Abstract
Background
Gleason scoring represents the standard for diagnosis of prostate cancer (PCa) and assessment of prognosis following radical prostatectomy (RP), but it does not account for patterns in neighboring normal-appearing benign fields that may be predictive of disease recurrence.
Objective
To investigate (1) whether computer-extracted image features within tumor-adjacent benign regions on digital pathology images could predict recurrence in PCa patients after surgery and (2) whether a tumor plus adjacent benign signature (TABS) could better predict recurrence compared with Gleason score or features from benign or cancerous regions alone.
Design, setting, and participants
We studied 140 tissue microarray cores (0.6 mm each) from 70 PCa patients following surgery between 2000 and 2004 with up to 14 yr of follow-up. Overall, 22 patients experienced recurrence (biochemical [prostate-specific antigen], local, or distant recurrence and cancer death) and 48 did not. Intervention: RP was performed in all patients.
Outcome measurements and statistical analysis
The top 10 features identified as most predictive of recurrence within both the benign and cancerous regions were combined into a 10-feature signature (TABS). Computer-extracted nuclear shape and architectural features from cancerous regions, adjacent benign fields, and TABS were evaluated via random forest classification accuracy and Kaplan-Meier survival analysis.
Results and limitations
Tumor-adjacent benign field features were predictive of recurrence (area under the receiver operating characteristic curve [AUC]: 0.72). Tumor-field nuclear shape descriptors and benign-field local nuclear arrangement were the predominant features found for TABS (AUC: 0.77). Combining TABS with Gleason sum further improved identification of recurrence (AUC: 0.81). All experiments were performed using threefold cross-validation without independent test set validation.
Conclusions
Computer-extracted nuclear features within cancerous and benign regions predict recurrence following RP. Furthermore, TABS was shown to provide added value to common predictors including Gleason sum and Kattan and Stephenson nomograms.
Patient summary
Future studies may benefit from evaluation of benign regions proximal to the tumor on surgically excised prostate cancer tissue for assessing risk of disease recurrence.
Keywords: Prostate cancer, Prognosis, Quantitative histomorphometry, Radical prostatectomy, Digital pathology, Field effect, Recurrence, Markers
1. Introduction
Each year, nearly 75 000 radical prostatectomies (RPs) are performed in the United States for the purpose of curing prostate cancer (PCa) [1]. Despite the effectiveness of RP, 20–40% of men will experience recurrence following surgery, manifested as biochemical, local, or distant cancer recurrence or cancer death [2–4]. There is an urgent need for improved methods of accurately predicting which men with PCa will have recurrence following surgery. Early identification of PCa patients at elevated risk for recurrence after surgery and prior to prostate-specific antigen (PSA) rising could help improve treatment management and monitoring.
Although high Gleason scores (ie, PCa with a Gleason score of 8–10) are typically associated with more aggressive disease and thus a higher risk of recurrence, its diagnosis is made solely based on the visual appearance of the morphology within the cancerous foci. There has been evidence to suggest that the microenvironment surrounding the prostate tumor may play a role in cancer progression [5,6], a phenomena known as the field effect. Epigenetic changes within the benign regions surrounding the tumor have been shown to be capable of initiating PCa [5]. This begs the question of whether morphometric attributes within tumor-adjacent benign regions can provide additional complementary features to Gleason scoring for better prediction of disease risk and recurrence.
There has been significant recent interest in investigating histomorphometric features of benign regions adjacent to the tumor and evaluating the association of these features with disease aggressiveness and outcome. Veltri et al [7] and Gann et al [6] showed that there are quantifiable morphometric attributes within tumor-adjacent benign regions that can provide additional information related to disease outcome. These studies, however, have been limited to the correlation of outcome and nuclear shape parameters alone. Although there has been substantial interest in computer-based evaluation of nuclear architecture in the context of developing better methods for automated Gleason scoring, there has been little to no work looking at the association with disease aggressiveness and outcomes.
Computer-based analysis of digital pathology images has allowed for extraction of image-based features from histologic tissue that have been shown to be able to predict biochemical recurrence following RP.
Graph-based algorithms allow for capturing the spatial architecture of nodes via connected edges. There has been recent interest in developing quantitative histomorphometry algorithms that use these graph-based approaches for assessing nuclear architecture. Quantitative measurements that can be extracted from these nuclear graphs include Voronoi and Delaunay tessellation graphs [8] and local cell cluster graphs [9]. These features summarize distance statistics between nuclei and have been shown to be useful in discriminating between different Gleason grades of PCa histopathology [10]. However, these features thus far have been evaluated within the tumor epithelium alone, whereas features from the tumor-adjacent benign regions have never been explicitly interrogated in the context of association with disease recurrence and outcome.
The primary objectives of this study were to identify (1) whether histomorphometric features relating to the nuclear architecture within the tumor-adjacent benign regions can predict recurrence following RP and (2) whether a combination of nuclear shape and architectural features from within the cancer and from adjacent benign regions can better predict disease progression compared with Gleason score.
In this work, we developed a tumor plus adjacent benign signature (TABS) for differentiating PCa patients who will develop recurrence from those who will not. The reported results represent a preliminary study in lieu of independent validation of TABS.
2. Materials and methods
2.1. Study population
Two tissue microarrays (TMAs; TMA681 and TMA682) were recruited for this study, each prepared using a Beecher MT1 manual arrayer (Beecher Instruments, Silver Spring, MD, USA) under the supervision of the Prostate Cancer Biorepository Network. The patients included in this TMA were selected independently of this work and based on an Early Detection Research Network grant to recruit a Gleason grade–stratified PCa cohort to study quantitative histomorphometry and molecular biomarkers.
