Abstract
Transcription activator-like effector nuclease (TALEN) is a programmable genome editing tool with wide applications. Since TALENs perform cleavage of DNA as heterodimers, a pair of TALENs must be synthesized for each target genome locus. Conventionally, TALEN pairs are either expressed on separate vectors or synthesized separately and then subcloned to the same vector. Neither approach allows high-throughput construction of TALEN libraries for large-scale applications. Here we present a single-step assembly scheme to synthesize and express a pair of TALENs in a single-transcript format with the help of a P2A self-cleavage sequence. Furthermore, we developed a fully automated platform to custom manufacture TALENs in a versatile biological foundry. 400 pairs of TALENs can be synthesized with over 96.2% success rate at a material cost of $2.1/pair. This platform opens the door to TALEN-based genome-wide studies.
Keywords: TALEN, DNA assembly, genome engineering, genetic screening, biofoundry, automation
Graphical Abstract
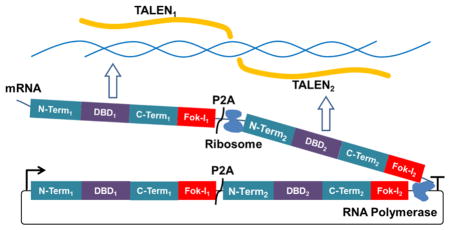
Transcription activator-like effector nuclease (TALEN) is a highly efficient and programmable genome editing tool, which has been applied in a wide range of organisms.1 A TALEN consists of a FokI DNA cleavage domain and a DNA binding domain (DBD) that has tandem repeats of a 33–35 amino acids (aa) motif. The 12th and 13th aa residue within each repeat is known as repeat-variable diresidue (RVD), and it determines the DNA binding specificity of the repeat. By assembling repeats with specific RVDs in order, a TAL effector DBD can bind to a specific DNA sequence.2 Because the FokI cleavage domain functions as a dimer, TALENs are typically used in tail-to-tail heterodimeric pairs to create double stranded breaks for genome editing.3 Such heterodimeric design generates high editing efficiency and improves specificity, but it also presents challenges in TALEN synthesis as well as usage.
A number of methods have been developed to synthesize TALEN expression DNA vectors.4 Taking advantage of an optimized set of 4-bp junctions as well as a preassembled direpeat part library, we developed a one-step assembly scheme based on the Golden Gate method.5 Custom TALEN vectors can be constructed in 24 h at 96% success rate and a material cost of $5. These methods, however, can only assemble vectors harboring a single TALE-FokI monomer. Since TALEN requires a heterodimer to make a cut, two monomers must be introduced into the host cells either on two separate vectors or a single subcloned vector with both monomers. Either option has significant drawbacks. For example, both of them will require twice as many vectors synthesized as the number of target sequences. When the monomers are on separate vectors, the number of cells transfected or transformed with both monomers can be reduced. More importantly, the dual vector scheme makes it very difficult to perform high-throughput genetic screening. Thanks to fluorescence-activated cell sorting (FACS) and next-generation sequencing, a large number of cells with different genotypes can be screened for phenotypes of interest and sequenced.6 As a precision genome editing tool, TALEN can potentially be used to generate a genomic knockout library. However, because the two monomers of each TALEN pair need to be introduced to the same cell, library transfection or transformation is not possible using a dual vector system. However, current methods to construct a single-vector TALEN require a lengthy and complicated subcloning procedure, which makes the synthesis process difficult to scale up. A high-throughput synthesis method for single-vector TALENs will open up new possibilities.
Building on our previously published “FairyTALE” protocol,5 we sought to assemble a pair of TALEN monomers onto a single vector in a one-step reaction. In a previous work, 2A self cleavage peptide7 was used to cotranscribe a pair of TALENs as one mRNA molecule but translated as separate functional proteins.8 We operationalized this coexpression strategy in a 15-insert one-pot assembly scheme, and assembled single-plasmid TALENs in one step at more than 87.7% fidelity. TALENs synthesized using this single-transcript design had comparable cleavage activity in mammalian cells as those synthesized using a two-plasmid design. We implemented the synthesis on iBioFAB (Illinois Biofoundry for Advanced Biomanufacturing), an integrated and versatile robotic system, to fully automate the synthesis process. 400 pairs of TALENs can be generated on a daily basis at a material cost of $2.1 per pair with minimal human intervention. We envision that genome-wide studies using TALENs can be scaled up to screen hundreds of loci in parallel with such a simplified design and automated synthesis.
