Abstract
In men with advanced penile squamous cell carcinoma receiving first-line chemotherapy, visceral metastases (VM) and Eastern Cooperative Oncology Group performance status ≥1 are poor prognostic factors for overall survival (OS). We hypothesized that tumor gene expression profiling may enhance prognostic stratification and identify potential therapeutic targets. In this retrospective study, RNA extracted from macrodissected tumors underwent profiling for the expression of 738 genes using NanoString. Univariate and multivariate analyses assessed the association of genes, VM, and performance status with OS. Tumors were available from 25 men who received first-line cisplatin-based chemotherapy. In univariate analysis, upregulated MAML2 (p = 0.004), KITLG (p ≤ 0.0001), and JAK1 (p = 0.029) genes were associated with poor OS, and upregulated FANCA was associated with better OS (p = 0.024). In stepwise multivariate analyses, VM (hazard ratio = 12.75, p = 0.0001) and MAML2 (hazard ratio = 10.411, p = 0.003) were associated with poor OS. The presence of none, one, and both of these poor risk factors was associated with significantly different median OS of 18.4 mo, 7.2 mo, and 2.1 mo, respectively. Unsupervised clustering demonstrated two major molecular subtypes with trend for different survivals (p = 0.052). Validation of results is necessary.
Keywords: Penile squamous cell carcinoma, Advanced, Cisplatin, Prognosis, Gene expression
Patient summary
The expression of the MAML2 gene in penile cancers from men receiving first-line cisplatin-based chemotherapy predicted overall survival independent of clinical factors.
Penile squamous cell carcinoma (PSCC) is an orphan disease in developed countries, although higher incidences are observed in less developed countries [1]. Unresectable and metastatic PSCC exhibits dismal outcomes using cisplatin-based combination chemotherapy [2]. A retrospective analysis of 140 patients with PSCC receiving first-line systemic therapy identified two prognostic factors: (1) visceral metastasis (VM), and (2) Eastern Cooperative Oncology Group (ECOG)-performance status (PS) ≥1 [3]. The incorporation of molecular panels may be hypothesized to enhance its prognostic ability.
Gene expressing profiling performed employing the NanoString technology appears promising and provides robust data even from formalin-fixed paraffin embedded tissue [4]. Multiple studies in other malignancies have utilized NanoString to subtype tumors, formulate prognostic signatures, or discover therapeutic targets. We performed tumor tissue NanoString analysis for a relevant panel of cancer promoting genes of patients with unresectable or metastatic PSCC receiving first-line cisplatin-based chemotherapy to identify a prognostic gene expression signature after controlling for VM and PS.
Patients with locally advanced unresectable primary tumor or regional bulky or fixed lymph nodes or metastatic PSCC who received first-line cisplatin-based systemic chemotherapy were identified. Baseline formalin-fixed paraffin embedded primary tumor tissue was required from a biopsy in addition to survival data, ECOG-PS, clinical stage, and sites of metastases. The study was approved by the Institutional Review Board at the University of Alabama, Birmingham (UAB).
The tissue underwent central pathology review at UAB and histologic macrodissection to separate tumor from benign tissue. RNA was harvested using the Qiagen RNAeasy kit (Qiagen, Valencia, CA, USA) and the quality of RNA was confirmed via the 260/280 ratio using nanodrop before inputting RNA to the nCounter NanoString platform (NanoString, Seattle, WA, USA). The nCounter GX PanCancer kit (NanoString) assays the gene expression of 730 cancer-related genes and 40 housekeeping genes (Supplementary Table 1; http://www.nanostring.com/products/gene_expression_panels) without the need for amplification of ≥100 ng of RNA and utilizes digital counting employing two ~50 hybridizing base probes per messenger RNA. Immunohistochemistry (IHC) studies were performed on tumor tissue to evaluate the expression of selected corresponding proteins for major overexpressed genes considered potentially actionable.
Individual patient level data were combined and patient, disease, and outcome characteristics summarized using descriptive statistics. Overall survival (OS) was calculated using the Kaplan-Meier method. A gene list was created by hierarchical clustering supervised for OS after false discovery p value correction [5]. For this analysis, nSolver text files (.csv) and NanoString.rcc files were imported into Partek Genomic Suite 6.6 (PGS, Saint Louis, MO, USA), and differential expressions were determined by analysis of variance. Cox proportional hazards regression analysis was conducted to examine the prognostic impact of VM, PS, and gene expression as continuous variables on OS. Also, unsupervised gene expression clustering was performed to identify major subtypes after importing gene expression to R package ConsensusClusterPlus (Bioconductor, Santa Cruz, CA, USA) to derive consensus-based clusters [6,7]. Genes and pathways differentially expressed between two clusters were identified [8,9]. All tests were two-sided and p ≤0.05 was considered statistically significant.
