Abstract
Arterial specification and differentiation are influenced by a number of regulatory pathways. While it is known that the Vegfa-Notch cascade plays a central role, the transcriptional hierarchy controlling arterial specification has not been fully delineated. To elucidate the direct transcriptional regulators of Notch receptor expression in arterial endothelial cells, we used histone signatures, DNaseI hypersensitivity and ChIP-seq data to identify enhancers for the human NOTCH1 and zebrafish notch1b genes. These enhancers were able to direct arterial endothelial cell-restricted expression in transgenic models. Genetic disruption of SoxF binding sites established a clear requirement for members of this group of transcription factors (SOX7, SOX17 and SOX18) to drive the activity of these enhancers in vivo. Endogenous deletion of the notch1b enhancer led to a significant loss of arterial connections to the dorsal aorta in Notch pathway-deficient zebrafish. Loss of SoxF function revealed that these factors are necessary for NOTCH1 and notch1b enhancer activity and for correct endogenous transcription of these genes. These findings position SoxF transcription factors directly upstream of Notch receptor expression during the acquisition of arterial identity in vertebrates.
KEY WORDS: Notch1, SoxF, Artery, Arterial enhancer, Endothelial cell, Transcriptional regulation, Zebrafish, Human, Mouse
Summary: Identification of novel enhancers in vertebrate Notch1 genes places SoxF transcription factors directly upstream of Notch signalling in controlling arterial specification and differentiation.
INTRODUCTION
Genetic specification of arterial fate has long been attributed to regulation downstream of the Vegfa and Notch pathways. Vegfa signalling is essential for arterial specification in both zebrafish and mammalian models, at least partially by stimulating the expression of components of the Notch pathway, while activation of Notch signalling can rescue defects in Vegfa-deficient zebrafish embryos (Lanahan et al., 2010; Lawson et al., 2002; Liu et al., 2003; Visconti et al., 2002). The Notch receptors Notch1 and Notch4 and the delta-like ligands Dll1, Dll4, Jag1 and Jag2 are expressed in endothelial cells, where they play crucial roles in both arteriogenesis and angiogenesis (Lawson et al., 2002; Liu et al., 2003; Phng and Gerhardt, 2009; Roca and Adams, 2007). Ligand binding to the Notch receptor releases the Notch intracellular domain (NICD), which translocates to the nucleus and forms a transcriptional activation complex with the otherwise repressive DNA-bound Rbpj [CSL, Su(H)] (Bray, 2006). Combined ablation of Notch1 and Notch4, which are both principally expressed in arterial endothelial cells during early vascular remodelling (Chong et al., 2011; Jahnsen et al., 2015), results in severe vascular remodelling defects (Krebs et al., 2000), as does ablation of the Notch downstream effector Rbpj or of Dll4, a Notch ligand specific within the vasculature to arteries (Duarte et al., 2004; Gale et al., 2004; Krebs et al., 2004). However, loss of Notch signalling does not fully recapitulate the arterial defects downstream of Vegfa ablation (Carmeliet et al., 1996; Krebs et al., 2000; Lawson et al., 2002), and the arterially restricted gene expression patterns of components of the Notch pathway do not fully overlap with activated Vegfa (Lawson, 2003), suggesting that additional factors are involved in the regulation of Notch-mediated arterial fate.
The SoxF group of transcription factors (Sox7, Sox17 and Sox18) are expressed in endothelial cells from early in development (Francois et al., 2010). While each SoxF member displays a subtly different endothelial expression pattern, all three factors are expressed early in arterial development (Corada et al., 2013; François et al., 2008; Zhou et al., 2015) and share considerable functional redundancy, complicating interpretation of the consequences of gene disruption (Hosking et al., 2009; Zhou et al., 2015). Combined loss of sox7 and sox18 in zebrafish, both by morpholino-based knockdown and genetic mutation, resulted in serious arteriovenous malformations similar to those seen after Notch ablation, suggesting that SoxF factors might genetically interact with the Notch pathway in endothelial cells (Cermenati et al., 2008; Hermkens et al., 2015; Herpers et al., 2008; Lawson et al., 2001). Further evidence was provided in mice, where endothelial-specific ablation of Sox17 caused arterial differentiation and remodelling defects (Corada et al., 2013), and conditional deletion of SoxF factors in the adult resulted in loss of major vessel identity in the retina (Zhou et al., 2015). Inhibition of Vegfa signalling can significantly impact SoxF expression and activity in both mouse and zebrafish models (Kim et al., 2016; Pendeville et al., 2008; Duong et al., 2014; Pennisi et al., 2000b), suggesting that members of the SoxF family lie downstream of Vegfa. Further, SoxF acts in a positive feed-forward loop to maintain Flk1 (Kdr; kdrl in zebrafish) expression (Kim et al., 2016). By contrast, the ablation of Notch signalling in the vasculature does not significantly impact the expression of SoxF (Abdelilah et al., 1996; Corada et al., 2013). Different experimental models have positioned SoxF genes both upstream (Corada et al., 2013) and downstream (Lee et al., 2014) of Notch signalling in endothelial cells, suggesting a complex relationship between these two pathways.
Analysis of the only known enhancers for the Notch pathway, namely the arterial-specific Dll4-12 (Sacilotto et al., 2013) and Dll4in3 enhancers (Sacilotto et al., 2013; Wythe et al., 2013), has demonstrated a crucial role for SoxF factors in the regulation of expression of the Notch ligand Dll4 in arteries, in combination with Rbpj/Notch binding and in the presence of Ets factors including Erg (Sacilotto et al., 2013; Wythe et al., 2013). Although this work clearly positioned the SoxF and Notch pathways as crucial regulators of arterial specification, and is supported by analysis placing Sox17 upstream of Notch signalling in mouse models (Corada et al., 2013), ablation of arterial marker expression only occurs after the removal of both SoxF factors and Notch signalling in combination (Sacilotto et al., 2013). Consequently, the precise transcriptional hierarchy of SoxF and Notch has yet to be fully established. Although studies of the Notch receptor genes Notch1 and Notch4 also identified SoxF binding motifs within putative promoter sequences (Corada et al., 2013; Lizama et al., 2015), a requirement for these SOX motifs in arterial-specific gene expression of Notch receptors has not been established. In this study, we have identified and characterised arterial-specific enhancers directing NOTCH1/notch1b gene expression in vivo, and used them to demonstrate a direct requirement for SoxF factors in the transcriptional regulation of Notch receptors during early arterial differentiation, positioning SoxF factors upstream of Notch in the acquisition of arterial cell identity.
RESULTS
Identification of an arterial-specific NOTCH1 intronic enhancer
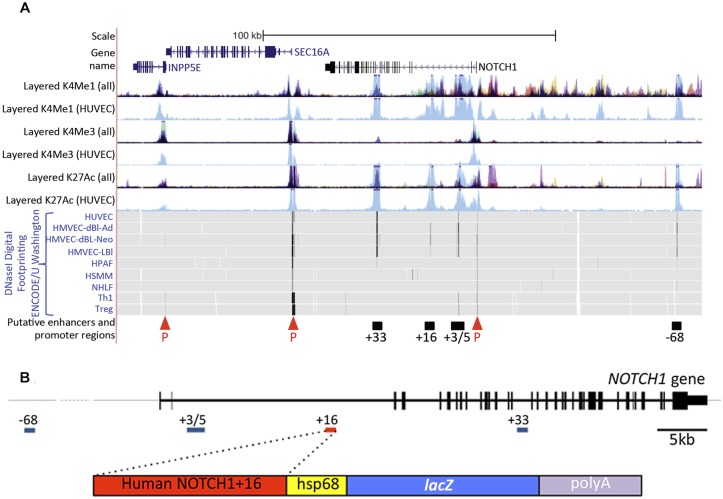
Previous studies into the transcriptional regulation of the Notch receptors have not extended to the identification or analysis of gene enhancers (cis-regulatory elements) (Corada et al., 2013; Lizama et al., 2015; Wu et al., 2005). We therefore conducted a detailed in silico analysis of the NOTCH1 locus with the aim of identifying novel, arterial-specific enhancers. This analysis focused on human NOTCH1 in order to take advantage of the wealth of publicly available information describing chromatin modifications in human endothelial cell lines. Using this information, we were able to pinpoint four regions of DNA rich in endothelial cell-specific H3K4me1 and H3K27ac histone modifications and DNaseI digital genomic footprints, all marks closely associated with enhancer activity (Heintzman and Ren, 2009; Sabo et al., 2004) (Fig. 1A, Fig. S1).
Fig. 1.
The human NOTCH1 locus contains multiple putative endothelial enhancers. (A) The human NOTCH1 locus from UCSC ENCODE Genome Browser (http://genome.ucsc.edu). Human umbilical vein endothelial cell (HUVEC)-specific H3me1 and H3K27ac (enhancer associated) and H3K4me3 (promoter associated) peaks are indicated in light blue [both in the separate HUVEC and combined tracks (denoted as ‘all’)]; other colours indicate H3K4me1, H3K4me3 and H3K4ac peaks specific to non-endothelial cell lines: GM12878 cells (red), H1-hESC cells (yellow), HSMM cells (green), K562 cells (purple) NHEK cells (lilac) and NHLF cells (pink). INPP5E and SEC16A are shown (blue text) in addition to NOTCH1 (black text). DNase I digital hypersensitive hotspots are indicated by black vertical lines on grey (HUVEC, HMVEC-dBl-Ad, HMVEC-dBl-Neo and HMVEC-LBl, which are all different endothelial cell types). The four NOTCH1 putative enhancer regions (black bars) were identified by high levels of HUVEC-specific H3K4me1 and H3K27ac associated with endothelial cell-specific DNaseI hypersensitivity hotspots. P, putative promoters identified by H3K4me3. (B) The human NOTCH1 gene (top, in 5′ to 3′ orientation with putative enhancers indicated) and the NOTCH1+16hsp transgene (bottom).
The putative enhancer regions were named NOTCH1+33, NOTCH1+16, NOTCH1+3/5 and NOTCH1−68 to reflect their distance from the transcription start site (TSS) in kb. Each enhancer region was cloned upstream of the silent hsp68 minimal promoter and the lacZ reporter gene (Fig. 1B) and tested for its ability to drive reporter gene expression specifically in arterial endothelial cells of transient transgenic mice at embryonic day (E) 12-13. Although each of the four putative enhancer regions was able to drive detectable levels of lacZ in transgenic mice, this expression was primarily neural, an expression pattern commonly seen when using the hsp68 minimal promoter (e.g. Becker et al., 2016; Sacilotto et al., 2013) (Table 1, Fig. S2). Only the NOTCH1+33 and NOTCH1+16 enhancers were able to direct expression in endothelial cells (Table 1, Fig. S2). In the case of NOTCH1+33, vascular expression was detected in only one of the five transgenic mice analysed. This expression was not restricted to the arterial endothelium but was pan-endothelial (Table 1, Fig. S2). This agrees with previous reports indicating that the mouse orthologue of this region, termed Notch1_enh1, also directs occasional vascular enhancer activity but did not show arterial-specific expression (Zhou et al., 2017). Conversely, the 274 bp NOTCH1+16 enhancer was able to robustly direct expression specifically to arterial endothelial cells within the vasculature at E12 in multiple independent transgenic embryos (Table 1, Fig. 2 and Fig. S2). This indicates that the NOTCH1+16 enhancer represents a novel, arterially restricted enhancer within the NOTCH1 locus. Analysis of a stable mouse line expressing the NOTCH1+16:lacZ transgene clearly demonstrated that this enhancer is strongly active from the very early stages of vascular development, mimicking the expression of endogenous Notch by becoming restricted to the arteries by late E9.5 and then maintaining an arterial endothelial cell-restricted expression pattern throughout embryonic development (Chong et al., 2011; Jahnsen et al., 2015) (Fig. 2).
