Abstract
Genetic changes are infrequent in acute myeloid leukemia (AML) compared to other malignancies and often involve epigenetic regulators, suggesting that an altered epigenome may underlie AML biology and outcomes. In 96 AML cases including 65 pilot samples selected for cured/not-cured, we found higher CpG island (CGI) promoter methylation in cured patients. Expanded genome-wide digital restriction enzyme analysis of methylation (DREAM) data revealed a CGI methylator phenotype independent of IDH1/2 mutations we term AML-CIMP (A-CIMP+). A-CIMP was associated with longer overall survival (OS) in this dataset (median OS, years: A-CIMP+ = Not reached, A-CIMP− =1.17; P=0.08). For validation we used 194 samples from The Cancer Genome Atlas interrogated with Illumina 450k methylation arrays where we confirmed longer OS in A-CIMP (median OS, years: A-CIMP+ =2.34, A-CIMP− =1.00; P=0.01). Hypermethylation in A-CIMP favored CGIs (OR: CGI/non-CGI=5.21), and while A-CIMP was enriched in CEBPA (P=0.002) and WT1 mutations (P=0.02), 70% of cases lacked either mutation. Hypermethylated genes in A-CIMP function in pluripotency maintenance, and a gene expression signature of A-CIMP was associated with outcomes in multiple datasets. We conclude that CIMP in AML cannot be explained solely by gene mutations (e.g. IDH1/2, TET2), and that curability in A-CIMP+ AML should be validated prospectively.
Keywords: AML, DNA methylation, CIMP, IDH1, IDH2
Introduction
Acute myeloid leukemia (AML) in adults is curable in a subset of cases that remain incompletely characterized. Certain molecular aberrations have been shown to associate with differential outcomes in AML1–4. For instance, curability is highest in younger patients with core binding factor (CBF) rearrangements between chromosomes 8 and 21, and within chromosome 16, while prognosis is poor in patients with autosomal monosomies2, 5–9. Despite these clinically useful cytogenetic risk associations, outcomes in AML remain heterogeneous and the biological mechanisms for diverse outcomes remain obscure10. Importantly, a number of patients with intermediate or poor risk cytogenetics can still be cured and may benefit from dose-intensive chemotherapy but their identification is uncertain, even in the era of whole genome mutational analysis1, 2, 10–12.
DNA methylation is an epigenetic process that is frequently altered in AML, either as a primary defect or secondary to mutations in regulators of DNA methylation such as TET2, DNMT3A and IDH1 or IDH213–20. Aberrant methylation at many CpG islands (CGIs) characterizes a subset of cancers of multiple primary origin that has been termed CGI methylator phenotype (CIMP)21. Interestingly, CIMP is often associated with a relatively better outcome. In colon cancer, CIMP is associated with microsatellite instability, and these patients often survive longer, perhaps as a result of enhanced anti-tumor immunity22–26. In gliomas, CIMP is an independent predictor for improved outcome27–29 and in breast cancer, CIMP is associated with a gene expression signature characteristic of good survival30, 31. Consistent with CIMP, Marcucci, et al. recently reported expression of seven genes that gain methylation in AML is associated with improved survival32. While these and other data suggest that CIMP exists in AML, its biological and clinical characteristics remain incompletely defined32–34. Observed mutations in IDH1/2 are a notable potential cause of CIMP. It has previously been shown that the R132H mutation in IDH1 and the R140Q or R172K mutations in IDH2 can cause aberrant enzymatic generation of the oncometabolite, 2-hydroxyglutarate, which inhibits normal TET-mediated DNA demethylation35–38. While IDH1/2 mutations can lead to hypermethylation, CIMP in colorectal and gastric cancers are not IDH1/2 associated; thus, other potential causes of CIMP remain unexplored.
In this study, we analyzed AML patient samples for DNA methylation status and identified two distinct CIMP phenotypes: an IDH1/2 mutation-associated CIMP (I-CIMP), and an IDH1/2 mutation-independent AML CIMP (A-CIMP). We found that the DNA methylation patterns, genetic backgrounds, and clinical characteristics between I-CIMP and A-CIMP are distinct, with an overall survival (OS) benefit for patients with A-CIMP+ disease.
Methods
Patient samples
For the pilot analysis we examined 65 bone marrow samples from AML patients who had been treated at MD Anderson Cancer Center (MDACC) from 1985 to 2004. The samples were selected for patients with short and long survival from a tissue bank. The expanded whole-genome methylation analysis was based on 42 of these 65 samples (selected by DNA availability) in addition to an unselected cohort of 59 consecutive samples from patients treated for AML at MDACC. We included only patients treated with MDACC-standard idarubicin + cytarabine based chemotherapy. None of the patients received treatment with hypomethylating agents. Standard diagnostic and remission criteria were used. DNA was extracted by standard methods. Cytogenetic risk groups were defined as follows: good (inv16), intermediate (normal karyotype; +8; +18; +6, +21), and poor (−5/del5q; −7; 11q23; t(6;11), +21; complex karyotype with 3 or more genetic abnormalities). For normal controls, peripheral blood leukocytes were obtained from 32 healthy volunteers (18–53 years of age). Patient characteristics for the expanded analysis are described in Table 1, and for the pilot study alone in Supplementary Table S1. The Institutional Review Boards at MDACC, and Temple University approved all protocols, and all patients gave informed consent for the collection of residual tissues as per institutional guidelines and in accordance with the Declaration of Helsinki. A summary of the different patient sample cohorts used and their DNA methylation status is provided in Supplementary Figure S1.
Table 1.
Clinical characteristics of MDACC cohort.
| A-CIMP | I-CIMP | CIMP-negative | P-value | |
|---|---|---|---|---|
| Samples in cluster | 15 | 10 | 71 | - |
| Age, years, median (range) | 43 (17–62) | 43 (20–75) | 53 (20–77) | 0.02 |
| Male sex, number (%) | 6 (40%) | 2 (20%) | 35 (49%) | 0.21 |
| Platelet count, × 10^9/L, median (range) |
40 (6–135) | 48 (13–126) | 57 (11–676) | 0.34 |
| WBC count, × 10^9/L, median (range) | 10.1 (0.8–312) | 39.7 (11.8–263) | 16.4 (0.8–271) | 0.34 |
| Peripheral blood blast percent, median (range) |
50 (4–97) | 91 (40–96) | 31 (0–95) | <0.01 |
| Bone marrow blast percent, median (range) |
65 (30–96) | 82 (42–94) | 64 (20–99) | 0.14 |
| Cytogenetic risk, number (%) | ||||
| Favorable | 0 (0%) | 0 (0%) | 5 (7%) | |
| Intermediate | 14 (93%) | 7 (70%) | 48 (68%) | 0.39 |
| Adverse | 1 (7%) | 3 (30%) | 18 (25%) |
Bisulfite-pyrosequencing
DNA was extracted, bisulfite-treated, and sequenced as previously reported13. In brief, bisulfite-treated DNA was amplified with gene-specific primers in a 2-step polymerase chain reaction (PCR). The second step of PCR was used to label the reverse DNA strand with biotin. DNA methylation was measured as the percentage of bisulfite-resistant cytosines at CpG sites by pyrosequencing. Assays close to transcription start sites were used for OSCP1, CDH13, CDKN2B, NPM2, OLIG2, SLC26A4, PGR, and SCGB3A1, as well as the LINE-1 repeat (GenBank accession number X58075) to approximate global methylation.
