ABSTRACT
Treatment of triple negative breast cancer (TNBC) has been a big challenge since it is defined. To date, platinum-based chemotherapy has played a significant role in the treatment of TNBC patients. However, some patients do not respond to platinum salts or gradually develop chemoresistance, resulting in little effect, or even some adverse effects. Here, we review numerous preclinical and clinical investigations to summarize possible mechanisms and potential predictive biomarkers of platinum in TNBC. The homologous recombination deficiency (HRD) resulting from the loss of BRCA function is the main rationale of platinum efficacy in TNBC. BRCA mutation and methylation have been demonstrated to be important potential biomarkers. Based on genome-wide effects, BRCA-like classifier can identify the functional loss of BRCA and work as the predictor. HRD score that is able to identify the “BRCAness” and predict the sensitivity of platinum is increasingly considered. Taken together, all findings suggest that HR deficiency profile encompassed by BRCA mutation and high HRD score could predict response to platinum, even to other DNA-damage inducing agents. p53 family members and molecular subtypes of TNBC are also important alternative considerations for predicting platinum response based on the preclinical trials. Currently, tumor infiltrating lymphocyte level and thrombocytopenia are emerging as predictive biomarkers.
KEYWORDS: BRCA, DNA damage, homologous recombination deficiency, platinum, predictive biomarker, triple negative breast cancer
Introduction
Triple negative breast cancer (TNBC), defined as lack of expression of estrogen receptor (ER) and progesterone receptor (PR), together with amplification of human epidermal growth factor receptor 2 (HER2) gene, accounts for approximately 20% of breast cancers. Compared with any other subtypes of breast cancer, TNBC follows a more aggressive clinical course and shows a higher risk of recurrence and death from disease in the first 3 to 5 y after diagnosis.1 TNBC patients cannot benefit from endocrine therapy or HER2-targeted agents because of absence of ER/PR/HER2. Furthermore, there are no available target agents for TNBC.2 Because TNBC is a highly heterogeneous disease composed of molecularly distinct subtypes.3 It's intriguing to observe that TNBC shows partial overlap with the basal-like breast cancer. In addition, although over 75% of BRCA1/2-mutated breast cancers show the TNBC phenotype, the majority of TNBC are still sporadic.1 To date, chemotherapy is the mainstay of systemic treatment of TNBC. And the use of platinum-based chemotherapy in TNBC patients has shown promise in increasing preclinical and clinical trials. Many retrospective studies have suggested an improved outcome for TNBC patients compared with non-TNBC patients treated with platinum-based chemotherapy, as shown in Table 1. Most commonly used platinum agents for TNBC are cisplatin and carboplatin.4-8 However, only a part of TNBC patients are extremely sensitive to platinum-based chemotherapy, while most of unselected TNBC patients get only substantial toxicities from platinum treatment.9,10 Hence, the predictive biomarkers are urgently needed to screen out the suitable patients among the unselected groups. Here, we will devote comprehensive attention to possible biomarkers of platinum salts.
Table 1.
TNBC and Non-TNBC patients Treated with platinum-based chemotherapy.
| Study | Study design | Setting | Outcome | TNBC | Non-TNBC | P value |
|---|---|---|---|---|---|---|
| Sirohi(2008)4 | Retrospective | Neo-adjuvant/Adjuvant | CRR | 88.0% | 51.0% | 0.005 |
| PFS | 6 m | 4 m | 0.05 | |||
| OS | 11 m | 7 m | 0.1 | |||
| Uhm(2009)6 | Retrospective | First- or second-line | RR | 37.5% | 39.5% | 0.926 |
| OS | 21 m | 56 m | 0.03 | |||
| Koshy(2010)7 | Retrospective | First-line | PFS | 5.3 m | 1.7 m | 0.058 |
| OS | 10.8 m | 4.3 m | 0.109 | |||
| Staudacher(2010)8 | Retrospective | First-line | ORR | 33.3% | 22% | 0.1 |
CRR: complete response rate. PFS: progression-free survival. OS: overall survival. RR: response rate ORR: overall response rate
Molecular mechanisms of platinum and homologous recombination deficiency in TNBC
Dasari et al has reviewed the main molecular mechanisms of platinum-based agents. Once platinum salts enter cells, their ligands are displaced by water and then the hydrolyzed products, performing as a electrophile, can covalently bind to DNA to form DNA-platinum adducts with various types of intra and inter-strand crosslinks.11 DNA-platinum adducts can lead to single-strand breaks (SSBs) and/or double-strand breaks (DSBs), ultimately resulting in DNA damage. It is worthy mentioning that inter-strand crosslinks can induce stalled replication fork.12 Once DNA is damaged, cell cycle checkpoints will be activated through activation of p53 either to repair the DNA damage or induce cell apoptosis. Whether cancer cells can repair platinum-induced DNA damage will decide the cells to survive or die,11,12 shown as Fig. 1A.
Figure 1.
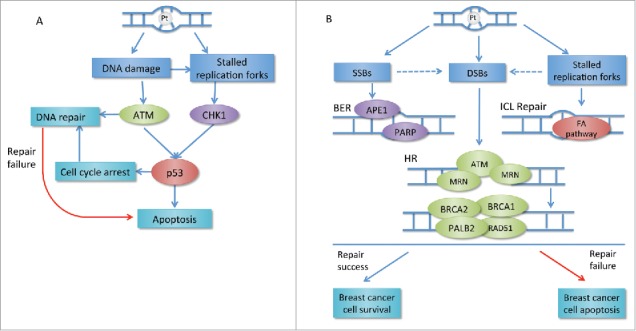
(A) Transduction of platinum-induced DNA damage: DNA repair or apoptosis. DNA-platinum adducts can result in DNA damage and replication fork stalling. This leads to the activation of PI3K-like kinases ATM and CHK, which induces activation of p53 resulting in cell cycle arrest or apoptosis. Cell cycle arrest and activation of ATM can initiate DAN damage repair. If the repair fails, the apoptosis will still happen. (B) Main repair pathways involved in platinum-induced DNA damage repair in breast cancer. DNA-platinum adducts can result in SSB and/or DSB breaks and stalled replication forks. MRN complex senses the DSBs and subsequently recruits ATM, which induces the following steps in error-free HR. BRCA1 and BRCA2 are main components in HR pathway. BRCA1 controls DSB resection and facilitates the transition from DSB resection to BRCA2-mediated RAD51 loading. SSBs are mainly repaired by BER pathway requiring PARP in breast cancer. FA pathway is important for replication-coupled repair of ICLs. If the repair is successful, the cancer cell will proliferate. If any part of these repair pathways changes, the repair will probably fail followed by apoptosis. Pt: platinum; CHK: checkpoint effector kinase; ATM: ataxia-telangiectasia mutated; SSB: single strand break; DSB: double strand break; BER: base excision repair; APE1: apurinic-apyrimidinic endonuclease 1; PARP: poly (ADP-ribose) polymerase; MRN: MRE11–RAD50–NBS1 complex; PALB2: partner and localizer of BRCA2; FA: Fanconi anaemia.