Table 1.
Reporter gene expression patterns in E12 mice transgenic for each putative NOTCH1 enhancer region and the effects of SOX motif mutation on NOTCH1+16 enhancer activity
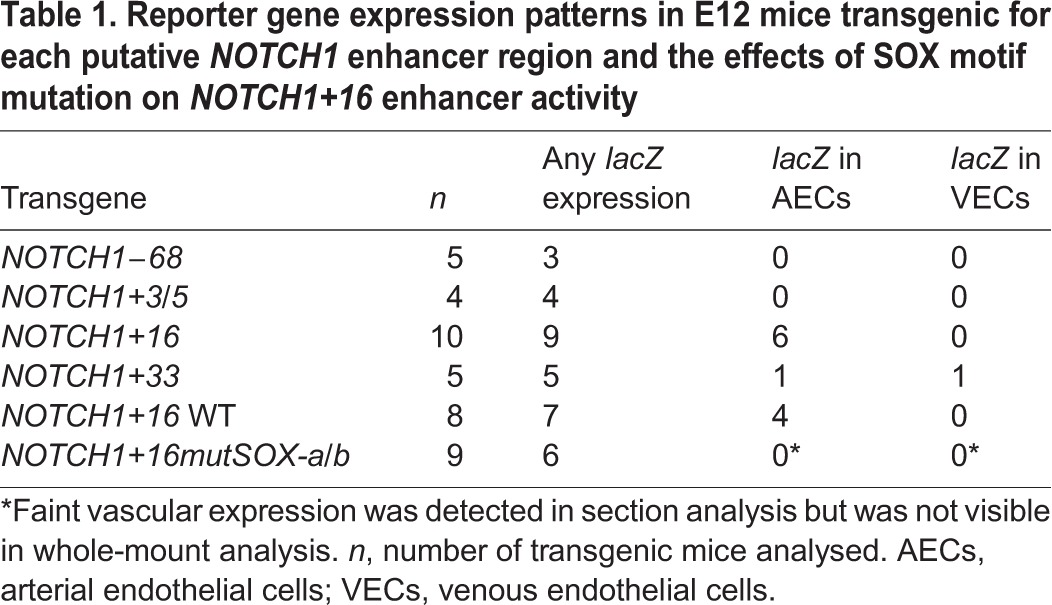
Fig. 2.
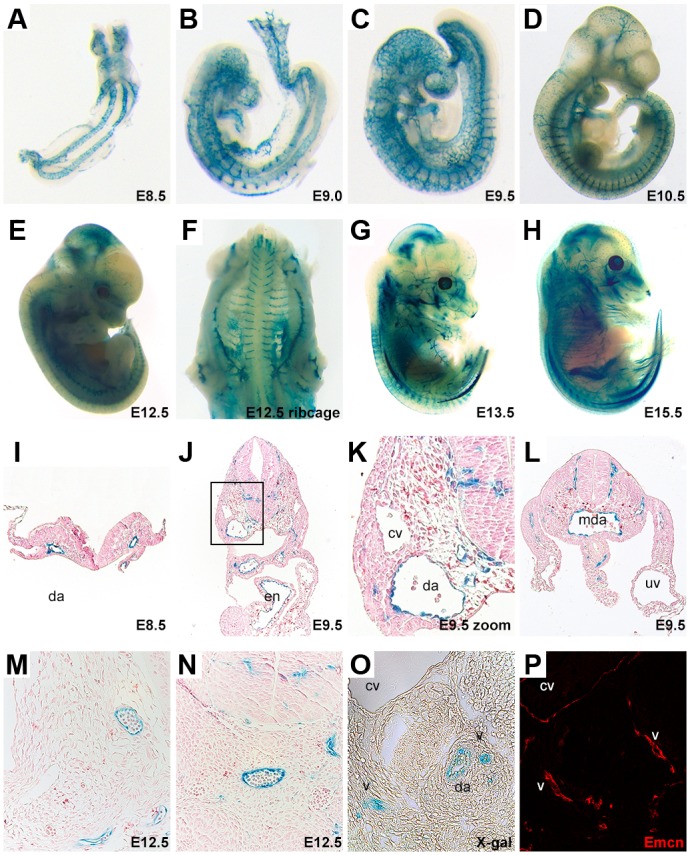
The NOTCH1+16 transgene directs arterial endothelial cell-restricted expression in transgenic mice. (A-N) Representative transgenic whole-mount embryos (A-H) and transverse sections (I-N) showing lacZ reporter gene expression (β-galactosidase detected by blue X-gal staining) in arterial endothelial cells throughout embryonic development. The boxed region in J is magnified in K. (O,P) E12 transverse section showing that expression of the venous marker endomucin (Emcn) does not overlap with lacZ reporter gene expression on the same section. cv, cardinal vein; da, dorsal aorta; Emcn, endomucin; en, endocardium; mda, midline dorsal aorta; uv,umbilical vein; v, vein.
The NOTCH1+16 enhancer is bound and regulated by SoxF factors
To identify the transcription factors that potentially regulate the NOTCH1+16 enhancer, we performed a ClustalW analysis of orthologous mammalian sequences (Fig. 3A). This analysis clearly identified nine conserved core consensus ETS binding motifs [GGAW (Hollenhorst et al., 2011)] and two consensus SOX binding motifs [WWCAAW (Mertin et al., 1999)] (Fig. 3A) within the 274 bp NOTCH1+16 enhancer. Because not all in silico consensus binding motifs are able to functionally bind the cognate protein, these motifs were then tested by electrophoretic mobility shift assay (EMSA). Both SOX motifs (termed hmSOX-a and hmSOX-b) were able to bind recombinant SOX7 and SOX18 proteins in EMSA (Fig. 3B), and three ETS motifs (termed hmETS-a, hmETS-b and hmETS-c) were able to bind the endothelial Ets protein ETV2 (Fig. 3C).
Fig. 3.
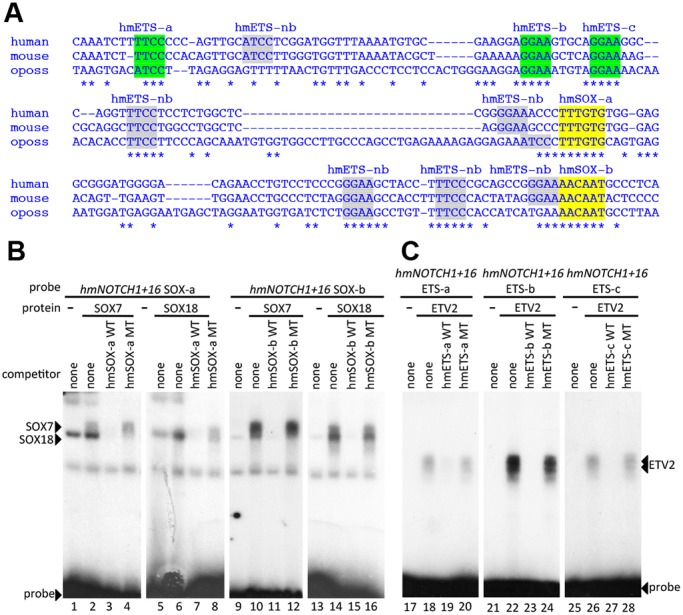
The NOTCH1+16 enhancer contains SOX and ETS binding motifs. (A) Multispecies alignment of the orthologous region of the NOTCH1+16 enhancer from human, mouse and opossum (oposs) using ClustalW. Coloured sequences are confirmed by EMSA; grey sequences are motifs identified in silico that did not bind in EMSA. (B) Radiolabelled oligonucleotide probes encompassing NOTCH1+16 hmSOX-a (lanes 1-8) and hmSOX-b (lanes 9-16) were bound to recombinant SOX7 (lanes 2-4 and 10-12) and SOX18 (lanes 6-8 and 14-16). Both proteins, which efficiently bound labelled probes (lanes 2, 6, 10 and 14), were competed by excess unlabelled self-probe (WT, lanes 3, 7, 11 and 15) but not by mutant self-probe (MT, lanes 4, 8, 12 and 16). (C) Radiolabelled oligonucleotide probes encompassing NOTCH1+16 hmETS-a (lanes 17-20), hmETS-b (lanes 21-24) and hmETS-c (lanes 25-28) were bound to recombinant ETV2 protein. ETV2, which efficiently bound to labelled probes (lanes 18, 22 and 26), was competed by excess unlabelled self-probe (WT, lanes 19, 23 and 27) but not by mutant self-probe (MT, lanes 20, 24 and 28).
ETS motifs are common to all endothelial-expressed gene enhancers (De Val and Black, 2009). Although the Ets factor Erg has been implicated in arterial specification (Wythe et al., 2013), previous studies have shown that ETS motifs were unable to direct expression of the arterial-specific Dll4 and Flk1 enhancers without additional transcription factor binding motifs (Becker et al., 2016; Sacilotto et al., 2013; Wythe et al., 2013), suggesting that Ets factors alone are unlikely to regulate the NOTCH1+16 enhancer.
To test whether the SOX motifs play a role in NOTCH1+16 enhancer activity, we mutated the core nucleotides of these motifs (see Materials and Methods for sequences) and tested the ability of the resultant NOTCH1+16mutSOX-a/b enhancer to drive reporter gene expression. Strikingly, enhancer mutation resulted in a dramatic reduction in reporter gene expression in endothelial cells in transgenic mice, although transgene expression was detected outside of the vascular system (Fig. 4, Table 1). This result differs notably from that reported for the Dll4 enhancers, where mutations in SOX motifs, or loss of SoxF factors, resulted in no detectable decrease in Dll4 expression unless accompanied by ablation of Notch signalling (Sacilotto et al., 2013).
Fig. 4.
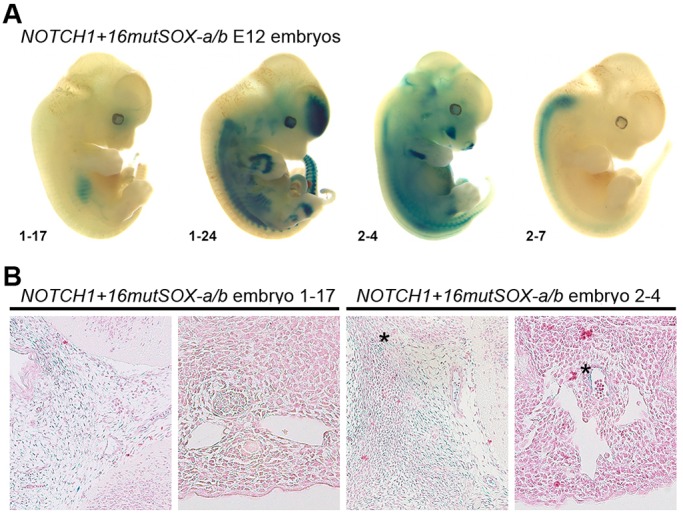
SoxF factors are required for NOTCH1+16 activity. (A) Four representative whole-mount E12 X-gal-stained embryos transgenic for the NOTCH1+16mutSOX-a/b construct. Numbers at the bottom left indicate the unique embryo identifier. (B) Transverse sections taken from two NOTCH1+16mutSOX-a/b embryos demonstrate the very limited endothelial expression detected in these embryos (asterisks). In each case, the section to the left is through the head region, the section to the right is through the upper torso region.