RESULTS AND DISCUSSION
Design of a single-transcript TALEN synthesis scheme
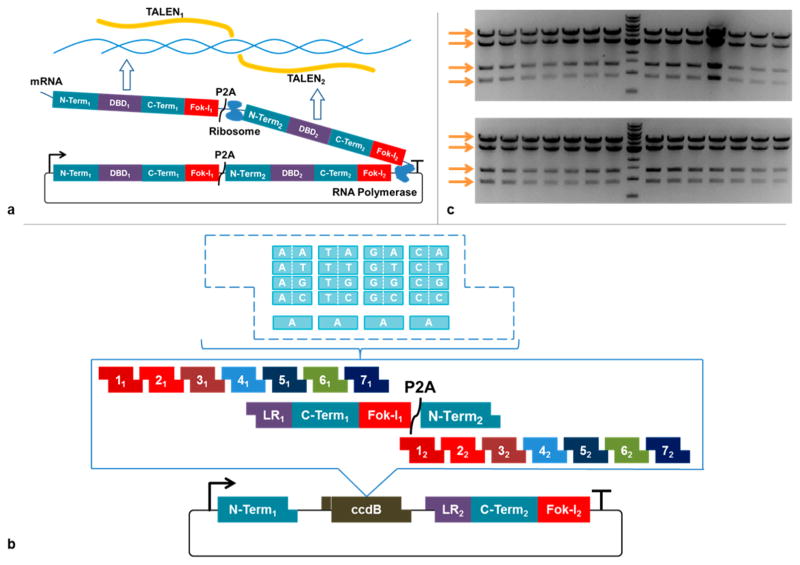
The TALEN architecture used in this work is based upon the AvrXa10 TALE from Xanthomonas oryzae pv oryzae, as previously reported.5 In brief, it utilizes a +207 aa N-terminus extension and a +63 aa C-terminus extension, which negates the 5′-T requirement and allows greater flexibility in target sequence design.9 Attached at the C-terminus is an engineered FokI cleavage domain that showed greater cleavage efficiency in yeast as well as human cells.10 The central repeat domains of the two TALENs are constructed from a library of direpeat substrates; that is, each substrate contains two TALE repeats that recognize two consecutive DNA bases. For this work, we used a library of 441 direpeat substrates, adapted from “FairyTALE”, divided into 17 groups according to their position in the assembly5 (Supplementary Table 1). In addition to the 4̂2 substrates to cover all possible DNA dibases at each assembly position, we included the option to use either NH or NN to code for guanine. To separate the two TALENs on the single plasmid, we employed a polycistronic format utilizing a P2A self-cleaving peptide sequence.7 Both TALENs are coded as a single transcript, but during translation, the P2A peptide will self-cleave the growing polypeptide to give two independent TALENs (Figure 1a).
Figure 1.
Design and preliminary test of single-transcript TALEN synthesis. (a) Overall design. Both TALENs were transcribed as one mRNA, but they were sliced to separate proteins in translation as a P2A sequence was inserted between the open reading frames. (b) Assembly scheme. A library of all possible combinations of single and dual TALE repeats were preassembled with standardized Golden Gate linkers for each position. Thus, each TALEN monomer can target 8 to 15 nucleotides with a mix of single and dual repeats. Repeats for both monomers as well as the LR-C-terminus-Fok-I-P2A-N-terminus fragment are assembled in a single Golden Gate assembly reaction. LR: last repeat. Term.: terminus. (c) Test assembly of a single-transcript TALEN pair. 28 independent clones were picked and digested by PvuI and StuI. All had correct digestion pattern. Arrows indicate the correct digestion pattern.
Using the set of optimized 4-bp junctions in the “fairyTALE” construction scheme, 2 sets of 7 direpeat substrates, with a P2A linker substrate in between, were ligated onto a TALEN receiver vector in a single step via Golden Gate assembly. The N-terminus extension of the first TALEN and the C-terminus extension of the second TALEN were carried by the vector, whereas the C-terminus extension of the first TALEN and the N-terminus extension of the second TALEN were carried by the linker substrate. Since the linker substrate and the receiver carried the last repeat of the two TALENs, 4 TALEN receivers and 4 linker substrates were created (Figure 1b). This construction scheme assembled 15 DNA fragments onto a 5 kb mammalian expression vector to create a single-plasmid TALEN pair that recognized a 30 bp DNA sequence.
One-pot assembly of TALENs
To fulfill the requirements of TALEN library creation, we optimized the reaction condition to maximize the assembly fidelity. For library creation applications, picking individual clones for verification would be an obvious throughput bottleneck, and we would therefore need to achieve high assembly fidelity to allow us to skip clonal isolation without drastically affecting the quality of the library. We picked 28 colonies from a single-transcript TALEN assembly, and assessed them by restriction digest followed by gel electrophoreses. As shown in Figure 1c, all 28 clones gave the correct digestion pattern. We then sequenced 4 of the clones, and they all appeared to be correct. This (28/28) corresponds to a fidelity of at least 87.7% based on binomial probability with 95% confidence.
