell's leading edge mediates chemotaxis. Curr. Biol. 18, 1034-1043. 10.1016/j.cub.2008.06.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein E., Vánky F., Ben-Bassat H., Neumann H., Ralph P., Zeuthen J. and Polliack A. (1976). Properties of the K562 cell line, derived from a patient with chronic myeloid leukemia. Int. J. Cancer 18, 421-431. 10.1002/ijc.2910180405 [DOI] [PubMed] [Google Scholar]
- Langenick J., Araki T., Yamada Y. and Williams J. G. (2008). A Dictyostelium homologue of the metazoan Cbl proteins regulates STAT signalling. J. Cell Sci. 121, 3524-3530. 10.1242/jcs.036798 [DOI] [PubMed] [Google Scholar]
- Lee D. H. and Goldberg A. L. (1998). Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8, 397-403. 10.1016/S0962-8924(98)01346-4 [DOI] [PubMed] [Google Scholar]
- Lee S., Comer F. I., Sasaki A., Mcleod I. X., Duong Y., Okumura K., Yates J. R. III, Parent C. A. and Firtel R. A. (2005). TOR complex 2 integrates cell movement during chemotaxis and signal relay in Dictyostelium. Mol. Biol. Cell 16, 4572-4583. 10.1091/mbc.E05-04-0342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C. J., Spiegelman G. B. and Weeks G. (2001). RasC is required for optimal activation of adenylyl cyclase and Akt/PKB during aggregation. EMBO J. 20, 4490-4499. 10.1093/emboj/20.16.4490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore T. M., Chubb J. R. and Saiardi A. (2016). Developmental accumulation of inorganic polyphosphate affects germination and energetic metabolism in Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA 113, 996-1001. 10.1073/pnas.1519440113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui E. L.-H., Ao C. K.-L., Li L., Khong M.-L. and Tanner J. A. (2016). Inorganic polyphosphate triggers upregulation of interleukin 11 in human osteoblast-like SaOS-2 cells. Biochem. Biophys. Res. Commun. 479, 766-771. 10.1016/j.bbrc.2016.09.137 [DOI] [PubMed] [Google Scholar]
- Luo H. R., Huang Y. E., Chen J. C., Saiardi A., Iijima M., Ye K., Huang Y., Nagata E., Devreotes P. and Snyder S. H. (2003). Inositol pyrophosphates mediate chemotaxis in Dictyostelium via pleckstrin homology domain-PtdIns(3,4,5)P3 interactions. Cell 114, 559-572. 10.1016/S0092-8674(03)00640-8 [DOI] [PubMed] [Google Scholar]
- Maeda Y. (2005). Regulation of growth and differentiation in Dictyostelium. Int. Rev. Cytol. 244, 287-332. 10.1016/S0074-7696(05)44007-3 [DOI] [PubMed] [Google Scholar]
- Marin F. T. (1976). Regulation of development in Dictyostelium discoideum: I. Initiation of the growth to development transition by amino acid starvation. Dev. Biol. 48, 110-117. 10.1016/0012-1606(76)90050-6 [DOI] [PubMed] [Google Scholar]
- Meili R., Ellsworth C., Lee S., Reddy T. B., Ma H. and Firtel R. A. (1999). Chemoattractant-mediated transient activation and membrane localization of Akt/PKB is required for efficient chemotaxis to cAMP in Dictyostelium. EMBO J. 18, 2092-2105. 10.1093/emboj/18.8.2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meili R., Ellsworth C. and Firtel R. A. (2000). A novel Akt/PKB-related kinase is essential for morphogenesis in Dictyostelium. Curr. Biol. 10, 708-717. 10.1016/S0960-9822(00)00536-4 [DOI] [PubMed] [Google Scholar]
- Mitin N., Rossman K. L. and Der C. J. (2005). Signaling interplay in Ras superfamily function. Curr. Biol. 15, R563-R574. 10.1016/j.cub.2005.07.010 [DOI] [PubMed] [Google Scholar]
- Mohanty S., Lee S., Yadava N., Dealy M. J., Johnson R. S. and Firtel R. A. (2001). Regulated protein degradation controls PKA function and cell-type differentiation in Dictyostelium. Genes Dev. 15, 1435-1448. 10.1101/gad.871101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey J. H., Choi S. H. and Smith S. A. (2012). Polyphosphate: an ancient molecule that links platelets, coagulation, and inflammation. Blood 119, 5972-5979. 10.1182/blood-2012-03-306605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray B. A., Yee L. D. and Loomis W. F. (1981). Immunological analysis of a glycoprotein (contact sites A) involved in intercellular adhesion of Dictyostelium discoideum. J. Supramol. Struct. Cell Biochem. 17, 197-211. 10.1002/jsscb.380170302 [DOI] [PubMed] [Google Scholar]
- Murray B. A., Niman H. L. and Loomis W. F. (1983). Monoclonal antibody recognizing gp80, a membrane glycoprotein implicated in intercellular adhesion of Dictyostelium discoideum. Mol. Cell. Biol. 3, 863-870. 10.1128/MCB.3.5.863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naujokat C. and Hoffmann S. (2002). Role and function of the 26S proteasome in proliferation and apoptosis. Lab. Invest. 82, 965-980. 10.1097/01.LAB.0000022226.23741.37 [DOI] [PubMed] [Google Scholar]