Zebrafish notch1b is directly transcriptionally regulated by SoxF factors via an evolutionarily non-conserved enhancer
The SoxF-dependent NOTCH1+16 enhancer robustly directed arterial-restricted expression in transgenic mouse models. However, this enhancer did not exhibit sequence conservation beyond mammals (Fig. S1B), leaving it unclear how relevant these observations are to Notch signalling during arteriovenous specification in zebrafish, an extremely well-studied model system (Gore et al., 2012). We therefore investigated whether SoxF factors were able to transcriptionally regulate the zebrafish orthologue of NOTCH1, notch1b, by examining the binding patterns of the zebrafish SoxF transcription factors around the notch1b locus. Zebrafish SoxF are expressed in early endothelial cells and implicated in arteriovenous differentiation (Cermenati et al., 2008; Hermkens et al., 2015; Herpers et al., 2008; Pendeville et al., 2008). To probe for SoxF (Sox7, Sox17 and Sox18) genome-wide binding locations we used an endothelial-specific SOX18Ragged overexpression line. The SOX18Ragged dominant-negative protein has been shown to interfere with the endogenous function of all three SoxF transcription factors (James et al., 2003; Pennisi et al., 2000b). Using 26-28 hours post fertilisation (hpf) embryos from the tg(fli1a:Gal4FF;10×UAS:Sox18Ragged-mCherry) zebrafish line, in which a tagged SOX18Ragged is expressed specifically in endothelial cells (Fig. S3A), ChIP-seq analysis identified a SoxF binding event 15 kb upstream of the notch1b first exon (Fig. 5A). This binding peak was enriched in the enhancer-associated histone modifications H3K4me1 and H3K27ac at 24 hpf (Bogdanović et al., 2012; Kent et al., 2002) (Fig. 5A), suggesting that it might represent a novel enhancer of notch1b. A 1219 bp zebrafish DNA fragment corresponding to the SOX18-bound region, termed the notch1b-15 enhancer, was cloned upstream of a silent gata2a promoter and GFP reporter gene within the zebrafish enhancer detection (ZED) vector (Bessa et al., 2009) (Fig. 5B) and used to generate the stable tg(notch1b-15:GFP) fish line (Fig. 5C). The GFP transcript was detected in the vascular cord around the midline from 19 hpf, and persisted in the vascular rod as it formed the dorsal aorta at 22 hpf (Fig. 5C). GFP expression continued to be restricted to the dorsal aorta and the segmental arteries in larvae from 24 hpf until 48 hpf. This arterial-restricted pattern of expression within the vasculature was similar to that of endogenous notch1b (Fig. S3B), indicating that the SOX18-bound notch1b-15 element is a bona fide notch1b enhancer and suggesting that SoxF factors directly transactivate Notch receptor transcription in arterial endothelial cells in zebrafish.
Fig. 5.
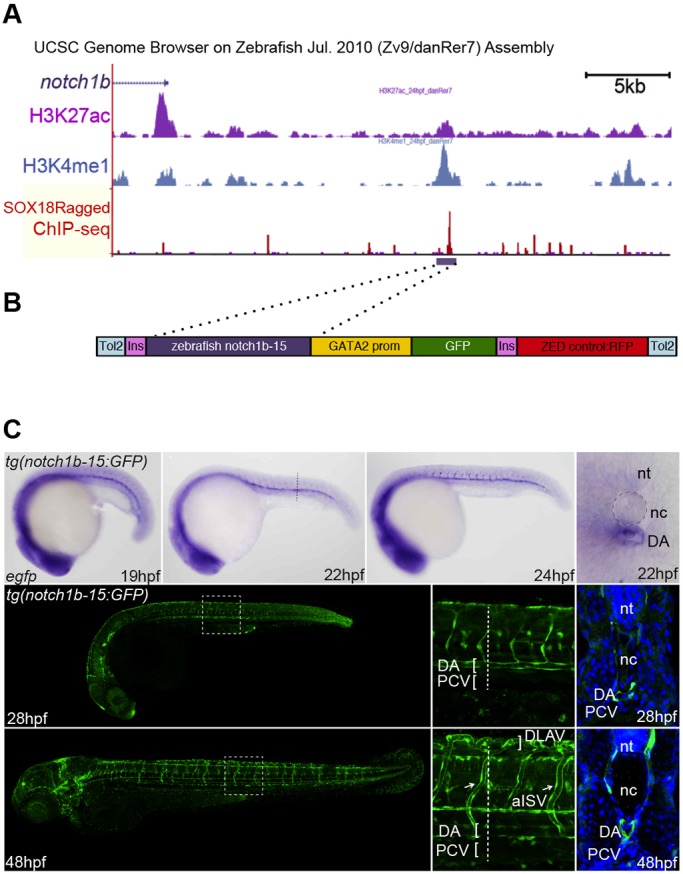
A SOX18-bound region within the notch1b locus represents a bona fide arterial-specific enhancer. (A) Part of the zebrafish notch1b locus from the UCSC ENCODE Genome Browser. The notch1b gene is in 3′ to 5′ orientation, H3K27ac peaks at 24 hpf are in purple, H3K4me1 peaks at 24 hpf are blue, SOX18Ragged ChIP-seq peaks are red, and the region encompassing the notch1b-15 enhancer is indicated by the purple horizontal bar. (B) The ZED notch1b-15:GFP transgene. Ins, insulator sequences; GATA2 prom, the silent GATA2 promoter; ZED control:RFP, the active cardiac actin enhancer/promoter construct fused to the RFP gene used as a positive control in the ZED vector. (C) The notch1b-15:GFP transgene directs arterial endothelial cell-specific expression in the zebrafish line tg(notch1b-15:GFP). Representative transgenic whole-mount embryos and transverse sections show reporter gene expression, as detected by in situ hybridisation (top row, blue) or GFP fluorescence (bottom rows, green) in arterial endothelial cells throughout embryonic development. nt, neural tube; nc, notochord; DA, dorsal aorta; PCV, posterior cardinal vein; aISV (arrows), arterial intersomitic vessel; DLAV, dorsal longitudinal anastomotic vessel.
ClustalW analysis comparing the orthologous enhancer sequences from fugu, stickleback and medaka revealed a remarkably similar pattern of conserved transcription factor motifs when compared with the NOTCH1+16 enhancer (Fig. 6A), with multiple ETS and two SOX binding motifs, termed zfSOX-a and zfSOX-b, confirmed by EMSA analysis (Fig. 6B,C). To establish whether the zfSOX-a and zfSOX-b binding motifs were required for notch1b-15 arterial enhancer function, we generated transient transgenic fish lines harbouring mutated SOX binding motifs (notch1b-15mutSOX-a/b:GFP) and compared the activity of the transgene with wild-type (WT) notch1b-15:GFP control transient transgenic animals (Fig. S4A,B). Simultaneous disruption of both zfSOX-a and zfSOX-b sites led to a reduction of arterial-specific GFP expression in endothelial cells. Although a minority of mutant fish still expressed GFP after SOX binding site mutation, the loss of expression was still much greater than that seen after SOX motif mutation in a previously published Dll4 enhancer in transgenic zebrafish, where vascular expression rates were unaffected by mutations of SoxF binding motifs (Sacilotto et al., 2013), supporting a key role for SoxF factors in notch1b activation. To further confirm our observation, we established stable transgenic notch1b-15mutSOX-a/b:GFP fish lines and compared them with the established WT notch1b-15:GFP lines. Analysis of the stably transgenic embryos (Fig. 7A) confirmed a GFP expression pattern in the dorsal aorta and segmental arteries for the WT transgene. By contrast, the tg(notch1b-15mutSOX-a/b:GFP) lines showed ectopic GFP expression in neurons and a significant decrease of GFP expression in the arterial endothelium (Fig. 7A). Quantitative analysis of GFP intensity in both dorsal aorta and segmental arteries showed lower expression in most tg(notch1b-15mutSOX-a/b:GFP) than in WT tg(notch1b-15:GFP) embryos (Fig. 7B,C, Fig. S4C). Taken together, these data clearly demonstrate that the zfSOX-a/b binding sites are required to guide notch1b-15-specific enhancer activity in vivo during arterial development.
Fig. 6.
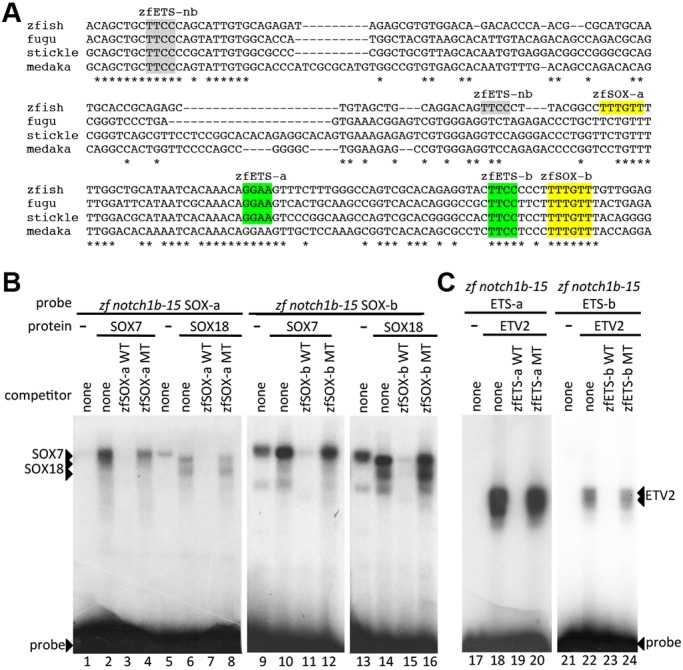
The notch1b-15 enhancer contains essential SoxF binding motifs. (A) Multispecies alignment of the orthologous regions of the notch1b-15 enhancer from zebrafish (zfish), fugu, stickleback (stickle) and medaka using ClustalW. Coloured sequences are confirmed by EMSA; grey sequences are motifs identified in silico that did not bind robustly in EMSA. (B) Radiolabelled oligonucleotide probes encompassing notch1b-15 zfSOX-a (lanes 1-8) and zfSOX-b (lanes 9-16) were bound by recombinant SOX7 (lanes 2-4 and 10-12) and SOX18 (lanes 6-8 and 14-16). Both proteins, which efficiently bound labelled probes (lanes 2, 6, 10 and 14), were competed by excess unlabelled self-probe (WT, lanes 3, 7, 11 and 15) but not by mutant self-probe (MT, lanes 4, 8, 12 and 16). (C) Radiolabelled oligonucleotide probes encompassing notch1b-15 zfETS-a (lanes 17-20) and zfETS-b (lanes 21-24) (see A) were bound by recombinant ETV2 proteins. ETV2, which efficiently bound to labelled probes (lanes 18 and 22), was competed by excess unlabelled self-probe (WT, lanes 19 and 23) but not by mutant self-probe (MT, lanes 20 and 24).
Fig. 7.