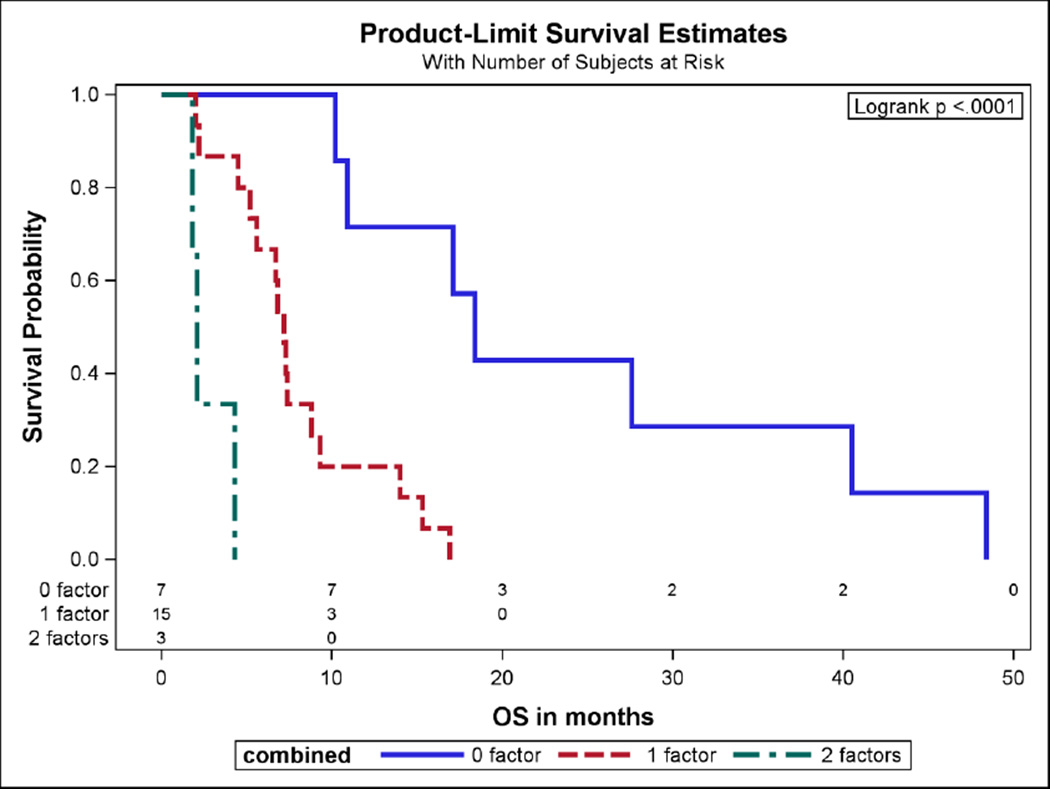
Patient data for 25 patients with PSCC treated between 2000 and 2012 with first-line cisplatin-based chemotherapy was captured (Supplementary Fig. 1) from Fondazione IRCCS Istituto Nazionale dei Tumori, Italy (n = 14 [56%]) and the British Columbia Cancer Agency, Vancouver, Canada (n = 11 [44%]). Correlative tumor studies were conducted at UAB Comprehensive Cancer Center. The median age was 60.5 yr (range, 47–76 yr). All patients had Stage 4 disease except for one patient who had Stage 3 disease. All of the 25 patients contributed to death events and no one was censored, which yielded a median OS of 7.4 mo (range, 1.8–48.4 mo). The ECOG-PS was 0 in 14 (56%) and ≥1 in 11 (44%) patients, and eight (32%) patients had visceral disease. In univariate analyses evaluating genes and supervised for association with OS, upregulated mastermind-like protein (MAML)-2 (p = 0.004), KIT-ligand (KITLG, p ≤ 0.0001), and JAK1 (p = 0.029) were associated with poor survival, and upregulated FANCA was associated with better OS (p = 0.024; Table 1, Supplementary Figs. 2 and 3). High BRCA1 gene expression exhibited a positive trend for association with OS (p = 0.088) and p16, a surrogate for human papilloma virus, was not associated with OS. In the multivariate analysis, stepwise selection approach was used to select the best two variable combinations. VM (hazard ratio [HR] = 12.75, 95% confidence interval [CI] = 3.438–47.282, p = 0.0001) and MAML2 (HR = 10.411, 95% CI = 2.967–36.539, p = 0.003) were significantly associated with poor OS. The presence of none, one, and both of these poor risk factors (VM present and MAML2 ≥ median) was associated with a median OS of 18.4 mo, 7.1 mo, and 2.1 mo, respectively (Fig. 1); the HR for one versus no factor was 9.870 (95% CI: 2.168–44.937, p = 0.003), the HR for two versus no factors was 136.182 (95% CI: 13.325–1391.797, p < 0.0001), and the HR for both versus one factor was 13.797 (95% CI: 2.228–85.449, p = 0.0048).