Single-transcript TALEN functionality in HEK293T and hESC cells
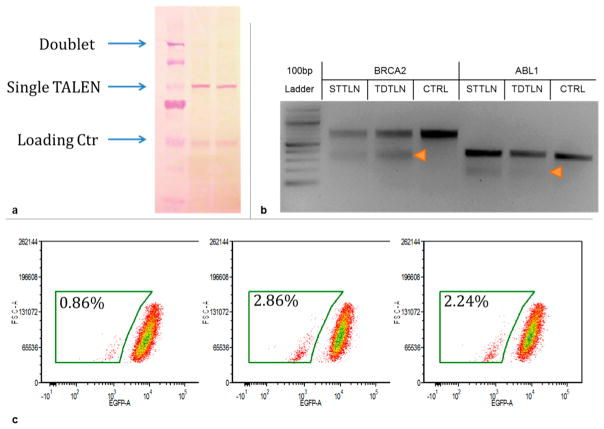
To ensure that P2A cleaves the protein effectively, we performed a Western blot analysis from the cell lysates of HEK293T that had been transfected with single-plasmid TALENs. As shown in Figure 2a, only TALEN monomer was detected and no dimer could be observed, suggesting that the P2A sequence cleaved the protein effectively in HEK293T cells.
Figure 2.
Functional test of single-transcript TALENs. (a) Single-transcript expression of a TALEN pair. Two distinctive TALEN pairs were expressed in HEK293T cells with the single-transcript design. TALEN monomers showed visible bands on Western blot while no band for the size of the uncleaved doublet was detected. (b) Genome editing in HEK293T cells. Single-transcript TALENs (STTLN) were compared against the traditional dual plasmid TALENs (TDTLN) by targeting BRCA2 as well as ABL1 sites in HEK293T cells. T7E1 assay was performed to detect the indel introduced by TALEN cleavage and NHEJ. The STTLN transfected samples showed comparable cleavage efficiency to TDTLN transfected samples. CTRL: sample with no TALEN transfection served as negative control. (c) Disruption of an Oct4 enhancer in H1 hESC. Flow cytometry was used to quantify the GFP expression level in H1-Oct4-GFP cells. The gated population had lower than normal GFP expression. Left: control population without enhancer disruption, middle: enhancer disrupted by traditional 2-plasmid TALEN, right: enhancer disrupted by single-transcript TALEN.
After confirming P2A functionality, we went on to compare the DNA cleavage efficiency of single-transcript TALENs against previously reported traditional two-plasmid TALENs. Two sites, ABL1 and BRCA2, were chosen for this comparison, and the experiments were performed in HEK293T cells. Cleavage efficiency was measured using the T7E1 nuclease assay, which detects indels introduced via NHEJ after TALEN induced double stranded breaks. As shown in Figure 2b, the cleavage efficiency of the two single-transcript TALENs was comparable to that of traditional TALENs. The 1P-TALEN used in this experiment used NH to recognize guanine, whereas the traditional TALENs used NN to recognize guanine. According to our observation and in agreement with that reported by others,11 when used in large number, NH RVD is detrimental to TALE binding. We therefore recommend using NN or a mix of NN and NH RVD when there are more than 4 guanine bases in the recognition site (Supplementary Figure 2).
We further compared the cleavage efficiency of single-transcript TALENs in H1 hESC cells that had an IRES-EGFP marker behind the endogenous Oct4 (H1 Oct4-EGFP, WiCell). We targeted OREG1393087, a site that is known to be an important enhancer for Oct4 expression, with either traditional two-plasmid TALEN or single-transcript TALEN, and monitored the Oct4 expression level in the stem cell population. As shown in Figure 2c, targeting the enhancer region using either TALEN produced an Oct4-reduced stem cell population. The activity produced by the single-transcript TALEN was comparable to that of the traditional TALEN.
