Abstract
Boundaries in the Bithorax complex (BX-C) of Drosophila delimit autonomous regulatory domains that drive parasegment-specific expression of homeotic genes. BX-C boundaries have two crucial functions: they must block crosstalk between adjacent regulatory domains and at the same time facilitate boundary bypass. The C2H2 zinc-finger protein Pita binds to several BX-C boundaries, including Fab-7 and Mcp. To study Pita functions, we have used a boundary replacement strategy by substituting modified DNAs for the Fab-7 boundary, which is located between the iab-6 and iab-7 regulatory domains. Multimerized Pita sites block iab-6↔iab-7 crosstalk but fail to support iab-6 regulation of Abd-B (bypass). In the case of Fab-7, we used a novel sensitized background to show that the two Pita-binding sites contribute to its boundary function. Although Mcp is from BX-C, it does not function appropriately when substituted for Fab-7: it blocks crosstalk but does not support bypass. Mutation of the Mcp Pita site disrupts blocking activity and also eliminates dCTCF binding. In contrast, mutation of the Mcp dCTCF site does not affect Pita binding, and this mutant boundary retains partial function.
KEY WORDS: Insulator, Chromatin organization, Zinc-finger transcription factors, Protein-protein interactions, Architectural proteins
Summary: The chromatin architectural protein Pita functions much like dCTCF in the organization of Drosophila BX-C, providing insight into how boundaries impact BX-C gene regulation and segment specification.
INTRODUCTION
Studies over the past few decades have substantially enhanced our understanding of chromosomal architecture in multicellular eukaryotes and have revealed an intimate connection between structure and function. High-resolution chromosome conformation capture techniques have shown that human, mouse and Drosophila chromosomes are partitioned into a series of loops called topologically associating domains (TADs) (Dixon et al., 2012; Sexton et al., 2012; Rao et al., 2014; Dekker and Mirny, 2016). Loop formation is driven largely by a special class of cis-acting elements called boundaries or insulators that pair with each other. These elements have binding sites for architectural proteins that mediate the boundary-boundary pairing interactions responsible for loop formation (Liang et al., 2014; Maksimenko and Georgiev, 2014; Bouwman and de Laat, 2015; Dekker and Mirny, 2016). In transgenic assays, boundaries can block regulatory interactions between enhancers/silencers and target genes; however, when appropriately arranged, boundaries can also bring distant regulatory elements in contact with promoters (Gohl et al., 2011; Chetverina et al., 2014; Kyrchanova and Georgiev, 2014; Matzat and Lei, 2014; Fujioka et al., 2016).
Although only one architectural DNA-binding protein, CTCF, has been as yet identified in mammals (Ali et al., 2016; Ghirlando and Felsenfeld, 2016; Merkenschlager and Nora, 2016), ongoing studies suggest that Drosophila probably has many architectural proteins (Maksimenko et al., 2015; Zolotarev et al., 2016a). This discrepancy likely reflects the relatively simple methods and assays that are available for studying boundaries/insulators in flies that are not readily feasible in higher eukaryotes. However, in spite of the experimental advantages offered by the fly model system, there is only a limited amount of information on the functional roles of architectural proteins in their native environment.
One context where it is readily possible to study the functioning of the endogenous fly boundaries is the Drosophila Bithorax complex. As is the case for the mammalian Hox gene complexes, many of the boundaries in the Drosophila Bithorax complex (BX-C) use a homolog of the mammalian CTCF protein, dCTCF (Moon et al., 2005; Holohan et al., 2007; Mohan et al., 2007; Perez-Lluch et al., 2008; Bonchuk et al., 2015; Magbanua et al., 2015). BX-C contains three homeotic genes, Ultrabithorax (Ubx), abdominal-A (abd-A) and Abdominal-B (Abd-B), which are responsible for specifying the parasegments (PS5 to PS13) that make up the posterior two-thirds of the fly segments (A) (Lewis, 1978; Kyrchanova et al., 2015; Maeda and Karch, 2015). Expression of each homeotic gene in the appropriate parasegment-specific pattern is controlled by a series of nine cis-regulatory domains, abx/bx, bxd/pbx and iab-2 to iab-9. In order to properly specify segment identity, these regulatory domains must be able to function autonomously. Genetic and molecular studies have shown that each of these regulatory domains is bracketed by boundary elements (Fab-3, Fab-4, Mcp, Fab-6, Fab-7, Fab-8 and AB-I; Fig. 1A) (Celniker et al., 1990; Gyurkovics et al., 1990; Galloni et al., 1993; Karch et al., 1994; Kyrchanova et al., 2008, 2011, 2016; McCall et al., 1994; Mihaly et al., 1997, 2006; Barges et al., 2000; Iampietro et al., 2010; Bender and Lucas, 2013; Bowman et al., 2014). BX-C boundaries have two important functions (Kyrchanova et al., 2015): one is to block crosstalk between adjacent regulatory domains; the other is bypass activity, i.e. enabling distal regulatory domains to bypass one or more intervening boundaries and contact their regulatory targets.
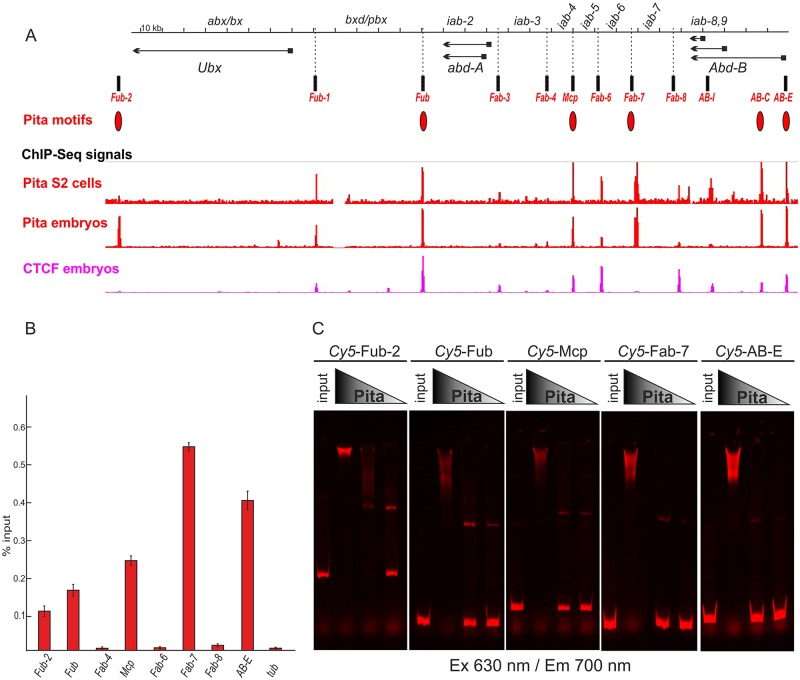
Fig. 1.
Boundaries of the BX-C coincide with dCTCF and Pita. (A) Distribution of ChIP signals for Pita and dCTCF across the BX-C. The BX-C is presented as a sequence coordinate line. Transcripts of Ubx, abd-A and Abd-B are marked by horizontal arrows. The mapped insulator elements are indicated under the coordinate map as vertical black bars. Red ovals denote sequence motifs for Pita found in the BX-C. Tracks show the binding profiles for Pita and dCTCF according to the ChIP-Seq data. (B) Binding of Pita at the boundaries of BX-C. Chromatin was isolated from pupae and treated with antibodies against Pita. Nonspecific IgG was used as a negative control (not shown, see Fig. S1 for comparison). The results of ChIPs are presented as a percentage of input DNA. The γTub37C (tub)-coding region (devoid of binding sites for Pita) was used as a negative control. Error bars indicate standard deviations of the triplicate PCR measurements from two independent biological samples of chromatin. (C) EMSA of recombinant zinc-finger domains of Pita with the DNA fragments from BX-C containing Pita-binding sites (Fub-2, Fub, Mcp, Fab-7 and AB-E). Zinc-finger domains of Pita fused with MBP were incubated with the Cy5-labeled DNA fragments. Specificity of the interaction was demonstrated by incubation of the DNA fragments with varying amounts of Pita protein, presented as a series of twofold dilutions. In parallel, the FAM-labeled DNA fragments with no binding sites for Pita were used as a negative control (data not shown).