Each TMA was composed of hematoxylin and eosin (H&E)–stained 0.6-mm samples of tumor, adjacent benign, and control regions. The formalin-fixed, paraffin-embedded RP PCa tissues and normal (benign) cancer-adjacent controls to be included in the two TMAs were selected and reviewed by a pathologist at Johns Hopkins University School of Medicine (J.I.E.). H&E-stained slides from all selected cases were reviewed by the pathologist: The tumor-adjacent normal-appearing regions along with staged and/or graded index tumor areas were identified and marked on the H&E slide for each case. Benign region selection was confined to that of normal-appearing prostatic glands and did not include any atypical benign pathology such as atrophy, basal cell hyperplasia, or high-grade prostate epithelial neoplasia.
The two TMAs included 80 unique PCa patients. Among the 80 patients, 5 from TMA681 and 5 from TMA682 were removed from this study due to lack of follow-up data following RP. This resulted in 70 patients remaining for this study identifying relationships between recurrence and features extracted from tumor-adjacent benign tissue.
All patients from this cohort underwent RP between 2000 and 2004 and were followed for PSA updates for up to 14 yr. Average time to biochemical recurrence after surgery was 6.6 yr. Moreover, 22 patients displayed recurrence (in the form of biochemical, local, or distant recurrence or cancer death), whereas 48 did not. Demographic information for the study cohort is summarized in Table 1.
Table1.
Summary of clinicopathological features of the studied prostate cancer cohort at the time of diagnosis, patient demographics of the studied prostate cancer cohort, and cases with recurrence and no recurrence within the studied prostate cancer cohort
| Clinicopathological cohort, n (%) | |||
|---|---|---|---|
| Recurrence | Nonrecurrence | Total | |
| No. of patients | 22 (31.4) | 48 (68.6) | 70 |
| Gleason score | |||
| 6 | 1 (11.1) | 8 (88.9) | 9 (12.9) |
| 7 (3 + 4) | 3 (17.7) | 14 (82.3) | 17 (24.3) |
| 7 (4 + 3) | 3 (16.7) | 15 (83.3) | 18 (25.7) |
| 8 | 8 (47.1) | 9 (52.9) | 17 (24.3) |
| 9 | 7 (77.8) | 2 (22.2) | 9 (12.9) |
| Primary tumor stage | |||
| pT2 | 2 (6.9) | 27 (93.1) | 29 (41.4) |
| pT3a | 15 (48.4) | 16 (51.6) | 31 (44.3) |
| pT3b | 5 (62.5) | 3 (37.5) | 8 (11.4) |
| Unknown | 0 (0) | 2 (100) | 2 (2.9) |
| Lymph node | |||
| Positive | 0 (0) | 1 (100) | 1 (1.4) |
| Negative | 22 (31.9) | 47 (68.1) | 69 (98.6) |
| Surgical margin | |||
| Positive | 20 (51.3) | 19 (48.7) | 39 (55.7) |
| Negative | 2 (6.4) | 29 (93.5) | 31 (44.3) |
| Presurgical PSA, ng/mL | |||
| 0–10 | 12 (25.0) | 36 (75.0) | 48 (68.6) |
| 10–20 | 6 (37.5) | 10 (62.5) | 16 (22.9) |
| 20–30 | 2 (66.7) | 1 (33.3) | 3 (4.3) |
| 30–40 | 2 (66.7) | 1 (33.3) | 3 (4.3) |
| Patient demographics, mean ± SD | |||
| Variable | Recurrence | Nonrecurrence | |
| Age, yr | 59.04 ± 6.57 | 58.82 ± 5.80 | |
| Gland weight, g | 51.94 ± 14.90 | 56.19 ± 21.38 | |
| PSA, ng/ml | 8.29 ± 5.29 | 13.17 ± 10.15 | |
| PSAD, ng/ml/g | 0.17 ± 0.11 | 0.25 ± 0.20 | |
| Time to event, yr | 5.06 ± 4.50 | 3.82 ± 2.36 | |
| Outcomes for recurrence and no recurrence, n | |||
| Recurrence | 22 | ||
| Died from prostate cancer | 2 | ||
| Died from another cause with recurrence | 1 | ||
| Distant metastasis | 1 | ||
| Increase in PSA (>0.2 ng/ml) | 14 | ||
| Local recurrence | 1 | ||
| Local recurrence and distant metastasis | 3 | ||
| No recurrence | 48 | ||
| Died from noncancer cause | 1 | ||
| No recurrence | 47 | ||
PSA = prostate-specific antigen; PSAD = prostate-specific antigen density; SD = standard deviation.