Table 1.
Univariate and multivariate analysis for association of factors with overall survival
| Variable | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p value | HR | 95% CI | p value | |
| Visceral metastasis | 3.547 | 1.364–9.225 | 0.006 | 12.75 | 3.438–47.282 | 0.0001 |
| ECOG-PS ≥ 1a | 1.826 | 0.797–4.184 | 0.15 | — | — | — |
| KITLGa | 1.014 | 1.007–1.021 | <0.0001 | |||
| JAK1a | 1.008 | 1.001–1.016 | 0.03 | |||
| MAML2 | 1.029 | 1.010–1.050 | 0.004 | 10.411 | 2.967–36.539 | 0.003 |
| FANCAa | 0.966 | 0.937–0.995 | 0.024 | |||
CI = confidence interval; ECOG-PS = Eastern Cooperative Oncology Group-Performance Status; HR = hazard ratio.
Not included in multivariate analysis since not significant in stepwise selection.
Fig. 1.
Overall survival (OS) based on visceral metastasis and/or gene expression of MAML2, or neither.
The presence of 0, 1, and both factors (visceral metastases and MAML2 gene expression ≥ median) was associated with a median overall survival of 18.4 mo, 7.1 mo, and 2.1 mo, respectively.
KITLG and JAK1 were selected for IHC based on their relevance as potentially actionable molecules and drugs available in the clinic to inhibit these pathways. IHC for KITLG was performed using Anti-KITLG EP665Y (Rabbit mAb, #TA300957, Origene, Rockville, MD, USA) and for JAK1 performed using Jak1 (6G4 Rabbit mAb, #3344, Cell Signaling Technology, Danvers, MA, USA) with overall scoring accounting for intensity of expression and percentage of cells showing expression (Supplementary Fig. 4). Following antigen unmasking using citrate buffer and following manufacturers protocol in 10 evaluable tumor samples, which showed a significant positive correlation between JAK1 gene expression and corresponding protein expression by IHC (Spearman’s rho = 0.66; p = 0.04), and a trend for KITLG gene expression and KITLG protein IHC expression (Spearman’s rho = 0.59; p = 0.074).
Unsupervised hierarchical clustering demonstrated two major clusters. Cluster 1 (16 patients [64%], three with VM) was characterized by the overexpression of the following pathways: Janus kinase/signal transducers and activators of transcription (JAK/STAT), mitogen-activated protein kinases (MAPK), gonadotropin-releasing hormone, hedgehog, and T-cell receptor signaling. Cluster 2 (nine patients [36%], five with VM) was characterized by MAPK, cell cycle, and focal adhesion signaling pathways. Cluster 1 exhibited a trend for improved OS compared with Cluster 2 (median OS = not reached vs 6.8 mo, p = 0.052; Supplementary Fig. 5).
The study is limited by the modest sample size. We focused on a homogeneous cohort of patients with locally advanced unresectable or metastatic disease receiving cisplatin-based chemotherapy. A larger and more heterogeneous cohort may have been considered, although the relative advantages of these strategies are unclear. This study is the first to describe a prognostic model incorporating both clinical and molecular factors for patients receiving first-line cisplatin-based systemic chemotherapy for locally advanced or advanced PSCC. Moreover, two major molecular clusters were demonstrated. The combination of one clinical factor, VM, and one molecular factor, tumor tissue MAML2 gene overexpression, was significantly associated with poor OS. In addition, KITLG, JAK1, and MAML2 may also warrant evaluation as therapeutic targets. KITLG binds the KIT membrane receptor, which activates downstream signaling through the PI3K, MAPK, and STAT pathways. JAK1 is a nonreceptor kinase, which signals downstream through STAT. MAML2 binds to the Notch receptors at intracellular and transcriptional activation domains. Indeed, agents targeting these molecules are already approved for other indications, for example, imatinib, sunitinib, regorafenib, and other kinase inhibitors inhibit KIT and ruxolitinib inhibits JAK1. NOTCH inhibitors are undergoing early phase 1 clinical investigation, but are not yet approved for clinical use. Other studies have preliminarily reported somatic gene alterations in EGFR, PIK3CA, NOTCH1, CDKN2A, CCND1, and BRCA2, but did not construct a prognostic model [10,11]. Unfortunately, there are no preclinical PSCC systems to validate these molecules as therapeutically actionable. Hence, validation of our data in a larger sample in conjunction with clinical trials targeting these molecules is justified. Despite the limitations of a small sample size, the present study is invaluable in improving our understanding of biology and management of this rare disease with substantial unmet needs.