The most harmful lesion DSBs are repaired by either homologous recombination (HR) or non-homologous end joining (NHEJ), depending on the phase of cell cycle. MRN complex (MRE11-RAD50-NBS1) senses the DSBs and subsequently recruits ATM (ataxia telangiectasia mutated) and its related ATR (ataxia-telangiectasia and Rad3-related). And the process will induce the activation of checkpoint and cell cycle arrest through p53. Subsequently, ATM phosphorylates histone H2AX resulting in the recruitment of p53 binding protein 1 (53BP1) and BRCA1 to the sites of damage. HR is the error-free DSB repair mechanism and BRCA family plays a very important role in HR pathway.13 During early DSB repair, BRCA1 controls DSB resection and facilitates the transition from DSB resection to BRCA2-mediated RAD51 loading.14 The previous study has demonstrated that approximately 20% of TNBC patients harbor a BRCA1 or BRCA2 mutation,15 which suggests that TNBC has deficiency in HR. HR deficiency leads to the activation of the error-prone NHEJ repair pathway which easily causes genomic instability and/or cell death.16,17 As platinum salts are able to form DNA-platinum adducts leading to DNA DSBs, the repair of platinum-induced DNA damage requires an intact HR repair capacity. Theoretically, TNBC with HR deficiency are supposed to be sensitive to the platinum.13
Sporadic TNBCs and BRCA-mutated breast cancers share many common molecular and clinical characteristics.1 A hypothesis is proposed that sporadic TNBCs may possess DNA repair defects similar to the BRCA-mutated tumors. The explanation could be “BRCAness,” or more accurately clarified as “BRCAlessness.” BRCAness is defined as patients without BRCA mutations, but with epigenetic inactivation of BRCA, mutations in other genes, or post-translational modifications of key proteins involved in HR and pharmacological inhibitors of the HR, resulting in impairment of HR.18,19 The BRCAness is identified in at least 25% of BRCA wild-type breast cancers and is more frequent in TNBC.20,21 The BRCA1 promoter CpG island methylation is in approximately 30% of TNBC patients22 and tumors with this methylation share the pattern of gene expression similar to inherited BRCA1-mutated breast cancers.23 Pan-histone deacetylase inhibitor and many other strategies have been reported to induce BRCAness and then enable TNBC patients sensitive to platinum-based regimens.24 Besides platinum salts, BRCAness patients can be also sensitive to other DNA damage-inducing regimens, such as Poly (ADP-ribose) polymerase (PARP) inhibitors, as a result of the deficiency in HR repair.
Lesions in other DNA repair pathways including base excision repair and inter-strand crosslink (ICL) repair also have the significantly roles in TNBC.25,26 The comprehensive illustration of main pathways involved in platinum-induced DNA damage repair in breast cancer is shown in Fig. 1B.11,12,17,27
The biomarkers from HR deficiency
As above, we know that patients with BRCA mutation or BRCAness do not have an intact HR repair capacity. This group of patients cannot impair platinum-induced DNA damage and are supposed to be sensitive to platinum.
BRCA expression
Dating back to 2000, it was recognized that BRCA-mutated cells were sensitive to cisplatin.28 Xenograft tumors derived from BRCA1-deficient TNBC cell lines also show cisplatin sensitivity.29 In a group of BRCA1-mutated breast cancers after neoadjuvant chemotherapy, pathologic complete response (pCR) was observed in 10 (83%) of 12 breast cancer patients treated with cisplatin.30 Many prospective studies of early-stage and metastatic TNBC have confirmed that BRCA-mutation status is a potential predictive biomarker.56,59-61 In addition, it has been validated that different BRCA mutation positions may be associated with different levels of HR deficiency and then lead to different sensitivities to platinum in TNBC and ovarian cancer patients.16,31-33 It has also been observed that sporadic TNBCs are sensitive to platinum-based chemotherapy3 and the gene methylation is one of the reasons. Whatever in vivo or vitro, the point that BRCA1 epigenetic inactivation can predict the sensitivity to platinum-based drugs in breast cancer has been proven.5,34 In conclusion, lower BRCA1 mRNA expression is significantly associated with better response in patients treated with platinum-based chemotherapy. However, loss of 53BP1 can allow cancer cells to tolerate BRCA1 deficiency and partly restore HR repair deficiency in breast cancer, potentially contributing to platinum resistance.35 Interestingly, loss of 53BP1 was reported to have higher frequency in BRCA1-deficient TNBC cells than other subtypes.16,36 Therefore, it is necessary to simultaneously evaluate 53BP1 and BRCA1 expression status to assess HR capacity.
Biomarkers based on genome-wide effects of functional loss of BRCA
How to identify signatures of BRCA functions has been in intense investigation for a long time, especially in TNBC.37 Recently, Vollebergh et al. developed a classifier based on array comparative genomic hybridization (aCGH), which could identify the functional loss of BRCA1 in breast cancer. The result showed that almost 60% of BRCA1-likeCGH tumors harbored either BRCA1 mutation or BRCA1 methylation. In this study, they found that these BRCA1-likeCGH patients were associated with better response to platinum-based adjuvant chemotherapy and more likely to be TNBC.38 Later, in a randomized controlled trial of HER2-negative breast cancer patients, they proposed that if tumors were with aCGH patterns resembling BRCA1 and/or BRCA2-mutated breast cancers, they would be defined as the BRCA-likeCGH ones. They verified the predictive value of BRCA-likeCGH for high-dose cyclophosphamide-thiotepa-carboplatin (HD-CTC) adjuvant chemotherapy in TNBC in the study. Compared to other chemotherapies, HD-CTC was associated with better OS in BRCA-likeCGH tumors, but not in non-BRCA-likeCGH tumors. Remarkably, almost 76% of TNBC patients showed a BRCA-likeCGH phenotype.39 Although the study has its limitations, aCGH platform has its potential to predict platinum response.
Biomarkers associated with genomic instability
Both BRCA1-mutated breast cancers and sporadic TNBCs are with high levels of genomic and chromosomal aberrations. Allelic imbalance (AI) is defined as a difference in expression levels of 2 alleles with or without changes in total DNA copy number.40 AI extending to the telomeric end of the chromosome (NtAI) can be applied as a method of evaluating subchromosomal abnormalities. NtAI was found to be an accurate biomarker of pathologic response to cisplatin-based preoperative chemotherapy in TNBC patients.41 In multiple tumors, the correlation between high NtAI grade and low BRCA1 expression indicated that this kind of genomic abnormality was able to represent HR deficiency.41 Large-scale state transition (LST) is defined as chromosomal break of at least 10 Mb between adjacent regions, while loss of heterozygosity (LOH) is defined as loss of parental copies of a heterozygous region of DNA. LST and LOH were positively correlated in TNBC patients.10 High numbers of LST were associated with BRCA1-inactivivation in basal-like breast cancer.42 LOH clustering could indicate BRCA1/2 dysfunction and sensitivity to platinum drugs in high-grade serous ovarian cancer.43
HR deficiency (HRD)-LOH score, HRD-LST score and HRD-NtAI score are formulated to evaluate genomic instability, or so-called genomic scar like signatures of aberrations resulting from HR repair defects by single-nucleotide polymorphism array-based sequencing, which are supposed to be related to BRCA functions and the platinum effect.44 Recently, a combined score called HRD score has been developed, which is calculated based on the quantitative sum of the 3 scores. HRD score equal to or over a predefined threshold 42 is considered as HR deficiency.45 In multivariable analyses of breast cancers, HRD score was able to detect significant BRCA1/2 deficiency not captured by the any single score.43 The predictive effects of HRD score and/or HR deficiency in early-stage TNBC have been illustrated by recent studies,45-47 as shown in Table 2. But in metastatic setting, the dichotomized HRD score was not found to be associated with sensitivity to carboplatin and the reason may be that nonmutated HRD-high cancers frequently reverse the homologous repair defect in metastatic setting.48 HRD assay determines the genomic instability score by formalin-fixed and paraffin-embedded (FFPE) tissues, which improves its clinical viability.37 Besides, a recent research found that HRD score was highly conserved because it was consistent across the multiple repeat biopsies from different regions of the same cancer tissue.49 This strategy for predicting the sensitivity of platinum-based chemotherapy focuses on general aspects of HRD rather than the function of a single gene. Now, whether HRD score is limited to the evaluation of the platinum salts is a new focus of further studies.
Table 2.
The different response rates of BRCA-wild TNBC patients treated with platinum-based chemotherapy between with high and with low HRD score.
| Author | Year | Study design | Responders | HRD score ≥ 42 | HRD score < 42 | P value |
|---|---|---|---|---|---|---|
| Richardson et al.45 | 2014 | Cisplatin-containing neoadjuvant chemotherapy | pPR | 10 (52.6%) | 2 (10.5%) | 0.0039 |
| pCR | 5 (26.3%) | 0 (0%) | 0.018 | |||
| Kaklamani et al. 47 | 2015 | Carboplatin-containing neoadjuvant chemotherapy | pCR | 6 (66.7%) | 2 (14.2%) | 0.0016 |
| Von Minckwitz et al.66 | 2015 | Carboplatin-containing neoadjuvant chemotherapy | pCR | 41 (49.4%) | 17 (30.9%) | 0.05 |
| Tutt et al.68 | 2015 | carboplatin monotherapy in metastatic setting | Response rate | 31(38.2%) | 33(29.2%) | NA |
NA: Not available. pCR: pathologic complete response.