The most thoroughly characterized BX-C boundary, Fab-7, is located between iab-6 and iab-7 (Fig. 1A). Fab-7 deletions fuse the iab-6 and iab-7 regulatory domains, and mutant flies exhibit a complex mixture of gain- and loss-of-function phenotypes in PS11 (Gyurkovics et al., 1990; Karch et al., 1994; Mihaly et al., 1997). The gain-of-function phenotypes arise because the iab-6 initiator inappropriately activates the iab-7 domain in PS11 (A6), whereas the loss-of-function phenotypes arise because repressive elements in iab-7 that are active in PS11 silence iab-6 in that parasegment. Deletions that remove not only the Fab-7 boundary, but also the iab-7 Polycomb response element (PREiab-7) exhibit an exclusively gain-of-function transformation, and A6 is completely missing in mutant males. A similar fusion of neighboring regulatory domains and a consequent misregulation of Abd-B is observed when Fab-6 and Fab-8 are deleted (Barges et al., 2000; Iampietro et al., 2008, 2010).
Unlike other BX-C boundaries, Fab-7 does not contain binding sites for dCTCF, and instead relies on other factors to confer boundary activity. Thus far, two developmentally regulated complexes have been shown to be important for Fab-7 boundary function: Elba and LBC (Aoki et al., 2012; Wolle et al., 2015). The Elba complex confers boundary function during early embryogenesis, whereas LBC functions later in development. Although several different lines of evidence indicate that both of these factors must contribute to Fab-7 activity, the boundary spans a DNA segment of over 1 kb and it is clear that other, partially redundant, factors are also important for boundary function (Wolle et al., 2015).
The studies reported here focus on a previously reported zinc-finger DNA-binding protein called Pita (Laundrie et al., 2003) or Spotted Dick (Page et al., 2005) that has two binding sites in Fab-7. pita is an essential gene and null alleles die during larval stages. The mutant larvae exhibit a range of phenotypes, including an accumulation of abnormal mitotic figures in larval brains, severely reduced endoreplication in salivary glands, melanotic tumors and small imaginal discs. Experiments by Page et al. (2005) indicate that the mitotic and endoreplication defects arise because pita is required for DNA replication; however, this replication requirement is indirect, as Pita is present throughout the cell cycle and does not localize to replication foci. Instead, RNAi knockdown experiments in tissue culture cells suggest that Pita functions as a transcription activator, and argue that its role in replication is to ensure the expression of a specific origin of replication protein Orc4.
More recent work points to another very different function for pita, namely chromosomal architecture (Maksimenko et al., 2015). Chromatin immunoprecipitation (ChIP) experiments show that Pita binds to several boundaries in BX-C including Fab-7 and Mcp. The Pita protein contains two recognizable domains: the N-terminal ZAD domain and a central cluster consisting of 10 C2H2 zinc-finger domains. Only part of the zinc-finger domains is responsible for DNA binding (Maksimenko et al., 2015). The N-terminal ZAD domain, together with an unstructured region located between the ZAD and zinc-finger domains, are thought to be involved in organizing long-distance interactions by the Pita protein (Zolotarev et al., 2016a,b). The ZAD domain is responsible for homodimer formation, whereas the unstructured region mediates interactions with the BTB domain of CP190. CP190 is a chromatin architectural protein that interacts with many DNA-binding proteins and is believed to promote loop formation by linking proteins bound to distant sequences (Vogelmann et al., 2014).
To better understand the role of Pita as a boundary factor and to elucidate how this activity might contribute to the regulation of BX-C homeotic gene expression, we have taken advantage of the Fab-7attP50 deletion (Wolle et al., 2015). The attP site in this allele can be used to introduce sequences of interest into Fab-7. Using this boundary replacement approach, we have explored the functioning of the Pita protein on its own and in the context of two previously characterized boundaries, Fab-7 and Mcp. These experiments provide new insights into how boundaries impact BX-C gene regulation and segment specification.
RESULTS
Pita sites in the Bithorax complex
Previous genome-wide ChIP experiments with samples from embryos and S2 tissue culture cells identified several Pita-bound regions in BX-C with a high confidence (Maksimenko et al., 2015; Zolotarev et al., 2016a). These Pita-bound regions coincide with known functional elements in BX-C, including the Fub-2, Fub, Mcp and Fab-7 boundaries, and the Abd-B [AB-E] and [AB-C] promoter regions (Fig. 1A and Fig. S1). Other known or suspected boundaries, Fub-1, Fab-3, Fab-4, Fab-6 and AB-I also scored positive for Pita, but with a lower confidence. As shown in Fig. 1B, we found that ChIPs of chromatin, isolated from pupae, confirmed Pita binding to the high-confidence sites, but not to the other, lower confidence sites.
To further validate the identification of the Pita-binding sites in Fub-2, Fub, Mcp, Fab-7 and AB-E, we tested for DNA binding in vitro using an electrophoretic mobility shift assay (EMSA) with fluorescently labeled DNA probes spanning the predicted Pita consensus-binding sites in each of these elements. For the Pita protein, we used an MBP fusion to the zinc-finger domain. As shown in Fig. 1C, the cluster of Pita zinc-finger domains fused with MBP binds to all five of these probes. To provide further evidence that Pita binds to the predicted consensus sequences and set the stage for functional studies, we mutated the Pita sites in the Fab-7 and Mcp probes. As shown in Fig. S2, the fusion of the cluster of Pita zinc-finger domains with MBP shifts the wild-type probes for both Fab-7 and Mcp, but not the probes in which the predicted Pita-binding sites are mutated.
Multimerized Pita-binding sites have blocking but not bypass activity
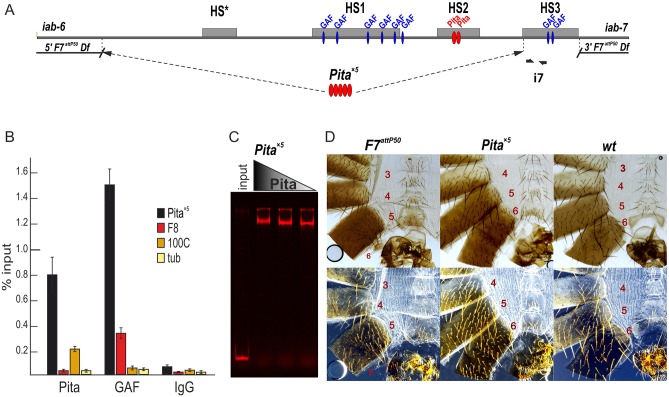
Next, we determined whether Pita has boundary activity on its own by replacing the Fab-7 boundary with multimerized Pita-binding sites (Pita×5). We used five Pita-binding sites because it was shown previously for Su(Hw), the best-studied insulator protein, that no fewer than four reiterated binding sites can display an enhancer-blocking activity in the transgenic assays (Scott et al., 1999). For replacement, we used the previously described Fab-7attP50 landing platform (Wolle et al., 2015). In this platform, the region spanning the Fab-7 nuclease-hypersensitive sites, HS*, HS1 and HS2, and the hypersensitive site associated with the PREiab-7, HS3, are deleted and replaced with an attP target site for the phiC31 recombination. The attP target site was used to insert a DNA fragment in which the Pita×5 multimer was fused to the PREiab-7 sequence (Fig. 2A). EMSA experiments show that Pita binds to the Pita×5 in vitro (Fig. 2C). ChIP experiments confirm that the multimerized Pita-binding sites recruit Pita to the Fab-7 region in vivo. Fig. 2B shows that Pita antibodies immunoprecipitate sequences from the very proximal end of the neighboring HS3 hypersensitive site (which corresponds to the PREiab-7). HS3 contains two GAGA factor (GAF) sites, and in the control experiments, GAF antibodies were found to immunoprecipitate the same sequence from the proximal end of HS3 (Schweinsberg et al., 2004).