Full automation of single-transcript TALEN assembly on a biological foundry
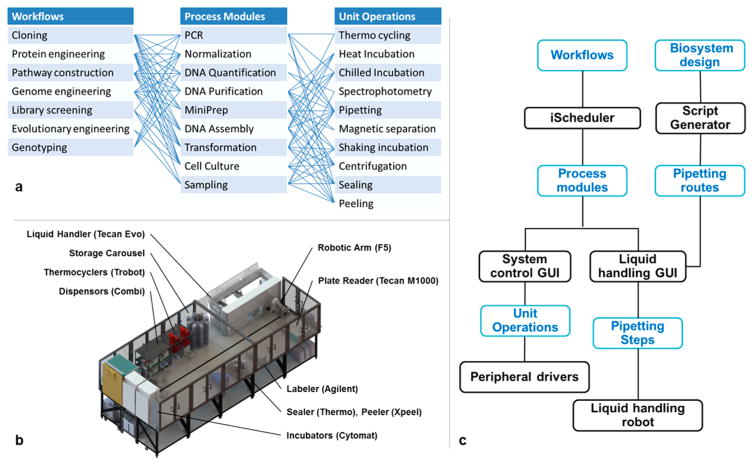
Automation has been used to accelerate biological engineering by either reducing human interventions in individual steps, or completely eliminating human intervention using integrated systems.12 The latter approach has demonstrated the great power of full automation by creating a large number of genetic variants in a short time period. To enable large-scale applications of TALENs, such as genetic screening, we sought to fully automate the synthesis process of TALENs. However, existing integrated platforms are extensively customized for specific tasks and difficult to reconfigure. It would not be efficient and economical to build a deeply customized system dedicated to TALEN synthesis. Instead, we applied a generalized Golden Gate assembly workflow implemented on iBioFAB.
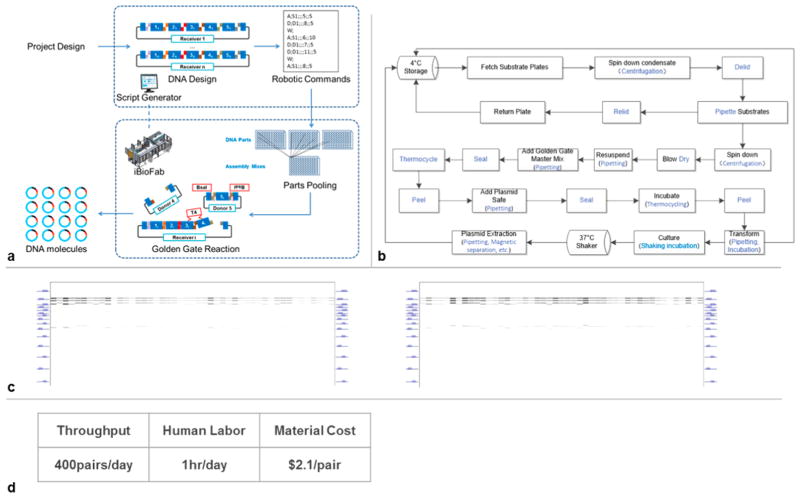
The iBioFAB consisted of component instruments, a central robotic platform, and a modular computational framework (Figure 3). Twenty devices, each in charge of a unit operation, such as pipetting and incubation, were linked by two robotic arms into various process modules, such as DNA assembly and transformation, and then further organized into workflows such as pathway construction and genome engineering (Figure 3a,b). An overall scheduler was developed to orchestrate the unit operations and allow hierarchical programming of the workflows (Figure 3c). The iBioFAB was configured to perform a generalized automatic DNA assembly workflow where various kinds of DNA constructs can be manufactured on-demand with the Golden Gate method.13 A sequence of unit operations was designed to implement this workflow (Figure 4a,b). To streamline the process, we developed Script Generator, a design tool that automatically converts DNA assembly designs to experimental routines of mix-and-matching arbitrary DNA parts. Script Generator then generates robotic commands for iBioFAB to conduct the complex pipetting work. The pipetting routes were also optimized to minimize tip and time consumption. The aspiration steps are combined as much as possible for the same substrate and dispensed into the corresponding destination. Tips are loaded on demand from the storage carousel to the liquid handling station.
Figure 3.
Overview of the iBioFAB system. (a) Breakdown of unit operations. (b) Layout of iBioFAB’s hardware. iBioFAB has two robotic arms. A centralized 6-degree-of-freedom arm on a 5-m track is used to transport labware between instruments. A 3-degree-of-freedom arm moves labware inside the liquid handling station. (c) Control hierarchy of iBioFAB. Process modules are developed in the system control GUI. iScheduler is in charge of workflow level control. Script Generator generates pipetting routes for the liquid handling GUI. Process modules can be quickly recombined to compose different workflows.
Figure 4.
Fully automated synthesis of TALEN libraries. (a) General workflow for the DNA assembly pipeline based on the Golden Gate method. Script Generator converted project design ideas, such as permutations of DNA parts, to assembly designs with appropriate extensions and further robotic commands for pipetting the stock plasmids to DNA mixes. In Golden Gate reactions, Type IIs restriction enzymes such as BsaI generated a set of standard precharacterized 4-bp single strained ends as linkers. The corresponding linkers annealed and were ligated by T4 ligase. (b) Process flow diagram for the build step. Unit operations employed were marked in blue. (c) Verification of single-transcript TALENs synthesized in high throughput. 94 samples were randomly selected from the 192 TALEN pairs synthesized in the full batch test. Each plasmid sample encoding a pair of TALENs was extracted from a polyclonal E. coli cell culture and restriction digested. The fragment sizes were analyzed by capillary electrophoresis. The digestion pattern was simulated. (d) Summary of efficiency, throughput, and cost.