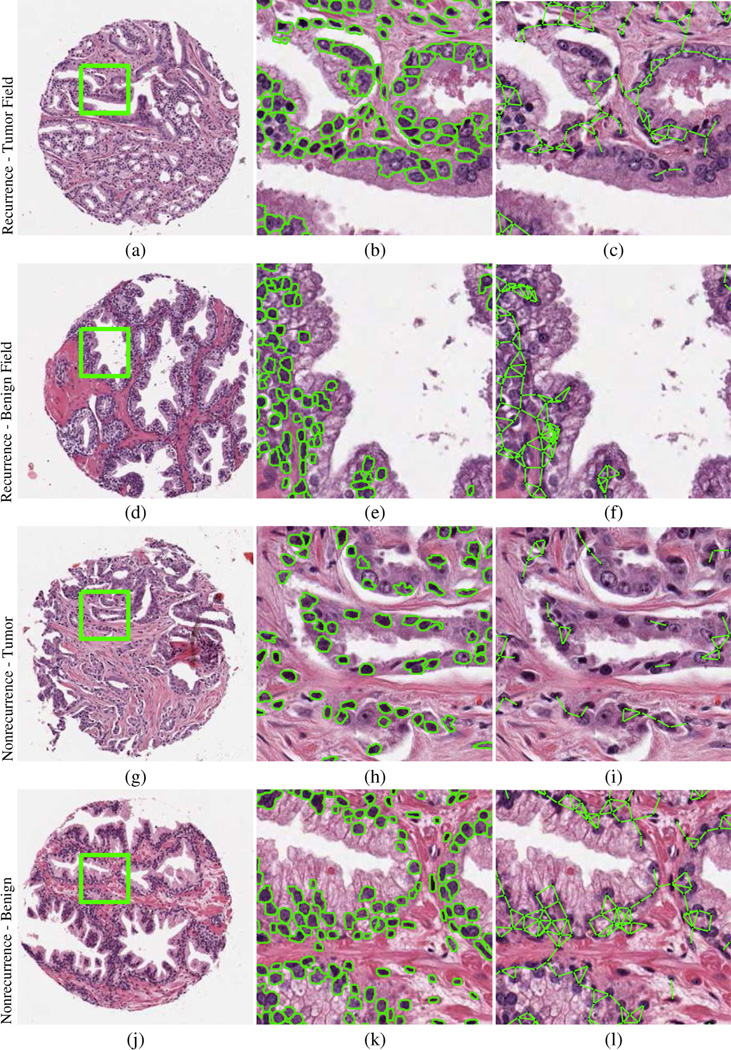
The TMA samples were scanned at ×20 magnification with a resolution of 0.5 µm per pixel using an Aperio whole-slide scanner (Leica, Wetzlar, Germany). The scanning resulted in a 1670 × 1670-pixel RGB color image for each TMA core, similar to the one shown in Figure 1a. For this study, a single randomly selected core was chosen from each of the tumor and benign sets to represent each patient.
Fig. 1.
Prostate tissue microarray cores (a, d, g, j) corresponding to patients who experienced (a–f) recurrence and (g–l) no recurrence. Automated segmentation defines nuclear boundaries and locations from tumor and benign field tissue cores, characterizing (b, e, h, k) nuclear shape and (c, f, i, l) nuclear subgraphs describing local nuclear architecture.
In total, we identified 140 fields of view from the surgically excised histopathology specimens of 70 PCa patients, corresponding to regions extracted from within tumor and benign regions on the surgical specimens. Characteristics of nuclear shape and global and local nuclear arrangement via Voronoi and Delaunay graphs, nuclear subgraph, and nuclear texture were extracted to quantify the tumor and benign field images.
2.2. Nuclear detection and segmentation
Prior to extracting nuclear shape features, the individual nuclei had to be automatically identified and segmented. Toward this end, we used a shape-based active-contour segmentation scheme [11,12] to detect and segment nuclei. Color deconvolution was used for preprocessing the RGB image and isolating the nuclear stain. A watershed segmentation method was used to initialize nuclear boundaries, and the centroids of these regions were used as seed points for the active contour. Both luminal and basal cells were identified in the segmentation process and considered equally.
2.3. Feature extraction
For our analysis, we extracted 199 features (Table 2) to quantify the cancerous and benign fields within the prostate pathology slide images. These features were grouped into six categories or feature types: nuclear shape, Voronoi diagram, Delaunay triangulation, nuclear density, nuclear subgraphs, and nuclear texture.
Table 2.
Summary of 199 quantitative histomorphometric features extracted from both the tumor fields and adjacent benign fields
| Feature type | No. | Description |
|---|---|---|
| Nuclear shape | 100 | Area ratio, distance ratio, SD of distance, variance of distance, distance ratio, perimeter ratio, smoothness, invariant moment 1–7, fractal dimension, Fourier descriptor 1–10: mean, SD, median, minimum/maximum of each |
| Voronoi diagram | 12 | Polygon area, perimeter, chord length: mean, SD, minimum/maximum ratio, disorder |
| Delaunay triangulation | 8 | Triangle side length, area: mean, SD, minimum/maximum ratio, disorder |
| Nuclear density | 27 | Density of nuclear centroids, distance to nearest nuclear centroid |
| Nuclear subgraphs | 26 | Eccentricity; clustering coefficients C, D, and E; largest connected component: mean, SD |
| Nuclear texture | 26 | Entropy, energy, intensity contrast: mean, SD |
SD = standard deviation.
Using the nuclear boundaries obtained via the automated nuclear segmentation (Fig. 1b, 1e, 1h, and 1k), a set of 100 shape features [13] were calculated from statistics of features such as nuclear area and smoothness. From within the segmented nuclei, 26 nuclear texture features were computed, as described by Doyle et al [13], to measure subvisual image intensity co-occurrence information. Graph tesselations of nuclei were used to describe the spatial organization of nuclei in prostate tissue. Global graph features (12 Voronoi and 8 Delaunay) were extracted to quantify the arrangement of cells present in the prostate section. This process involved identifying the centroids of the automatically identified nuclei as nodes in a graph. The individual nodes were then connected via a different set of rules (Voronoi, Delaunay) to create different graphic arrangements. Statistics of the Voronoi polygon area and Delaunay edge length were extracted from the resulting graphs.