Fig. 2.
The phenotypic effects of Fab-7 replacement by multimerized Pita-binding sites. (A) A schematic presentation of the Fab-7 boundary. Hypersensitive sites are shown as gray boxes. The proximal and distal deficiency endpoints of the F7attP50 deletions are shown. GAF- and Pita-binding sites are designated as blue and red ovals, respectively. In this experiment, HS*, HS1 and HS2 are replaced by five Pita sites, whereas HS3 (PREiab-7) is retained. Bold black half-arrows (marked i7) indicate primers that were used for ChIP experiments. (B) Binding of Pita and GAF at the Pita×5 fragment in Pita×5 pupae. Nonspecific IgG was used as a negative control. The results of ChIP are presented as a percentage of input DNA. The Fab-8 (F8) region was used as a positive control for GAF binding, the 100C region was used as a positive control for Pita binding. The γTub37C-coding region (tub), devoid of binding sites for Pita and GAF, was used as a negative control. Error bars indicate standard deviations of triplicate PCR measurements from two independent biological samples of chromatin. (C) EMSA of the recombinant zinc-finger domains of Pita with a Pita×5 DNA fragment. Zinc-finger domains of Pita fused with MBP were incubated with a Cy5-labeled DNA fragment. In parallel, the FAM-labeled DNA fragment with no binding site for Pita was used as negative control (data not shown). (D) Morphology of the 4th to 6th male abdominal segments (numbered) is determined by the Abd-B cis-regulatory regions. F7attP50 males have the classic gain-of-function transformation of A6 (PS11) into A7 (PS12) seen in mutations that remove both the Fab-7 boundary and the PREiab-7, HS3. Pita×5 demonstrates a strong loss-of-function phenotype: A6 sternite in males is quadrilateral and covered in bristles, just like the sternite in A5; whole A6 tergite is covered with trichomes, similar to the A5 tergite. Wild-type (wt) males have pigmented A5 and A6 tergites. The A6 sternite is recognizable by the absence of bristles and a specific form; A7 does not contribute to any visible cuticle structures. Trichomes are visible in the dark-field images (bottom row) and cover all the surface of A5 tergite, and only a thin stripe along the anterior and ventral edges of the A6 tergite.
We next examined the function of Pita×5 in BX-C. In wild type, cells in A6 (PS11) and A7 (PS12) abdominal segments have different fates in adult males. The former establish distinct cuticular structures (tergites and sternites) and internal tissues of the A6 abdominal segment, whereas the latter are lost during morphogenesis. In Fab-7attP50 mutant males, iab-7 is ectopically activated in all A6 (PS11) cells and they assume an A7 (PS12) identity. Consequently, these males lack both the A6 and A7 segments (Fig. 2D). Phenotypic analysis of the cuticle of adult male Pita×5 flies indicates that the functioning of the multimerized Pita sites in the context of Fab-7 resembles that seen for the multimerized su(Hw)- or dCTCF-binding sites (Kyrchanova et al., 2016). Just like these other multimers, Pita×5 blocks crosstalk between iab-6 and iab-7, but does not permit regulatory interactions between the distal iab-6 regulatory domain and Abd-B. As a result, the cuticle in A6 resembles A5. In wild-type males, the A6 sternite has a distinct elongated banana shape and is devoid of bristles, whereas in A5 the sternite has a more compact quadrilateral shape and is covered in bristles. As can be seen in Fig. 2D, the A6 sternite in Pita×5 males is compact and covered in bristles, just like the sternite in A5. A similar loss-of-function transformation is evident in the tergite. Whereas only the anterior and ventral edges of the A6 tergite have trichomes in wild-type flies, the A6 tergite in Pita×5 males is completely covered in trichomes, just like A5.
Pita contributes to Fab-7 boundary function
To further explore the functional properties of the Pita protein, we asked whether it contributes to the Fab-7 boundary activity. The Fab-7 boundary spans a DNA sequence of ∼1.2 kb and contains three nuclease-hypersensitive sites: HS*, HS1 and HS2 (Fig. 3A). The Fab-7 Pita-binding sites are located in the hypersensitive site HS2. Next to HS2 is HS3, which corresponds to the major PRE in the iab-7 regulatory domain. Mutagenesis experiments, both in transgenes and in the context of BX-C, point to a considerable degree of redundancy in the cis-acting elements (Schweinsberg et al., 2004; Schweinsberg and Schedl, 2004; Wolle et al., 2015; Kyrchanova et al., 2016). Because of this redundancy, mutations in the binding sites for the factors that contribute to boundary activity can have little or no phenotypic effects. In this respect, Fab-7 differs from the neighboring boundary, Fab-8, where mutations in the two dCTCF sites abrogate boundary activity (Moon et al., 2005; Kyrchanova et al., 2016). Because of this redundancy, we used two different contexts to test for Pita function. In the first, we introduced Pita mutations into a Fab-7 replacement that includes HS1, HS2 and HS3. Although this replacement lacks HS*, it appears to be fully functional in an otherwise wild-type background. In the second, we introduced the Pita mutations into a Fab-7 replacement that includes HS1 and HS2, but not HS3. As described further below, the removal of HS3 perturbs boundary function, giving a ‘sensitized’ background. As anticipated, redundancy compensates for the Pita mutations and we observed obvious phenotypic effects in only the sensitized HS1+HS2 replacement.
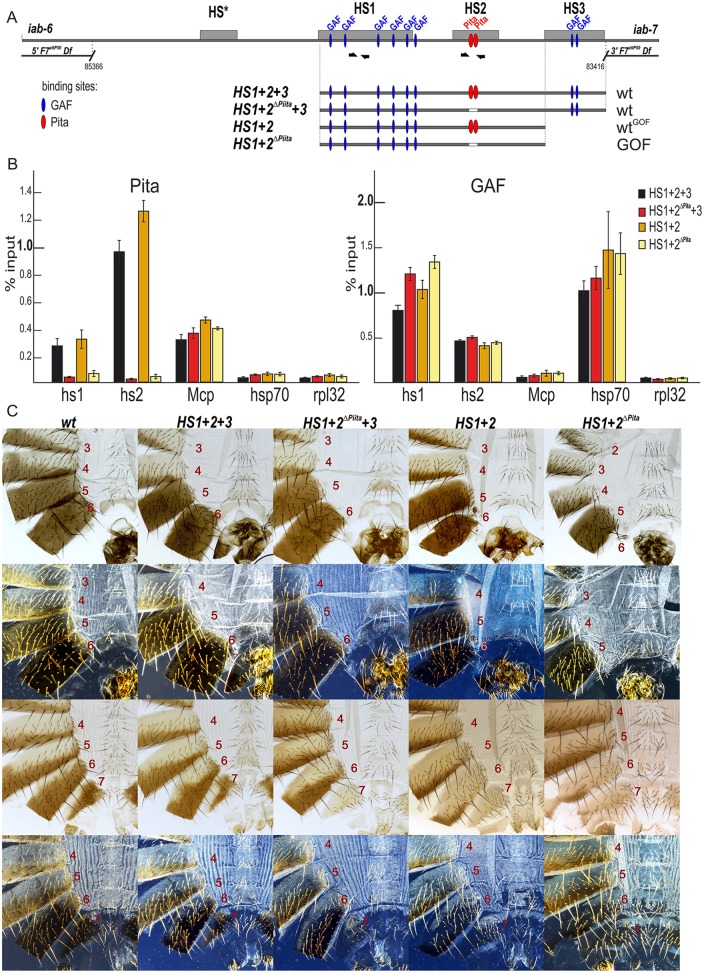
Fig. 3.