DREAM interrogation of genome-wide methylation
DREAM analysis was performed on 96 AML samples and 32 normal blood controls as previously described39. In brief genomic DNA extracted from AML samples was sequentially cut with two enzymes recognizing CCCGGG sites in DNA. SmaI does not cut methylated sites, and leaves blunt ends. XmaI can cleave methylated sites and leaves a 5’ overhang sequence. Thus, specific signatures are created for methylated and unmethylated sites. Enzyme-treated DNA was then used to generate sequencing libraries according to Illumina protocols, and run on an Illumina HiSeq 2000 or 2500. Sequencing data were mapped to CCCGGG sites in the human genome (hg19) and methylation was calculated as the fraction of total CCCGGG site sequencing reads that mapped to the methylated signature. For quality control, these data were filtered to include sites with at least 100 reads in 75% of samples, giving 11,499 CpG sites for bioinformatic analysis. For clustering analyses missing values in each sample were imputed as the median of all non-missing values for that site. In order to select for cancer specific hypermethylation patterns we selected CpG sites with average methylation and standard deviation <20% in normal blood, and standard deviation >12% in AML (1210 sites).
Targeted mutational analysis
Targeted next-generation sequencing was performed on DNA extracted from 88 AML samples using the Illumina TruSight Myeloid Sequencing Panel (54 genes) with library preparation done according to manufacturer instructions (Illumina, Inc.). In brief, genomic DNA was hybridized to region-specific upstream and downstream oligonucleotide pairs. Unbound oligonucleotides were washed and hybridized pairs were joined by a DNA polymerase and ligated. Captured target regions were PCR amplified with multiplexing index sequences added and pooled for sequencing. Paired-end sequencing was performed on an Illumina HiSeq 2500 which generated 78, and 85 million total aligned reads across two rapid run lanes. Over 95% of aligned reads had sequencing quality scores greater than 30, and mean coverage across all interrogated genes for all samples was approximately 5,000×. Because there were no normal control tissues available for these AML cases, we used Illumina BaseSpace TrueSeq Amplicon app and BaseSpace Variant Studio 2.2 (Illumina, Inc.) in conjunction with variant filtering criteria to make likely distinctions between somatic and germline alterations. To this end we required that non-synonymous variants with a minimum allelic ratio of 10% be detected by at least 50 reads with quality scores > 50, and that the variants be present in the COSMIC database with a hematologic cancer association. Based on these criteria, we detected 212 likely somatic mutations in our DREAM cohort.
TCGA and microarray validation data
Level 1 Illumina HumanMethylation450k methylation array data for 194 AML patients were downloaded from the TCGA data portal40. The raw data were pre-processed using functional normalization in the minfi R package41. For normal blood controls profiled using the Illumina HumanMethylation450k platform we used a publicly available dataset (GSE51388)42. Analysis was restricted to only those CpG sites with non-NA values for all samples (375,324 sites). To select for cancer-specific hypermethylation, and to remove potential age-related sites we selected for CpG sites unmethylated and non-variable in normal blood (beta-value average < 20%, standard deviation < 5%) and with variable methylation in AML (beta-value standard deviation > 20%). To confirm the overlap in A-CIMP targets of hypermethylation in the high-throughput datasets (DREAM and 450k) we examined normally unmethylated (< 20%) promoter CpG islands hypermethylated by at least 10% in 30% or more A-CIMP+ AML cases versus normal blood and found significant overlap (1.34E-49) across the two platforms (Supplementary Figure S2, Supplementary Table S2). RNA-seq data was also downloaded for the 176 available cases. Differential expression analysis was performed on read counts using the edgeR package in R, and hierarchical clustering was performed using z-score transformed RPKM values. For validation of differential gene expression, we downloaded datasets composed of 461, and 52 well-annotated AML cases from GEO with available outcomes data (GSE6891, and GSE23312-GPL10107)43, 44. Cluster analysis was performed using normalized probe intensity data for all interrogated genes which were down-regulated and hypermethylated in TCGA A-CIMP (318 genes). For visual clarity, heatmap of GSE6891 microarray data display only genes in the 80th percentile by standard deviation. Comparisons of average z-score gene expression for up and down –regulated genes were done using the non-parametric Kruskal-Wallis test followed by the Dunn post-hoc test implemented in R.
Statistics
Unsupervised hierarchical clustering was performed in R using Ward’s method (Ward.D) as implemented by the hclust function45. Differential methylation between clusters of AML cases was evaluated using the student t-test which was corrected for multiple hypothesis testing using the FDR method implemented in R45, 46. Clinical characteristics were compared between groups using single factor ANOVA and Fisher’s Exact Test. Unless otherwise stated, two-tailed p-values ≤ 0.05 were considered significant. For the preliminary pyrosequencing analysis we calculated methylation z-score by subtracting the mean methylation from each individual methylation level and dividing the difference by the standard deviation. This allows for comparison of a small number of methylation sites with differing variance across the samples. Cox regression analysis and Kaplan-Meier curves were generated in R using the survival package47.
Data Access
The DREAM data have been submitted to the Gene Expression Omnibus (GEO) repository (GSE92254). TCGA data used for validation and extended analyses are publicly available through the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/). Methylation data for normal blood on the Illumina 450k platform are available on GEO (GSE51388). Affymetrix array data used for validation of the A-CIMP gene expression signature are also available on GEO (GSE6891, GSE23312-10107).
Results
AML patients with long survival have increased CpG island DNA methylation
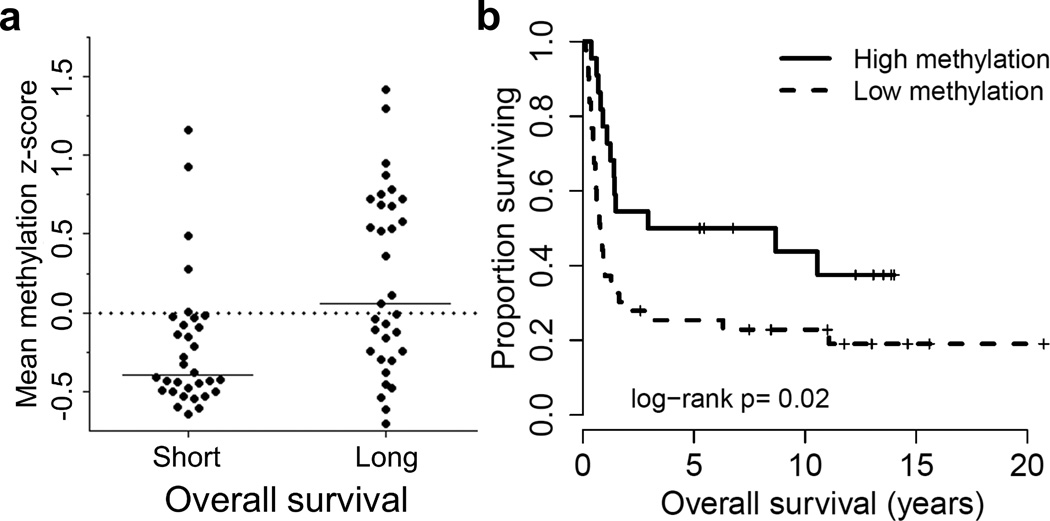
To study a potential association between DNA methylation and curability in AML, we selected patients based on long and short survival as well as sample availability in the MDACC leukemia sample bank. Using bisulfite pyrosequencing, DNA methylation was compared between 32 patients with OS less than one year after diagnosis (short survivors, median OS =6.9 months) and 33 AML patients with long OS (long survivors, median OS =89.8 months). Patient characteristics for this pilot cohort are described in Supplementary Table S1. We compared DNA methylation at promoter CGIs of the genes listed in Supplementary Table S3 (selected from a previous study13), and found that patients in the long survival group had significantly more aberrant DNA methylation than the short survivors at multiple genes including SCGB3A1, NPM2, CDKN2B, and OSCP1 (Supplementary Table S3). In order to account for the heterogeneous nature of a small number of CpG sites, we computed z-scores to normalize methylation by the standard deviation for each respective locus. When we compared average z-scores across all sites interrogated between long and short survivors, we found that the long survivor group had significantly higher average z-scores (Figure 1a; P=0.002), and Kaplan-Meier analysis demonstrated that patients with more methylation at the interrogated CpG sites had longer OS (median OS, years: High methylation =5.8, Low methylation =0.78, P=0.02; Figure 1b). Furthermore, when other prognostic factors, including cytogenetics, age, blast percentage, and FLT3 mutations, were included in a multivariate Cox regression, the average methylation z-score retained independent significance (Supplementary Table S4), suggesting that CIMP may be an independent prognostic factor in AML.