A total of 27 nuclear density features were also incorporated to describe the clustering of nuclei in the image [14]. These features described the number of nuclei present within various distances from each other and are shown in Table 2.
Analysis of local subgraphs [15] allowed for the quantification of interactions within localized neighborhoods by creating edge connections between nearby nuclear centroids. Edges were created between these nuclei to generate sparse graphs from which 26 subgraph features were used to characterize local cell organization, including eccentricity and connected component size. These local graphs are shown in Figure 1c, 1f, 1i, and 1l.
2.4. Experimental design
2.4.1. Experiment 1: Univariable and multivariable methods for predicting recurrence
To establish a baseline for our cohort, we investigated the most common univariable (Gleason sum, primary tumor stage, surgical margin, PSA) and multivariable models (nomograms developed by Kattan et al [16] and Stephenson et al [17]) for predicting recurrence of postsurgical PCa.
2.4.2. Experiment 2: Adjacent benign field features for predicting recurrence
We identified specific features from specific regions of interest for predicting recurrence using a univariable Cox regression model [18], demonstrating separation of nonrecurrence and recurrence cases for each feature. This results in feature sets CT, extracted from the tumor, CAB for adjacent benign regions, and a joint feature set called CTABS. This process is described in detail below.
2.4.2.1. CT: Extraction of tumor field features
To evaluate tumor field features, we extracted 199 features from the tumor field core images for each TMA (Table 2). A univariable Cox regression model was used to select the top 10 most predictive features for recurrence (Supplementary Table 1a). The top 10 features for CT identified by the univariable Cox regression model were used to train a classifier for predicting recurrence.
2.4.2.2. CAB: Extraction of adjacent benign field features
To evaluate adjacent benign field features, we extracted 199 features from the adjacent benign field core images for each TMA (Table 2). A univariable Cox regression model was used to select the top 10 most predictive features for recurrence (Supplementary Table 1a). The top 10 features for CAB identified by the univariable Cox regression model were used to train a classifier for predicting recurrence.
2.4.2.3. CTABS: Extraction of tumor plus adjacent benign signature
To evaluate TABS, a univariable Cox regression model was used to select the top 10 most predictive features for recurrence (Supplementary Table 1a) from the combination of CT and CAB. The top 10 features for CTABS identified by the univariable Cox regression model were used to train a classifier for predicting recurrence.
2.4.3. Experiment 3: Tumor plus adjacent benign signature for predicting recurrence
To evaluate TABS for predicting recurrence, each patient was characterized by the 199 features (Table 2) extracted from the tumor core and the 199 features extracted from the adjacent benign core. Each feature set (CT, CAB, CTABS) was used to train a random forest classifier [19], a powerful methodology utilizing a combination of decision tree models. Random forests were shown to be most likely to provide the highest classification accuracy in a study of 121 public data sets [20].
We performed threefold cross-validation for 100 trials, selecting a random two-thirds of the data set for training and one-third for testing. The area under the receiver operating characteristic curve (AUC) [21] values and 95% confidence intervals associated with each feature set were calculated.
Kaplan-Meier analysis was used to compare recurrence-free survival time between positive and negative control groups following RP. In this study, the two predicted groups (recurrence and nonrecurrence) were determined by the prediction of 100 runs of random forests on each patient. The quantitative difference between the survival outcome was determined by the log-rank test, in which lower p values denoted greater significance between the survival distributions.
The CT, CAB, and CTABS feature sets can be compared using AUC and Kaplan-Meier analysis, as described in section 3.2. Similarly, the Gleason sum feature set (CGS) and feature combinations with Gleason sum are also demonstrated.
3. Results
3.1. Experiment 1: Univariable and multivariable methods for predicting recurrence
Table 3 provides a baseline measure of the performance of univariable methods for predicting recurrence in postsurgical PCa patients. Table 3 illustrates the predictive value of Gleason sum compared with quantitative histomorpho-metric features automatically extracted from cancerous and cancer-adjacent benign fields, respectively. Table 3 also provides the baseline of multivariable predictors (Kattan and Stephenson nomograms) for predicting recurrence. Supplementary Table 2 shows the Spearman correlation between each univariable and multivariable predictor with the TABS feature set.
Table 3.
Mean area under the curve values show the ability of univariable predictors, comparisons of computer-extracted features with Gleason sum, and multivariable Kattan and Stephenson nomograms with TABS for predicting recurrence in prostate cancer patients following radical prostatectomy for 100 runs of threefold random forest classification
| Gleason sum | Tumor stage | Surgical margin | PSA | |
|---|---|---|---|---|
| 0.72 (0.68–0.76) | 0.68 (0.68–0.69) | 0.71 (0.70–0.72) | 0.63 (0.59–0.67) | |
| Tumor QH | Benign QH | TABS | ||
| Without Gleason sum | 0.68 (0.61–0.75) | 0.72 (0.64–0.80) | 0.77 (0.69–0.85) | |
| With Gleason sum | 0.73 (0.65–0.80) | 0.78 (0.67–0.84) | 0.81 (0.72–0.87) | |
| Kattan | Stephenson | |||
| Without TABS | 0.84 (0.77–0.89) | 0.80 (0.71–0.85) | ||
| With TABS | 0.90 (0.83–0.94) | 0.89 (0.84–0.92) |
PSA = prostate-specific antigen; QH = quantitative histomorphometry; TABS = tumor plus adjacent benign signature.
Data are shown as mean area under the curve (95% confidence interval). Best classification performance metrics are highlighted in bold.