Role of Pita-binding sites in Fab-7 boundary function. (A) A schematic of the Fab-7 boundary replacement constructs at the F7attP50 site. Hypersensitive sites are shown as gray boxes. The proximal and distal deficiency endpoints of the F7attP50 deletions are shown. GAF- and Pita-binding sites are designated as blue and red ovals, respectively. Bold black half-arrows (marked i7) indicate primers that were used for ChIP experiments. (B) Binding of Pita and GAF at the HS1 and HS2 regions in pupae of tested transgenic lines (HS1+2+3, HS1+2ΔPita +3, HS1+2 and HS1+2ΔPita). The results of ChIP are presented as a percentage of the input DNA. The Mcp region was used as a positive control for Pita binding, the Hsp70 region was used as a positive control for GAF binding and the RpL32-coding region was used as a negative control for binding. Error bars indicate standard deviations of triplicate PCR measurements from two independent biological samples of chromatin. (C) All abdominal segments in HS1+2+3 and HS1+2ΔPita +3 males (first row, bright field; second row, dark field) and females (third row, bright field; fourth row, dark-field images) have essentially a wild-type identity. HS1+2 males have a mixed wild-type and gain-of-function phenotype: an almost complete A6 tergite in combination with an absence of the A6 sternite. HS1+2ΔPita males and females have a strong gain-of-function phenotype with small spots of cells with loss-of-function features.
For the HS1+2+3 replacement series, ChIP experiments show that Pita is bound to the HS2 region in the wild-type version, whereas it is substantially reduced in the HS1+2ΔPita +3 mutant (Fig. 3B). In spite of the loss of the Pita protein in the HS1+2ΔPita +3 replacement, there was no detectable phenotypic effect, and like the starting HS1+2+3 replacement, the mutant flies were phenotypically wild type (Fig. 3C). Thus, in this context the two Pita-binding sites are not needed for boundary function.
Very different results were obtained for the HS1+2 replacement. The replacement itself has properties that have not been observed in any of the previously described Fab-7 boundary mutations. First, the activity of the HS1+2 replacement is differentially compromised in a rather striking tissue-specific pattern. As shown in the male HS1+2 fly in Fig. 3C, the A6 sternite on the ventral side is completely absent, as would be expected for a gain-of-function transformation from PS11 (A6) to PS12 (A7) identity. Although the vast majority of HS1+2 males lack an A6 sternite, infrequently (∼10%) we observe a remaining small patch of sternite tissue (see Fig. S7). If a similar gain-of-function transformation occurred in the cells giving rise to the dorsal tergite, this tissue should also be absent in HS1+2 males. However, as evident from Fig. 3C, all HS1+2 males have an almost complete A6 tergite. Although there are often regions along the edges of the A6 tergite that are deformed or irregular, the tergite in the majority of the HS1+2 males is close to the size of the A6 tergite in wild type and, judging from the unique pattern of trichomes in A6, is properly specified. A preferential reduction in boundary activity in the ventral tissues is also evident in female flies. Whereas the A6 tergite in the HS1+2 females is properly specified and resembles that in wild type, this is not true for the sternite. Instead, the morphology of the A6 sternite indicates that there is a gain-of-function transformation in identity, albeit incomplete. Unlike wild type, the HS1+2 A6 sternite has hairs pointing inward rather than outward, and has a partially abnormal inverted U-like shape. All of these morphological features are normally observed in A7, not in A6. It is worth noting that the specification of cell identity in A6 in HS1+2 males and females differs from that observed in the standard Fab-7 deletions. In standard deletions, A6 cells either activate iab-7 and assume an A7 identity or silence A6 and assume and A5 identity. In contrast, in the HS1+2 flies, the A6 cells either activate iab-7 and assume an A7 identity or block iab-6↔iab-7 crosstalk and assume the proper A6 identity. Second, all of the previously described Fab-7 mutants are dominant, and their phenotypic effects are evident in flies that are heterozygous for the mutation. In contrast, the HS1+HS2 replacement is recessive, and there are no detectable phenotypes in HS1+HS2/+ flies.
ChIP experiments show that Pita is bound to the HS2 region in HS1+2, but not in the HS1+2ΔPita mutant (Fig. 3B). However, unlike the HS1+2+3 series, loss of Pita binding in this already sensitized background leads to a further disruption of boundary function. Fig. 3C shows that HS1+2ΔPita males lack the entire A6 segment (both sternite and tergite) as would be expected for a complete transformation of PS11 into PS12. This is confirmed by the morphology of the A6 segment in HS1+2ΔPita females. A6 is transformed into a copy of A7. These findings indicate that Pita contributes to the boundary activity of Fab-7.
Pita is essential for blocking activity of the Mcp boundary
In previous studies, Fab-7 has been replaced not only by multimerized binding sites for insulator proteins such as dCTCF, but also by the endogenous D. melanogaster boundary elements, scs, gypsy su(Hw) and Fab-8 (Hogga et al., 2001; Kyrchanova et al., 2016). Of these three endogenous boundary elements, the only one that fully substitutes for Fab-7 is the neighboring Fab-8 boundary element (Kyrchanova et al., 2016). For this boundary element, rescuing activity is orientation dependent. When Fab-8 is inserted in the same (forward) orientation, as the endogenous Fab-8 boundary element, it rescues the Fab-7attP50 deletion. It blocks crosstalk between iab-6 and iab-7 and is also able to facilitate boundary bypass. By contrast, when Fab-8 is inserted in the reverse orientation, it still blocks crosstalk, but is no longer able to promote boundary bypass. scs inserted in both orientations resembles Fab-8R in that it blocks crosstalk and also prevents bypass. A similar result is observed for gypsy su(Hw) replacement in the epidermis; however, in the embryonic CNS, the gypsy su(Hw) replacement is inactive and fails to block iab-6↔iab-7 crosstalk.
One interpretation of these findings is that BX-C boundaries have unique properties that enable them to prevent interactions between adjacent regulatory domains, but at the same time facilitate bypass (Kyrchanova et al., 2015). Supporting this idea, Iampietro et al. (2008) found that Fab-7 could substitute for Fab-8. To test this hypothesis further and at the same time probe the functioning of the Pita protein, we selected the BX-C Mcp boundary, as it shares insulator proteins with both Fab-7 and Fab-8. Like Fab-7, it has a Pita-binding site, and like Fab-8, it has a site for dCTCF (Fig. 4A). Although both sites are important for the Mcp-blocking activity in transgene assays (Kyrchanova et al., 2007), it is not known whether they are functional in the context of BX-C.
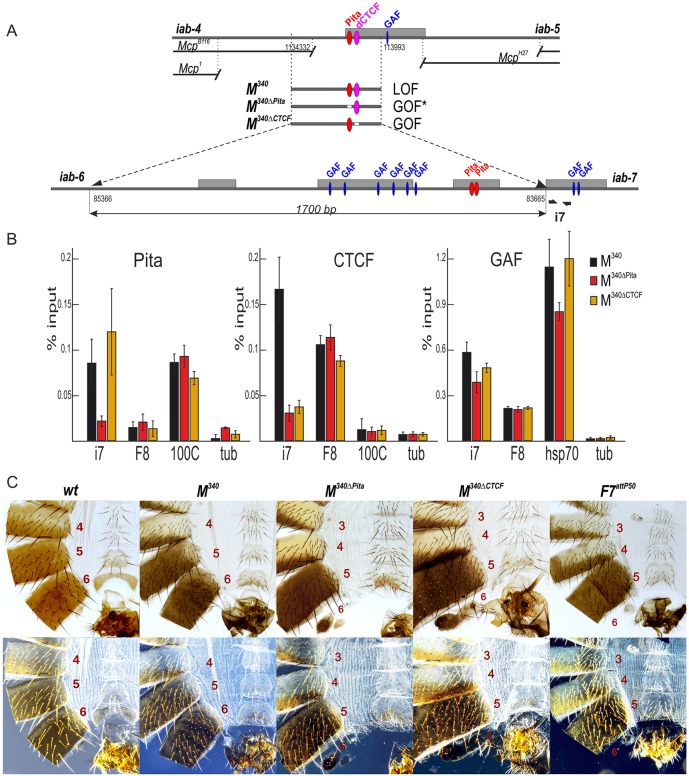
Fig. 4.