In this work, we adapted this DNA assembly workflow for synthesizing single-transcript TALENs. An extension that automated the DNA assembly design specifically for TALENs was added to Script Generator. Using such a pipeline, the operator only needs to input the target DNA sequence to Script Generator, and iBioFAB would perform the rest of TALEN synthesis with minimal human intervention. It only requires the operator to load reagents and consumables on a daily basis. Any arbitrary number between 1 to 192 TALEN pairs can be synthesized in each batch.
High-throughput synthesis of 192 single-transcript TALENs
To test the high-throughput synthesis pipeline, we fed 192 different human genomic target loci to Script Generator. iBioFAB performed 3648 pipetting steps from 444 different DNA parts, and reagents within 17 h at a material cost of $2.1 per TALEN pair (Supplementary Table 2). By staggering batches, over 400 TALENs can be generated in a single day (Supplementary Figure 3).
To evaluate the success rate of the synthesis, 94 randomly selected constructs were verified by polyclonal restriction digestion. All samples showed the correct digestion pattern (Figure 4c), which corresponds to a success rate of at least 96.2% with 95% confidence based on binomial probability. For activity verification, we randomly selected 22 TALENs for T7E1 assay in HEK293T cells.14 Fifteen of 22 samples showed cleavage activity. Since cleavage activity was known to be sequence dependent, the lack of activity for some sites was not unexpected.15 To eliminate the possibility of misassembly, we sequenced all the constructs that did not show cleavage activity. All sequencing reads were aligned to the intended TALEN designs, indicating that the TALENs were correctly assembled (Supplementary Table 3).
Besides TALEN, CRISPR-Cas9 is another popular technology used in genome editing applications.16 As opposed to using a specific protein to recognize DNA sequences, CRISPR utilizes RNA to perform the recognition through base pairing. Using a nucleic acid for targeting has many advantages, but most importantly, through the use of microarray DNA synthesis, a large nucleic acid library is readily accessible. As such, even though TALEN had a two-year head start over CRISPR, multiple targeting and genetic screening were both first achieved using CRISPR.6,17 However, due to its relatively short recognition sequence, 20 bp, the off-target effect is a significant problem in CRISPR.18 In a genetic screening that targets structural genes, the off-target effect can be compensated by targeting multiple sites within the same gene, so that a high-confidence hit can be identified by looking for the enrichment of a set of sites instead of any single site. However, in the case where the functional DNA element is very small, e.g., a transcriptional enhancer, or a miRNA gene, there is simply not enough length to fit in multiple targeting sites. Furthermore, in the case of an enhancer, the target cut sites are transcription factor binding sites that are typically around 10 bp. Given the limited range for target selection, CRISPR may not be able to find a site that is sufficiently unique in the genome. Furthermore, given the small number of selectable sites for such screens, the level of confidence for any resultant hits will be low. A TALEN library, with a different off-target profile, can be used in conjunction with a CRISPR library to improve the confidence of any potential hits.
In conclusion, we have developed a scheme to synthesize TALEN pairs on a single vector in a one-pot reaction, which has substantially simplified the synthesis of TALENs while achieving an outstanding success rate. An automated process was developed accordingly, and the resulting pipeline makes it possible to create large TALEN libraries at a reasonable cost and in a reasonable time frame.
METHODS
iBioFAB
iBioFAB consists of a F5 robotic arm on a 5-m track (Fanuc, Oshino-mura, Japan), an Evo200 liquid handling robot (Tecan, Mannedorf, Switzerland), two shaking temperature controlled blocks (Thermo Scientific, Waltham, MA), a M1000 microplate reader (Tecan, Mannedorf, Switzerland), a Cytomat 6000 incubator (Thermo Scientific, Waltham, MA), two Cytomat 2C shaking incubators (Thermo Scientific, Waltham, MA), three Multidrop Combi reagent dispensers (Thermo Scientific, Waltham, MA), four Trobot thermocyclers (Biometra, Gottingen, Germany), Vspin plate centrifuge (Agilent, Santa Clara, CA), a storage carousel (Thermo Scientific, Waltham, MA), a delidding station (Thermo Scientific, Waltham, MA), an Alps plate sealer (Thermo Scientific, Waltham, MA), a WASP plate sealer (Thermo Scientific, Waltham, MA), a Xpeel seal pealer (Brooks, Chelmsford, MA), and a label printer (Agilent, USA). The liquid handling robot was equipped with an 8-channel independent pipetter, a robotic manipulation arm, a 96-channel pipetter, six Peltier temperature controlled blocks (Torrey Pine, Carlsbad, CA), two shakers (Q.Instruments, Jena, Germany), a light box, and a camera for colony picking (Scirobotics, Kfar Saba, Israel).