3.2. Experiment 2: Adjacent benign field features for predicting recurrence
Figure 2 shows box plots for features extracted from the benign field and the resulting difference in the feature distribution in recurrent and nonrecurrent patients. Supplementary Table 1a shows the representation of feature types described in Table 2 for the CT, CAB, and CTABS feature sets. Supplementary Table 1b shows the top features identified from CTABS sorted by univariable Cox regression model p values for discriminating between recurrent and nonrecurrent patients.
Fig. 2.
Box plots of nuclear subgraph features in benign fields of view show the separation in prostate cancer patients with recurrence versus nonrecurrence following radical prostatectomy. The p values were obtained using the Wilcoxon rank sum test between cases of recurrence and nonrecurrence.
3.3. Experiment 3: Tumor plus adjacent benign signature for predicting recurrence
Table 3 displays the classification AUC for classifiers built from features CT, CAB, and CTABS as well as the AUC by combining those respective features with Gleason sum (CGS).
The Spearman correlation was calculated between each clinical variable (gland weight, Gleason sum, tumor stage, and PSA) with TABS probability scores compared across the 70 patients studied. The probability scores were generated from the predicted probability of the CTABS-based classifier.
4. Discussion
Current methods of predicting recurrence risk after surgery rely heavily on the Gleason scoring system, which describes only the tumor region, and elevated PSA ≥0.2 ng/mL and have shown only a 71% classification rate in predicting biochemical recurrence [22,23]. The Kattan [16] and Stephenson [17,24] nomograms, Cancer of the Prostate Risk Assessment (CAPRA) risk scores [25], and genomic predictors [26] are among the many solutions for predicting recurrence following RP, but none of these measures explicitly extracts information from the adjacent benign regions. We provided head-to-head comparisons as possible (lack of biopsy data and genomic information available provided limitations for analysis) to demonstrate the value added by TABS.
4.1. Experiment 1: Univariable and multivariable methods for predicting recurrence
Table 3 suggests that Gleason sum is arguably the strongest single predictor for predicting biochemical recurrence. However, it can be seen in Table 3 that the TABS feature set outperformed each individual univariable predictor, most notably Gleason sum. Along with the low correlations between TABS and any individual predictor shown in Supplementary Table 2, these tables demonstrate the value added by investigating tumor and benign field features.
We also demonstrated in comparison with the multivarate models that adding TABS resulted in a better AUC (Table 3) and greater stratification in recurrence-free survival outcomes (Fig. 3). This may to be due to low correlations between TABS and current models (shown in Supplementary Table 2), owing to the use of an independent source of data—benign field features—that was not previously considered. For comparison, Gleason sum showed a Spearman correlation of r = 0.59 with the Kattan score and r = 0.62 with the Stephenson score. Respectively, Kattan and Stephenson scores yielded r = 0.75 and r = 0.68, respectively, with Primary Tumor Stage; r = 0.76 and r = 0.69, respectively, with surgical margins; and r = 0.32 and r = 0.21, respectively, with PSA. These results appear to suggest that TABS provides information that is largely independent (r < 0.35) of the factors used in extant prognostic models. In addition, our results appear to suggest that TABS can potentially improve the ability to predict recurrence in PCa patients over current multivariable nomogram models alone.
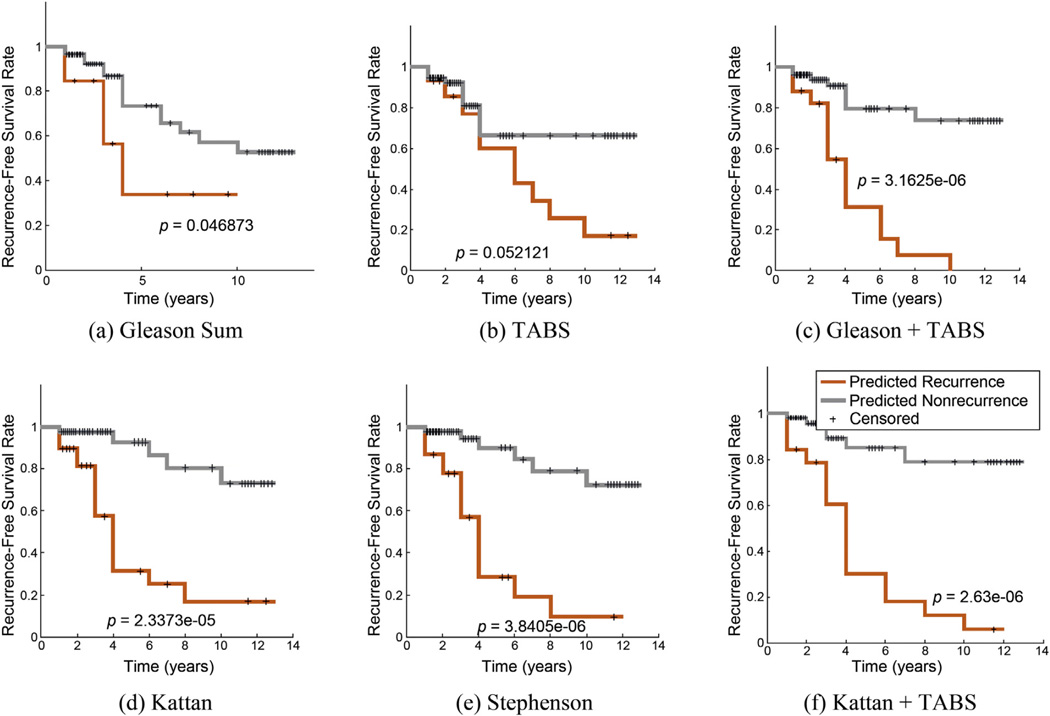
Fig. 3.