Figure 1. Long surviving AML patients have increased DNA methylation.
a) Average methylation z-score is plotted for patients with long survival (>12 months) versus short survival (<12 months). b) Overall survival (OS) of patients with high methylation (average methylation z-score > 0.0) and low methylation (average methylation z-score ≤ 0.0).
Genome-wide methylation analysis identifies an IDH1/2-independent hypermethylator phenotype
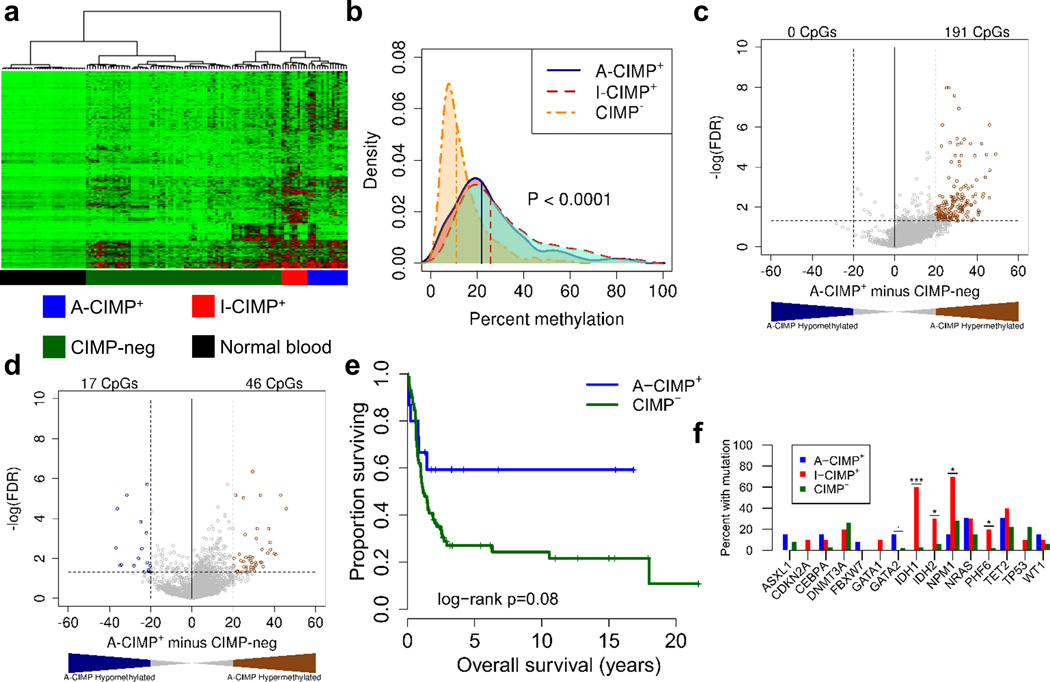
To validate and extend our findings, we performed genome-wide DREAM analysis on 96 AML samples and 32 normal blood controls. DREAM interrogates DNA methylation at thousands of CpGs across the genome and is highly quantitative39. Our preliminary and previously published data indicated that the best CIMP markers are those that show the most cancer-specific hypermethylation patterns48, 49. To enrich for those we applied filtering criteria as described in the methods section; this included removal of age-related methylation changes by excluding CpG sites which are highly variable in normal blood. Hierarchical clustering of the AML samples based on DNA methylation revealed at least two distinct hypermethylator phenotypes (Figure 2a). Ten cases clustered together tightly in a group we termed IDH-CIMP (I-CIMP+) because 7/10 (70%) of these cases harbor IDH1/2 mutations, a previously described cause for aberrant hypermethylation. Fifteen other cases clustered together in a separate group we termed AML-CIMP (A-CIMP+), of which none harbored IDH1 or IDH2 mutations. Because A-CIMP was not previously characterized we focused on exploring and validating its unique biology.
Figure 2. DREAM analysis identifies A-CIMP in AML.
a) Hierarchical clustering of 96 AML patient samples and 32 normal blood controls on quantitative DNA methylation levels. b) The distribution of methylation values across all 1,210 selected CpG sites from Figure 2a were stratified by cluster. Vertical lines correspond to the median of average methylation values across all CpG sites for each respective cluster. P-value was computed using the non-parametric Kruskal-Wallis test. c, d) Volcano plot differential methylation analysis comparing A-CIMP+ to CIMP− AML for CGI sites (c), and non-CGI sites (d). e) Kaplan-Meier survival analysis of A-CIMP+ compared to CIMP− AML. f) Genetic mutations associated with A-CIMP, and I-CIMP. * P < 0.05, ** P < 0.01, *** P < 0.001.
From an epigenetic perspective A-CIMP+ was distinct. Comparing the distribution of DNA methylation levels across each leukemia cluster revealed that both CIMP+ groups had significantly higher median methylation compared to CIMP− cases (Kruskal-Wallis P<0.001; Figure 2b). To explore the targets of hypermethylation we performed separate differential methylation analyses of CGI and non-CGI sites between A-CIMP+ and CIMP− leukemia (Figure 2c, d).
Compared to CIMP− cases, A-CIMP+ cases hypermethylated CGIs preferentially. Overall hypermethylation was observed (defined as FDR<0.05 and average methylation difference > 20%) in 3% of detected CGI sites and 1% of non-CGI sites suggesting a significant preference for CGI hypermethylation (odds ratio CGI/non-CGI =3.54; 95% CI: 2.55–5.01; P<0.001). In contrast, I-CIMP+ cases favored non-CGI hypermethylation (Supplementary Figure S3a, b). Thus, the methylation targets of A-CIMP were distinct.
From a clinical perspective the A-CIMP+ patients were younger than CIMP− patients (median age, years: A-CIMP+ = 43, CIMP− = 53, P=0.02; Table 1), and importantly, A-CIMP+, but not I-CIMP+ AML was associated with longer OS compared to CIMP− (median OS, years: A-CIMP+ = Not reached, versus CIMP− = 1.17, P = 0.08; Figure 2e; median OS I-CIMP+ = 3.35, P = 0.50 compared to CIMP−). Because A-CIMP is a relatively rare phenotype in AML we could not prove statistical independence from age in this dataset, however, an analysis restricted to younger patients (<60 years) revealed a trend for prolonged survival within this group (Supplementary Figure S4). We then interrogated the AML cases from a genetic mutational perspective and also identified distinct backgrounds. By targeted high-throughput sequencing of a hematologic cancer gene panel (54 genes), we identified a significant enrichment for IDH1 and IDH2 mutations in I-CIMP+ cases, however, A-CIMP appeared to lack a genetic definition (Figure 2f, Supplementary Figure S5).