Momentum (Thermo Scientific, Waltham, MA) was used to communicate with the peripheral devices, control the central robotic arm, and program process modules. Process modules defined the unit operations and sample transportation routes between unit operations. Freedom Evoware (Tecan, Mannedorf, Switzerland) was used to control the liquid handling robot and program pipetting modules. Pipetting modules specifically defined the general procedure of pipetting on the liquid handling robot, such as labware fetching from the central robotic arm, DNA part dispensing, reagent dispensing, and temperature controls. iScheduler and ScriptGenerator are programed in Visual Basic. iScheduler executes process modules by sending commands in Extensible Markup Language to Momentum. The ScriptGenerator converted user defined DNA assembly as permutations of parts to source and destination locations based on preloaded parts storage plate layouts. The corresponding pipetting routes were optimized by queueing the destination locations from the same source. Pipetting worklists were compiled accordingly and sent to Freedom Evoware to control aspiration, dispense, as well as tip change actions. Defined amount of each DNA part was aspirated and multidispensed without contacting the liquid in the destination. Tips were reused as much as possible and changed when all destinations for the same source were dispensed. Constraints such as tip volume and maximum number of aspirations with each tip were also imposed in the algorithm.
Plasmids
Based on the RVDs parts used in the previous work,5 a new library of TALEN stock plasmids were developed for the single plasmid design. The group for position 6 was replaced with LR_N-term_FokI_P2A+C-term constructs (Supplementary Figure 1a). Dual and single RVD parts with NN were supplemented into the stock library. The RVD and P2A fragments were inserted to a receiver plasmid (Supplementary Figure 1b) with human CMV promoter as well as last repeat, N terminus, and FokI domain for the second TALEN monomer.
Golden gate assembly and verification
Golden Gate DNA assembly was performed with the methods described in the previous work.5 Competent E. coli HST08 strain (Clontech, Mountain View, CA) was prepared with Mix & Go E. coli Transformation Buffer Set (Zymo Research, Irvine, CA). 2.5 μL of Golden Gate reaction products were first mixed with E. coli competent cells on a Peltier block held at 0 °C and incubated for 30 min. The cell plate was then transferred to a second Peltier block held at 42 °C by the plate manipulation arm. After 1 min heat shock, the cell plate was transferred back to the 0 °C block and chilled for 2 min. The transformants were recovered in LB broth (Becton, Dickinson and Company, Franklin Lakes, NJ) for 1 h. The recovered cell suspensions were either plated on LB agar media with 100 μg/mL of ampicillin or used to inoculate polyclonally LB liquid media supplemented with 200 μg/mL of carbenicillin. Plasmids were extracted from the polyclonal cultures with MagJET Plasmid DNA Kit (Thermo Scientific, Waltham, MA) and restriction digested by EcoRI-HF (New England Biolabs, Ipswich, MA). The digestion products were analyzed by 1% agarose gel in low throughput or Fragment Analyzer (Advanced Analytical Technologies, Ankeny, IA) in high throughput. Selected plasmids were also verified by Sanger sequencing reactions (ACGT, Wheeling, IL) with 4 primers. The binomial probability confidence interval for assembly success rate was calculated with Clopper-Pearson method.19
Mammalian gene knockout and verification
Human embryonic kidney (HEK) cell line HEK293T was transfected with randomly selected TALENs plasmids. HEK293T cells were used as they are easy to cultivate and transfect. Although no cell authentication or mycoplasma contamination tests were performed, we reason that the results of T7E1 assay is relatively insensitive to the cell line background. Cells were maintained in Dulbecco’s modified Eagle’s Medium (DMEM) (Corning Life Sciences, Tewksbury, MA) supplemented with 10% heat inactivated fetal bovine serum (Life Technologies, Carlsbad, CA) at 37 °C and 5% CO2 incubation. One day prior to transfection, 293T cells were seeded into 12-well BioCoat Collagen-I coated plates (Corning Life Sciences, Tewksbury, MA) at a confluency of ~50%. Transfections were performed with FuGENE HD Transfection Reagent (Promega, Madison, WI) according to the manufacturer’s protocols. Briefly, for each well of the 12-well plate, 1 μg of clonally purified TALEN plasmid was first diluted in Opti-MEM (Life Technologies, Carlsbad, CA) to a total volume of 100 μL. After addition of 3μL Fugene HD reagent and incubation at room temperature for 5 min, the mix was added onto the cells. Cells were harvested at 60 h post-transfection. The genomic DNA was extracted with QuickExtract DNA Extraction Solution (Epicenter, Madison, WI).