Role of Pita- and CTCF-binding sites in Mcp insulator function. (A) Schematic of the Mcp boundary and replacement fragments used for the insertion at F7attP50. The dCTCF-binding site is shown as magenta ovals. Bold black half-arrows (marked i7) below the map indicate primers that were used for ChIP experiments. Hypersensitive sites are shown as gray boxes. GAF- and Pita-binding sites are designated as blue and red ovals, respectively. (B) Binding of Pita, dCTCF and GAF at the PREiab-7 region in pupae of tested transgenic lines (M340, M340ΔPita and M340ΔCTCF). The results of ChIPs are presented as percentages of the input DNA. The 100C region was used as a positive control for Pita and hsp70 for GAF binding, the F8 region was used as a positive control for dCTCF and GAF binding, and the γTub37C-coding region was used as a negative control for binding. Error bars indicate standard deviations of triplicate PCR measurements from two independent biological samples of chromatin. (C) Morphology of the abdominal segments of the M340 mutant males (bright field, top row; dark field, bottom row). In M340 homozygous males, A6 is completely transformed into A5. The phenotypic effects are the same as in the case of Pita×5. M340ΔPita and M340ΔCTCF males demonstrate a mixed gain- and loss-of-function transformation of A6 segment. Phenotypes of M340 mutant females can be seen in Fig. S3.
To explore these issues, we used the Fab-7attP50 platform to introduce the 340 bp Mcp boundary either in the same, forward orientation (M340) as it is in BX-C, or in the reverse orientation, fused with the PREiab-7 (Fig. 4A). The results for the forward orientation are shown in Fig. 4C. The same results were obtained for the reverse orientation (not shown).
Like the Pita×5 replacement, M340 blocks crosstalk between iab-6 and iab-7 but does not permit boundary bypass. Whereas the gain-of-function transformation in Fab-7attP50 deletion almost completely eliminates A6, this segment is restored in the Mcp replacement. However, instead of assuming an A6 (PS11) identity, the morphology of the cuticle in this segment is typical of the immediately anterior segment A5. In wild type, the A6 sternite has a banana shape and is devoid of bristles. In contrast, the A6 sternite in the male M340 fly, shown in Fig. 4C, resembles the sternite in A5: it has a quadrilateral shape and is covered in bristles. Similarly, the A6 tergite is completely covered in trichome hairs, just like the A5 tergite of wild type. Thus, unlike Fab-8, Mcp cannot substitute for Fab-7.
We next used Mcp fragments, which have either the Pita or dCTCF sites deleted, for the replacement experiments. Both mutations compromised the blocking activity of the Mcp replacement. The most straightforward is the deletion of the Pita site. As illustrated by the male fly in Fig. 4C, the M340ΔPita mutant boundary has a mixed gain-of-function and loss-of-function phenotype, like that observed in classical Fab-7 boundary deletions that retain the PREiab-7, HS3. In the fly shown here, the A6 sternite is absent, whereas the size of the tergite is greatly reduced. These phenotypes are indicative of a gain-of-function transformation. Likewise, as expected for a loss-of-function transformation, the rudimentary tergite has trichome hairs distributed throughout, which is characteristic of A5.
Whereas a mixed gain-/loss-of-function phenotype (like that seen for M340ΔPita) is expected for mutations that inactivate boundary function, this is not what was observed for M340ΔCTCF. Instead, the A6 (PS11) phenotypes in M340ΔCTCF flies are a more complicated mixture of not only gain- and loss-of-function but also what appears to be wild type. A typical M340ΔCTCF male fly is shown in Fig. 4C. Like most M340ΔPita males, the sternite is absent, as would be expected for a loss-of-function transformation of A6 (PS11) into A7 (PS12). However, in other respects it differs from the typical M340ΔPita male. Although the size of tergite is clearly reduced, it is larger than the residual tergite in most M340ΔPita males. In fact, as illustrated in Fig. S5, in about 10% of the M340ΔCTCF males the size of A6 tergite is between one quarter and one half that of the wild-type A6 tergite. The other even more unusual feature is that the posterior side of the residual tergite is typically devoid of trichomes. As shown in the dark-field images in Fig. 4C and Fig. S5, this is most readily seen in M340ΔCTCF males that have larger residual tergites. This morphological feature is characteristic of A6 (PS11), indicating that these cells have assumed a proper PS11 identity. However, it should be noted that there are also some males in which the residual tergite is covered in trichome hairs, indicating that the cells have assumed an A5 (PS10) identity. This is expected of the loss-of-function phenotype of boundary mutants. An equivalent loss-of-function phenotype can also be seen in the few males that have retained a residual sternite.
An important question is why does the deletion of the M340ΔCTCF site lead to a mixture of gain- and loss-of-function, and wild-type phenotypes in A6, whereas the Pita site deletion gives the expected mixture of gain-/loss-of-function where the remnant cuticle is transformed to A5? One possible hint comes from ChIP experiments on the Mcp replacements M340, M340ΔPita and M340ΔCTCF (Fig. 4B). As expected, both Pita and dCTCF are present in ChIPs of the wild-type M340 replacement. However, the changes in Pita and dCTCF binding in the M340ΔPita and M340ΔCTCF mutant replacements are not equivalent. For the M340ΔPita, not only Pita but also dCTCF are reduced, indicating that dCTCF association is dependent on the presence of Pita. By contrast, for M340ΔCTCF, although the phenotype is unconventional, the effects of the mutation in the dCTCF site on protein association are completely conventional: dCTCF association is greatly reduced, while Pita binding is unaffected. Thus, one plausible explanation for the distinct phenotypes of the M340ΔPita and M340ΔCTCF would be the difference in protein association. In the case of M340ΔPita, loss of both Pita and dCTCF would be equivalent to deleting the Fab-7 boundary in terms of residual Mcp boundary activity. By contrast, in the M340ΔCTCF mutant, Pita protein remains bound, and partial boundary function is retained. Significantly, as evidenced by the appropriately specified cuticle in A6, this partially functional Mcp boundary seems to have acquired bypass activity.
DISCUSSION
Previous studies on the Pita (also known as Spotted Dick) protein suggested that it is a transcriptional activator and showed that the replication defects in pita mutants and in RNAi knockdowns were due to a reduction in the expression of the replication origin protein Orc4 (Page et al., 2005). The experiments presented here, together with our previous studies (Maksimenko et al., 2015), indicate that pita has an additional, if not an entirely different, function, which is chromosome architecture. Below, we detail the evidence in favor of this conclusion, and also discuss the implications of our findings for boundary function in the context of BX-C.
Pita has ‘classic’ boundary activity
Our boundary replacement experiments provide compelling evidence that the zinc-finger protein Pita functions just like other insulator/architectural proteins. When placed in the context of Fab-7, multimerized Pita-binding sites block crosstalk between iab-6 and iab-7, but are not permissive for the regulatory interactions between iab-6 and the Abd-B gene. In this respect, the functioning of the multimerized Pita-binding sites is similar to that observed when multimerized sites for ‘canonical’ boundary factors, dCTCF and Su(Hw), are substituted for the Fab-7 boundary. In the context of Fab-7, they also block crosstalk between iab-6 and iab-7, but do not support bypass (Hogga et al., 2001; Kyrchanova et al., 2016).