A refined classifier of A-CIMP in TCGA data
In order to validate our findings and extend our analysis we used AML samples from TCGA40. These Illumina 450k array-based data interrogated methylation ~480,000 sites in 194 AML patients. We also used publicly available normal blood data analyzed on the same Illumina 450k methylation platform (GSE51388)42. After pre-processing, we initially used filtering criteria to enrich for cancer-specific CpG sites similar to those described earlier (see methods section). Hierarchical clustering of the 6,843 filtered CpG sites revealed a pattern consistent with our DREAM data; two distinct hypermethylator phenotypes were evident with one being highly enriched (9/10) in IDH1 and IDH2 mutation-positive cases (Supplementary Figure S6). To refine this classification, we used volcano plot analysis to identify 603 CpG sites showing A-CIMP-specific hypermethylation (Supplementary Figure S7). Reclustering the TCGA cases using the 603 A-CIMP-specific sites revealed a group of 43 A-CIMP+ patients (Figure 3a).
Figure 3. Characterization of A-CIMP+ AML in the TCGA cohort.
a) Hierarchical clustering of 194 AML patient samples and 24 normal blood controls on the basis of 603 CpG sites. b) Kaplan-Meier survival analysis of 194 cases based on the clusters derived in Figure 3a. c) Clinical characteristics associated with A-CIMP+ versus A-CIMP− AML.
From a clinical perspective A-CIMP+ patients had significantly longer median OS compared to A-CIMP− patients (median OS, years: A-CIMP+ =2.34, A-CIMP− =1.00, P=0.01; Figure 3b). The TCGA A-CIMP+ patients also tended to be younger than A-CIMP− (median age, years A-CIMP+ =43, A-CIMP− =60, P<0.001; Figure 3c), and this group had relatively more favorable risk cytogenetic aberrations (P=0.004, Figure 3c; Supplementary Table S5). Analysis of I-CIMP using the same methods confirmed a lack of prognostic benefit and a significant enrichment for IDH mutations (Supplementary Figure S8; Supplementary Table S6).
The distinct methylation landscape of A-CIMP+ AML
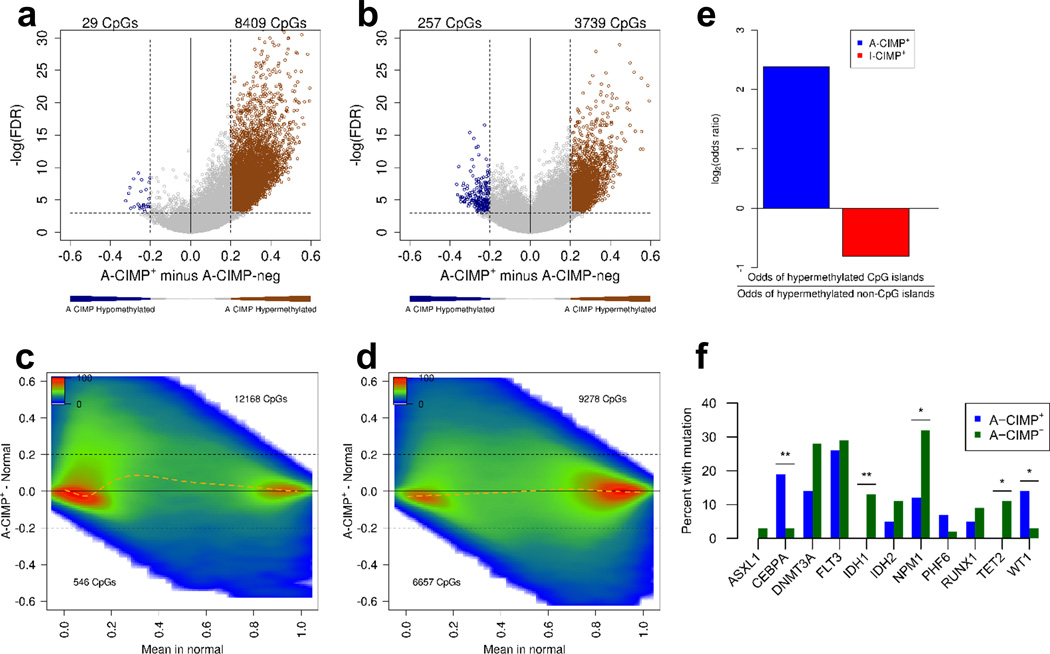
After refining a classification of A-CIMP, we examined these cases from an epigenetic perspective. Differential methylation analysis using volcano plots comparing A-CIMP+ to A-CIMP− AML across all interrogated CpG sites confirmed that significant hypermethylation was disproportionately at CGIs, and relatively less prevalent at non-CGIs (odds ratio CGI/non-CGI =5.21; 95% CI: 5.01–5.42; P<0.001; Figure 4a, b, e). As expected, we found similar results when comparing A-CIMP+ to normal blood instead of to A-CIMP− (Figure 4c and d). Examination of I-CIMP confirmed the preference for distinct non-CGI hypermethylation (Figure 4e, Supplementary Figure S9a–d).
Figure 4. A-CIMP+ AML is defined by CGI enriched hypermethylation.
Volcano plot and density plot differential methylation analysis between A-CIMP+ and A-CIMP− for CGI sites (a), and non-CGI sites (b); and between A-CIMP+ AML and normal blood for CGI sites (c), and non-CGI sites (d). Numbers in each volcano plot correspond to CpG sites with methylation beta-value differences greater than 0.2, and FDR<0.001. Numbers in each density plot correspond to CpG sites with methylation beta-value differences compared to normal blood greater than 0.2. Orange dashed lines in density plots represent LOWESS regression of the CpG density data. The enrichment of CGI sites hypermethylated in A-CIMP is distinct from the non-CGI preference seen in I-CIMP, as reflected by odds ratios of hypermethylated CpGs (e). f) Plots of somatic mutations associated with A-CIMP. Despite widespread epigenetic changes in A-CIMP, there is not a dominant genetic mutational definition for this subset of AML. * P < 0.05, ** P < 0.01, *** P < 0.001.
Examining the overlap in differentially methylated sites revealed that many CpGs are hypermethylated in any AML versus normal blood, regardless of CIMP status (Supplementary Figure S9e, f). However, a large fraction of CGIs is specifically hypermethylated in A-CIMP+ (Supplementary Figure S9e), and a large fraction of non-CGIs is specifically hypermethylated in I-CIMP+ (Supplementary Figure S9f), again confirming the distinct methylation pattern of each phenotype. Most of the hypomethylated CpG sites were shared between CIMP+ and CIMP− AML, with many more non-CGI sites losing methylation compared to normal blood (Supplementary Figure S9g and h for CGI and non-CGI sites, respectively).
In order to dissect possible functional relationships with the observed methylation patterns in AML, we performed Gene Set Enrichment Analysis (GSEA), and Ingenuity Pathway Analysis (IPA). Both analyses revealed that promoter CGI sites characteristic of A-CIMP+ AML were significantly enriched for genes involved in maintaining pluripotency (Supplementary Tables S7–9), with the top upstream regulators including OCT4, SOX2, and POU5F1. Taken together, these data suggest A-CIMP+ is a biologically distinct entity with divergent pathway dysregulation compared to A-CIMP− AML.