Kaplan-Meier curves for (a) Gleason sum, (b) tumor plus adjacent benign signature (TABS), (c) TABS plus Gleason sum, (d) Kattan score, (e) Stephenson score, and (f) combination of Kattan and TABS demonstrate the ability of each feature set to stratify patients with different recurrence-free survival times in radical prostatectomy cases.
TABS = tumor plus adjacent benign signature.
4.2. Experiment 2: Adjacent benign field features for predicting recurrence
A univariable Cox regression model of the nuclear subgraph and nuclear shape feature set (Supplementary Table 1a) revealed the most important features for extracting prognostic information from each tissue cohort. In benign tissue, nuclear subgraph features were found to be the most representative, as 4 of the 10 selected features in CTABS were nuclear subgraph features. This suggests a that nuclear architecture in adjacent benign fields can be used to predict recurrence.
Figure 2a describes the distribution of nuclear subgraph diameter, suggesting that in PCa patients with recurrence, there exist larger nuclear graphs compared with nonrecurrent tissue. This difference may be related to the presence of benign glands being fused in the case of tissue from recurrent patients, resulting in graphs that span multiple benign glands.
Figure 2b and 2c may illustrate more local nuclear architectural differences between recurrent and nonrecurrent benign field tissue. These differences are probably manifest by shorter internuclear distance in the benign tissue of nonrecurrent PCa patients, manifesting as thicker layers of tightly packed epithelial cells. In contrast, larger internuclear distances in the benign tissue of recurrent patients could be a reflection of the more sparse distribution of nuclei, resulting in thinner epithelial tissue layers.
The univariable Cox regression model of the nuclear subgraph and nuclear shape feature set (Supplementary Table 1a) revealed the most important features for TABS. Nuclear shape was found to be the most informative, making up 8 of the 10 informative features, and supporting previous work on the shape of cancerous nuclei [7,27].
The feature types represented in TABS illustrate that it is the combination of features extracted from both the tumor region and the benign region together that contribute to better prediction. It can be seen in Supplementary Table 1b that the features extracted from the benign areas make up 8 of the 10 most important features in TABS. The remaining two features are from the tumor region. We can surmise that local nuclear architecture appears to be important in the adjacent benign regions and is captured by the nuclear subgraph features. Nuclear shape of cancerous cells appears to be able to distinguish between recurrent and nonrecurrent patients (p = 0.0119) with the low Fourier descriptor, which describes the complexity of the nuclear boundary.
In Table 3 and Supplementary Table 2, we demonstrated the predictive value of adjacent benign field features, showing a mean AUC of 0.72 in predicting recurrence cases. In summary, we found (1) that both tumor and benign regions contain features that are capable of distinguishing recurrent from nonrecurrent patients, (2) that the arrangement of nuclei in the benign regions provides a majority of the contribution in CAB, and (3) that the size and shape of nuclei in tumor regions are primary components of CT.
The subgraph features identified describe long graphic constructs stemming from the individual nuclei. This is suggestive of a high degree of nuclear clustering in the image. This pattern may manifest in two biologically informed patterns: central zone morphology and prostate intraepithelial neoplasia (PIN)–like morphology.
In central zone tumors, glands are arranged in a more compact manner with less stroma between them. In contrast, additional stromal content in the peripheral zone separating the epithelial regions should lead to shorter nuclear subgraph lengths. Furthermore, central zone tumors have been shown to have higher rates of biochemical recurrence compared with peripheral or transition zone tumors [28], results that are consistent with our findings associated with these patterns.
The morphology of tumor-adjacent benign prostate tissue could suggest a path for disease progression or metastasis. Malignant disease requires cancer cells to invade, and the morphology of adjacent tissue surrounding the tumor may provide clues about the potential invasion. Longer nuclear subgraph edges suggest a clustering of glands, particularly those with thick epithelial walls. This clustering of glands could be associated with cribriform patterns, which are known to be associated with more malignant and aggressive disease. These PIN-like patterns displayed features similar to PIN including thick epithelial layers containing large clusters of nuclei; however, the tissue patterns lacked enlarged nuclei and prominent nucleoli, which tend to be typical of high-grade PIN.
A contextual pattern such as central zone gland morphology or PIN-like morphology typically will not be present in tumor morphology but can manifest in features of tumor-adjacent benign tissue. These contextual clues may be important for identifying those PCa patients at risk of recurrence; however, we note and reiterate that the cohort size has limitations with regard to the statistical power of the study. Consequently, these results represent preliminary work.
4.3. Experiment 3: Tumor plus adjacent benign signature for predicting recurrence
In the first row of Table 3, classification AUC associated with CT, CAB, and CTABS illustrate the improvement afforded by combining the top features of CT and CAB, as shown in Supplementary Table 1b. Although investigation of CT showed a predictive AUC of 0.68, adding features from CAB improved AUC to 0.73.
Similarly, a Kaplan-Meier analysis demonstrated greater separation using the joint benign and cancer feature set CTABS (p = 0.052) compared with cancer features alone (CT; p = 0.094). These results support the use of TABS to improve current methods of quantitative histomorphometry for predicting recurrence.