Pita sites in HS2 contribute to Fab-7 boundary function
The boundary functions of the Pita protein are also supported by experiments testing its activity in a native context. For Fab-7, there are two Pita-binding sites in the HS2 hypersensitive region. Since previous studies have shown that HS1 is sufficient for full boundary activity, when the iab-7 PRE (HS3) is present, it is clear that Pita function is redundant (Wolle et al., 2015). We confirmed this by introducing mutations in the two Pita-binding sites in a Fab-7 boundary, HS1+2+3, that lacks the ‘*’ nuclease-hypersensitive site, but contains the iab-7 PRE. However, a different result was obtained in the context of a sensitized replacement, HS1+2, in which the iab-7 PRE (HS3) is deleted. In this sensitized background, the two Pita-binding sites in HS2 are essential for boundary activity.
Interestingly, the sensitized HS1+2 Fab-7 replacement has unprecedented properties. Unlike previously described Fab-7 mutations, which are dominant, the boundary defects of HS1+HS2 can be fully complemented by a wild-type boundary in trans. Additionally, as a homozygote, it has differential effects on the specification of dorsal and ventral tissues. The A6 (PS11) sternite is missing in HS1+2 males. This gain-of-function transformation indicates that boundary activity is disrupted in the cells that give rise to this ventral cuticular structure. By contrast, the A6 tergite is not only nearly normal in size, but is also properly specified. This finding means that boundary activity is largely retained in the PS11 cells that give rise to dorsal cuticle structures.
It is also worth noting that HS1+2 is very different from mutations that delete the iab-7 PRE (HS3) but retain the entire Fab-7 boundary (Mihaly et al., 1997). First, the vast majority of homozygous iab-7 PRE (HS3) deletion males are indistinguishable from wild type, arguing that the HS3 deletion retains full boundary function. Second, in a few of the males (∼2.5%), small sections of the dorsal A6 tergite are missing. This phenotype is most readily explained by a loss of PRE silencing, and consequent gain-of-function transformation, in a subset of the cells that give rise to the dorsal cuticle (Mihaly et al., 1997). As the HS1+2 replacement differs from all of the iab-7 PRE (HS3) deletions isolated by Mihaly et al. in that it lacks ‘HS*’, it would appear that this part of the boundary contains binding motifs for factors that are important for boundary function specifically in ventral tissues.
This would not be the only Fab-7 boundary factor that has ‘developmentally’ restricted activity. The two large complexes known to be important for Fab-7 HS1 boundary function, Elba and LBC, are active at different stages of development; the former in early embryos and the latter from mid-embryogenesis onwards. The fact that there is likely to be yet another boundary factor whose activity is developmentally restricted, fits with the idea that boundary function in flies can be subject to stage- and/or tissue-specific regulation (Magbanua et al., 2015).
The Mcp boundary, Pita and dCTCF
One of the paradoxes posed by the BX-C boundaries is that six of the nine regulatory domains in the complex are separated from their homeotic target genes by one or more boundaries. Consequently, these boundaries must, on the one hand, block regulatory interactions between adjacent domains and, on the other, facilitate boundary bypass. One of models to explain these two contradictory activities is that BX-C boundaries have unique properties, i.e. they are designed to block interactions between enhancers/silencers, but not between enhancers/silencers and promoters (Kyrchanova et al., 2015). This model gained currency from replacement experiments, which showed that the BX-C boundary Fab-8 can substitute for Fab-7, while two heterologous boundaries cannot. A prediction of the model is that other BX-C boundaries could also substitute for Fab-7. However, contrary to this prediction, our experiments indicate that Mcp340 boundary behaves like the heterologous fly boundaries – it blocks both crosstalk and bypass.
Our analysis of the effects of mutations in the Pita and dCTCF sites of the Mcp340 boundary suggest that there is a complicated relationship between blocking crosstalk and blocking or enabling bypass. Although mutations in the Pita and dCTCF disrupt the functioning of Mcp340 replacement, the actual consequences of each mutation are quite distinct. In the case of the Pita mutation, we found that loss of Pita binding leads to a substantial reduction in the binding of dCTCF. This means that for this particular boundary, Pita association is required to recruit dCTCF. The requirement is not, however, reciprocal: deleting the Mcp dCTCF site has no effect on Pita association.
Correlated with the differential effects on protein binding, the M340ΔPita and M340ΔCTCF mutants have quite different phenotypes. The phenotype (mixed gain and loss of function) of the former resembles a classic Fab-7 boundary deletion in which the iab-7 PRE (HS3) is retained (Mihaly et al., 1997). In contrast, the phenotype of the latter is a mixture of gain and loss of function, together with cuticle that has morphological features identical to that in A6 (PS11) of wild-type flies. The presence of cuticle that has the proper PS11 identity argues that M340ΔCTCF retains residual boundary function that, in a subset of cells, is sufficient to not only block crosstalk between iab-6 and iab-7, but is also able to facilitate iab-6 bypass.
A simple interpretation of this finding is that the Pita protein differs from dCTCF in that it blocks crosstalk but can facilitate bypass. However, this simple model is not supported by other findings. First, as noted above, just like multimerized dCTCF sites, multimerized Pita sites block both crosstalk and bypass when substituted for Fab-7. Second, the two Pita sites in Fab-7 are not in themselves sufficient for blocking crosstalk. Third, the Fab-8 boundary, which has two dCTCF sites, but no sites for Pita, has both blocking and bypass activity when substituted for Fab-7 (Kyrchanova et al., 2016). Moreover, these dCTCF sites appear to contribute to the bypass activity of the Fab-8 replacement. Thus, a more likely hypothesis is that there are other, as yet unidentified, factors that are bound to Mcp340 and contribute to the blocking and (newly acquired) bypass activities of the M340ΔCTCF mutant, in addition to the Pita protein. Taken together with the finding that reversing Fab-8 eliminates bypass activity (Kyrchanova et al., 2016), our experiments with Mcp suggest that there may not be a common mechanism for generating both blocking and bypass activity. Rather, each BX-C boundary would appear to deploy distinct mechanisms that are adapted for their specific context within the complex.
Is Pita also a transcriptional activator?
Based on their RNAi knockdown experiments in S2 cells, Page et al. (2005) suggested that Pita is a transcriptional activator and that it could play a crucial role in coordinating S phase progression. This idea was supported by their experiments showing that the replication defects induced by Pita depletion are caused by a reduction in Orc4 expression. However, only 32 genes are downregulated (and 10 upregulated) after Pita RNAi, and most appear to have nothing to do with replication. Furthermore, as there are several thousand Pita sites in the genome, the number of affected genes is surprisingly low. In this light, an obvious question is whether blocking, instead of transcriptional activation, might account for the effects of Pita depletion on the Orc4 transcription? Although we did not investigate how Pita functions in the S2 cells, there are reasons to think that this is a distinct possibility. ChIP experiments by Page et al. showed that Pita binds to a region upstream of the Orc4 gene in S2 cells. ModEncode ChIP experiments indicate that there is a large PcG silenced domain just beyond this Pita site (Fig. S6). Thus, an alternative possibility is that Orc4 expression is reduced when Pita is depleted, because the gene is silenced by the PcG spreading. Several of the other Pita transcriptional targets are also close to the PcG domains, and could be silenced in a similar manner (Fig. S6).
MATERIALS AND METHODS
Antibodies
Antibodies against Pita (amino acids 550-683), dCTCF (amino acids 612-818) and GAF (full length) were raised in rabbits and purified from the sera by the ammonium sulfate fractionation followed by affinity purification on the CNBr-activated Sepharose (GE Healthcare) according to standard protocols and have been previously described (Maksimenko et al., 2015).
Chromatin immunoprecipitation (ChIP)
Chromatin for the subsequent immunoprecipitations was prepared from 12-24 h embryos and mid-late pupae as described previously (Maksimenko et al., 2015). Aliquots of chromatin were incubated with rabbit antibodies against Pita (1:500), GAF (1:200) and dCTCF (1:500), or with nonspecific rabbit IgG (control). At least two independent biological replicates were made for each chromatin sample. The results of the ChIP experiments are presented as a percentage of the input genomic DNA after triplicate PCR measurements. The RpL32- or γTub37C-coding region (devoid of binding sites for the test proteins) was used as a negative control; Mcp, F8, 100C and hsp70 regions were used as positive controls. The sequences of used primers are presented in Table S1.