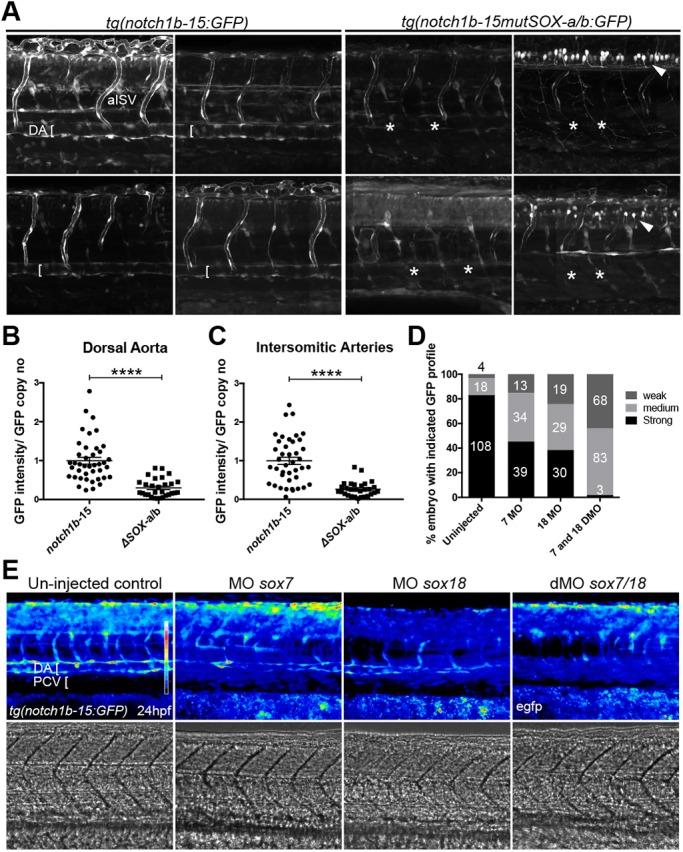
SoxF factors are required for notch1b-15 activity. (A) Confocal projection of stably transgenic WT tg(notch1b-15:GFP) (left) and tg(notch1b-15mutSOX-a/b:GFP) (right), where both zfSOX-a and zfSOX-b sites have been simultaneously mutated. Representative larvae from four independent founders are shown. Asterisks indicate the reduced GFP expression in the dorsal aorta. Arrowheads show the ectopic neuronal expression observed in tg(notch1b-15mutSOX-a/b:GFP). All panels show composite images from tile scan acquisition. (B,C) The intensity of GFP expression in the dorsal aorta and arterial intersomitic vessels of the stably transgenic tg(notch1b-15:GFP) and tg(notch1b-15mutSOX-a/b:GFP) at 2 dpf. Expression is normalised to GFP genomic copy number. Fish were pooled from three or four separate founders. Mean±s.e.m. tg(notch1b-15), n=41; tg(notch1b-15mutSOX-a/b:GFP), n=29. ****P<0.0001 (Mann–Whitney U-test). (D) Levels of GFP expression in tg(notch1b-15:GFP) zebrafish embryos after MO injection. Number of fish for each condition is indicated. (E) Representative examples of sox7/18 double-morphant tg(notch1b-15:GFP) zebrafish at 24 hpf as compared with uninjected controls. Double morphants (dMO) demonstrated reduced EGFP expression in the dorsal aorta and intersomitic vessels. DA, dorsal aorta; PCV, posterior cardinal vein; aISV, arterial intersomitic vessel.
We next investigated the consequences of morpholino (MO)-based knockdown of SoxF on tg(notch1b-15:GFP) fish. sox7/sox18 double morphants exhibit a severe vascular phenotype (Fig. S5), including fusions and shunts between the dorsal aorta and cardinal vein (Cermenati et al., 2008; Herpers et al., 2008; Pendeville et al., 2008), phenotypes that are shared with sox7;sox18 double-mutant fish (Hermkens et al., 2015). MO-induced transcript depletion of sox7 or sox18 resulted in downregulation of notch1b-15:GFP expression, while sox7/sox18 double-morphant tg(notch1b-15:GFP) fish demonstrated a near-complete loss of reporter gene expression (Fig. 7D,E).
Taken together, these results indicate that SoxF proteins directly modulate the activity of arterial-specific enhancers for both the mammalian NOTCH1 and zebrafish Notch1b receptors, positioning SoxF transcription factors directly upstream of Notch signalling during early arterial differentiation in both mammals and zebrafish.
Loss of endogenous notch1b-15 enhancer activity perturbs notch1b transcription and causes arteriovenous defects
To assess whether the endogenous notch1b-15 regulatory element is functionally relevant during arteriovenous differentiation in vivo, we deleted the endogenous notch1b-15 enhancer in zebrafish. The resultant notch1b-15uq1mf allele was generated using two guide RNAs to drive rapid genome editing using the CRISPR/Cas9 system (Fig. 8A), resulting in excision of a 203 bp fragment overlapping the notch1b-15 enhancer. Analysis of endogenous notch1b expression in the F2 generation demonstrated lower notch1b expression levels in both dorsal aorta and arterial intersomitic vessel (aISV) of the notch1b-15uq1mf/uq1mf homozygous embryos as compared with their sibling controls, whereas no change was observed in the neural tube (Fig. 8B). Next, we assessed the notch1b transcript levels in purified flt1-positive arterial endothelial cell populations from the F3 generation of homozygous fish (F2 notch1b-15uq1mf homozygous in-cross) compared with WT control larvae (F2 notch1b-15+/+ × flt1:YFP;lyve1:dsRed) at 24-28 hpf (Fig. 8C). As expected, notch1b transcripts were significantly downregulated in the homozygous animals, strongly supporting a role for the notch1b-15 enhancer in the transcription of endogenous notch1b in arterial endothelial cells.
Fig. 8.
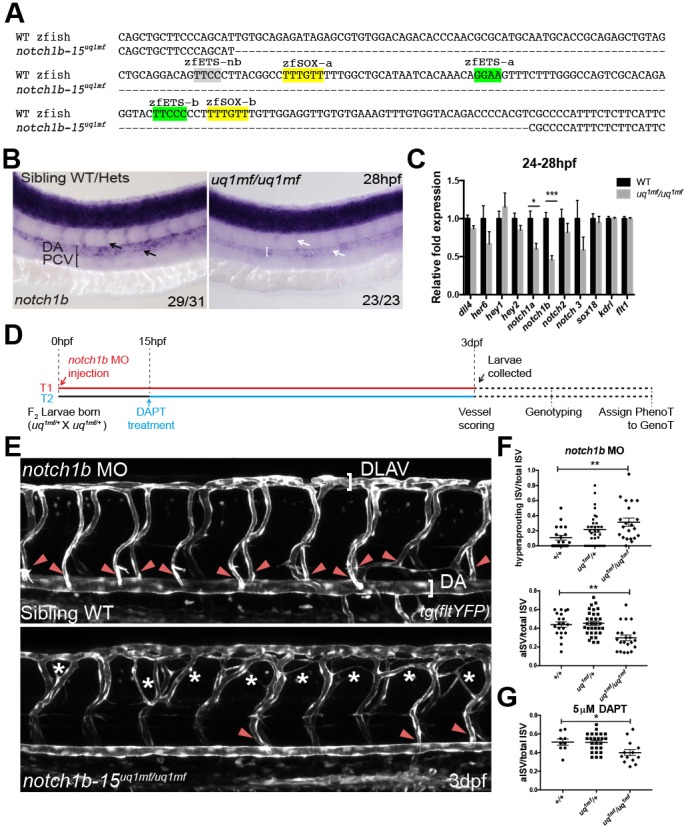
Loss of endogenous notch1b-15 compromises artery formation and reduces the endogenous notch1b transcript level. (A) The deleted region (dashed line) of notch1b-15 mutant allele uq1mf, which includes both the zfSOX-a and zfSOX-b sites (yellow). (B) F2 notch1b-15uq1mf/uq1mf has reduced notch1b expression in dorsal aorta and intersomitic vessels (white arrows) as compared with sibling WT and heterozygotes (black arrows) at 26-28 hpf. The number of embryos showing the illustrated phenotype among the total examined is indicated. (C) Quantitative PCR on FACS-sorted endothelial populations at 24-28 hpf, showing that F3 notch1b-15uq1mf/uq1mf fish have lower notch1b expression than WT fish. Expression is relative to kdrl and flt1. Mean±s.e.m. n=6 (uq1mf/uq1mf) and n=8 (WT) independent sorts, where each sort was pooled from 60-100 larvae;. *P<0.05, ***P<0.001 (t-test). (D) The treatment regime conducted to characterise the vascular phenotype of the uq1mf/+ cross. In treatment 1 (T1, red), notch1b MO was injected at the 1-2 cell stage. The developing vasculature of each embryo was then analysed blindly at 3 dpf. After scoring, genotypes were assigned to each larva. In treatment 2 (T2, blue), larvae from the uq1mf/+ cross were treated with or without DAPT (5 µM) from 15-16 hpf until 3 dpf. Vessels of these treated larvae were blindly scored prior to genotyping, as reported for T1. (E) At 3 dpf, notch1b-15uq1mf/uq1mf notch1b morphants frequently showed ectopic sprouting in between the intersomitic vessels (asterisks) as compared with sibling WT notch1b morphants. Mutants also show loss of arterial connections (red arrowheads) between the dorsal longitudinal anastomotic vessel (DLAV) and dorsal aorta (DA), as indicated by the loss of YFP expression in the tg(flt1:YFP) background. (F) (Top) Quantification of hypersprouting ISV number in individual notch1b morphants labelled by tg(flt1:YFP) at 3 dpf. Mean±s.e.m. Sibling WT (+/+), n=20; heterozygote (uq1mf/+), n=32; homozygous mutant (uq1mf/uq1mf), n=21. (Bottom) Quantification of YFP-positive intersomitic vessels that connect between DLAV and DA in individual notch1b morphants labelled by tg(flt1:YFP) at 3 dpf. Mean±s.e.m. Sibling WT, n=20; heterozygote, n=32; homozygous mutant, n=21. **P<0.005 (Mann–Whitney U-test). (G) Quantification of YFP-positive intersomitic vessels that connect between DLAV and DA in individual embryos treated with 5 μM DAPT at 3 dpf. Vessels are labelled by tg(flt1:YFP). Mean±s.e.m. Sibling WT, n=9; heterozygote, n=25; homozygous mutant, n=14. *P<0.05 (Mann–Whitney U-test).
To assess the phenotypic outcome of notch1b-15 loss of function, we took advantage of the tg(notch1b-15uq1mf/+;flt1:YFP;lyve1:dsRed) line to analyse the developing vasculature after notch1b-15 enhancer deletion in both the F2 and F3 generations. Surprisingly, given the reduced levels of notch1b (Fig. 8B), no overt vascular phenotype was detected in F2 notch1b-15uq1mf/uq1mf homozygous zebrafish (F1 heterozygous notch1b-15uq1mf/+ in-cross) (Fig. S6A), suggesting partial enhancer redundancy. Such redundancy, which is potentially explained by the pervasiveness of redundant, or ʻshadow', enhancers around developmental genes (Cannavò et al., 2016), has previously been well documented in key endothelial genes, with examples including the Dll4, Flk1 and Tal1 loci (Cannavò et al., 2016). By contrast, we detected a subpopulation (20-30%) of larvae from the F3 generation (F2 homozygous notch1b-15uq1mf/uq1mf in-cross) that displayed a phenocopy of the notch1b loss-of-function phenotype (Fig. S6B). This increase in the phenotypic severity in the F3 generation suggests that, in the context of the notch1b-15uq1mf/+ cross, maternal mRNA deposition is likely to help compensate for the disruption of notch1b transcription caused by deletion of the notch1b enhancer, a compensation that is reduced in a purer notch1b-15uq1mf/uq1mf genetic background (Harvey et al., 2013).