Supplementary Material
In men with advanced penile squamous cell carcinoma receiving first-line cisplatin-based chemotherapy, visceral metastases and tumor tissue MAML2 gene expression were independently associated with poor overall survival. KITLG, JAK1, and MAML2 may also warrant evaluation as therapeutic targets.
Acknowledgments
Funding/Support and role of the sponsor: Guru Sonpavde received a University of Alabama, Birmingham Urologic Oncology startup fund for use in the management of the data. Sooryanarayana Varambally is supported by NIH R01CA154980 and R01CA157845.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: Guru Sonpavde had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Necchi, Eigl, Yang, Sonpavde.
Acquisition of data: Necchi, Eigl, Yang, Varambally, Sonpavde.
Analysis and interpretation of data: Necchi, Eigl, Yang, Varambally, Sonpavde.
Drafting of the manuscript: Necchi, Eigl, Yang, Varambally, Sonpavde.
Critical revision of the manuscript for important intellectual content: Necchi, Eigl, Yang, Bae, Chandrashekar, Chen, Naik, Mehta, Giannatempo, Colecchia, Gordetsky, Wei, Cooper, Varambally, Sonpavde.
Statistical analysis: Bae, Chandrashekar, Chen.
Obtaining funding: Sonpavde.
Administrative, technical, or material support: Necchi, Eigl, Yang, Bae, Chandrashekar, Chen, Naik, Mehta, Giannatempo, Colecchia, Gordetsky, Wei, Cooper, Varambally, Sonpavde.
Supervision: Necchi, Eigl, Yang, Varambally, Sonpavde.
Other: None.
Financial disclosures: Guru Sonpavde certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: Sonpavde: research support from Onyx, Bayer, Boehringer-Ingelheim, and consultant for Pfizer, Novartis, Merck, Genentech, Sanofi, Argos, Agensys, and Bayer;Yang: research support from Eli Lilly and Company, Advisory Board for Bayer, Nanostring Technologies; Necchi: research grants from Amgen, Millennium-Takeda, GlaxoSmithKline, MerckSharp&Dohme (MSD), Astra Zeneca, advisor for MSD, consultant for Roche, Celgene, MSD, Pierre Fabre, Bristol Myers Squibb, Astra Zeneca, and Pfizer; Eigl: research support from Janssen, Advisory Board for Roche, Astellas, and Pfizer.
Presented in part at the American Society Of Clinical Oncology annual conference in June 2015.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Sonpavde G, Pagliaro LC, Buonerba C, Dorff TB, Lee RJ, Di Lorenzo G. Penile cancer: Current therapy and future directions. Ann Oncol. 2013;24:1179–1189. doi: 10.1093/annonc/mds635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pond GR, Di Lorenzo G, Necchi A, et al. Prognostic risk stratification derived from individual patient level data for men with advanced penile squamous cell carcinoma receiving first-line systemic therapy. Urol Oncol. 2014;32:501–508. doi: 10.1016/j.urolonc.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Geiss GK, Bumgarner RE, Birditt B, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 5.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 6.Wilkerson MD, Hayes DN. ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mortensen MM, Hoyer S, Lynnerup AS, et al. Expression profiling of prostate cancer tissue delineates genes associated with recurrence after prostatectomy. Sci Rep. 2015;5:16018. doi: 10.1038/srep16018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 10.McDaniel AS, Hovelson DH, Cani AK, et al. Genomic profiling of penile squamous cell carcinoma reveals new opportunities for targeted therapy. Cancer Res. 2015;75:5219–5227. doi: 10.1158/0008-5472.CAN-15-1004. [DOI] [PubMed] [Google Scholar]
- 11.Ali SM, Pal SK, Wang K, et al. Comprehensive genomic profiling of advanced penile carcinoma suggests a high frequency of clinically relevant genomic alterations. Oncologist. 2016;21:33–39. doi: 10.1634/theoncologist.2015-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.