The cleavage efficiency was evaluated by T7E1 assay.14 DNA amplicons were designed to have a length of 400–1000bp flanking the nominal cleavage site by a custom developed Visual Basic script. It searches the genome sequence within a given range for a pair of primer binding sites to avoid off-targets, long stretches of GC, AT, or any single type of nucleotide. End nucleotides, GC contents, and melting temperatures were optimized. The relevant genome sequences were downloaded by querying UCSC DAS server (https://genome.ucsc.edu/cgi-bin/das/) while off-target check was performed by querying GGGenome server (https://gggenome.dbcls.jp/). The PCR amplification was conducted with Q5 polymerase (New England Biolabs, Ipswich, MA) and annealing temperature touchdown (65–55 °C for 10 cycles, 55 °C for 20 cycles). In the cleavage assay, 200 ng of purified amplicon in 10 μL NEB Buffer 2 was first denatured and renatured (95 °C, 5 min; 95–85 °C at −2 °C/s; 85–25 °C at −0.1 °C/s; hold at 4 °C). 10U of T7 Endonuclease I (New England Biolabs, Ipswich, MA) was added and incubated at 37 °C for 15 min. The reaction was stopped by adding 1 μL of 0.5 M EDTA. The digestion products were analyzed by Fragment Analyzer (Advanced Analytical Technologies, Ankeny, IA).
Oct4 down-regulation assay
TALEN constructs under evaluation were transfected into H1-Oct4-EGFP stem cells (WiCell, Madison, WI) by nucleofection according to manufacturer’s recommendations. After optimization, we settled on the P4 Primary Cell 4D-Nucleofector Kit, and program CA-137 on the 4D-Nucleofector (Lonza, Cologne, Germany). Cells were passaged 1 day after nucleofection, and were harvested on the fourth day after nucleofection. After harvest, the cells were counted and stained using Alexa Fluor 647 conjugated SSEA4 antibody (Life Technologies, Carlsbad, CA) at a concentration of 5 × 105 cells in 50 μL PBS with 2% BSA and 2.5 μL SSEA4 antibody. The cells were stained in the dark at room temperature for 30 min, and washed 3 times in PBS before flow cytometry analysis. During analysis, the stem cell population was first selected by gating for the SSEA4 positive cells. Within this population, we then look at the spread of EGFP expression, and gate for the EGFP-reduced population.
Supplementary Material
Acknowledgments
This work was supported by the Roy J. Carver Charitable Trust, Carl R. Woese Institute for Genomic Biology at the University of Illinois at Urbana–Champaign (UIUC), Defense Advanced Research Projects Agency, and National Institutes of Health (1U54DK107965). R.C. acknowledges fellowship support from 3M Corporation. T.S. acknowledges postdoctoral fellowship support from Carl R. Woese Institute for Genomic Biology (UIUC). The authors acknowledge Vasundhara Vigh for her help in iBioFAB maintenance and Han Xiao, Xiong Xiong, and Zehua Bao for preparing substrates for TALEN synthesis. We also thank Mark Band and Mary Majewski at Roy J. Carver Biotechnology Center (UIUC) for help with DNA capillary electrophoresis. We give special thanks to Christopher V. Rao for insightful discussions and suggestions.
ABBREVIATIONS
- TALEN
transcription activator-like effector nucleases
- CRISPR
clustered regulatory interspaced short palindromic repeat
- iBioFAB
Illinois Biological Foundry for Advanced Bioengineering
Footnotes
Author Contributions
R.C. and J.L. contributed equally. R.C., J.L., and H.Z. designed and conceived the study. R.C. developed the iBioFAB automation system and automated the DNA assembly workflow. J.L. developed the P2A-based TALEN synthesis scheme, performed initial tests, and selected genome editing targets. I.T. performed TALEN functional tests in human cells. L.J. helped with preparation of TALEN plasmids and verification of genome editing efficiency. H.Z. supervised the research. R.C., J.L., and H.Z. wrote the manuscript with important input from T.S. and I.T.
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acssynbio.6b00293.