Biomarkers based on p53 family
Although p53 family plays an important role in DNA damage repair, there is no valid evidence that p53 mutation is associated with response to platinum-based treatment. However, there exists a correlation between p53 nonsense or frameshift mutation and cisplatin response in TNBC.5 Also, this special type of p53 mutation has been found associated with BRCA1 mutation, which might function as an alternative marker for BRCA1 deficiency.50
p63 and p73 which are the p53 family members, are sequence-specific DNA-binding factors mediating transcription of pivotal down-stream target genes.51 Under normal conditions, p63 protein isoform ΔNp63 can repress the proapoptotic activity of p73 isoform TAp73 by direct physical interaction. Platinum salts can promote TAp73 to dissociate from ΔNp63 by phosphorylation, leading to the activation of TAp73-dependent proapoptotic pathway followed by apoptosis.5 The result in vitro found that TNBC cells co-expressing TAp73 and ΔNp63 showed sensitivity to cisplatin, but not to other common chemotherapy agents. In other words, co-expression of ΔNp63 and TAp73 in TNBC could predict cisplatin sensitivity. Notably, the ΔNp63/TAp73 pathway appeared in both BRCA1-related and sporadic TNBC.52 One clinical trial provided a positive conclusion that ΔNp63/TAp73 ratio > 2 was probably associated with sensitivity to cisplatin in early-stage TNBC.5 However, another study did not observe a significant association of ΔNp63/TAp73 ratio with clinical response of platinum salts in advanced TNBC.10 The conflicting result indicates that the predictive role of ΔNp63/TAp73 ratio is not explicit.
Molecular subtype associated biomarkers
In PAM50 gene expression analysis, the majority of TNBCs are basal-like breast cancer (BLBC) with the proliferation gene signatures.53 Retrospective trials have found that molecular profiles of TNBC have clinical utility in predicting chemotherapy response.29,54 In a clinical research of 1055 patients with TNBC, BLBC or both, only when TNBC can be stratified into BLBC subtype could patients be identified with notably better response after chemotherapy.55 Previous studies show that BLBC harbors the hallmarks of BRCAness.21 In conclusion, BLBC is an important consideration for prediction of platinum efficacy in TNBC.
Lehmann et al. have classified TNBC into 6 subtypes based on their gene expression profiles. Among these subtypes, cell lines representative of basal-like (BL1 and BL2) subtypes were significantly more sensitive to cisplatin than other subtypes.3 The possible mechanism may be that genes of the basal-like subtypes encompass elevated DNA damage response (ATR/BRCA) pathways and increased cell-cycle checkpoint loss. Further exploration revealed that BL1/2 cells had relatively higher sensitivity to cisplatin than other TNBC subtypes irrelevant to BRCA1 mutation status,3 and it suggested that there were probably some other potential HR defects in BL1/2 TNBC cells with intact BRCA1. However, the predictive role of molecular subtypes needs further investigation in clinical trials. Besides, identification of the molecular subtypes should be based on the gene expression profile, which limits its clinical application.
Emerging biomarkers
Nowadays, emerging predictive biomarkers have been investigated in the context of platinum-based therapy in TNBC. Although tumor-infiltrating lymphocytes (TILs) have been proposed for many years, whether they are able to predict the response of platinum in TNBC has just received attention.56 Another emerging biomarker is thrombocytopenia, an adverse effect of platinum regimens.57
Increasing data have showed that TIL is associated with prognosis in several types of tumors, especially in TNBC. TIL has been confirmed to be highly present in TNBC and related to increased pCR rate and improved disease-free survival (DFS) and OS after chemotherapy.58-60 In neoadjuvant GeparSixto and PrECOG0105 trials, TIL levels are positively associated with pCR rate among TNBC patients. LPBC is defined as breast cancer with lymphocytes in 50% or more of tumor or stroma, which was also proved to be associated with pathologic response in GeparSixto trial.56,61 TIL is assessed by the routine hemotoxylin and eosin slides, other than molecular analysis, which makes it more feasible in clinical setting. The possible mechanism of predictive role of TIL is that effective chemotherapy can facilitate an antitumor immune response and induce immunogenic tumor cell death by activating the adaptive immunity.56 In the ECOG and BIG 02–98 studies, higher levels of TILs in TNBC patients receiving anthracycline-based chemotherapy were also linked with higher pCR rate,59,62 suggesting that TILs may not be a distinct predictor for platinum but a common one for different chemotherapy in TNBC.
In the recent SABCAS meeting, a research emphasized that Fanconi anemia pathway might have certain relevance to the response of platinum in TNBC patients. Fanconi anemia pathway not only plays an important role in HR repair, but also in maintaining haematopoietic stem cell (HSC) function. The impaired DNA DSB repair can also lead to HSC and progenitor cell dysfunction. Therefore, it is possible that platinum salts may induce significant hematological toxicity in patients with HR repair dysfunction. A prospective trial evaluating the association between hematological toxicity and response to neoadjuvant carboplatin treatment in 49 TNBC patients found that pCR rates in BRCA-wild type patients with thrombocytopenia (Tp) and without Tp were respectively 85% and 47% (p = 0.013). The conclusion was that Tp was correlated with an improved pCR in BRCA wild-type TNBC. The exact mechanism requires comprehensive assessment of HR repair defects beyond BRCA mutation.57
Besides TILs level and Tp status, there are studies assessing the predictive values of circulating tumor cell (CTC) status and circulating tumor DNA (ctDNA) in breast cancer after chemotherapy.63
Platinum salts in early-stage TNBC
In 2010, Silver et.al conducted a prospective clinical trial of evaluating efficacy of neoadjuvant cisplatin in 28 TNBC patients and a pCR was observed in 6 patients (21%) in this trial. The results also demonstrated that tumors with BRCA1 promoter methylation were more likely to respond to neoadjuvant cisplatin (pCR rate: 75% vs 27%; p = 0.04) and decreased BRCA1 mRNA expression was significantly associated with better response.5 Randomized phase II GeparSixto study was conducted to demonstrate the value of addition of carboplatin to neoadjuvant chemotherapy in stage II-III TNBC and Her-2 positive breast cancer patients. This study found that the addition of carboplatin to anthracylines and taxanes improved pCR rate from 37% to 58% (p = 0.005) in TNBC patients, but not HER2-positive breast cancer patients.64 And the findings are consistent with the results from the similar CALGB 40603 study.65 Exploratory results of GeparSixto showed that 70.5% of TNBC patients were with HR deficiency and 60.3% of TNBC patients were with high HRD score without BRCA mutation. HR deficient tumors had better response to carboplatin-containing chemotherapy compared with HR non-deficient ones. Among patients without BRCA mutations, a higher HRD score was associated with better response to the treatment.66 GeparSixto trial also assessed the predictive value of TILs. In the complete cohort of TNBC and HER-positive breast cancer, the pCR rate was 59.9% in LPBC, compared with 33.8% in non-LPBC (p = 0.001). And the result that the pCR rate was 75% in TNBC with a LPBC phenotype suggested that TIL was a significant predictor for platinum response particularly in TNBC. Besides, mRNA expressions of key modulators of immune reactions were also found to be associated with platinum-based chemotherapy response in this research.61 PrECOG0105 study was a small and single arm clinical trial assessing gemcitabine, carboplatin, and iniparib as neoadjuvant therapy for triple-negative and BRCA1/2 mutation–associated breast cancer. The results of PrECOG0105 indicated that mean HRD-LOH score was higher in patients with response to carboplatin-based treatment than those without response67 and increased probability of pCR was associated with the high level of TILs among 70 TNBC patients.56 Another recent single arm phase II clinical trial evaluating the efficacy and safety of neoadjuvant treatment with carboplatin and eribulin in early-stage TNBC patients suggested the pCR rate was 43.3% (13/30). And further study revealed that HRD score (P = 0.0024) and HR deficiency status (P = 0.0012) significantly predicted pCR. More clinical trials about efficacy and predictive biomarkers of platinum in TNBC patients are shown in Table 3 and Table 4.