The results of the second row of Table 3 point to the following conclusions. First, both CT- and CAB-derived classifiers show performance improvement by incorporating Gleason sum. In fact, combining all features improved the predictive AUC of CTABS from 0.77 to 0.81, a statistically significant improvement over Gleason sum alone (AUC: 0.72), supporting the utility of combining CTABS and Gleason sum. Kaplan-Meier analysis in Figure 3c shows similar findings, illustrating the predictive value of combining Gleason sum with CTABS to yield a p value of 3.16 × 10−6, which is superior to CGS, CT, and CTABS. Furthermore, we found TABS to show statistically significantly better AUC performance (p < 0.001) compared with any individual marker in Table 3. Second, combining TABS with Kattan and Stephenson scores yielded statistically significant (p < 0.001) improvements in classification AUC (Supplementary Table 2) compared with the nomograms without TABS.
The value added by TABS can also be demonstrated by the potential reclassification caused by incorporating TABS. We found, using the Cox regression model, that TABS reclassified 24 patients differently compared with Gleason sum alone but provided no net benefit alone; however, by combining TABS with Gleason sum, 22 patients in the cohort were reclassified compared with Gleason sum alone. Overall, 12 additional patients were correctly classified compared with Gleason sum alone. Moreover, by combining TABS and the Kattan and Stephenson nomograms, we found that six additional patients were correctly classified compared with the Kattan or Stephenson nomogram alone.
We acknowledge the limitations of this preliminary study, which did not include an independent validation set. In future work, we plan to recruit additional cases to create an independent validation set to allow for the evaluation of the reproducibility of the results generated. Furthermore, the current study did not aim to evaluate specific end points such as biochemical recurrence, metastasis, or PCa-specific mortality individually but rather opted to predict poor outcomes associated with RP. In this study, the most common poor outcome was biochemical recurrence.
5. Conclusions
In this study, we aimed to investigate (1) the role of benign field features in predicting recurrence in RP cases and (2) the predictive value of TABS for predicting recurrence. We found that nuclear subgraphs appear to have predictive value in predicting recurrence from benign tissue fields. Furthermore, the CTABS feature set performed better than features derived from the cancerous field and benign fields individually while improving the classification AUC for the best clinical predictors in Gleason sum (AUC: 0.72) and Kattan (AUC: 0.84) and Stephenson (AUC: 0.80) nomograms to AUCs of 0.81, 0.90, and 0.89, respectively. We acknowledge that our findings represent preliminary work, and we plan to identify an independent validation cohort to evaluate TABS in future work.
Supplementary Material
Acknowledgments
Funding/Support and role of the sponsor: Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award numbers K01ES026841, R01CA136535-01, R01CA140772-01, R21CA167811-01, R21CA179327-01; the National Institute of Diabetes and Digestive and Kidney Diseases under award number R01DK098503-02, the DOD Prostate Cancer Synergistic Idea Development Award (PC120857); the DOD Lung Cancer Idea Development New Investigator Award (LC130463), the Ohio Third Frontier Technology development Grant, the CTSC Coulter Annual Pilot Grant, and the Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering at Case Western Reserve University. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Author contributions: George Lee had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Lee, Veltri, Madabhushi.
Acquisition of data: Veltri, Zhu, Epstein.
Analysis and interpretation of data: Lee, Ali.
Drafting of the manuscript: Lee, Madabhushi.
Critical revision of the manuscript for important intellectual content: Lee, Veltri, Zhu, Epstein, Madabhushi.
Statistical analysis: Lee.
Obtaining funding: Veltri, Madabhushi.
Administrative, technical, or material support: Lee, Veltri, Zhu, Madabhushi.
Supervision: Lee, Veltri, Madabhushi.
Other (specify): None.
Financial disclosures: George Lee certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.euf.2016.05.009.
References
- 1.Tyson MD, 2nd, Andrews PE, Ferrigni RF, Humphreys MR, Parker AS, Castle EP. Radical prostatectomy trends in the United States: 1998 to 2011. Mayo Clin Proc. 2016;91:10–16. doi: 10.1016/j.mayocp.2015.09.018. [DOI] [PubMed] [Google Scholar]
- 2.Paller CJ, Antonarakis ES. Management of biochemically recurrent prostate cancer after local therapy: evolving standards of care and new directions. Clin Adv Hematol Oncol. 2013;11:14–23. [PMC free article] [PubMed] [Google Scholar]
- 3.Boorjian SA, Thompson RH, Tollefson MK, et al. Long-term risk of clinical progression after biochemical recurrence following radical prostatectomy: the impact of time from surgery to recurrence. Eur Urol. 2011;59:893–899. doi: 10.1016/j.eururo.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 4.Trock BJ, Han M, Freedland SJ, et al. Prostate cancer–specific survival following salvage radiotherapy vs observation in men with biochemical recurrence after radical prostatectomy. JAMA. 2008;299:2760–2769. doi: 10.1001/jama.299.23.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zong Y, Huang J, Sankarasharma D, et al. Stromal epigenetic dysregulation is sufficient to initiate mouse prostate cancer via paracrine Wnt signaling. Proc Natl Acad Sci U S A. 2012;109:E3395–E3404. doi: 10.1073/pnas.1217982109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gann PH, Deaton R, Amatya A, et al. Development of a nuclear morphometric signature for prostate cancer risk in negative biopsies. PLoS One. 2013;8:e69457. doi: 10.1371/journal.pone.0069457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veltri RW, Khan MA, Miller MC, et al. Ability to predict metastasis based on pathology findings and alterations in nuclear structure of normal-appearing and cancer peripheral zone epithelium in the prostate. Clin Cancer Res. 2004;10:3465–3473. doi: 10.1158/1078-0432.CCR-03-0635. [DOI] [PubMed] [Google Scholar]