To bypass potential rescue effects of shadow enhancers or maternally deposited transcripts, we also investigated arteriovenous differentiation and sprouting angiogenesis in F2 notch1b-15uq1mf/uq1mf zebrafish after low-level depletion of notch1b mRNA. A splice notch1b MO was injected into eggs from a notch1b-15uq1mf/+ in-cross in the tg(flt1:YFP;lyve1:dsRed) background. The notch1b MO was used at suboptimal concentration (5 ng/embryo), which is known to result in minimal phenotypes (Sacilotto et al., 2013). The developing vasculature of each resulting embryo was analysed blindly at 3 dpf, and genotypes were assigned to embryos after image acquisition (Fig. 8D, red). Whereas most WT siblings had normal ISV development, we observed an increased number of hypersprouting ISVs across the notch1b-15uq1mf/+ and notch1b-15uq1mf/uq1mf population (Fig. 8E, asterisks; Fig. 8F, top) in a gene dosage-dependent manner, similar to the phenotype described previously in high MO concentration notch1b morphants and Notch signalling-deficient embryos (Geudens et al., 2010; Siekmann and Lawson, 2007). Further, mutant embryos also demonstrated loss of arterial connections between the dorsal aorta and dorsal longitudinal anastomotic vessel, similar to those described in Notch signalling-deficient zebrafish (Geudens et al., 2010; Quillien et al., 2014), while the notch1b-depleted notch1b-15+/+ and notch1b-15uq1mf/+ morphant embryos demonstrated an equal proportion of arterial and venous ISVs as previously reported (Bussmann et al., 2010) (Fig. 8E,F bottom).
To further confirm that interfering with notch1b-15 enhancer activity is additive to notch1b transcript depletion, we chemically treated the notch1b-15uq1mf/+ cross with DAPT, a well characterised Notch signalling inhibitor, over the course of endothelial differentiation (15- to 16-somite stage through to 3 dpf) (Fig. 8D, blue). All embryos treated with a suboptimal concentration of DAPT (5 µM) showed a straight body axis, indicating that somitogenesis (and therefore Notch activity) was not significantly compromised (Fig. S7A). Interestingly, despite this lack of morphological defects, F2 fish homozygous for the notch1b-15uq1mf allele displayed a lower arterial-to-total ISV ratio (Fig. 8G, Fig. S7A). By contrast, fish homozygous for the notch1b-15uq1mf allele treated with DMSO vehicle alone had a comparable aISV ratio to both notch1b-15+/+ and notch1b-15uq1mf/+ siblings (Fig. S7B), similar to the untreated control. This suggests that the observed loss of aISV is specific to an additive effect of DAPT treatment and notch1b-15 enhancer activity disruption. Overall, these data suggest a functional role of the notch1-15 enhancer in the endothelial-specific initiation of notch1b transcription to promote the acquisition of arterial cell identity.
SoxF factors are required for endogenous Notch1/notch1b expression
Our results have clearly implicated SoxF factors as direct upstream regulators of arterial Notch enhancers, and therefore suggest a considerably greater role for SoxF in the regulation of the Notch receptors than of the Notch ligands. However, since the notch1b-15 enhancer is partially redundant with other notch1b shadow enhancers, we wished to establish whether SoxF regulation is required for endogenous notch1b expression itself, not just enhancer activity. Further, our results so far do not entirely rule out the possibility of SoxF/Rbpj combinatorial regulation of notch1b, as was previously shown for Dll4 enhancers (Sacilotto et al., 2013). Although neither the human NOTCH1+16 nor the zebrafish notch1b-15 enhancer contains conserved consensus Rbpj/Notch binding motifs, transcription factors can bind non-consensus motifs, and not all transcription factors necessarily bind conserved motifs (Wong et al., 2015). The nature of the SoxF/Notch combinatorial regulation of Dll4, where the SOX or RBPJ binding motifs play functionally interchangeable roles, indicates potential direct interactions between these two proteins, such that only a single SOX binding motif might be necessary for SoxF/Rbpj synergy. We therefore investigated the consequences of SoxF depletion on endogenous Notch1/notch1b expression in vivo.
Although Sox17 is robustly expressed in arterial endothelial cells (Corada et al., 2013; Hosking et al., 2009), compound Sox7;Sox18 deletion in mice resulted in a reduction of Notch1 mRNA levels in the trunk dorsal aorta and primitive heart cavities of E8.5 embryos (Fig. 9A, Fig. S8A). These results concur with observations in the mouse retina, where the vascular phenotype after Sox7;Sox17;Sox18 endothelial-specific triple deletion closely resembled defects caused by loss of Notch signalling (Zhou et al., 2015). Strikingly, both MO-induced gene knockdown and compound mutation of sox7 and sox18 in zebrafish embryos also led to a near-complete loss of notch1b transcript expression specifically in endothelial cells, as shown by in situ hybridisation analysis (Fig. 9B,C, Fig. S8B). These results further establish an essential role for SoxF transcription factors in the induction of Notch1/notch1b gene expression, and position SoxF proteins at the top of the transcriptional hierarchy regulating arterial specification.
Fig. 9.
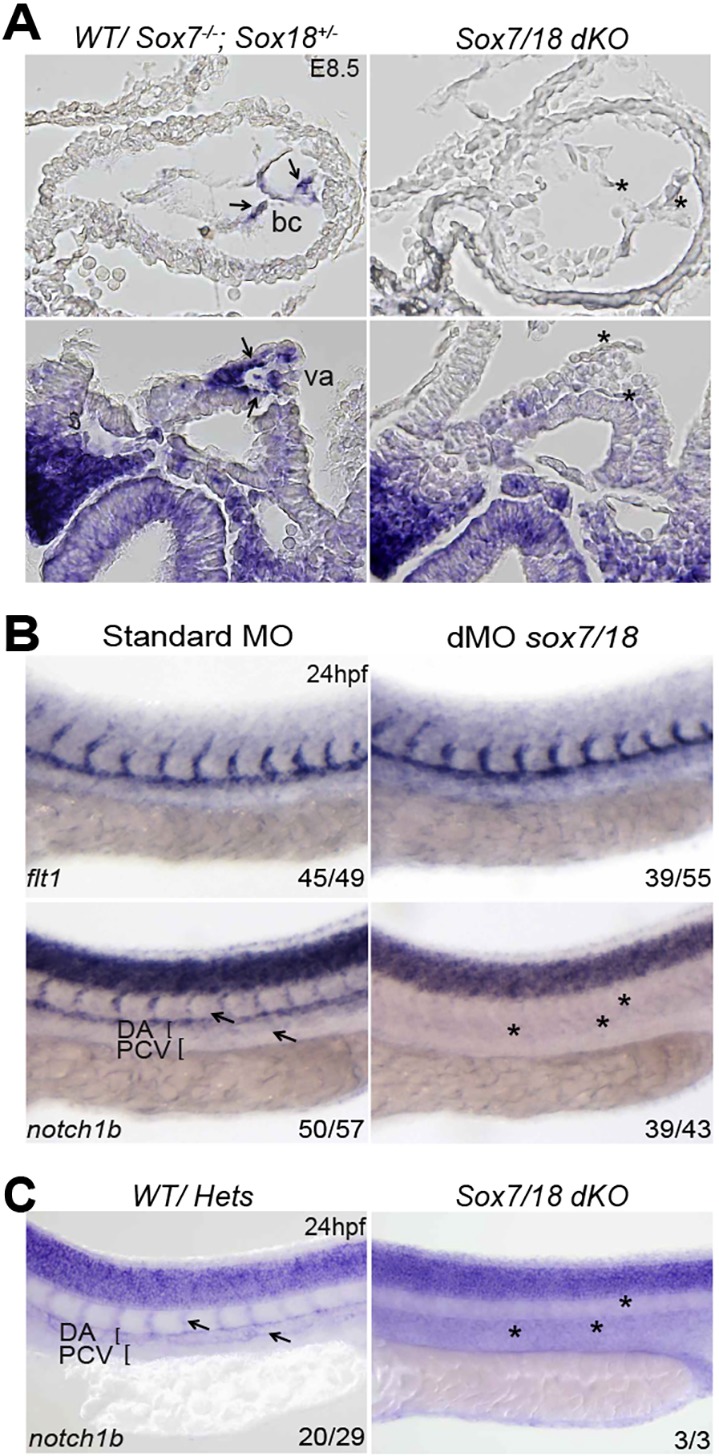
Mouse and zebrafish arterial Notch1 expression is dependent on SOX7/18 activity. (A) Transverse sections of whole-mount in situ hybridisation for Notch1 transcript on E8.5 mouse embryos shows a reduction of Notch1 expression (asterisks) in the bulbus cordis (bc) region of the primitive heart and vitelline artery (va) of Sox7/Sox18 double knockouts. (B) At 24 hpf notch1b expression is significantly downregulated in the dorsal aorta (asterisks) of sox7/sox18 double-morphant zebrafish, whereas its signal is unaffected in the neural tube. flt1 expression is comparable between controls and sox7/sox18 double morphants, indicating that the dorsal aorta is correctly formed. (C) notch1b was barely detectable in the dorsal aorta and ISVs of sox7/18 double-knockout zebrafish (asterisks), as compared with the WT, sox7 or sox18 heterozygotes (arrows). The number of embryos showing the illustrated phenotype among the total examined is indicated. DA, dorsal aorta; PCV, posterior cardinal vein.
DISCUSSION
Recent work has implicated SoxF, Ets and Rbpj, the Notch transcriptional effector, in the regulation of the Notch ligand Dll4 and many other key arterial genes (Corada et al., 2013; Lizama et al., 2015; Sacilotto et al., 2013; Wythe et al., 2013), but has not established the hierarchical arrangement of these diverse factors in arterial specification and differentiation. In this study, we demonstrate that arterial expression of the Notch receptor Notch1/notch1b, a key player in arterial specification, is directly downstream of SoxF regulation in both fish and mouse. Unlike other key arterial specification markers, including Dll4, Efnb2a and Dlc (Sacilotto et al., 2013), ablation of Notch1/Notch1b expression after depletion of SoxF factors occurred without concurrent inhibition of Notch signalling. Therefore, this work positions SoxF factors directly above Notch signalling in the transcriptional hierarchy initiating arterial development, and suggests that SoxF factors might initiate a feed-forward loop directing arterial identity. In this model, SoxF factors would first activate Notch signalling via the transcriptional activation of Notch receptors in combination with weak activation of Notch ligands (Sacilotto et al., 2013). This early SoxF-mediated activity would then be boosted by the initiation of Notch signalling, resulting in the sustained activation of other downstream genes, eventually activating the full cohort of genes necessary to acquire and maintain arterial endothelial cell identity.