Genetic backgrounds in epigenetically defined AML subtypes
The distinct biology underlying A-CIMP+ AML was also evident in the set of genetic aberrations detected (Figure 4f, Supplementary Figure S10). From a genetic perspective, A-CIMP+ cases demonstrated associations with WT1 (6/43; 14%) and CEBPA (8/43; 19%) mutations (Fisher’s Exact Test P =0.02, and P =0.002, respectively; Figure 4f, Supplementary Figure S10a). These mutations were mostly mutually exclusive, with only one case being positive for both. However, most A-CIMP+ cases (30/43; 70%) lacked either WT1 or CEBPA mutations. A-CIMP+ also demonstrated significantly fewer IDH1, NPM1, and TET2 mutations compared to A-CIMP− AML (Fisher’s Exact Test P =0.009, P =0.01, and P =0.02, respectively; Figure 4f). In contrast, I-CIMP+ cases were mostly defined by IDH1 and IDH2 mutations (75% with IDH1 or IDH2; Fisher’s Exact Test P<0.001), although they also demonstrated significant enrichments for RUNX1 and PHF6 mutations, and a relative lack of FLT3 mutations (Fisher’s Exact Test P =0.01, P =0.04, and P=0.02, respectively; Supplementary Figure S8d, S10b). Notably, the 25% of I-CIMP+ cases with wild-type IDH1/2 were not enriched for any other genetic mutations. These observations suggest that the distinct DNA methylation profiles observed in AML are associated with specific genetic backgrounds, but that unlike, I-CIMP+, most A-CIMP+ cases do not have a defining mutational signature.
An A-CIMP-associated gene expression program is prognostic in multiple datasets
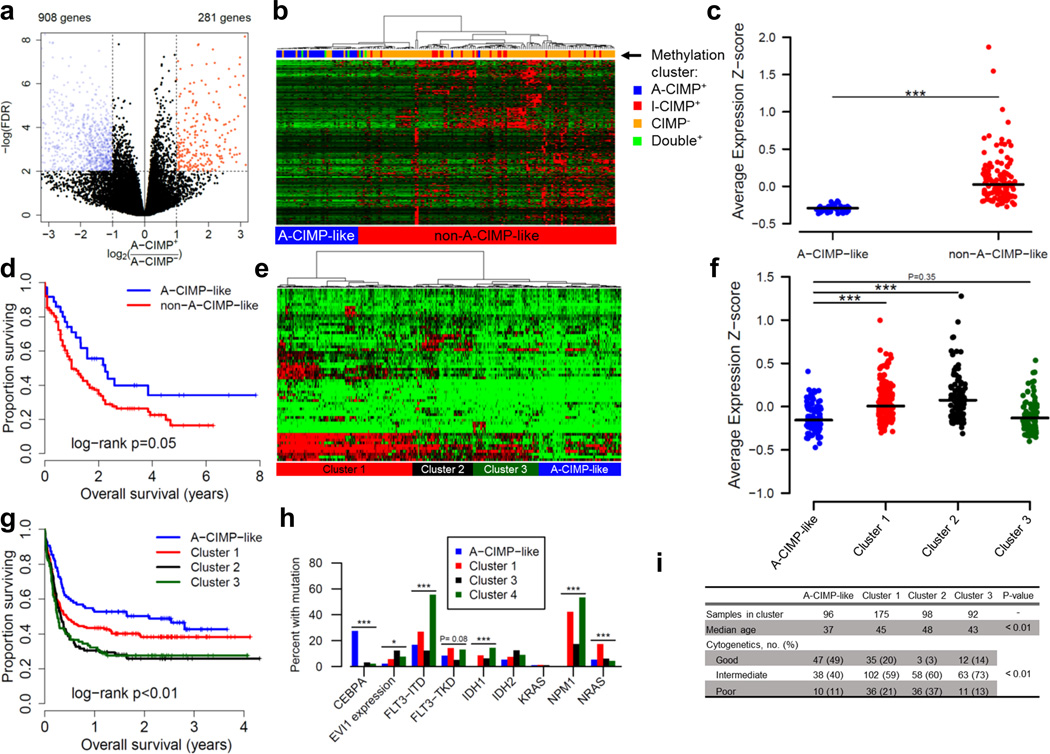
We hypothesized that, by virtue of differences in DNA methylation, A-CIMP+ AML would have a unique gene expression signature. To test this we performed a differential expression analysis comparing A-CIMP+ to A-CIMP−. By using the edgeR package with subsequent volcano plot analyses we observed 1,189 differentially expressed genes (FDR<0.01, Fold-change>2), of which 908 (76%) were down-regulated in A-CIMP+ (Figure 5a). Among the down-regulated genes, there were promoter CGI methylation data on 403 of them, and we found significant hypermethylation (FDR<0.05) in A-CIMP+ versus A-CIMP− AML in 318/403 (79%). A pathway enrichment analysis on these genes confirmed significant representation of functions maintaining human embryonic stem cell pluripotency. Some of the relevant down-regulated hypermethylated genes include BMPR2, WNT3A, FZD3, and FZD8, among others. We identified significant negative Spearman correlation coefficients between methylation and expression for 240 of these genes (60% of down-regulated genes with methylation data on the 450k array; Supplementary Figure S11, Supplementary Table S10).
Figure 5. Gene expression program characteristic of A-CIMP is prognostic in multiple datasets.
a) Volcano plot of differential expression analysis comparing TCGA A-CIMP+ to A-CIMP− AML defined by methylation. b) Hierarchical clustering of TCGA cases on RNA-seq data for down-regulated and hypermethylated genes identified in (a). c) Average expression z-scores for clusters in (b). Lines correspond to median z-score values. d) Kaplan-Meier analysis of gene expression clusters. e) Hierarchical clustering of 461 cases from GSE6891 based on down-regulated and hypermethylated genes identified in (a). For clarity, heatmap shows only genes with 80th percentile standard deviation, however clustering was done using all genes. f) Average expression z-scores for clusters in (e). g) Kaplan-Meier analysis of gene expression clusters in (e). h) Molecular characteristics of gene expression clusters in (e). i) Other clinical characteristics associated with gene expression clusters from (e). * P < 0.05, ** P < 0.01, *** P < 0.001.
Hierarchical clustering of TCGA cases on the expression of all 318 down-regulated and hypermethylated genes revealed a group of cases highly enriched for A-CIMP+ AML by methylation status (A-CIMP-like; Figure 5b). As expected, average z-score transformed RPKM values were lowest for the A-CIMP-like cluster (Figure 5c). In addition, Kaplan-Meier survival analysis of gene expression-based clusters recapitulated the prognostic advantage associated with A-CIMP methylation (median OS, years: A-CIMP-like = 2.25, Cluster 1 = 1.00, P=0.05; Figure 5d). To validate the prognostic importance of the A-CIMP transcriptional signature, we used published Affymetrix expression array data (GSE6891) to cluster 461 well-annotated AML cases on A-CIMP-down-regulated and hypermethylated genes (Figure 5e)43, 50. This analysis revealed an A-CIMP-like cluster of cases with the lowest average expression z-score for the A-CIMP signature genes (Figure 5f). Importantly, the A-CIMP-like cluster demonstrated significantly improved survival compared to other gene expression clusters (median OS, years: A-CIMP-like = 2.10, Cluster 1 = 0.44, Cluster 2 = 0.32, Cluster 3 = 0.26, log-rank P<0.01; Figure 5g). We also examined the genetic and clinical characteristics associated with the A-CIMP-like cluster in GSE6891, and found an enrichment for favorable cytogenetics, younger age, presence of CEBPA mutations, and a lack of IDH1/2 and NPM1 mutations, all of which are consistent with the genetic background of A-CIMP identified in our previous analyses (Figure 5h, i). We next performed the same analysis on one more independent gene expression microarray dataset (GSE23312-GPL10107; Supplementary Figure S12). We clustered AML cases on A-CIMP-down-regulated and hypermethylated genes and identified a low-expression cluster with improved overall survival (median OS, years: A-CIMP-like = Not reached, Cluster 1 =0.29, Cluster 2 =1.83, Cluster 3 =1.27, log-rank P=0.13; Supplementary Figure S12a–c). These results support the clinical importance of an altered transcriptional program related to DNA methylation status specific to A-CIMP+ AML.