Table 3.
TNBC patients treated by chemotherapy with or without platinum.
| Study | Study design | Setting | Outcome | With platinum | Without platinum | P value |
|---|---|---|---|---|---|---|
| Fan(2012)76 | Phase II | Metastatic | ORR | 63.0% | 15.4% | 0.001 |
| PFS | 10.9 m | 4.8 m | < 0.001 | |||
| OS | 32.8 m | 21.5 m | 0.027 | |||
| TBCRC001/Carey(2012)77 | Phase II | Metastatic | ORR | 17.0% | 6.0% | NA |
| PFS | 2.1 m | 2.6 m | NA | |||
| OS | 10.4 m | 7.5 m | NA | |||
| CALGB 40603/Sikov (2015)65 | Phase II | Neo-adjuvant | pCR | 60.0% | 44.0% | 0.0018 |
| GeparSixto/VonMinckwitz(2014)64 | Phase II | Neo-adjuvant | pCR | 53.2% | 36.9% | 0.005 |
| Villarreal-Garza(2014)78 | Retrospective | Adjuvant | OS | 14.5 m | 10 m | 0.041 |
| TnT/Tutt(2015)69 | Phase III | Adjuvant | ORR | 31.4% | 35.6% | 0.44 |
| PFS | 3.1 m | 4.5 m | 0.29 | |||
| OS | 12.4 m | 12.3 m | 0.31 | |||
| WSG-ADAPT/Gluz(2015)79 | Phase II | Neo-adjuvant | pCR | 49.2% | 25.0% | 0.006 |
| CBCSG006/Hu(2015)68 | Phase III | Adjuvant | PFS | 7.73 m | 6.47 m | NA |
NA: Not available. ORR: overall response rate. PFS: progression-free survival. OS: overall survival. pCR: pathologic complete response.
Table 4.
Clinical studies assessing predictive biomarkers in relation to the activity or efficacy of platinum in TNBC.
| Study | Study design | Setting | Markers | Positively associated with clinical endpoints |
|---|---|---|---|---|
| Silver(2010)5 | Phase I/II | Neoadjuvant | Age,BRCA1 mutation status,BRCA1 mRNA Expression,BRCA1 Promoter Methylation,ΔNp63/TAp73,p53 Mutation, Array Mining | Young age, low BRCA1 mRNA expression, BRCA1 promoter methylation, p53 nonsense or frameshift mutations, and a gene expression signature of E2F3 activation |
| Vollebergh(2011)38 | Prospective | Adjuvant | BRCA1-likeCGH classifier | BRCA1-likeCGH profile |
| Birkbak(2012)41 | Retrospective | Neoadjuvant | NtAI | Higher levels of NtAI |
| Vollebergh(2014)39 | Retrospective | Adjuvant | BRCA-likeCGH classifier | BRCA-likeCGH profile |
| Richardson(2014)45 | Retrospective | Neoadjuvant | BRCA1 mutation status, HRD score | BRCA1 mutation and HRD score ≥ 42 |
| Telli(2015)67 | Phase II | Neoadjuvant | BRCA1 mutation status, HRD-LOH | BRCA1 mutation and higher HRD-LOH score |
| Isakoff(2015)10 | Phase II | First/second-line | BRCA1/2 mutation status,HRD-LOH/HRD-LST score,p63/p73,PAM50 gene expression subtypes,p53 and PIK3CA mutation status | BRCA1 mutation and higher HRD-LOH/HRD-LST score |
| Kaklamani(2015)47 | Phase II | Neoadjuvant | HRD score, BRCA1/2 mutation status, protein-based biomarkers (Ki67, TP53, androgen receptor, Cyclin E, CDK2, Cyclin D, CDK4, Pin1 and Smad3) | HRD score ≥ 42, BRCA1/2 mutation and cytoplasmic CDK2 |
| Telli(2015)46 | Phase II | Neoadjuvant | HRD score, BRCA1/2 mutation status | HRD score ≥ 42 and BRCA1/2 mutation |
| GeparSixto/Von Minckwitz (2015)66 and Denkert(2015)61 | Phase II | Neoadjuvant | HRD score, Stromal TILs, mRNA expression of immunoactivating factors (CXCL9, CCL5, CD8A, CD80, CXCL13, IGKC, CD21), immunosuppressive factors (IDO1, PD-1, PD-L1, CTLA4, FOXP3) | HRD score ≥ 42, Increased levels of stromal TILs and all 12 immune mRNA markers |
| Tutt(2015)69 | Phase III | First line | BRCA1/2 mutation status,HRD score | BRCA1/2 mutation |
| Sharma(2015)57 | Prospective | Neoadjuvant | Hematological toxicity | Thrombocytopenia |
ORR: overall response rate. PFS: progression-free survival. OS: overall survival. pCR: pathologic complete response. CGH: comparative genomic hybridization. NtAI: allelic imbalance extending to the telomere. HRD: homologous recombination deficiency. LDH: loss of heterozygosity. LST: large-scale state transitions. RR: response rate.
Platinum salts in metastatic TNBC
In a recent phase III trial, the CBCSG006 study, 240 Chinese advanced TNBC patients were randomly assigned to cisplatin plus gemcitabine or paclitaxel plus gemcitabine group. The cisplatin-containing group had better response compared with paclitaxel-containing group, with a median progression-free survival (PFS) of 7.73 months versus 6.47 months.68 The exploratory assessment of biomarkers of platinum is still being investigated. Another randomized phase III TNT trial that compared carboplatin with taxanes in 376 advanced triple-negative or BRCA1/2-mutation breast cancer patients showed that carboplatin showed no superior activity in unselected TNBC patients, but had better effect in BRCA-mutation patients. The median PFS was 3.1 months in the carboplatin arm and 4.5 months in the docetaxel arm (P = 0.29) among unselected arm. In carboplatin treatment group, BRCA-mutation patients had a median PFS of 6.8 months, compared with 3.1 months in patients with no BRCA mutations, and the differences were not seen in docetaxel group. In this trial, although patients with high HRD score seemed to have better response than those patients with low HRD score when treated with carboplatin, high HRD score could not select carboplatin-sensitive patients among all patients.69 TBCRC009 trial was a single-arm phase II clinical trial of platinum monotherapy for metastatic TNBC with biomarker analysis. Results from TBCRC009 trial indicated that TNBC patients with BRCA1/2 mutation had better response compared with non-mutation ones and HRD-LOH/HRD-LST scores might help identify the patients who are more likely to benefit from platinum-based therapy.10 Further randomized clinical trials evaluating platinum salts in TNBC patients are ongoing.70,71
Discussion and conclusion
Recently, increasing studies have shown that TNBC patients treated by platinum-based chemotherapy will have better response. Considering the heterogeneity of TNBC and several substantial toxicities after chemotherapy, platinum treatment should incorporate some distinct biomarkers for patient selection. Now, personalized treatment is the core principle of cancer therapy and the predictive biomarkers of platinum in TNBC will promote this project. In the Table 4 and Fig. 2, we make a comprehensive summary of any potential predictive biomarkers to give some inspiration to the oncologists.
Figure 2.