Understanding the function of SoxF factors through mutational analysis has presented significant challenges. Strain-specific variations in mice after depletion of individual SoxF genes and varying levels of compensation from other SoxF factors have resulted in some contradictory reports (Corada et al., 2013; Lee et al., 2014), as is also the case for zebrafish morphant analysis (Cermenati et al., 2008; Herpers et al., 2008; Pendeville et al., 2008). Nonetheless, the results described here agree with an increasingly convincing body of work suggesting that SoxF factors influence Notch signalling yet are unaffected by Notch ablation. For example, overexpression of Sox17 upregulates components of the Notch pathway (Corada et al., 2013; Lizama et al., 2015), loss of functional SoxF factors results in defects similar to those observed after Notch inhibition in mice and fish (Corada et al., 2013; Sakamoto et al., 2007; Zhou et al., 2015), while Notch ablation results in little alteration to the endothelial expression of SoxF factors in mice and fish (Abdelilah et al., 1996; Corada et al., 2013). Data reported here combine with these reports to strongly support a role for SoxF factors as part of the initial transcriptional machinery that instructs arterial specification events.
However, some questions remain. In particular, it is notable that sox7;sox18 double-mutant fish, although exhibiting severe arteriovenous defects very similar to those seen in Notch-deficient fish (Lawson et al., 2001), do not fully recapitulate the effects of Vegfa depletion on arterial specification (Lawson et al., 2002). While this difference may in part be attributed to weak expression of zebrafish sox17, which is expressed in some arterial endothelial cells (Hermkens et al., 2015), it is also expected that the Vegfa pathway has a wider effect on arterial endothelial cells more generally, away from SoxF-mediated activation of Notch signalling. SoxF factors are also influenced by signalling pathways beyond Vegfa. The diverse nature of Vegfa roles in the vasculature, including the regulation of both sprouting angiogenesis and arteriogenesis, processes that inevitably involve different cohorts of downstream targets, make it necessary that multiple regulatory pathways interact with Vegfa during vascular development. While recent work has shown that Vegfa signalling increases the nuclear translocation of SoxF (Duong et al., 2014), and inhibition of Vegfa results in the loss of vascular sox7 in fish, sox18 is still expressed in the absence of intact Vegfa signalling (Pendeville et al., 2008). Additionally, loss of the Vegf co-receptor Nrp1 has little effect on SoxF expression in mouse retinal vasculature (Zhou et al., 2015), pointing to other upstream influences on SoxF function in endothelial cells. Other upstream effectors of SoxF function are likely to include canonical Wnt signalling and Vegfd, both of which have been shown to influence SoxF nuclear localisation (Corada et al., 2013; Duong et al., 2014; Zhou et al., 2015).
Recent evidence has also implicated a role for a coordinated Vegf-Mapk-Ets pathway in the induction of Notch signalling components and early arterial differentiation (Wythe et al., 2013). Notably, in addition to SoxF motifs, both the Notch1 and Dll4 enhancers share a number of highly conserved consensus motifs for the Ets family of transcription factors. While ETS motifs are common to all vascular enhancer elements, including many that are not preferentially expressed in the arterial vasculature (De Val and Black, 2009), the Ets factor Erg has been specifically implicated in arterial-specific regulation of Dll4 (Wythe et al., 2013). It is therefore likely that Vegfa-mediated activation of Ets factors may contribute to the transcriptional activity of Notch downstream effectors, and thus may influence arterial establishment independently of SoxF factors. However, it is notable that Vegfa-mediated activation of Ets transcription factors alone does not appear to be sufficient for arterial gene expression. Dll4 enhancers lacking SOX and RBPJ motifs but retaining all ETS motifs were unable to drive any transgene expression (Sacilotto et al., 2013), nor were Notch enhancers lacking SOX motifs (Figs 2-4). Similar results were found in other delineated arterial enhancers, including the Ece1 upstream enhancer (Robinson et al., 2014) and the Flk1 intron 10 enhancer, where loss of Rbpj-mediated repression resulted in expansion of enhancer activity into venous cells without alterations to ETS motif binding (Becker et al., 2016). Combined with recent observations demonstrating that Erg also plays a crucial role in venous specification through activation of Aplnr (Lathen et al., 2014), it is therefore likely that the role of Erg, and of other Ets factors, downstream of Vegfa in arterial-restricted gene expression occurs in co-operation with additional essential, arterial-specifying transcription factors. The data presented in this work, combined with the analysis of further arterial enhancers, including those of Dll4 and Ece1 (Robinson et al., 2014; Sacilotto et al., 2013; Wythe et al., 2013), increasingly suggest that SoxF may fulfil this role.
MATERIALS AND METHODS
Cloning
The 10×UAS:Sox18Ragged-mCherry plasmid was generated using the full-length mouse Sox18Ragged cDNA sequence, tagged with 10×UAS and mCherry and cloned into pDestTol2CG2 (Kwan et al., 2007). notch1b-15:GFP WT was generated by cloning a 1219 bp PCR fragment from zebrafish genomic DNA together with gata2a promoter and GFP reporter gene into the zebrafish enhancer detection (ZED) vector (Bessa et al., 2009). notch1b-15mutSOX-a/b was generated by site-directed PCR mutagenesis of the WT construct. The NOTCH1−68, NOTCH1+3/5 and NOTCH1+33 enhancers were generated by PCR from human genomic DNA, NOTCH1+16 WT and NOTCH1+16mutSOX-a/b enhancers were generated as custom-made, double-stranded linear DNA fragments (GeneArt Strings, Life Technologies). All mammalian fragments were cloned into the hsp68-lacZ Gateway vector (provided by N. Ahituv) (De Val et al., 2004). Primers and sequences for DNA fragments are listed in the supplementary Materials and Methods.
Transgenic animals and genome editing
Animal procedures were approved by local ethical review and licensed by the UK Home Office or conformed to institutional guidelines of the University of Queensland Animal Ethics Committee. Transgenic mice were generated by oocyte microinjection and analysed as detailed in the supplementary Materials and Methods (De Val et al., 2004). Compound Sox7−/−;Sox18−/− (C57BL/6) mouse embryos were generated on the C57BL/6 background through crossing heterozygous Sox7:tm1 to Sox18:tm1 generating Sox7+/−;Sox18+/− mice, which were subsequently in-crossed (Pennisi et al., 2000a).
Transgenic zebrafish embryos were generated using the Tol2 system in conjunction with the ZED vector (Bessa et al., 2009). The sox7hu5626; sox18hu10320 double-homozygous mutant zebrafish have been described previously (Hermkens et al., 2015). The tg(fli1a:Gal4FF,10×UAS:Sox18Ragged-mCherry) zebrafish line was generated by crossing 10×UAS:Sox18Ragged-mCherry with fli1a:Gal4FF, 4×UAS Utrophin GFP. MO-mediated knockdown was performed as previously described (Herpers et al., 2008). CRISPR genome editing for notch1b-15 was performed as described by Gagnon et al. (2014) using the primers listed in the supplementary Materials and Methods to generate notch1b-15uq1mf (203 bp deletion) allele.
The F2 notch1b-15uq1mf/uq1mf was generated by in-crossing notch1b-15uq1mf/+, while F3 notch1b-15uq1mf/uq1mf was generated from the F2 notch1b-15uq1mf/uq1mf in-cross, both in the tg(flt1:YFP;lyve1:dsRed) background.
Chromatin immunoprecipitation (ChIP)
Positive tg(fli1a:Gal4FF;10×UAS:Sox18Ragged-mCherry) fish larvae were collected at 26-28 hpf and processed as described in supplementary Materials and Methods (Mohammed et al., 2013). DNA amplification was performed using the TruSeq ChIP-seq Kit (Illumina, IP-202-1012) following immunoprecipitation. The library was quantified using the KAPA library quantification kit for Illumina sequencing platforms (KAPA Biosystems, KK4824) and 50 bp single-end reads were sequenced on a HiSeq 2500 (Illumina) following the manufacturer's protocol. FASTQ files were mapped to GRCz10/danRer10 genome assembly using bowtie (Langmead, 2010), and peaks were called using MACS version 2.1.0. using input as a reference. To avoid false-positive peaks calling due to the mCherry epitope, ChIP-seq with the mCherry epitope only was performed in parallel to SOX18Ragged-mCherry ChIP-seq and peaks called in these experimental conditions were subtracted from the peaks called in the SOX18Ragged-mCherry conditions. For details, see the supplementary Materials and Methods.
Motif identification and EMSA
Sequences were analysed for consensus sequence motifs by eye and using TRANSFAC (BIOBASE; http://genexplain.com/transfac/) (Matys et al., 2006). EMSAs were performed as previously described (De Val et al., 2004), as outlined in the supplementary Materials and Methods.
Morpholinos and drug treatment
MO-mediated knockdown was performed as previously described (Duong et al., 2014; Cermenati et al., 2008). ATG MOs against sox7 and sox18 were injected into the tg(notch1b-15:GFP) stable line at the 1-2 cell stage at 5 ng/embryo (Herpers et al., 2008). To assess the effect of sox7/18 knockdown on endogenous notch1b transcripts, sox7 and sox18 MOs were injected into WT zebrafish larvae at 1 ng/embryo in parallel with a standard control MO (std-MO) injected at 2 ng/embryo (Cermenati et al., 2008). To characterise notch1b-15uq1mf, notch1b MO was injected into the notch1b-15uq1mf/+ cross at 5 ng/embryo. For MO sequences, see the supplementary Materials and Methods.
N-[(3,5-difluorophenyl)acetyl]-L-alanyl-2-phenyl]glycine-1,1-dimethylethyl ester (DAPT; Sigma-Aldrich) was used at 5 μM dissolved in 1% DMSO. Fish were treated from the 15- to 16-somite stage to 3 dpf and the medium containing DAPT was refreshed daily.
In situ hybridisation and immunofluorescence staining
Whole-mount in situ hybridisation in zebrafish larvae was performed as described (Coxam et al., 2015; Duong et al., 2014). Section and whole-mount in situ hybridisation in mouse was performed as described (Metzis et al., 2013; Fowles et al., 2003). The Notch1 probe was generated by PCR from mouse embryo cDNA pool at E14.5, and reverse transcribed with T7 polymerase. Whole-mount immunohistochemistry for anti-GFP was performed as described (Koltowska et al., 2015). For details, see the supplementary Materials and Methods.
Quantification and data analysis
To characterise the vasculature in Fig. 8E-G and Figs S6, S7, intersomitic vessels (20-22 ISVs) expressing tg(flt1:YFP) were analysed across 10-11 somites through a z-stack using ImageJ (NIH) after image acquisition by confocal microscopy. Intersomitic vessels connecting the dorsal longitudinal anastomotic vessel to the dorsal aorta expressing YFP were assigned as arterial ISVs. Vessels were also scored for ectopic sprouting. The proportion of aISVs or hypersprouts among the total number of ISVs was analysed by two-tailed Mann–Whitney U-test. To quantify the GFP intensity of tg(notch1b-15:GFP) and tg(notch1b-15mutSOX-a/b:GFP) (Fig. 7A), two to three ISVs across five to six somites in the trunk region were analysed using ImageJ. A region of interest (ROI) covering a single ISV was selected and mean pixel intensity for each ISV was quantified from each individual stack across three z-sections. This value was further corrected by subtracting the background value. Average ISV GFP intensity (quantified from two to three ISVs) for each fish larva was subsequently corrected for its genomic GFP copy number. A similar method was used to quantify GFP intensity in the endothelial lining along the dorsal aorta.
Fluorescence-activated cell sorting (FACS) and expression analysis
Flt1:YFP-positive endothelial cells were isolated from WT and F3 notch1b-15uq1mf/uq1mf at 24-28 hpf. RNA was extracted, amplified and cDNA was synthesized as previously described (Coxam et al., 2014; Picelli et al., 2014). Primer sequences and details of the quantitative PCR analysis are provided in the supplementary Materials and Methods.