Electrophoretic mobility shift assays
Recombinant proteins for the binding assays were expressed and purified as described previously (Maksimenko et al., 2015). Fluorescently labeled DNA fragments were generated by PCR amplification with the corresponding FAM- or Cy5-labeled primers. Aliquots of purified recombinant proteins (10-15 μg) were incubated with the fluorescently labeled DNA fragments in the presence of nonspecific binding competitor poly(dI-dC). Incubations were performed in PBS (pH 8.0) containing 5 mM MgCl2, 0.1 mM ZnSO4, 1 mM DTT, 0.1% NP-40 and 10% glycerol at room temperature for 30 min. The mixtures were then resolved by nondenaturing 5% PAGE in 0.5× TBE buffer at 5 V/cm. Signals were detected using the Kodak Image System for the FAM-labeled fragments at Ex 500 nm/Em 535 nm and for the Cy5-labeled fragments at the Ex 630 nm/Em 700 nm.
Generation of the Fab-7attP50 replacement lines
The strategy of the creation of the Fab-7attP50 landing platform and generation of the Fab-7 replacement lines is described in detail elsewhere (Wolle et al., 2015; Kyrchanova et al., 2016). DNA fragments used for the replacement experiments were generated by PCR amplification and verified by sequencing. The sequences of the used fragments are shown in Fig. S4.
Cuticle preparations
Adult abdominal cuticles of homozygous eclosed 3- to 4-day-old flies were prepared essentially as described previously (Kyrchanova et al., 2016) and mounted in Hoyer's solution. Photographs in the bright- or dark-field were taken on the Nikon SMZ18 stereomicroscope using Nikon DS-Ri2 digital camera, processed with ImageJ 1.50c4 and Fiji bundle 2.0.0-rc-46, and assembled using Impress of LibreOffice 5.2.3.3.
Acknowledgements
We are grateful to F. Karch for replacement constructs in the Fab-7 region.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: O.K., P.S., P.G.; Methodology: O.K., N.Z., V.M., O.M., P.S., P.G.; Validation: O.K., N.Z., V.M., O.M.; Formal analysis: N.Z., O.M.; Investigation: O.K., N.Z., V.M., O.M., P.G.; Resources: P.S., P.G.; Data curation: N.Z., V.M., O.M.; Writing - original draft: O.K., P.S., P.G.; Writing - review & editing: O.K., V.M., P.S., P.G.; Visualization: O.K., N.Z., V.M., O.M.; Supervision: P.S., P.G.; Project administration: P.G.; Funding acquisition: P.S., P.G.
Funding
This study was supported by the Russian Science Foundation (project 14-24-00166 to P.G.), by a Ministry of Education and Science of the Russian Federation grant to the Institute of Gene Biology (14.B25.31.0 022 to P.S.) and by the National Institutes of Health (GM043432 to P.S.). Equipment belonging to the IGB RAS Shared Equipment Center, supported by the Ministry of Science and Education of the Russian Federation, was used in this study. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.149815.supplemental
References
- Ali T., Renkawitz R. and Bartkuhn M. (2016). Insulators and domains of gene expression. Curr. Opin. Genet. Dev. 37, 17-26. 10.1016/j.gde.2015.11.009 [DOI] [PubMed] [Google Scholar]
- Aoki T., Sarkeshik A., Yates J. and Schedl P. (2012). Elba, a novel developmentally regulated chromatin boundary factor is a hetero-tripartite DNA binding complex. Elife 1, e00171 10.7554/eLife.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barges S., Mihaly J., Galloni M., Hagstrom K., Muller M., Shanower G., Schedl P., Gyurkovics H. and Karch F. (2000). The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127, 779-790. [DOI] [PubMed] [Google Scholar]
- Bender W. and Lucas M. (2013). The border between the ultrabithorax and abdominal-A regulatory domains in the Drosophila bithorax complex. Genetics 193, 1135-1147. 10.1534/genetics.112.146340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonchuk A., Maksimenko O., Kyrchanova O., Ivlieva T., Mogila V., Deshpande G., Wolle D., Schedl P. and Georgiev P. (2015). Functional role of dimerization and CP190 interacting domains of CTCF protein in Drosophila melanogaster. BMC Biol. 13, 163 10.1186/s12915-015-0168-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman B. A. M. and de Laat W. (2015). Architectural hallmarks of the pluripotent genome. FEBS Lett. 589, 2905-2913. 10.1016/j.febslet.2015.04.055 [DOI] [PubMed] [Google Scholar]
- Bowman S. K., Deaton A. M., Domingues H., Wang P. I., Sadreyev R. I., Kingston R. E. and Bender W. (2014). H3K27 modifications define segmental regulatory domains in the Drosophila bithorax complex. Elife 3, e02833 10.7554/eLife.02833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker S. E., Sharma S., Keelan D. J. and Lewis E. B. (1990). The molecular genetics of the bithorax complex of Drosophila: cis-regulation in the Abdominal-B domain. EMBO J. 9, 4277-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetverina D., Aoki T., Erokhin M., Georgiev P. and Schedl P. (2014). Making connections: Insulators organize eukaryotic chromosomes into independent cis-regulatory networks. BioEssays 36, 163-172. 10.1002/bies.201300125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J. and Mirny L. (2016). The 3D genome as moderator of chromosomal communication. Cell 164, 1110-1121. 10.1016/j.cell.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J. R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J. S. and Ren B. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376-380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M., Mistry H., Schedl P. and Jaynes J. B. (2016). Determinants of Chromosome Architecture: Insulator Pairing in cis and in trans. PLoS Genet. 12, e1005889 10.1371/journal.pgen.1005889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloni M., Gyurkovics H., Schedl P. and Karch F. (1993). The bluetail transposon: evidence for independent cis-regulatory domains and domain boundaries in the bithorax complex. EMBO J. 12, 1087-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirlando R. and Felsenfeld G. (2016). CTCF: making the right connections. Genes Dev. 30, 881-891. 10.1101/gad.277863.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohl D., Aoki T., Blanton J., Shanower G., Kappes G. and Schedl P. (2011). Mechanism of chromosomal boundary action: roadblock, sink, or loop? Genetics 187, 731-748. 10.1534/genetics.110.123752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyurkovics H., Gausz J., Kummer J. and Karch F. (1990). A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J. 9, 2579-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogga I., Mihaly J., Barges S. and Karch F. (2001). Replacement of Fab-7 by the gypsy or scs insulator disrupts long-distance regulatory interactions in the Abd-B gene of the bithorax complex. Mol. Cell 8, 1145-1151. 10.1016/S1097-2765(01)00377-X [DOI] [PubMed] [Google Scholar]
- Holohan E. E., Kwong C., Adryan B., Bartkuhn M., Herold M., Renkawitz R., Russell S. and White R. (2007). CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet. 3, e112 10.1371/journal.pgen.0030112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iampietro C., Cleard F., Gyurkovics H., Maeda R. K. and Karch F. (2008). Boundary swapping in the Drosophila Bithorax complex. Development 135, 3983-3987. 10.1242/dev.025700 [DOI] [PubMed] [Google Scholar]
- Iampietro C., Gummalla M., Mutero A., Karch F. and Maeda R. K. (2010). Initiator elements function to determine the activity state of BX-C enhancers. PLoS Genet. 6, e1001260 10.1371/journal.pgen.1001260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch F., Galloni M., Sipos L., Gausz J., Gyurkovics H. and Sched P. (1994). Mcp and Fab-7: molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res. 22, 3138-3146. 10.1093/nar/22.15.3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O. and Georgiev P. (2014). Chromatin insulators and long-distance interactions in Drosophila. FEBS Lett. 588, 8-14. 10.1016/j.febslet.2013.10.039 [DOI] [PubMed] [Google Scholar]