- Ozeki N., Hase N., Yamaguchi H., Hiyama T., Kawai R., Kondo A., Nakata K. and Mogi M. (2015). Polyphosphate induces matrix metalloproteinase-3-mediated proliferation of odontoblast-like cells derived from induced pluripotent stem cells. Exp. Cell Res. 333, 303-315. 10.1016/j.yexcr.2015.01.007 [DOI] [PubMed] [Google Scholar]
- Prabhu Y. and Eichinger L. (2006). The Dictyostelium repertoire of seven transmembrane domain receptors. Eur. J. Cell Biol. 85, 937-946. 10.1016/j.ejcb.2006.04.003 [DOI] [PubMed] [Google Scholar]
- Rao N. N., Gómez-García M. R. and Kornberg A. (2009). Inorganic polyphosphate: essential for growth and survival. Annu. Rev. Biochem. 78, 605-647. 10.1146/annurev.biochem.77.083007.093039 [DOI] [PubMed] [Google Scholar]
- Rock K. L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D. and Goldberg A. L. (1994). Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78, 761-771. 10.1016/S0092-8674(94)90462-6 [DOI] [PubMed] [Google Scholar]
- Sasaki A. T. and Firtel R. A. (2006). Regulation of chemotaxis by the orchestrated activation of Ras, PI3k, and TOR. Eur. J. Cell Biol. 85, 873-895. 10.1016/j.ejcb.2006.04.007 [DOI] [PubMed] [Google Scholar]
- Schmidt M. and Finley D. (2014). Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta 1843, 13-25. 10.1016/j.bbamcr.2013.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwebs D. J. and Hadwiger J. A. (2015). The Dictyostelium MAPK ERK1 is phosphorylated in a secondary response to early developmental signaling. Cell. Signal. 27, 147-155. 10.1016/j.cellsig.2014.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. A., Mutch N. J., Baskar D., Rohloff P., Docampo R. and Morrissey J. H. (2006). Polyphosphate modulates blood coagulation and fibrinolysis. Proc. Natl. Acad. Sci. USA 103, 903-908. 10.1073/pnas.0507195103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza G. M., Lu S. and Kuspa A. (1998). Yaka, a protein kinase required for the transition from growth to development in Dictyostelium. Development 125, 2291-2302. [DOI] [PubMed] [Google Scholar]
- Suess P. M. and Gomer R. H. (2016). Extracellular polyphosphate inhibits proliferation in an autocrine negative feedback loop in Dictyostelium discoideum. J. Biol. Chem. 291, 20260-20269. 10.1074/jbc.M116.737825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundström C. and Nilsson K. (1976). Establishment and characterization of a human histiocytic lymphoma cell line (U-937). Int. J. Cancer 17, 565-577. 10.1002/ijc.2910170504 [DOI] [PubMed] [Google Scholar]
- Tang Y. and Gomer R. H. (2008). A protein with similarity to PTEN regulates aggregation territory size by decreasing cyclic AMP pulse size during Dictyostelium discoideum development. Eukaryot. Cell 7, 1758-1770. 10.1128/EC.00210-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya S., Yamabe M., Yamaguchi Y., Kobayashi Y., Konno T. and Tada K. (1980). Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int. J. Cancer 26, 171-176. 10.1002/ijc.2910260208 [DOI] [PubMed] [Google Scholar]
- Veltman D. M., Keizer-Gunnik I. and Van Haastert P. J. M. (2008). Four key signaling pathways mediating chemotaxis in Dictyostelium discoideum. J. Cell Biol. 180, 747-753. 10.1083/jcb.200709180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Fraley C. D., Faridi J., Kornberg A. and Roth R. A. (2003). Inorganic polyphosphate stimulates mammalian Tor, a kinase involved in the proliferation of mammary cancer cells. Proc. Natl. Acad. Sci. USA 100, 11249-11254. 10.1073/pnas.1534805100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. and Janetopoulos C. (2013). Systematic analysis of gamma-aminobutyric acid (GABA) metabolism and function in the social amoeba Dictyostelium discoideum. J. Biol. Chem. 288, 15280-15290. 10.1074/jbc.M112.427047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C., Anjard C., Riemann K., Konzok A. and Nellen W. (2000). gdt1, a new signal transduction component for negative regulation of the growth-differentiation transition in Dictyostelium discoideum. Mol. Biol. Cell 11, 1631-1643. 10.1091/mbc.11.5.1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler-Heitbrock H. W. L., Thiel E., Futterer A., Herzog V., Wirtz A. and Riethmüller G. (1988). Establishment of a human cell line (Mono Mac 6) with characteristics of mature monocytes. Int. J. Cancer 41, 456-461. 10.1002/ijc.2910410324 [DOI] [PubMed] [Google Scholar]