Acknowledgements
We thank M. Shipman for help with imaging. The ZED vector was kindly provided by F. Tessadori (Bakkers lab, Hubrecht Institute). The notch1b plasmid used to generate the probe was kindly provided by N. D. Lawson (University of Massachusetts Medical School).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: I.K.-N.C., M.Frit., S.D.V., M.Fran.; Methodology: I.K.-N.C., M.Frit., S.D.V., M.Fran.; Formal analysis: K.H., J.C.; Investigation: I.K.-N.C., M.Frit., C.P.-T., A.N., K.H., A.L., J.O., D.D., A.O., D.H., E.L., K.L., I.R., M.C., B.H.; Resources: A.L., G.B.-G., J.C., S.S.-M., M.B.; Data curation: K.H., J.C.; Writing - original draft: I.K.-N.C., S.D.V., M.Fran.; Writing - review & editing: I.K.-N.C., B.H., M.B., S.D.V., M.Fran.; Visualization: I.K.-N.C., S.D.V., M.Fran.; Supervision: G.B.-G., J.C., E.D., B.H., M.B., S.D.V., M.Fran.; Project administration: S.D.V., M.Fran.; Funding acquisition: S.D.V., M.B., M.Fran.
Funding
This work was supported by the National Health and Medical Research Council of Australia (NHMRC) (APP1107643); The Cancer Council Queensland (1107631) (M.Fran.); the Australian Research Council Discovery Project (DP140100485) and a Career Development Fellowship (APP1111169) (M.Fran.); the Ludwig Institute for Cancer Research (M.Frit., A.N., I.R., S.D.V.); the Medical Research Council (MR/J007765/1) (K.L., G.B.-G., S.D.V.); the Fondazione Cariplo (2011-0555) (M.B., B.H., M.Fran.); and the Biotechnology and Biological Sciences Research Council (BB/L020238/1) (A.N., K.L., G.B.-G., S.D.V.). Deposited in PMC for release after 6 months.
Data availability
ChIP-seq data are available in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) with accession number E-MTAB-5843.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.146241.supplemental
References
- Abdelilah S., Mountcastle-Shah E., Harvey M., Solnica-Krezel L., Schier A. F., Stemple D. L., Malicki J., Neuhauss S. C., Zwartkruis F., Stainier D. Y. et al. (1996). Mutations affecting neural survival in the zebrafish Danio rerio. Development 123, 217-227. [DOI] [PubMed] [Google Scholar]
- Becker P. W., Sacilotto N., Nornes S., Neal A., Thomas M. O., Liu K., Preece C., Ratnayaka I., Davies B., Bou-Gharios G. et al. (2016). An intronic Flk1 enhancer directs arterial-specific expression via RBPJ-mediated venous repression. Arterioscler. Thromb. Vasc. Biol. 36, 1209-1219. 10.1161/ATVBAHA.116.307517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessa J., Tena J. J., de la Calle-Mustienes E., Fernández-Miñán A., Naranjo S., Fernández A., Montoliu L., Akalin A., Lenhard B., Casares F. et al. (2009). Zebrafish enhancer detection (ZED) vector: a new tool to facilitate transgenesis and the functional analysis of cis-regulatory regions in zebrafish. Dev. Dyn. 238, 2409-2417. 10.1002/dvdy.22051 [DOI] [PubMed] [Google Scholar]
- Bogdanović O., Fernández-Miñán A., Tena J. J., de la Calle-Mustienes E., Hidalgo C., van Kruysbergen I., van Heeringen S. J., Veenstra G. J. C. and Gómez-Skarmeta J. L. (2012). Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res. 22, 2043-2053. 10.1101/gr.134833.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray S. J. (2006). Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678-689. 10.1038/nrm2009 [DOI] [PubMed] [Google Scholar]
- Bussmann J., Bos F. L., Urasaki A., Kawakami K., Duckers H. J. and Schulte-Merker S. (2010). Arteries provide essential guidance cues for lymphatic endothelial cells in the zebrafish trunk. Development 137, 2653-2657. 10.1242/dev.048207 [DOI] [PubMed] [Google Scholar]
- Cannavò E., Khoueiry P., Garfield D. A., Geeleher P., Zichner T., Gustafson E. H., Ciglar L., Korbel J. O. and Furlong E. E. M. (2016). Shadow enhancers are pervasive features of developmental regulatory networks. Curr. Biol. 26, 38-51. 10.1016/j.cub.2015.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P., Ferreira V., Breier G., Pollefeyt S., Kieckens L., Gertsenstein M., Fahrig M., Vandenhoeck A., Harpal K., Eberhardt C. et al. (1996). Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature 380, 435-439. 10.1038/380435a0 [DOI] [PubMed] [Google Scholar]
- Cermenati S., Moleri S., Cimbro S., Corti P., Del Giacco L., Amodeo R., Dejana E., Koopman P., Cotelli F. and Beltrame M. (2008). Sox18 and Sox7 play redundant roles in vascular development. Blood 111, 2657-2666. 10.1182/blood-2007-07-100412 [DOI] [PubMed] [Google Scholar]
- Chong D. C., Koo Y., Xu K., Fu S. and Cleaver O. (2011). Stepwise arteriovenous fate acquisition during mammalian vasculogenesis. Dev. Dyn. 240, 2153-2165. 10.1002/dvdy.22706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corada M., Orsenigo F., Morini M. F., Pitulescu M. E., Bhat G., Nyqvist D., Breviario F., Conti V., Briot A., Iruela-Arispe M. L. et al. (2013). Sox17 is indispensable for acquisition and maintenance of arterial identity. Nat. Commun. 4, 2609 10.1038/ncomms3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coxam B., Sabine A., Bower N. I., Smith K. A., Pichol-Thievend C., Skoczylas R., Astin J. W., Frampton E., Jaquet M., Crosier P. S. et al. (2014). Pkd1 regulates lymphatic vascular morphogenesis during development. Cell Rep. 7, 623-633. 10.1016/j.celrep.2014.03.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coxam B., Neyt C., Grassini D. R., Le Guen L., Smith K. A., Schulte-Merker S. and Hogan B. M. (2015). carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase (cad) regulates Notch signaling and vascular development in zebrafish. Dev. Dyn. 244, 1-9. 10.1002/dvdy.24209 [DOI] [PubMed] [Google Scholar]
- De Val S. and Black B. L. (2009). Transcriptional control of endothelial cell development. Dev. Cell 16, 180-195. 10.1016/j.devcel.2009.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Val S., Anderson J. P., Heidt A. B., Khiem D., Xu S.-M. and Black B. L. (2004). Mef2c is activated directly by Ets transcription factors through an evolutionarily conserved endothelial cell-specific enhancer. Dev. Biol. 275, 424-434. 10.1016/j.ydbio.2004.08.016 [DOI] [PubMed] [Google Scholar]
- Duarte A., Hirashima M., Benedito R., Trindade A., Diniz P., Bekman E., Costa L., Henrique D. and Rossant J. (2004). Dosage-sensitive requirement for mouse Dll4 in artery development. Genes Dev. 18, 2474-2478. 10.1101/gad.1239004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong T., Koltowska K., Pichol-Thievend C., Le Guen L., Fontaine F., Smith K. A., Truong V., Skoczylas R., Stacker S. A., Achen M. G. et al. (2014). VEGFD regulates blood vascular development by modulating SOX18 activity. Blood 123, 1102-1112. 10.1182/blood-2013-04-495432 [DOI] [PubMed] [Google Scholar]
- Fowles L. F., Bennetts J. S., Berkman J. L., Williams E., Koopman P., Teasdale R. D. and Wicking C. (2003). Genomic screen for genes involved in mammalian craniofacial development. Genesis 35, 73-87. 10.1002/gene.10165 [DOI] [PubMed] [Google Scholar]
- François M., Caprini A., Hosking B., Orsenigo F., Wilhelm D., Browne C., Paavonen K., Karnezis T., Shayan R., Downes M. et al. (2008). Sox18 induces development of the lymphatic vasculature in mice. Nature 456, 643-647. 10.1038/nature07391 [DOI] [PubMed] [Google Scholar]
- Francois M., Koopman P. and Beltrame M. (2010). SoxF genes: Key players in the development of the cardio-vascular system. Int. J. Biochem. Cell Biol. 42, 445-448. 10.1016/j.biocel.2009.08.017 [DOI] [PubMed] [Google Scholar]
- Gagnon J. A., Valen E., Thyme S. B., Huang P., Akhmetova L. Pauli A., Montague T. G., Zimmerman S., Richter C. and Schier A. F. (2014). Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS ONE 9, e98186 10.1371/journal.pone.0098186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale N. W., Dominguez M. G., Noguera I., Pan L., Hughes V., Valenzuela D. M., Murphy A. J., Adams N. C., Lin H. C., Holash J. et al. (2004). Haploinsufficiency of delta-like 4 ligand results in embryonic lethality due to major defects in arterial and vascular development. Proc. Natl. Acad. Sci. USA 101, 15949-15954. 10.1073/pnas.0407290101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geudens I., Herpers R., Hermans K., Segura I., Ruiz de Almodovar C., Bussmann J., De Smet F., Vandevelde W., Hogan B. M., Siekmann A. et al. (2010). Role of delta-like-4/Notch in the formation and wiring of the lymphatic network in zebrafish. Arterioscler. Thromb. Vasc. Biol. 30, 1695-1702. 10.1161/ATVBAHA.110.203034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore A. V., Monzo K., Cha Y. R., Pan W. and Weinstein B. M. (2012). Vascular development in the zebrafish. Cold Spring Harb. Perspect. Med. 2, a006684-a006684 10.1101/cshperspect.a006684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey S. A., Sealy I., Kettleborough R., Fényes F., White R., Stemple D. and Smith J. C. (2013). Identification of the zebrafish maternal and paternal transcriptomes. Development 140, 2703-2710. 10.1242/dev.095091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N. D. and Ren B. (2009). Finding distal regulatory elements in the human genome. Curr. Opin. Genet. Dev. 19, 541-549. 10.1016/j.gde.2009.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermkens D. M. A., van Impel A., Urasaki A., Bussmann J., Duckers H. J. and Schulte-Merker S. (2015). Sox7 controls arterial specification in conjunction with hey2 and efnb2 function. Development 142, 1695-1704. 10.1242/dev.117275 [DOI] [PubMed] [Google Scholar]
- Herpers R., van de Kamp E., Duckers H. J. and Schulte-Merker S. (2008). Redundant roles for Sox7 and Sox18 in arteriovenous specification in zebrafish. Circ. Res. 102, 12-15. 10.1161/CIRCRESAHA.107.166066 [DOI] [PubMed] [Google Scholar]
- Hollenhorst P. C., McIntosh L. P. and Graves B. J. (2011). Genomic and biochemical insights into the specificity of ETS transcription factors. Annu. Rev. Biochem. 80, 437-471. 10.1146/annurev.biochem.79.081507.103945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosking B., François M., Wilhelm D., Orsenigo F., Caprini A., Svingen T., Tutt D., Davidson T., Browne C., Dejana E. et al. (2009). Sox7 and Sox17 are strain-specific modifiers of the lymphangiogenic defects caused by Sox18 dysfunction in mice. Development 136, 2385-2391. 10.1242/dev.034827 [DOI] [PubMed] [Google Scholar]