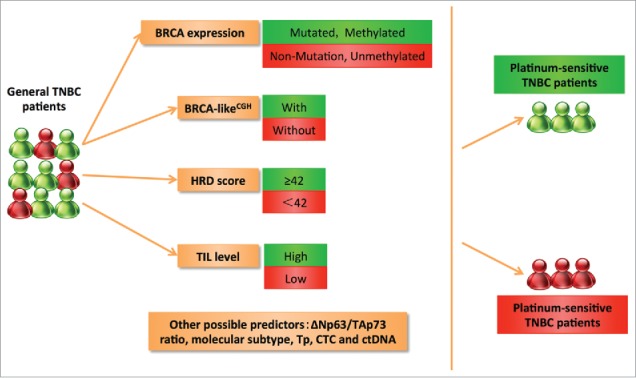
The possible predictive biomarkers for TNBC patients treated with platinum-based chemotherapy based on clinical trials. BRCA mutation status and methylation status are the important potential biomarkers for platinum therapy. Based on genome-wide effects, BRCA-likeCGH classifier may identify the functional loss of BRCA and then work as the predictor. HRD score≥ 42 is related to the chemosensitivity to platinum. Besides, high TILs level seems to be associated with better platinum response. p53 family and molecular subtype are also important alternative considerations. Tp, CTC and ctDNA are emerging biomarkers. TIL: Tumor infiltrating lymphocyte; Tp: thrombocytopenia; CTC: circulating tumor cell; ctDNA: circulating tumor DNA.
As BRCA mutation predisposes to breast cancer development, it also sensitizes cancer cells to DNA-damaging agents based on the fact that BRCA-associated HR repair is the only error-free repair pathway of inter-strand crosslinks.72 BRCA mutation status has been demonstrated to be an important potential biomarker for platinum therapy in almost all researches.10,45,67,69
Because the number of genes related to HR repair is increasing, and more importantly, HR repair pathway is complex and dynamic during tumor development, it may be inaccurate to identify HR deficiency by the single gene alteration.72 In the clinical context, inactivation of HR in BRCA-wild TNBC patients may also induce hypersensitivity to platinum.47 The most compelling problem is how to define the “BRCAness." Vollebergh and colleagues proposed aCGH platform as a predictive marker, but its clinical application had not been reached. Because it was unclear whether the benefit observed in the BRCA-likeCGH patients was mainly a result of the platinum or the intensified alkylating agents or both.39 NtAI, LOH score, LSH score and HRD score can evaluate genomic instability related to BRCA dysfunction to serve as a marker of HRD and response to platinum-based chemotherapy in TNBC.10,47,67 Many researches suggest HR deficiency encompassed by BRCA mutation and high HRD score has shown the most potential promise in predicting the response of platinum agents.47 However, these studies are retrospective or single-arm, which do not provide enough evaluation of a predictive biomarker. It has been recognized that the proper biomarker trial design requires control group.44 It is sure that BRCA-associated HRD biomarkers are potential predictors, but there is no definitive data of their clinical utility. And the preliminary results of phase III TNT tail did not show the significant evidence that high HRD score was associated with the better efficacy of carboplatin compared with docetaxel.69 Hence the role of these genomic instability assays needs further clinical researches. When appropriately validated, the measures assessing HR status will capture more available cancer patients who are likely to benefit from platinum agents or other DNA-damaging agents.
Besides the predictors from HR repair pathway, there are biomarkers from other biomolecules. As the correlation between ΔNp63 and clinical tumor response to cisplatin has been investigated for more than 10 years,73 the predictive value of ΔNp63/TAp73 ratio is intriguing and worth exploring. And the existing clinical trials have not gotten the positive results with statistical significance, possibly as a result of methodology. While the prognostic implications of TILs in multiple tumors, including TNBC, are highly consistent, the predictive effect of TILs has its limitations. The methods for TIL evaluation usually have measure errors in multicenter context. And as TILs are a group of heterogeneous immune cells, identification of a specific subtype of TILs closely related to platinum response will improve their clinical value as the biomarker.61
Actually, platinum agents have been applied to treat a wide range of tumor types for decades. In high-grade serous ovarian cancer (HG-OSC) which is similar to TNBC in molecular and clinical characteristics,16 somatic hypermutation is significantly associated with platinum sensitivity.74 Besides, a multiparameter flow cytometric assay has been investigated as a potential biomarker for HR deficiency in HG-OSC.75
More prospective and large researches should be conducted to identify the value of these biomarkers and confirm their clinical utility. Combining these biomarkers as a composite marker that thoroughly evaluates the properties of TNBC can more accurately predict drug efficacy. For example, HRD score together with TILs level will reflect both tumor and microenvironment properties. Further comprehension of the complexity of DDRs, particularly HR, will improve the process of identifying some practical predictive biomarker tests that facilitate the clinical application of platinum-based chemotherapy in TNBC.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by National Natural Science Foundation of China (No. 81470357) and a Foundation for Clinical Medicine Science and Technology Special Project of the Jiangsu Province, China (No. BL2014071) (to X. G).
Reference
- 1.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med 2010; 363:1938-48; PMID:21067385; https://doi.org/ 10.1056/NEJMra1001389 [DOI] [PubMed] [Google Scholar]
- 2.Bramati A, Girelli S, Torri V, Farina G, Galfrascoli E, Piva S, Moretti A, Dazzani MC, Sburlati P, La Verde NM. Efficacy of biological agents in metastatic triple-negative breast cancer. Cancer Treat Rev 2014; 40:605-13; PMID:24529896; https://doi.org/ 10.1016/j.ctrv.2014.01.003 [DOI] [PubMed] [Google Scholar]
- 3.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011; 121:2750; PMID:21633166; https://doi.org/ 10.1172/JCI45014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sirohi B, Arnedos M, Popat S, Ashley S, Nerurkar A, Walsh G, Johnston S, Smith IE. Platinum-based chemotherapy in triple-negative breast cancer. Ann Oncol 2008; 19(11):1847-52; PMID:18567607; https://doi.org/ 10.1093/annonc/mdn395 [DOI] [PubMed] [Google Scholar]
- 5.Silver DP, Richardson AL, Eklund AC, Wang ZC, Szallasi Z, Li Q, Juul N, Leong CO, Calogrias D, Buraimoh A, et al.. Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J Clin Oncol 2010; 28:1145-53; PMID:20100965; https://doi.org/ 10.1200/JCO.2009.22.4725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uhm JE, Park YH, Yi SY, Cho EY, Choi YL, Lee SJ, Park MJ, Lee SH, Jun HJ, Ahn JS, et al.. Treatment outcomes and clinicopathologic characteristics of triple‐negative breast cancer patients who received platinum‐containing chemotherapy. Int J Cancer 2009; 124:1457-62; PMID:19065658; https://doi.org/ 10.1002/ijc.24090 [DOI] [PubMed] [Google Scholar]
- 7.Koshy N, Quispe D, Shi R, Mansour R, Burton GV. Cisplatin–gemcitabine therapy in metastatic breast cancer: Improved outcome in triple negative breast cancer patients compared to non-triple negative patients. The Breast 2010; 19:246-8; PMID:20227277; https://doi.org/ 10.1016/j.breast.2010.02.003 [DOI] [PubMed] [Google Scholar]