Finally, a previous study found a gene expression-based prognostic signature by also selecting for hypermethylated promoter CGI sites associated with outcome32. We compared this signature to the A-CIMP classifier. We found that the previously published signature – Marcucci et al. CIMP (M-CIMP) – is prognostic in the TCGA methylation dataset (median OS, years: M-CIMP+ =2.25, M-CIMP− =1.00, P=0.04; Supplementary Figure S13a, b); however, it identifies a mix of A-CIMP+ and I-CIMP+ cases, and in this dataset we could not determine which epigenetic classifier performs better (Supplementary Figure S13c). Notably, FAM92A1 and SCRN1 from the M-CIMP signature were hypermethylated in both A-CIMP and I-CIMP in our classification of the TCGA cases, and appeared to drive their M-CIMP status identification (i.e. by magnitude of change they were the most hypermethylated genes in M-CIMP+ cases). The other genes in this signature (VWA8, CD34, RHOC, and F2RL1) were not found to be differentially methylated or expressed in our analyses of A-CIMP in the TCGA data (miR-155 was also part of this signature, however, we did not have data for this miRNA). In the GSE6891 microarray expression dataset M-CIMP gene expression was prognostic, and in a multivariate analysis, it was independent of A-CIMP gene expression status (Supplementary Figure S13d–g).
Discussion
In this study we interrogated DNA methylation in AML and identified two hypermethylation patterns, A-CIMP and I-CIMP. An important implication of these two CIMP phenotypes is that they affect different genomic compartments, and must arise via distinct mechanisms. A-CIMP is a CGI-favoring process, while I-CIMP targets are more often non-CGIs. A further important observation is that A-CIMP+ cancers lack mutations in epigenetic regulators known to cause aberrant DNA hypermethylation defects, such as IDH1/2, or TET1/2/3. Conversely, I-CIMP+ AML was predominantly characterized by known mutations in IDH1 or IDH2.
The causes of IDH1/2-independent CIMP remain unknown, but some possibilities are lncRNA/miRNA mutations, epigenetic changes affecting epigenetic regulators, and exogenous factors (e.g. infectious agents) leading to metabolites affecting DNA methylation. Mutations in non-coding RNA species may function by preventing proper recruitment of epigenetic regulators to their genomic targets, as has recently been described51, 52. Down-regulation of TET family members, or down-regulation of their interacting partners either through DNA methylation or repressive histone marks may also explain mutation-independent hypermethylation53. Finally, it has been shown that colorectal cancers harboring high amounts of fusobacterium are enriched for CIMP, and EBV is associated with CIMP in gastric cancer54, 55. It is possible that metabolites and/or inflammation related to chronic infections may affect DNA methylation in other disease contexts, and it is worth exploring these possibilities in AML.
The specific targets of A-CIMP hypermethylation were to a large degree, normally unmethylated CGIs which were significantly enriched for pluripotency maintenance genes. This association is also intriguing in light of the genetic background of A-CIMP+ AML which has a relatively high frequency of mutations in CEBPA and WT1. Recent data have reported interactions between wild-type TET2 and both WT1, and CEBPα56–60. Di Stefano, et al. found that CEBPα poises B cells for transformation into pluripotent stem cells, and induces the expression of TET2, while Sinha, et al. found that mutant WT1 can cause DNA hypermethylation at PRC2 targets59, 60. Both observations are consistent with our description of A-CIMP, and may represent one possible contributing mechanism in a subset of these patients who harbor such mutations. It is important to note, however, that TET2 mutant AML cases do not phenocopy methylation in WT1 or CEBPA mutant cases, suggesting that these genes have additional effects, perhaps on TET1 and/or TET3. We speculate that TET1 and TET3 may be more specific to CGIs because they contain a conserved CXXC domain that allows them to bind DNA and protect CGIs from methylation. TET2 lacks a CXXC domain, and thus its specificity is likely dependent on other factors, including its various interacting partners.
From an epigenetic mechanistic perspective, we find these alternative explanations for hypermethylation attractive given the very small number of patients in our analyses who were classified as harboring both hypermethylator profiles (3%). Presumably A-CIMP and I-CIMP may be redundant, but since we do not know the cause of A-CIMP, we can only speculate that the two phenotypes very likely arise via different mechanisms. It is also notable that in our analyses, true epigenetic instability either manifesting in A-CIMP or I-CIMP, comprises only about 30% of AML cases. The remaining CIMP-negative leukemias are not devoid of epigenetic aberrations, however, many of the DNA methylation changes one could observe are likely age-related, and do not reflect distinct cancer phenotypes61. This phenomenon has been shown in colorectal cancer, where the majority of cases show aberrant methylation at age-related loci, but are CIMP-negative at disease phenotype-specific targets62. Similarly, while we do observe epigenetic aberrations in CIMP-negative AML, disease specific hypermethylation is limited to the relatively small subset of A-CIMP and I-CIMP leukemias.
A final important implication of our data is the clinical relevance of CIMP in AML management. Our analysis revealed that only A-CIMP is associated with a favorable prognosis, and that this phenotype tends to occur more frequently in younger patients. Because A-CIMP is relatively rare, we could not definitively prove prognostic independence from age, however, we observed trends hinting at independence. Given the functional nature of A-CIMP+ hypermethylation targets it is plausible that these patients’ improved survival is related to a lack of full dedifferentiation in their leukemia, possibly making them more chemosensitive. Because the targets of hypermethylation in I-CIMP+ patients are quite distinct, and their outcomes are poor compared to A-CIMP+, this subgroup does not seem to respond well to chemotherapy, but may potentially benefit from treatment with hypomethylating agents (e.g. decitabine, azacitidine). This possibility is further alluded to by recent data suggesting gliomas harboring IDH mutations favor hypermethylation of CTCF binding sites and demonstrate reduce expression of the oncogenic driver, PDGFRA, upon administration of azacitidine63. Ongoing clinical trials in AML may reveal whether such a treatment strategy benefits patients with an identifiable I-CIMP+ epigenomic signature.
In summary, we present evidence of multiple hypermethylator phenotypes in AML. Using high-throughput methylation profiling of clinical AML specimens we identified three epigenetic phenotypes defined by distinct DNA methylation patterns: A-CIMP+, I-CIMP+, and CIMP−. These epigenetic states are associated with differential outcomes and gene expression in AML, and were validated using TCGA data. Future studies should focus on both further characterizing the transcriptional changes and aberrant epigenetic mechanisms associated with these observed methylation patterns in AML, and investigating the use of novel epigenetic biomarkers as clinical tools in AML management.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants CA158112 and CA100632. J-P.J.I. is an American Cancer Society Clinical Research professor supported by a generous gift from the F. M. Kirby Foundation.
Footnotes
Conflict of Interest
The authors declare that there are no competing financial interests in relation to the results of this study.