Additional figures and tables as described in the text (PDF)
References
- 1.Sun N, Abil Z, Zhao H. Recent advances in targeted genome engineering in mammalian systems. Biotechnol J. 2012;7(9):1074–87. doi: 10.1002/biot.201200038. [DOI] [PubMed] [Google Scholar]
- 2.Boch J. TALEs of genome targeting. Nat Biotechnol. 2011;29(2):135–6. doi: 10.1038/nbt.1767. [DOI] [PubMed] [Google Scholar]
- 3.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–8. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 4.(a) Briggs AW, Rios X, Chari R, Yang L, Zhang F, Mali P, Church GM. Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res. 2012;40(15):e117. doi: 10.1093/nar/gks624. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30(5):460–5. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Ding Q, Lee YK, Schaefer EA, Peters DT, Veres A, Kim K, Kuperwasser N, Motola DL, Meissner TB, Hendriks WT, Trevisan M, Gupta RM, Moisan A, Banks E, Friesen M, Schinzel RT, Xia F, Tang A, Xia Y, Figueroa E, Wann A, Ahfeldt T, Daheron L, Zhang F, Rubin LL, Peng LF, Chung RT, Musunuru K, Cowan CA TALEN, A. genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013;12(2):238–51. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Kim Y, Kweon J, Kim A, Chon JK, Yoo JY, Kim HJ, Kim S, Lee C, Jeong E, Chung E, Kim D, Lee MS, Go EM, Song HJ, Kim H, Cho N, Bang D, Kim JS. A library of TAL effector nucleases spanning the human genome. Nat Biotechnol. 2013;31(3):251–8. doi: 10.1038/nbt.2517. [DOI] [PubMed] [Google Scholar]; (e) Schmid-Burgk JL, Schmidt T, Kaiser V, Honing K, Hornung V. A ligation-independent cloning technique for high-throughput assembly of transcription activator-like effector genes. Nat Biotechnol. 2013;31(1):76–81. doi: 10.1038/nbt.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang J, Chao R, Abil Z, Bao ZH, Zhao HM. FairyTALE: A high-throughput TAL effector synthesis platform. ACS Synth Biol. 2014;3(2):67–73. doi: 10.1021/sb400109p. [DOI] [PubMed] [Google Scholar]
- 6.(a) Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–7. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic Screens in Human Cells Using the CRISPR-Cas9 System. Science. 2014;343(6166):80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Ryan MD, Gani D, Flint M, Monaghan S, Donnelly MLL. The cleavage activities of aphthovirus and cardiovirus 2A proteins. J Gen Virol. 1997;78:13–21. doi: 10.1099/0022-1317-78-1-13. [DOI] [PubMed] [Google Scholar]; (b) Radcliffe PA, Mitrophanous KA. Multiple gene products from a single vector: ‘self-cleaving’ 2A peptides. Gene Ther. 2004;11(23):1673–1674. [Google Scholar]; (c) Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY, Kim MK, Shin BA, Choi SY. High Cleavage Efficiency of a 2A Peptide Derived from Porcine Teschovirus-1 in Human Cell Lines, Zebrafish and Mice. PLoS One. 2011;6:4. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Cermak T, Baltes NJ, Cegan R, Zhang Y, Voytas DF. High-frequency, precise modification of the tomato genome. Genome Biol. 2015;16:232. doi: 10.1186/s13059-015-0796-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Mariano A, Xu L, Han R. Highly efficient genome editing via 2A-coupled co-expression of two TALEN monomers. BMC Res Notes. 2014;7:628. doi: 10.1186/1756-0500-7-628. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Xu L, Zhao P, Mariano A, Han R. Targeted Myostatin Gene Editing in Multiple Mammalian Species Directed by a Single Pair of TALE Nucleases. Mol Ther–Nucleic Acids. 2013;2:e112. doi: 10.1038/mtna.2013.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun N, Liang J, Abil Z, Zhao HM. Optimized TAL effector nucleases (TALENs) for use in treatment of sickle cell disease. Mol BioSyst. 2012;8(4):1255–1263. doi: 10.1039/c2mb05461b. [DOI] [PubMed] [Google Scholar]
- 10.Sun N, Bao Z, Xiong X, Zhao H. SunnyTALEN: a second-generation TALEN system for human genome editing. Biotechnol Bioeng. 2014;111(4):683–91. doi: 10.1002/bit.25154. [DOI] [PubMed] [Google Scholar]
- 11.Streubel J, Blucher C, Landgraf A, Boch J. TAL effector RVD specificities and efficiencies. Nat Biotechnol. 2012;30(7):593–5. doi: 10.1038/nbt.2304. [DOI] [PubMed] [Google Scholar]
- 12.(a) Esvelt KM, Carlson JC, Liu DR. A system for the continuous directed evolution of biomolecules. Nature. 2011;472(7344):499–503. doi: 10.1038/nature09929. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460(7257):894–8. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engler C, Kandzia R, Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008;3(11):e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mashal RD, Koontz J, Sklar J. Detection of mutations by cleavage of DNA heteroduplexes with bacteriophage resolvases. Nat Genet. 1995;9(2):177–83. doi: 10.1038/ng0295-177. [DOI] [PubMed] [Google Scholar]
- 15.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39(12):e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–55. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, Hsu PD, Wu XB, Jiang WY, Marraffini LA, Zhang F. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–6. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clopper CJ, Pearson ES. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika. 1934;26:404–413. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.