- 8.Staudacher L, Cottu PH, Dieras V, Vincent-Salomon A, Guilhaume MN, Escalup L, Dorval T, Beuzeboc P, Mignot L, Pierga JY. Platinum-based chemotherapy in metastatic triple-negative breast cancer: The Institut Curie experience. Ann Oncol 2010; 22(4):848-56; PMID:20924076; https://doi.org/ 10.1093/annonc/mdq461 [DOI] [PubMed] [Google Scholar]
- 9.Grady SO, Finn SP, Cuffe S, Richard DJ, O'Byrne KJ, Barr MP. The role of DNA repair pathways in cisplatin resistant lung cancer. Cancer Treat Rev 2014; 40(10):1161-70; PMID:25458603; https://doi.org/ 10.1016/j.ctrv.2014.10.003 [DOI] [PubMed] [Google Scholar]
- 10.Isakoff SJ, Mayer EL, He L, Traina TA, Carey LA, Krag KJ, Rugo HS, Liu MC, Stearns V, Come SE, et al.. TBCRC009: A multicenter phase II clinical trial of platinum monotherapy with biomarker assessment in metastatic triple-negative breast cancer. J Clin Oncol 2015; 33(17):1902-9; PMID:25847936; https://doi.org/ 10.1200/JCO.2014.57.6660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discovery 2005; 4:307-20; PMID:15789122; https://doi.org/ 10.1038/nrd1691 [DOI] [PubMed] [Google Scholar]
- 12.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer 2008; 8:193-204; PMID:18256616; https://doi.org/ 10.1038/nrc2342 [DOI] [PubMed] [Google Scholar]
- 13.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature 2012; 481:287-94; PMID:22258607; https://doi.org/ 10.1038/nature10760 [DOI] [PubMed] [Google Scholar]
- 14.Carvalho JFS, Kanaar R. Targeting homologous recombination-mediated DNA repair in cancer. Expert Opin Ther Targets 2014; 18:427-58; PMID:24491188; https://doi.org/ 10.1517/14728222.2014.882900 [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez-Angulo AM, Timms KM, Liu S, Chen H, Litton JK, Potter J, Lanchbury JS, Stemke-Hale K, Hennessy BT, Arun BK, et al.. Incidence and outcome of BRCA mutations in unselected patients with triple receptor-negative breast cancer. Clin Cancer Res 2011; 17:1082-9; PMID:21233401; https://doi.org/ 10.1158/1078-0432.CCR-10-2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vollebergh MA, Jonkers J, Linn SC. Genomic instability in breast and ovarian cancers: Translation into clinical predictive biomarkers. Cell Mol Life Sci 2012; 69:223-45; PMID:21922196; https://doi.org/ 10.1007/s00018-011-0809-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bouwman P, Jonkers J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat Rev Cancer 2012; 12:587-98; PMID:22918414; https://doi.org/ 10.1038/nrc3342 [DOI] [PubMed] [Google Scholar]
- 18.Michels J, Vitale I, Saparbaev M, Castedo M, Kroemer G. Predictive biomarkers for cancer therapy with PARP inhibitors. Oncogene 2014; 33:3894-907; PMID:24037533; https://doi.org/ 10.1038/onc.2013.352 [DOI] [PubMed] [Google Scholar]
- 19.Chalasani P, Livingston R. Differential chemotherapeutic sensitivity for breast tumors with “BRCAness:" A review. Oncologist 2013; 18:909-16; PMID:23881989; https://doi.org/ 10.1634/theoncologist.2013-0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turner N, Tutt A, Ashworth A. Hallmarks of'BRCAness' in sporadic cancers. Nat Rev Cancer 2004; 4:814-9; PMID:15510162; https://doi.org/ 10.1038/nrc1457 [DOI] [PubMed] [Google Scholar]
- 21.De Summa S, Pinto R, Sambiasi D, Petriella D, Paradiso V, Paradiso A, Tommasi S. BRCAness: A deeper insight into basal-like breast tumors. Ann Oncol 2013; 24:viii13-21; PMID:24131964; https://doi.org/ 10.1093/annonc/mdt306 [DOI] [PubMed] [Google Scholar]
- 22.Xu Y, Diao L, Chen Y, Liu Y, Wang C, Ouyang T, Li J, Wang T, Fan Z, Fan T, et al.. Promoter methylation of BRCA1 in triple-negative breast cancer predicts sensitivity to adjuvant chemotherapy. Ann Oncol 2013; 24(6):1498-505; PMID:23406733; https://doi.org/ 10.1093/annonc/mdt011 [DOI] [PubMed] [Google Scholar]
- 23.Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi OP, et al.. Gene-expression profiles in hereditary breast cancer. N Engl J Med 2001; 344:539-48; PMID:11207349; https://doi.org/ 10.1056/NEJM200102223440801 [DOI] [PubMed] [Google Scholar]
- 24.Ha K, Fiskus W, Choi DS, Bhaskara S, Cerchietti L, Devaraj SGT, Shah B, Sharma S, Chang JC, Melnick AM, et al.. Histone deacetylase inhibitor treatment induces ‘BRCAness’ and synergistic lethality with PARP inhibitor and cisplatin against human triple negative breast cancer cells. Oncotarget 2014; 5:5637; PMID:25026298; https://doi.org/ 10.18632/oncotarget.2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill SJ, Clark AP, Silver DP, Livingston DM. BRCA1 pathway function in basal-like breast cancer cells. Mol Cell Biol 2014; 34:3828-42; PMID:25092866; https://doi.org/ 10.1128/MCB.01646-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alli E, Sharma VB, Sunderesakumar P, Ford JM. Defective repair of oxidative dna damage in triple-negative breast cancer confers sensitivity to inhibition of poly (ADP-ribose) polymerase. Cancer Res 2009; 69:3589-96; PMID:19351835; https://doi.org/ 10.1158/0008-5472.CAN-08-4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosoya N, Miyagawa K. Targeting DNA damage response in cancer therapy. Cancer Sci 2014; 105:370-88; PMID:24484288; https://doi.org/ 10.1111/cas.12366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem 2000; 275:23899-903; PMID:10843985; https://doi.org/ 10.1074/jbc.C000276200 [DOI] [PubMed] [Google Scholar]
- 29.Lehmann BD, Pietenpol JA. Identification and use of biomarkers in treatment strategies for triple‐negative breast cancer subtypes. J Pathol 2014; 232:142-50; PMID:24114677; https://doi.org/ 10.1002/path.4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Byrski T, Gronwald J, Huzarski T, Grzybowska E, Budryk M, Stawicka M, Mierzwa T, Szwiec M, Wisniowski R, Siolek M, et al.. Pathologic complete response rates in young women with BRCA1-positive breast cancers after neoadjuvant chemotherapy. J Clin Oncol 2010; 28:375-9; PMID:20008645; https://doi.org/ 10.1200/JCO.2008.20.7019 [DOI] [PubMed] [Google Scholar]
- 31.Thompson D, Easton D, Breast Cancer Linkage C . Variation in BRCA1 cancer risks by mutation position. Cancer Epidemiol Biomarkers Prev 2002; 11:329-36; PMID:11927492 [PubMed] [Google Scholar]
- 32.David YB, Chetrit A, Hirsh-Yechezkel G, Friedman E, Beck BD, Beller U, Ben-Baruch G, Fishman A, Levavi H, Lubin F, et al.. Effect of BRCA mutations on the length of survival in epithelial ovarian tumors. J Clin Oncol 2002; 20:463-6; PMID:11786575; https://doi.org/ 10.1200/JCO.2002.20.2.463 [DOI] [PubMed] [Google Scholar]
- 33.Frolova M, Ignatova E, Glazkova E, Petrovsky A, Tjulandin S. Neoadjuvant chemotherapy with dose-dense doxorubicin, cisplatin, and paclitaxel in patients with early triple-negative breast cancer (TNBC). ASCO Meeting Abstracts 2015; 33:1074; https://doi.org/ 10.1200/jco.2015.33.15_suppl.1074 [DOI] [Google Scholar]