- Kyrchanova O., Toshchakov S., Parshikov A. and Georgiev P. (2007). Study of the functional interaction between Mcp insulators from the Drosophila bithorax complex: effects of insulator pairing on enhancer-promoter communication. Mol. Cell. Biol. 27, 3035-3043. 10.1128/MCB.02203-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O., Toshchakov S., Podstreshnaya Y., Parshikov A. and Georgiev P. (2008). Functional interaction between the Fab-7 and Fab-8 boundaries and the upstream promoter region in the Drosophila Abd-B gene. Mol. Cell. Biol. 28, 4188-4195. 10.1128/MCB.00229-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O., Ivlieva T., Toshchakov S., Parshikov A., Maksimenko O. and Georgiev P. (2011). Selective interactions of boundaries with upstream region of Abd-B promoter in Drosophila bithorax complex and role of dCTCF in this process. Nucleic Acids Res. 39, 3042-3052. 10.1093/nar/gkq1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O., Mogila V., Wolle D., Magbanua J. P., White R., Georgiev P. and Schedl P. (2015). The boundary paradox in the Bithorax complex. Mech. Dev. 138, 122-132. 10.1016/j.mod.2015.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O., Mogila V., Wolle D., Deshpande G., Parshikov A., Cléard F., Karch F., Schedl P. and Georgiev P. (2016). Functional Dissection of the Blocking and Bypass Activities of the Fab-8 Boundary in the Drosophila Bithorax Complex. PLoS Genet. 12, e1006188 10.1371/journal.pgen.1006188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laundrie B., Peterson J. S., Baum J. S., Chang J. C., Fileppo D., Thompson S. R. and McCall K. (2003). Germline cell death is inhibited by P-element insertions disrupting the dcp-1/pita nested gene pair in Drosophila. Genetics. 165, 1881-1888. http://www.genetics.org/content/165/4/1881.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E. B. (1978). A gene complex controlling segmentation in Drosophila. Nature 276, 565-570. 10.1038/276565a0 [DOI] [PubMed] [Google Scholar]
- Liang J., Lacroix L., Gamot A., Cuddapah S., Queille S., Lhoumaud P., Lepetit P., Martin P. G., Vogelmann J., Court F. et al. (2014). Chromatin immunoprecipitation indirect peaks highlight long-range interactions of insulator proteins and pol II pausing. Mol. Cell. 53, 672-681. 10.1016/j.molcel.2013.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda R. K. and Karch F. (2015). The open for business model of the bithorax complex in Drosophila. Chromosoma. 124, 293-307. 10.1007/s00412-015-0522-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magbanua J. P., Runneburger E., Russell S. and White R. (2015). A variably occupied CTCF binding site in the ultrabithorax gene in the Drosophila bithorax complex. Mol. Cell. Biol. 35, 318-330. 10.1128/MCB.01061-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimenko O. and Georgiev P. (2014). Mechanisms and proteins involved in long-distance interactions. Front Genet. 5, 28 10.3389/fgene.2014.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimenko O., Bartkuhn M., Stakhov V., Herold M., Zolotarev N., Jox T., Buxa M. K., Kirsch R., Bonchuk A., Fedotova A. et al. (2015). Two new insulator proteins, Pita and ZIPIC, target CP190 to chromatin. Genome Res. 25, 89-99. 10.1101/gr.174169.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzat L. H. and Lei E. P. (2014). Surviving an identity crisis: a revised view of chromatin insulators in the genomics era. Biochim. Biophys. Acta 1839, 203-214. 10.1016/j.bbagrm.2013.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall K., O'Connor M. B. and Bender W. (1994). Enhancer traps in the Drosophila bithorax complex mark parasegmental domains. Genetics 138, 387-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M. and Nora E. P. (2016). CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annu. Rev. Genomics Hum. Genet. 17, 17-43. 10.1146/annurev-genom-083115-022339 [DOI] [PubMed] [Google Scholar]
- Mihaly J., Hogga I., Gausz J., Gyurkovics H. and Karch F. (1997). In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development 124, 1809-1820. [DOI] [PubMed] [Google Scholar]
- Mihaly J., Barges S., Sipos L., Maeda R., Cleard F., Hogga I., Bender W., Gyurkovics H. and Karch F. (2006). Dissecting the regulatory landscape of the Abd-B gene of the bithorax complex. Development 133, 2983-2993. 10.1242/dev.02451 [DOI] [PubMed] [Google Scholar]
- Mohan M., Bartkuhn M., Herold M., Philippen A., Heinl N., Bardenhagen I., Leers J., White R. A. H., Renkawitz-Pohl R., Saumweber H. et al. (2007). The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J. 26, 4203-4214. 10.1038/sj.emboj.7601851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon H., Filippova G., Loukinov D., Pugacheva E., Chen Q., Smith S. T., Munhall A., Grewe B., Bartkuhn M., Arnold R. et al. (2005). CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 6, 165-170. 10.1038/sj.embor.7400334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A. R., Kovacs A., Deak P., Torok T., Kiss I., Dario P., Bastos C., Batista P., Gomes R., Ohkura H., Russell S. and Glover D. M. (2005). Spotted-dick, a zinc-finger protein of Drosophila required for expression of Orc4 and S phase. EMBO J. 24, 4304-4315. 10.1038/sj.emboj.7600890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Lluch S., Cuartero S., Azorin F. and Espinas M. L. (2008). Characterization of new regulatory elements within the Drosophila bithorax complex. Nucleic Acids Res. 36, 6926-6933. 10.1093/nar/gkn818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S. S. P., Huntley M. H., Durand N. C., Stamenova E. K., Bochkov I. D., Robinson J. T., Sanborn A. L., Machol I., Omer A. D., Lander E. S. et al. (2014). A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665-1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinsberg S. E. and Schedl P. (2004). Developmental modulation of Fab-7 boundary function. Development 131, 4743-4749. 10.1242/dev.01343 [DOI] [PubMed] [Google Scholar]
- Schweinsberg S., Hagstrom K., Gohl D., Schedl P., Kumar R. P., Mishra R. and Karch F. (2004). The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics 168, 1371-1384. 10.1534/genetics.104.029561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K. C., Taubman A. D. and Geyer P. K. (1999). Enhancer blocking by the Drosophila gypsy insulator depends upon insulator anatomy and enhancer strength. Genetics 153, 787-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., Hoichman M., Parrinello H., Tanay A. and Cavalli G. (2012). Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458-472. 10.1016/j.cell.2012.01.010 [DOI] [PubMed] [Google Scholar]
- Vogelmann J., Le Gall A., Dejardin S., Allemand F., Gamot A., Labesse G., Cuvier O., Negre N., Cohen-Gonsaud M., Margeat E. et al. (2014). Chromatin insulator factors involved in long-range DNA interactions and their role in the folding of the Drosophila genome. PLoS Genet. 10, e1004544 10.1371/journal.pgen.1004544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolle D., Cleard F., Aoki T., Deshpande G., Schedl P. and Karch F. (2015). Functional requirements for Fab-7 boundary activity in the bithorax complex. Mol. Cell. Biol. 35, 3739-3752. 10.1128/MCB.00456-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolotarev N., Fedotova A., Kyrchanova O., Bonchuk A., Penin A. A., Lando A. S., Eliseeva I. A., Kulakovskiy I. V., Maksimenko O. and Georgiev P. (2016a). Architectural proteins Pita, Zw5,and ZIPIC contain homodimerization domain and support specific long-range interactions in Drosophila. Nucleic Acids Res. 44, 7228-7241. 10.1093/nar/gkw371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolotarev N. A., Maksimenko O. G., Georgiev P. G. and Bonchuk A. N. (2016b). ZAD-domain is essential for nuclear localization of insulator proteins in Drosophila melanogaster. Acta Naturae 8, 97-102. [PMC free article] [PubMed] [Google Scholar]