References
- 1.Bienz M, Ludwig M, Leibundgut EO, Mueller BU, Ratschiller D, Solenthaler M, et al. Risk assessment in patients with acute myeloid leukemia and a normal karyotype. Clin Cancer Res. 2005 Feb;11(4):1416–1424. doi: 10.1158/1078-0432.CCR-04-1552. [DOI] [PubMed] [Google Scholar]
- 2.Breems DA, Van Putten WL, De Greef GE, Van Zelderen-Bhola SL, Gerssen-Schoorl KB, Mellink CH, et al. Monosomal karyotype in acute myeloid leukemia: a better indicator of poor prognosis than a complex karyotype. J Clin Oncol. 2008 Oct;26(29):4791–4797. doi: 10.1200/JCO.2008.16.0259. [DOI] [PubMed] [Google Scholar]
- 3.Döhner K, Schlenk RF, Habdank M, Scholl C, Rücker FG, Corbacioglu A, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood. 2005 Dec;106(12):3740–3746. doi: 10.1182/blood-2005-05-2164. [DOI] [PubMed] [Google Scholar]
- 4.Schlenk RF, Döhner K, Krauter J, Fröhling S, Corbacioglu A, Bullinger L, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008 May;358(18):1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 5.Campbell LJ, Challis J, Fok T, Garson OM. Chromosome 16 abnormalities associated with myeloid malignancies. Genes Chromosomes Cancer. 1991 Jan;3(1):55–61. doi: 10.1002/gcc.2870030110. [DOI] [PubMed] [Google Scholar]
- 6.Cheng FJ, Yang AD, Fei HB, Tian H. Clinical and prognostic investigations on M2/t(8;21) acute nonlymphocytic leukemia. J Tongji Med Univ. 1993;13(4):218–220. doi: 10.1007/BF02888013. [DOI] [PubMed] [Google Scholar]
- 7.Dastugue N, Payen C, Lafage-Pochitaloff M, Bernard P, Leroux D, Huguet-Rigal F, et al. Prognostic significance of karyotype in de novo adult acute myeloid leukemia. The BGMT group. Leukemia. 1995 Sep;9(9):1491–1498. [PubMed] [Google Scholar]
- 8.Jung HA, Maeng CH, Park S, Kim SJ, Kim K, Jang JH, et al. Prognostic factor analysis in core-binding factor-positive acute myeloid leukemia. Anticancer Res. 2014 Feb;34(2):1037–1045. [PubMed] [Google Scholar]
- 9.Numata A, Fujimaki K, Aoshima T, Onizuka M, Hagihara M, Miyazaki K, et al. Retrospective analysis of treatment outcomes in 70 patients with t(8;21) acute myeloid leukemia. Rinsho Ketsueki. 2012 Jul;53(7):698–704. [PubMed] [Google Scholar]
- 10.O'Donnell MR, Tallman MS, Abboud CN, Altman JK, Appelbaum FR, Arber DA, et al. Acute myeloid leukemia, version 2.2013. J Natl Compr Canc Netw. 2013 Sep;11(9):1047–1055. doi: 10.6004/jnccn.2013.0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coenen EA, Raimondi SC, Harbott J, Zimmermann M, Alonzo TA, Auvrignon A, et al. Prognostic significance of additional cytogenetic aberrations in 733 de novo pediatric 11q23/MLL-rearranged AML patients: results of an international study. Blood. 2011 Jun;117(26):7102–7111. doi: 10.1182/blood-2010-12-328302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baldus CD, Mrózek K, Marcucci G, Bloomfield CD. Clinical outcome of de novo acute myeloid leukaemia patients with normal cytogenetics is affected by molecular genetic alterations: a concise review. Br J Haematol. 2007 Jun;137(5):387–400. doi: 10.1111/j.1365-2141.2007.06566.x. [DOI] [PubMed] [Google Scholar]
- 13.Kroeger H, Jelinek J, Estécio MR, He R, Kondo K, Chung W, et al. Aberrant CpG island methylation in acute myeloid leukemia is accentuated at relapse. Blood. 2008 Aug;112(4):1366–1373. doi: 10.1182/blood-2007-11-126227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toyota M, Kopecky KJ, Toyota MO, Jair KW, Willman CL, Issa JP. Methylation profiling in acute myeloid leukemia. Blood. 2001 May;97(9):2823–2829. doi: 10.1182/blood.v97.9.2823. [DOI] [PubMed] [Google Scholar]
- 15.Galm O, Wilop S, Lüders C, Jost E, Gehbauer G, Herman JG, et al. Clinical implications of aberrant DNA methylation patterns in acute myelogenous leukemia. Ann Hematol. 2005 Dec;84(Suppl 1):39–46. doi: 10.1007/s00277-005-0005-0. [DOI] [PubMed] [Google Scholar]
- 16.Damm F, Markus B, Thol F, Morgan M, Göhring G, Schlegelberger B, et al. TET2 mutations in cytogenetically normal acute myeloid leukemia: Clinical implications and evolutionary patterns. Genes Chromosomes Cancer. 2014 Jun; doi: 10.1002/gcc.22191. [DOI] [PubMed] [Google Scholar]
- 17.Kroeze LI, Aslanyan MG, van Rooij A, Koorenhof-Scheele TN, Massop M, Carell T, et al. Characterization of acute myeloid leukemia based on levels of global hydroxymethylation. Blood. 2014 Jul; doi: 10.1182/blood-2013-08-518514. [DOI] [PubMed] [Google Scholar]
- 18.Tian X, Xu Y, Yin J, Tian H, Chen S, Wu D, et al. TET2 gene mutation is unfavorable prognostic factor in cytogenetically normal acute myeloid leukemia patients with NPM1(+) and FLT3-ITD (−) mutations. Int J Hematol. 2014 Jul;100(1):96–104. doi: 10.1007/s12185-014-1595-x. [DOI] [PubMed] [Google Scholar]
- 19.Tie R, Zhang T, Fu H, Wang L, Wang Y, He Y, et al. Association between DNMT3A Mutations and Prognosis of Adults with De Novo Acute Myeloid Leukemia: A Systematic Review and Meta-Analysis. PLoS One. 2014;9(6):e93353. doi: 10.1371/journal.pone.0093353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hájková H, Marková J, Haškovec C, Sárová I, Fuchs O, Kostečka A, et al. Decreased DNA methylation in acute myeloid leukemia patients with DNMT3A mutations and prognostic implications of DNA methylation. Leuk Res. 2012 Sep;36(9):1128–1133. doi: 10.1016/j.leukres.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 21.Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004 Dec;4(12):988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 22.Shiovitz S, Bertagnolli MM, Renfro LA, Nam E, Foster NR, Dzieciatkowski S, et al. CpG Island Methylator Phenotype is Associated With Response to Adjuvant Irinotecan-Based Therapy for Stage 3 Colon Cancer. Gastroenterology. 2014 May; doi: 10.1053/j.gastro.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dumenil TD, Wockner LF, Bettington M, McKeone DM, Klein K, Bowdler LM, et al. Genome-wide DNA methylation analysis of formalin-fixed paraffin embedded colorectal cancer tissue. Genes Chromosomes Cancer. 2014 Jul;53(7):537–548. doi: 10.1002/gcc.22164. [DOI] [PubMed] [Google Scholar]
- 24.Nazemalhosseini Mojarad E, Kuppen PJ, Aghdaei HA, Zali MR. The CpG island methylator phenotype (CIMP) in colorectal cancer. Gastroenterol Hepatol Bed Bench. 2013;6(3):120–128. [PMC free article] [PubMed] [Google Scholar]