- 34.Stefansson OA, Villanueva A, Vidal A, Martí L, Esteller M. BRCA1 epigenetic inactivation predicts sensitivity to platinum-based chemotherapy in breast and ovarian cancer. Epigenetics 2012; 7:1225-9; PMID:23069641; https://doi.org/ 10.4161/epi.22561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bunting SF, Callén E, Wong N, Chen H-T, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al.. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 2010; 141:243-54; PMID:20362325; https://doi.org/ 10.1016/j.cell.2010.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, et al.. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol 2010; 17:688-95; PMID:20453858; https://doi.org/ 10.1038/nsmb.1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stecklein SR, Sharma P. Tumor homologous recombination deficiency assays: Another step closer to clinical application? Breast Cancer Res 2014; 16:409; PMID:25928813; https://doi.org/ 10.1186/s13058-014-0409-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vollebergh MA, Lips EH, Nederlof PM, Wessels LFA, Schmidt MK, Van Beers EH, Cornelissen S, Holtkamp M, Froklage FE, de Vries EG, et al.. An aCGH classifier derived from BRCA1-mutated breast cancer and benefit of high-dose platinum-based chemotherapy in HER2-negative breast cancer patients. Ann Oncol 2010; 22(7):1561-70; PMID:21135055; https://doi.org/ 10.1093/annonc/mdq624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vollebergh MA, Lips EH, Nederlof PM, Wessels LF, Wesseling J, Vd Vijver MJ, de Vries EG, van Tinteren H, Jonkers J, Hauptmann M, et al.. Genomic patterns resembling BRCA1-and BRCA2-mutated breast cancers predict benefit of intensified carboplatin-based chemotherapy. Breast Cancer Res 2014; 16:R47; PMID:24887359; https://doi.org/ 10.1186/bcr3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Loo P, Nordgard SH, Lingjærde OC, Russnes HG, Rye IH, Sun W, Weigman VJ, Marynen P, Zetterberg A, Naume B, et al.. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A 2010; 107:16910-5; PMID:20837533; https://doi.org/ 10.1073/pnas.1009843107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Birkbak NJ, Wang ZC, Kim J-Y, Eklund AC, Li Q, Tian R, Bowman-Colin C, Li Y, Greene-Colozzi A, Iglehart JD, et al.. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov 2012; 2:366-75; PMID:22576213; https://doi.org/ 10.1158/2159-8290.CD-11-0206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Popova T, Manié E, Rieunier G, Caux-Moncoutier V, Tirapo C, Dubois T, Delattre O, Sigal-Zafrani B, Bollet M, Longy M, et al.. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res 2012; 72:5454-62; PMID:22933060; https://doi.org/ 10.1158/0008-5472.CAN-12-1470 [DOI] [PubMed] [Google Scholar]
- 43.Watkins JA, Irshad S, Grigoriadis A, Tutt AN. Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Res 2014; 16:211; PMID:25093514; https://doi.org/ 10.1186/bcr3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schouten PC, Linn SC. Challenges in the use of DNA repair deficiency as a biomarker in breast cancer. J Clin Oncol 2015; 33(17):1867-9; PMID:25918281; https://doi.org/ 10.1200/JCO.2014.60.5501 [DOI] [PubMed] [Google Scholar]
- 45.Richardson AL, Silver DP, Szallasi Z, Birkbak NJ, Wang ZC, Iglehart JD, et al.. Abstract P3-06-11: Homologous recombination deficiency (HRD) assay predicts response to cisplatin neoadjuvant chemotherapy in patients with triple negative breast cancer. Cancer Res 2015; 75:P3-06-11; https://doi.org/ 10.1158/1538-7445.SABCS14-P3-06-11 [DOI] [Google Scholar]
- 46.Telli ML, Timms K, Reid JE, Neff C, Abkevich V, Gutin A, et al.. Combined Homologous Recombination Deficiency (HRD) scores and response to neoadjuvant platinum-based chemotherapy in triple-negative and/or BRCA1/2 mutation-associated breast cancer. J Clin Oncol (Meeting Abstracts) 2015; 33:1018; https://doi.org/ 10.1200/jco.2015.33.15_suppl.1018 [DOI] [Google Scholar]
- 47.Kaklamani VG, Jeruss JS, Hughes E, Siziopikou K, Timms KM, Gutin A, et al.. Phase II neoadjuvant clinical trial of carboplatin and eribulin in women with triple negative early-stage breast cancer (NCT01372579). Breast Cancer Research and Treatment 2015; 151:629-38: PMID:26006067; https://doi.org/ 10.1007/s10549-015-3435-y [DOI] [PubMed] [Google Scholar]
- 48.Andrew Tutt PE, Kilburn L, Gilett C, Pinder S, Abraham J, Barrett S, Barrett-Lee P, Chan S, Cheang M, Dowsett M, et al.. Catherine Harper-Wynne. TNT: A randomized phase III trail of carboplatin (C) compared with docetaxel (D) for patients with metastatic or recurrent locally advanced triple negative or BRCA1/2 breast cancer (CRUK/07/012). 2014 San Antonio Breast Cancer Symposium. San Antonio: American Association for Cancer Research, 2014; https://doi.org/ 10.1158/1538-7445.SABCS14-S3-01 [DOI] [Google Scholar]
- 49.Timms K, Chagpar AB, Wali V, Bossuyt V, Reid JE, Gutin A, et al.. Reproducibility of homologous recombination deficiency (HRD) scores in biopsies of triple negative breast cancer (TNBC) tumors. ASCO Meeting Abstracts 2015; 33:1091; https://doi.org/ 10.1200/jco.2015.33.15_suppl.1091 [DOI] [Google Scholar]
- 50.Holstege H, Joosse SA, van Oostrom CTM, Nederlof PM, de Vries A, Jonkers J. High incidence of protein-truncating TP53 mutations in BRCA1-related breast cancer. Cancer Res 2009; 69:3625-33; PMID:19336573; https://doi.org/ 10.1158/0008-5472.CAN-08-3426 [DOI] [PubMed] [Google Scholar]
- 51.Carroll DK, Carroll JS, Leong C-O, Cheng F, Brown M, Mills AA, Brugge JS, Ellisen LW. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat Cell Biol 2006; 8:551-61; PMID:16715076; https://doi.org/ 10.1038/ncb1420 [DOI] [PubMed] [Google Scholar]
- 52.Leong C-O, Vidnovic N, DeYoung MP, Sgroi D, Ellisen LW. The p63/p73 network mediates chemosensitivity to cisplatin in a biologically defined subset of primary breast cancers. J Clin Invest 2007; 117:1370; PMID:17446929; https://doi.org/ 10.1172/JCI30866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al.. Molecular portraits of human breast tumours. Nature 2000; 406:747-52; PMID:10963602; https://doi.org/ 10.1038/35021093 [DOI] [PubMed] [Google Scholar]
- 54.Masuda H, Baggerly KA, Wang Y, Zhang Y, Gonzalez-Angulo AM, Meric-Bernstam F, Valero V, Lehmann BD, Pietenpol JA, Hortobagyi GN, et al.. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin Cancer Res 2013; 19:5533-40; PMID:23948975; https://doi.org/ 10.1158/1078-0432.CCR-13-0799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prat A, Lluch A, Albanell J, Barry WT, Fan C, Chacon JI, Parker JS, Calvo L, Plazaola A, Arcusa A, et al.. Predicting response and survival in chemotherapy-treated triple-negative breast cancer. Br J Cancer 2014; PMID:25101563; https://doi.org/ 10.1038/bjc.2014.444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tung NM, Winer EP. Tumor-Infiltrating Lymphocytes and Response to Platinum in Triple-Negative Breast Cancer. J Clin Oncol 2015; 33(9):969-71; PMID:25559817; https://doi.org/ 10.1200/JCO.2014.59.6031 [DOI] [PubMed] [Google Scholar]
- 57.Sharma P, Powers BC, Kimler BF, Ward C, Klemp JR, Connor CS, et al.. Abstract P3-06-16: Thrombocytopenia is associated with pathological complete response to neoadjuvant carboplatin/docetaxel chemotherapy in BRCA wild type triple negative breast cancer. Cancer Res 2015; 75:P3-06-16; https://doi.org/ 10.1158/1538-7445.SABCS14-P3-06-16 [DOI] [Google Scholar]
- 58.West NR, Milne K, Truong PT, Macpherson N, Nelson BH, Watson PH. Tumor-infiltrating lymphocytes predict response to anthracycline-based chemotherapy in estrogen receptor-negative breast cancer. Breast Cancer Res 2011; 13:R126; PMID:22151962; https://doi.org/ 10.1186/bcr3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Loi S, Sirtaine N, Piette F, Salgado R, Viale G, Van Eenoo F, Rouas G, Francis P, Crown JP, Hitre E, et al.. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J Clin Oncol 2013; 31(7):860-7; PMID:23341518; https://doi.org/ 10.1200/JCO.2011.41.0902 [DOI] [PubMed] [Google Scholar]