- Jahnsen E. D., Trindade A., Zaun H. C., Lehoux S., Duarte A. and Jones E. A. V. (2015). Notch1 is pan-endothelial at the onset of flow and regulated by flow. PLoS ONE 10 10.1371/journal.pone.0122622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James K., Hosking B., Gardner J., Muscat G. E. and Koopman P. (2003). Sox18 mutations in the ragged mouse alleles ragged-like and opossum. Genesis 36, 1-6. 10.1002/gene.10190 [DOI] [PubMed] [Google Scholar]
- Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M. and Haussler D. (2002). The human genome browser at UCSC. Genome Res. 12, 996-1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Kim I.-K., Yang J. M., Lee E., Koh B. I., Song S., Park J., Lee S., Choi C., Kim J. W. et al. (2016). SoxF transcription factors are positive feedback regulators of VEGF signaling. Circ. Res. 119, 839-852. 10.1161/CIRCRESAHA.116.308483 [DOI] [PubMed] [Google Scholar]
- Koltowska K., Paterson S., Bower N. I., Baillie G. J., Lagendijk A. K., Astin J. W., Chen H., François M., Crosier P. S., Taft R. J. et al. (2015). mafba is a downstream transcriptional effector of Vegfc signaling essential for embryonic lymphangiogenesis in zebrafish. Genes Dev. 29, 1618-1630. 10.1101/gad.263210.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs L. T., Xue Y., Norton C. R., Shutter J. R., Maguire M., Sundberg J. P., Gallahan D., Closson V., Kitajewski J., Callahan R. et al. (2000). Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 14, 1343-1352. [PMC free article] [PubMed] [Google Scholar]
- Krebs L. T., Shutter J. R., Tanigaki K., Honjo T., Stark K. L. and Gridley T. (2004). Haploinsufficient lethality and formation of arteriovenous malformations in Notch pathway mutants. Genes Dev. 18, 2469-2473. 10.1101/gad.1239204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan K. M., Fujimoto E., Grabher C., Mangum B. D., Hardy M. E., Campbell D. S., Parant J. M., Yost H. J., Kanki J. P. and Chien C. B. (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088-3099. 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- Lanahan A. A., Hermans K., Claes F., Kerley-Hamilton J. S., Zhuang Z. W., Giordano F. J., Carmeliet P. and Simons M. (2010). VEGF receptor 2 endocytic trafficking regulates arterial morphogenesis. Dev. Cell 18, 713-724. 10.1016/j.devcel.2010.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. (2010). Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinformatics 11, 11.7 10.1002/0471250953.bi1107s32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathen C., Zhang Y., Chow J., Singh M., Lin G., Nigam V., Ashraf Y. A., Yuan J. X., Robbins I. M. and Thistlethwaite P. A. (2014). ERG-APLNR axis controls pulmonary venule endothelial proliferation in pulmonary Veno-occlusive disease. Circulation 130, 1179-1191. 10.1161/CIRCULATIONAHA.113.007822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson N. D. (2003). phospholipase C gamma-1 is required downstream of vascular endothelial growth factor during arterial development. Genes Dev. 17, 1346-1351. 10.1101/gad.1072203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson N. D., Scheer N., Pham V. N., Kim C. H., Chitnis A. B., Campos-Ortega J. A. and Weinstein B. M. (2001). Notch signaling is required for arterial-venous differentiation during embryonic vascular development. Development 128, 3675-3683. [DOI] [PubMed] [Google Scholar]
- Lawson N. D., Vogel A. M. and Weinstein B. M. (2002). sonic hedgehog and vascular endothelial growth factor act upstream of the Notch pathway during arterial endothelial differentiation. Dev. Cell 3, 127-136. 10.1016/S1534-5807(02)00198-3 [DOI] [PubMed] [Google Scholar]
- Lee S.-H., Lee S., Yang H., Song S., Kim K., Saunders T. L., Yoon J. K., Koh G. Y. and Kim I. (2014). Notch pathway targets proangiogenic regulator Sox17 to restrict angiogenesis. Circ. Res. 115, 215-226. 10.1161/CIRCRESAHA.115.303142 [DOI] [PubMed] [Google Scholar]
- Liu Z.-J., Shirakawa T., Li Y., Soma A., Oka M., Dotto G. P., Fairman R. M., Velazquez O. C. and Herlyn M. (2003). Regulation of Notch1 and Dll4 by vascular endothelial growth factor in arterial endothelial cells: implications for modulating arteriogenesis and angiogenesis. Mol. Cell. Biol. 23, 14-25. 10.1128/MCB.23.1.14-25.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizama C. O., Hawkins J. S., Schmitt C. E., Bos F. L., Zape J. P., Cautivo K. M., Borges Pinto H., Rhyner A. M., Yu H., Donohoe M. E. et al. (2015). Repression of arterial genes in hemogenic endothelium is sufficient for haematopoietic fate acquisition. Nat. Commun. 6, 7739 10.1038/ncomms8739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V., Kel-Margoulis O. V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K. et al. (2006). TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34, D108-D110. 10.1093/nar/gkj143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertin S., McDowall S. G. and Harley V. R. (1999). The DNA-binding specificity of SOX9 and other SOX proteins. Nucleic Acids Res. 27, 1359-1364. 10.1093/nar/27.5.1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzis V., Courtney A. D., Kerr M. C., Ferguson C., Rondon Galeano M. C., Parton R. G., Wainwright B. J. and Wicking C. (2013). Patched1 is required in neural crest cells for the prevention of orofacial clefts. Hum. Mol. Genet. 22, 5026-5035. 10.1093/hmg/ddt353 [DOI] [PubMed] [Google Scholar]
- Mohammed H., D'Santos C., Serandour A. A., Ali H. R., Brown G. D., Atkins A., Rueda O. M., Holmes K. A., Theodorou V., Robinson J. L. L. et al. (2013). Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 3, 342-349. 10.1016/j.celrep.2013.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendeville H., Winandy M., Manfroid I., Nivelles O., Motte P., Pasque V., Peers B., Struman I., Martial J. A. and Voz M. L. (2008). Zebrafish Sox7 and Sox18 function together to control arterial-venous identity. Dev. Biol. 317, 405-416. 10.1016/j.ydbio.2008.01.028 [DOI] [PubMed] [Google Scholar]
- Pennisi D., Bowles J., Nagy A., Muscat G. and Koopman P. (2000a). Mice null for Sox18 are viable and display a mild coat defect. Mol. Cell. Biol. 20, 9331-9336. 10.1128/MCB.20.24.9331-9336.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi D., Gardner J., Chambers D., Hosking B., Peters J., Muscat G., Abbott C. and Koopman P. (2000b). Mutations in Sox18 underlie cardiovascular and hair follicle defects in ragged mice. Nat. Genet. 24, 434-437. 10.1038/74301 [DOI] [PubMed] [Google Scholar]
- Phng L.-K. and Gerhardt H. (2009). Angiogenesis: a team effort coordinated by notch. Dev. Cell 16, 196-208. 10.1016/j.devcel.2009.01.015 [DOI] [PubMed] [Google Scholar]
- Picelli S., Faridani O. R., Björklund A. K., Winberg G., Sagasser S. and Sandberg R. (2014). Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171-181. 10.1038/nprot.2014.006 [DOI] [PubMed] [Google Scholar]
- Quillien A., Moore J. C., Shin M., Siekmann A. F., Smith T., Pan L., Moens C. B., Parsons M. J. and Lawson N. D. (2014). Distinct Notch signaling outputs pattern the developing arterial system. Development 141, 1544-1552. 10.1242/dev.099986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A. S., Materna S. C., Barnes R. M., De Val S., Xu S.-M. and Black B. L. (2014). An arterial-specific enhancer of the human endothelin converting enzyme 1 (ECE1) gene is synergistically activated by Sox17, FoxC2, and Etv2. Dev. Biol. 395, 379-389. 10.1016/j.ydbio.2014.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca C. and Adams R. H. (2007). Regulation of vascular morphogenesis by Notch signaling. Genes Dev. 21, 2511-2524. 10.1101/gad.1589207 [DOI] [PubMed] [Google Scholar]
- Sabo P. J., Hawrylycz M., Wallace J. C., Humbert R., Yu M., Shafer A., Kawamoto J., Hall R., Mack J., Dorschner M. O. et al. (2004). Discovery of functional noncoding elements by digital analysis of chromatin structure. Proc. Natl. Acad. Sci. USA 101, 16837-16842. 10.1073/pnas.0407387101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacilotto N., Monteiro R., Fritzsche M., Becker P. W., Sanchez-del-Campo L., Liu K., Pinheiro P., Ratnayaka I., Davies B., Goding C. R. et al. (2013). Analysis of Dll4 regulation reveals a combinatorial role for Sox and Notch in arterial development. Proc. Natl. Acad. Sci. USA 110, 11893-11898. 10.1073/pnas.1300805110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto Y., Hara K., Kanai-Azuma M., Matsui T., Miura Y., Tsunekawa N., Kurohmaru M., Saijoh Y., Koopman P. and Kanai Y. (2007). Redundant roles of Sox17 and Sox18 in early cardiovascular development of mouse embryos. Biochem. Biophys. Res. Commun. 360, 539-544. 10.1016/j.bbrc.2007.06.093 [DOI] [PubMed] [Google Scholar]
- Siekmann A. F. and Lawson N. D. (2007). Notch signalling limits angiogenic cell behaviour in developing zebrafish arteries. Nature 445, 781-784. 10.1038/nature05577 [DOI] [PubMed] [Google Scholar]
- Visconti R. P., Richardson C. D. and Sato T. N. (2002). Orchestration of angiogenesis and arteriovenous contribution by angiopoietins and vascular endothelial growth factor (VEGF). Proc. Natl. Acad. Sci. USA 99, 8219-8224. 10.1073/pnas.122109599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E. S., Thybert D., Schmitt B. M., Stefflova K., Odom D. T. and Flicek P. (2015). Decoupling of evolutionary changes in transcription factor binding and gene expression in mammals. Genome Res. 25, 167-178. 10.1101/gr.177840.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Iwata F., Grass J. A., Osborne C. S., Elnitski L., Fraser P., Ohneda O., Yamamoto M. and Bresnick E. H. (2005). Molecular determinants of NOTCH4 transcription in vascular endothelium. Mol. Cell. Biol. 25, 1458-1474. 10.1128/MCB.25.4.1458-1474.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wythe J. D., Dang L. T. H., Devine W. P., Boudreau E., Artap S. T., He D., Schachterle W., Stainier D. Y. R., Oettgen P., Black B. L. et al. (2013). ETS factors regulate Vegf-dependent arterial specification. Dev. Cell 26, 45-58. 10.1016/j.devcel.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Williams J., Smallwood P. M. and Nathans J. (2015). Sox7, Sox17, and Sox18 cooperatively regulate vascular development in the mouse retina. PLoS ONE 10, e0143650 10.1371/journal.pone.0143650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Gu F., Zhang L., Akerberg B. N., Ma Q., Li K., He A., Lin Z., Stevens S. M., Zhou B. et al. (2017). Mapping cell type-specific transcriptional enhancers using high affinity, lineage-specific Ep300 bioChIP-seq. eLife 6, e22039 10.7554/eLife.22039 [DOI] [PMC free article] [PubMed] [Google Scholar]