- 25.Yagi K, Akagi K, Hayashi H, Nagae G, Tsuji S, Isagawa T, et al. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res. 2010 Jan;16(1):21–33. doi: 10.1158/1078-0432.CCR-09-2006. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Long Y, Xu Y, Guan Z, Lian P, Peng J, et al. Prognostic and predictive value of CpG island methylator phenotype in patients with locally advanced nonmetastatic sporadic colorectal cancer. Gastroenterol Res Pract. 2014;2014:436985. doi: 10.1155/2014/436985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bady P, Sciuscio D, Diserens AC, Bloch J, van den Bent MJ, Marosi C, et al. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. 2012 Oct;124(4):547–560. doi: 10.1007/s00401-012-1016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010 May;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohka F, Natsume A, Motomura K, Kishida Y, Kondo Y, Abe T, et al. The global DNA methylation surrogate LINE-1 methylation is correlated with MGMT promoter methylation and is a better prognostic factor for glioma. PLoS One. 2011;6(8):e23332. doi: 10.1371/journal.pone.0023332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Branham MT, Marzese DM, Laurito SR, Gago FE, Orozco JI, Tello OM, et al. Methylation profile of triple-negative breast carcinomas. Oncogenesis. 2012;1:e17. doi: 10.1038/oncsis.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang F, Turcan S, Rimner A, Kaufman A, Giri D, Morris LG, et al. Breast cancer methylomes establish an epigenomic foundation for metastasis. Sci Transl Med. 2011 Mar;3(75):75ra25. doi: 10.1126/scitranslmed.3001875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marcucci G, Yan P, Maharry K, Frankhouser D, Nicolet D, Metzeler KH, et al. Epigenetics meets genetics in acute myeloid leukemia: clinical impact of a novel seven-gene score. J Clin Oncol. 2014 Feb;32(6):548–556. doi: 10.1200/JCO.2013.50.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jelinek J, Mannari R, Issa J-P. Identification of 41 Novel Promoter-Associated CpG Islands Methylated in Leukemias. ASH Annual Meeting Abstracts. 2004 Nov 16;104(11):1126-. 2004. [Google Scholar]
- 34.Kroeger H, Jelinek J, Kornblau SM, Bueso-Ramos CE, Issa J-P. Increased DNA Methylation Is Associated with Good Prognosis in AML. ASH Annual Meeting Abstracts. 2007 Nov 16;110(11):595-. 2007. [Google Scholar]
- 35.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009 Dec;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010 Mar;17(3):225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010 Dec;18(6):553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011 Jan;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jelinek J, Liang S, Lu Y, He R, Ramagli LS, Shpall EJ, et al. Conserved DNA methylation patterns in healthy blood cells and extensive changes in leukemia measured by a new quantitative technique. Epigenetics. 2012 Dec;7(12):1368–1378. doi: 10.4161/epi.22552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013 May;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fortin JP, Labbe A, Lemire M, Zanke BW, Hudson TJ, Fertig EJ, et al. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014;15(12):503. doi: 10.1186/s13059-014-0503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013 Jan;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Jonge HJ, Valk PJ, Veeger NJ, ter Elst A, den Boer ML, Cloos J, et al. High VEGFC expression is associated with unique gene expression profiles and predicts adverse prognosis in pediatric and adult acute myeloid leukemia. Blood. 2010 Sep;116(10):1747–1754. doi: 10.1182/blood-2010-03-270991. [DOI] [PubMed] [Google Scholar]
- 44.Gaidzik VI, Bullinger L, Schlenk RF, Zimmermann AS, Röck J, Paschka P, et al. RUNX1 mutations in acute myeloid leukemia: results from a comprehensive genetic and clinical analysis from the AML study group. J Clin Oncol. 2011 Apr;29(10):1364–1372. doi: 10.1200/JCO.2010.30.7926. [DOI] [PubMed] [Google Scholar]
- 45.R Core Development Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 46.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B. 1995;57(1):289–300. [Google Scholar]
- 47.Therneau T. A Package for Survival Analysis in S. R package version 2.37-7 ed. 2014 [Google Scholar]
- 48.Maegawa S, Gough SM, Watanabe-Okochi N, Lu Y, Zhang N, Castoro RJ, et al. Age-related epigenetic drift in the pathogenesis of MDS and AML. Genome Res. 2014 Apr;24(4):580–591. doi: 10.1101/gr.157529.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999 Jul;96(15):8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verhaak RG, Wouters BJ, Erpelinck CA, Abbas S, Beverloo HB, Lugthart S, et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica. 2009 Jan;94(1):131–134. doi: 10.3324/haematol.13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao Y, Sun H, Wang H. Long noncoding RNAs in DNA methylation: new players stepping into the old game. Cell Biosci. 2016;6:45. doi: 10.1186/s13578-016-0109-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013 Nov;503(7476):371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiong J, Zhang Z, Chen J, Huang H, Xu Y, Ding X, et al. Cooperative Action between SALL4A and TET Proteins in Stepwise Oxidation of 5-Methylcytosine. Mol Cell. 2016 Oct; doi: 10.1016/j.molcel.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 54.Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014 Mar;74(5):1311–1318. doi: 10.1158/0008-5472.CAN-13-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He D, Zhang YW, Zhang NN, Zhou L, Chen JN, Jiang Y, et al. Aberrant gene promoter methylation of p16, FHIT, CRBP1, WWOX, and DLC-1 in Epstein-Barr virus-associated gastric carcinomas. Med Oncol. 2015 Apr;32(4):92. doi: 10.1007/s12032-015-0525-y. [DOI] [PubMed] [Google Scholar]
- 56.Rampal R, Alkalin A, Madzo J, Vasanthakumar A, Pronier E, Patel J, et al. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep. 2014 Dec;9(5):1841–1855. doi: 10.1016/j.celrep.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y, Xiao M, Chen X, Chen L, Xu Y, Lv L, et al. WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol Cell. 2015 Feb;57(4):662–673. doi: 10.1016/j.molcel.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kallin EM, Rodríguez-Ubreva J, Christensen J, Cimmino L, Aifantis I, Helin K, et al. Tet2 facilitates the derepression of myeloid target genes during CEBPα-induced transdifferentiation of pre-B cells. Mol Cell. 2012 Oct;48(2):266–276. doi: 10.1016/j.molcel.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Di Stefano B, Sardina JL, van Oevelen C, Collombet S, Kallin EM, Vicent GP, et al. C/EBPα poises B cells for rapid reprogramming into induced pluripotent stem cells. Nature. 2014 Feb;506(7487):235–239. doi: 10.1038/nature12885. [DOI] [PubMed] [Google Scholar]
- 60.Sinha S, Thomas D, Yu L, Gentles AJ, Jung N, Corces-Zimmerman MR, et al. Mutant WT1 is associated with DNA hypermethylation of PRC2 targets in AML and responds to EZH2 inhibition. Blood. 2015 Jan;125(2):316–326. doi: 10.1182/blood-2014-03-566018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maegawa S, Hinkal G, Kim HS, Shen L, Zhang L, Zhang J, et al. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010 Mar;20(3):332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.An B, Kondo Y, Okamoto Y, Shinjo K, Kanemitsu Y, Komori K, et al. Characteristic methylation profile in CpG island methylator phenotype-negative distal colorectal cancers. Int J Cancer. 2010 Nov;127(9):2095–2105. doi: 10.1002/ijc.25225. [DOI] [PubMed] [Google Scholar]
- 63.Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016 Jan;529(7584):110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.