- 60.Denkert C, Loibl S, Noske A, Roller M, Müller BM, Komor M, Budczies J, Darb-Esfahani S, Kronenwett R, Hanusch C, et al.. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol 2010; 28:105-13; PMID:19917869; https://doi.org/ 10.1200/JCO.2009.23.7370 [DOI] [PubMed] [Google Scholar]
- 61.Denkert C, von Minckwitz G, Brase JC, Sinn BV, Gade S, Kronenwett R, Pfitzner BM, Salat C, Loi S, Schmitt WD, et al.. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. J Clin Oncol 2015; 33(9):983-91; PMID:25534375; https://doi.org/ 10.1200/JCO.2014.58.1967 [DOI] [PubMed] [Google Scholar]
- 62.Adams S, Gray RJ, Demaria S, Goldstein L, Perez EA, Shulman LN, Martino S, Wang M, Jones VE, Saphner TJ, et al.. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J Clin Oncol 2014; 32:2959-66; PMID:25071121; https://doi.org/ 10.1200/JCO.2013.55.0491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Mattos-Arruda L, Cortes J, Santarpia L, Vivancos A, Tabernero J, Reis-Filho JS, Seoane J. Circulating tumour cells and cell-free DNA as tools for managing breast cancer. Nat Rev Clin Oncol 2013; 10:377-89; PMID:23712187; https://doi.org/ 10.1038/nrclinonc.2013.80 [DOI] [PubMed] [Google Scholar]
- 64.von Minckwitz G, Schneeweiss A, Loibl S, Salat C, Denkert C, Rezai M, Blohmer JU, Jackisch C, Paepke S, Gerber B, et al.. Neoadjuvant carboplatin in patients with triple-negative and HER2-positive early breast cancer (GeparSixto; GBG 66): A randomised phase 2 trial. Lancet Oncol 2014; 15:747-56; PMID:24794243; https://doi.org/ 10.1016/S1470-2045(14)70160-3 [DOI] [PubMed] [Google Scholar]
- 65.Sikov WM, Berry DA, Perou CM, Singh B, Cirrincione CT, Tolaney SM, Kuzma CS, Pluard TJ, Somlo G, Port ER, et al.. Impact of the addition of carboplatin and/or bevacizumab to neoadjuvant once-per-week paclitaxel followed by dose-dense doxorubicin and cyclophosphamide on pathologic complete response rates in stage II to III triple-negative breast cancer: CALGB 40603 (Alliance). J Clin Oncol 2015; 33:13-21; PMID:25092775; https://doi.org/ 10.1200/JCO.2014.57.0572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Von Minckwitz G, Timms K, Untch M, Elkin EP, Fasching PA, Schneeweiss A, et al.. Prediction of pathological complete response (pCR) by Homologous Recombination Deficiency (HRD) after carboplatin-containing neoadjuvant chemotherapy in patients with TNBC: Results from GeparSixto. J Clin Oncol (Meeting Abstracts) 2015; 33:1004; https://doi.org/ 10.1200/jco.2015.33.15_suppl.1004 [DOI] [Google Scholar]
- 67.Telli ML, Jensen KC, Vinayak S, Kurian AW, Lipson JA, Flaherty PJ, Timms K, Abkevich V, Schackmann EA, Wapnir IL, et al.. Phase II Study of Gemcitabine, Carboplatin, and Iniparib As Neoadjuvant Therapy for Triple-Negative and BRCA1/2 Mutation–Associated Breast Cancer With Assessment of a Tumor-Based Measure of Genomic Instability: PrECOG 0105. J Clin Oncol 2015; 33(17):1895-901; PMID:25847929; https://doi.org/ 10.1200/JCO.2014.57.0085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu X-C, Zhang J, Xu B-H, Cai L, Ragaz J, Wang Z-H, Wang BY, Teng YE, Tong ZS, Pan YY, et al.. Cisplatin plus gemcitabine versus paclitaxel plus gemcitabine as first-line therapy for metastatic triple-negative breast cancer (CBCSG006): A randomised, open-label, multicentre, phase 3 trial. Lancet Oncol 2015; 16:436-46; PMID:25795409; https://doi.org/ 10.1016/S1470-2045(15)70064-1 [DOI] [PubMed] [Google Scholar]
- 69.Tutt A, Ellis P, Kilburn L, Gilett C, Pinder S, Abraham J, et al.. Abstract S3-01: The TNT trial: A randomized phase III trial of carboplatin (C) compared with docetaxel (D) for patients with metastatic or recurrent locally advanced triple negative or BRCA1/2 breast cancer (CRUK/07/012). Cancer Res 2015; 75:S3-01; https://doi.org/ 10.1158/1538-7445.SABCS14-S3-01 [DOI] [Google Scholar]
- 70.Yardley DA, Brufsky A, Coleman RE, Conte PF, Cortes J, Gluck S, et al.. tnAcity: A phase II/III trial of weekly nab-paclitaxel (nab-P) plus gemcitabine (gem) or carboplatin (carbo) versus gem/carbo as first-line treatment for metastatic triple-negative breast cancer (mTNBC). J Clin Oncol (Meeting Abstracts) 2014; 32:TPS1146; https://doi.org/ 10.1200/jco.2014.32.15_suppl.tps1146 [DOI] [Google Scholar]
- 71.Gluz O, Nitz U, Christgen M, Grischke E-M, Forstbauer H, Braun MW, et al.. Efficacy of 12 weeks neoadjuvant nab-paclitaxel combined with carboplatinum vs. gemcitabine in triple-negative breast cancer: WSG-ADAPT TN randomized phase II trial. J Clin Oncol (Meeting Abstracts) 2015; 33:1032; https://doi.org/ 10.1200/jco.2015.33.15_suppl.1032 [DOI] [Google Scholar]
- 72.Jiang P, Du W, Yang X. p53 and regulation of tumor metabolism. J Carcinog 2013; 12:21; PMID:24379735; https://doi.org/ 10.4103/1477-3163.122760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zangen R, Ratovitski E, Sidransky D. DeltaNp63alpha levels correlate with clinical tumor response to cisplatin. Cell Cycle 2005; 4:1313-5; PMID:16123597; https://doi.org/ 10.4161/cc.4.10.2066 [DOI] [PubMed] [Google Scholar]
- 74.Sohn I, Jung WY, Sung CO. Somatic hypermutation and outcomes of platinum based chemotherapy in patients with high grade serous ovarian cancer. Gynecol Oncol 2012; 126:103-8; PMID:22484402; https://doi.org/ 10.1016/j.ygyno.2012.03.050 [DOI] [PubMed] [Google Scholar]
- 75.Lee J-M, Gordon N, Trepel JB, Lee M-J, Yu M, Kohn EC. Development of a multiparameter flow cytometric assay as a potential biomarker for homologous recombination deficiency in women with high-grade serous ovarian cancer. J Transl Med 2015; 13:239; PMID:26198537; https://doi.org/ 10.1186/s12967-015-0604-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fan Y, Xu BH, Yuan P, Ma F, Wang JY, Ding XY, Zhang P, Li Q, Cai RG. Docetaxel–cisplatin might be superior to docetaxel–capecitabine in the first-line treatment of metastatic triple-negative breast cancer. Ann Oncol 2012; 24(5):1219-25; PMID:23223332; https://doi.org/ 10.1093/annonc/mds603 [DOI] [PubMed] [Google Scholar]
- 77.Carey LA, Rugo HS, Marcom PK, Mayer EL, Esteva FJ, Ma CX, Liu MC, Storniolo AM, Rimawi MF, Forero-Torres A, et al.. TBCRC 001: Randomized phase II study of cetuximab in combination with carboplatin in stage IV triple-negative breast cancer. J Clin Oncol 2012; 30:2615-23; PMID:22665533; https://doi.org/ 10.1200/JCO.2010.34.5579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Villarreal-Garza C, Khalaf D, Bouganim N, Clemons M, Peña-Curiel O, Baez-Revueltas B, Kiss A, Kassam F, Enright K, Verma S, et al.. Platinum-based chemotherapy in triple-negative advanced breast cancer. Breast Cancer Res Treat 2014; 146:567-72; PMID:25001611; https://doi.org/ 10.1007/s10549-014-3033-4 [DOI] [PubMed] [Google Scholar]
- 79.Gluz O, Nitz U, Christgen M, Grischke E-M, Forstbauer H, Braun MW, et al.. Efficacy of 12 weeks neoadjuvant nab-paclitaxel combined with carboplatinum vs. gemcitabine in triple-negative breast cancer: WSG-ADAPT TN randomized phase II trial. ASCO Meeting Abstracts 2015; 33:1032; https://doi.org/ 10.1200/jco.2015.33.15_suppl.1032 [DOI] [Google Scholar]