- 8.Christens-Barry W, Partin A. Quantitative grading of tissue and nuclei in prostate cancer for prognosis prediction. Johns Hopkins Apl Tech Digest. 1997;18:226–233. [Google Scholar]
- 9.Ali S, Veltri R, Epstein JI, Christudass C, Madabhushi A. Cell cluster graph for prediction of biochemical recurrence in prostate cancer patients from tissue microarrays [abstract] Proc SPIE. 2013;8676:86760H. [Google Scholar]
- 10.Doyle S, Hwang M, Shah K, Madabhushi A, Feldman M, Tomaszeweski J. Automated grading of prostate cancer using architectural and textural image features. Presented at: IEEE International Symposium on Biomedical Imaging; Arlington, VA. April 12–15.2007. [Google Scholar]
- 11.Ali S, Madabhushi A. An integrated region-, boundary-, shape-based active contour for multiple object overlap resolution in histological imagery. IEEE Trans Med Imaging. 2012;31:1448–1460. doi: 10.1109/TMI.2012.2190089. [DOI] [PubMed] [Google Scholar]
- 12.Lee G, Ali S, Veltri R, Epstein JI, Christudass C, Madabhushi A. Cell orientation entropy (core): predicting biochemical recurrence from prostate cancer tissue microarrays. In: Mori K, Sakuma I, Sato Y, Barillot C, Navab N, editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2013. New York, NY: Springer; 2013. pp. 396–403. [DOI] [PubMed] [Google Scholar]
- 13.Doyle S, Feldman MD, Shih N, Tomaszewski J, Madabhushi A. Cascaded discrimination of normal, abnormal, and confounder classes in histopathology: Gleason grading of prostate cancer. BMC Bioinformatics. 2012;13:282. doi: 10.1186/1471-2105-13-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee G, Singanamalli A, Wang H, et al. Supervised multi-view canonical correlation analysis (sMVCCA): integrating histologic and proteomic features for predicting recurrent prostate cancer. IEEE Trans Med Imaging. 2015;34:284–297. doi: 10.1109/TMI.2014.2355175. [DOI] [PubMed] [Google Scholar]
- 15.Bilgin C, Demir C, Nagi C, Yener B. Cell-graph mining for breast tissue modeling and classification. Conf Proc IEEE Eng Med Biol Soc. 2007:5311–5314. doi: 10.1109/IEMBS.2007.4353540. [DOI] [PubMed] [Google Scholar]
- 16.Kattan MW, Wheeler TM, Scardino PT. Postoperative nomogram for disease recurrence after radical prostatectomy for prostate cancer. J Clin Oncol. 1999;17:1499. doi: 10.1200/JCO.1999.17.5.1499. [DOI] [PubMed] [Google Scholar]
- 17.Stephenson AJ, Scardino PT, Eastham JA, et al. Postoperative nomogram predicting the 10-year probability of prostate cancer recurrence after radical prostatectomy. J Clin Oncol. 2005;23:7005–7012. doi: 10.1200/JCO.2005.01.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tibshirani RJ. Univariate shrinkage in the Cox model for high dimensional data. Stat Appl Genet Mol Biol. 2009;8:1–18. doi: 10.2202/1544-6115.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 20.Fernández-Delgado M, Cernadas E, Barro S, Amorim D. Do we need hundreds of classifiers to solve real world classification problems? J Machine Learning Res. 2014;15:3133–3181. [Google Scholar]
- 21.Zweig MH, Campbell G. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem. 1993;39:561–577. [PubMed] [Google Scholar]
- 22.Cordon-Cardo C, Kotsianti A, Verbel DA, et al. Improved prediction of prostate cancer recurrence through systems pathology. J Clin Invest. 2007;117:1876–1883. doi: 10.1172/JCI31399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shariat SF, Karakiewicz PI, Roehrborn CG, Kattan MW. An updated catalog of prostate cancer predictive tools. Cancer. 2008;113:3075–3099. doi: 10.1002/cncr.23908. [DOI] [PubMed] [Google Scholar]
- 24.Stephenson AJ, Kattan MW, Eastham JA, et al. Prostate cancer-specific mortality after radical prostatectomy for patients treated in the prostate-specific antigen era. J Clin Oncol. 2009;27:4300–4305. doi: 10.1200/JCO.2008.18.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooperberg MR, Freedland SJ, Pasta DJ, et al. Multiinstitutional validation of the UCSF cancer of the prostate risk assessment for prediction of recurrence after radical prostatectomy. Cancer. 2006;107:2384–2391. doi: 10.1002/cncr.22262. [DOI] [PubMed] [Google Scholar]
- 26.Lalonde E, Ishkanian AS, Sykes J, et al. Tumour genomic and microenvironmental heterogeneity for integrated prediction of 5-year biochemical recurrence of prostate cancer: a retrospective cohort study. Lancet Oncol. 2014;15:1521–1532. doi: 10.1016/S1470-2045(14)71021-6. [DOI] [PubMed] [Google Scholar]
- 27.Veltri RW, Isharwal S, Miller MC, Epstein JI, Partin AW. Nuclear roundness variance predicts prostate cancer progression, metastasis, and death: a prospective evaluation with up to 25 years of follow-up after radical prostatectomy. Prostate. 2010;70:1333–1339. doi: 10.1002/pros.21168. [DOI] [PubMed] [Google Scholar]
- 28.Cohen RJ, Shannon BA, Phillips M, Moorin RE, Wheeler TM, Garrett KL. Central zone carcinoma of the prostate gland: a distinct tumor type with poor prognostic features. J Urol. 2008;179:1762–1767. doi: 10.1016/j.juro.2008.01.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.