Abstract
The PIM kinase family (PIM1, 2 and 3) play a central role in integrating growth and survival signals, and are expressed in a wide range of solid and hematological malignancies. We now confirm that PIM2 is overexpressed in multiple myeloma (MM) patients, and within MM group it is overexpressed in the high-risk MF subset (activation of proto-oncogenes MAF/MAFB). This is consistent with our finding of PIM2’s role in key signaling pathways (IL-6, CD28 activation) that confer chemotherapy resistance in MM cells. These studies have identified a novel PIM2-selective non-ATP competitive inhibitor (JP11646) that has a 4 to 760-fold greater suppression of MM proliferation and viability than ATP-competitive PIM inhibitors. This increased efficacy is due not only to the inhibition of PIM2 kinase activity, but also to a novel mechanism involving specific downregulation of PIM2 mRNA and protein expression not seen with the ATP competitive inhibitors. Treatment with JP11646 in xenogeneic myeloma murine models demonstrated significant reduction in tumor burden and increased median survival. Altogether our findings suggest the existence of previously unrecognized feedback loop(s) where PIM2 kinase activity regulates PIM2 gene expression in malignant cells, and that JP11646 represents a novel class of PIM2 inhibitors that interdicts this feedback.
Introduction
The PIM (Proviral Insertion site in Moloney Murine Leukemia Virus (MMLV)) family of serine/threonine kinases (PIM1, PIM2 and PIM31) play a central role in integrating growth signals that regulate a number of cellular pathways2–7. PIMs are constitutively active, transcriptionally and translationally regulated8, 9 and they are upregulated in a wide range of solid and hematological malignancies2, 10, 11. PIM2 specifically is upregulated in acute myeloid leukemia (AML)12 and the plasma cell dyscrasia multiple myeloma (MM)10, 11, 13–15. In myeloma, stromally-derived factors like IL-616 or the TNF family of cytokines13, via NFκB signaling13 regulate PIM2 and its key role in MM cell survival and adaptation to stress (e.g. to chemotherapy treatment)13, 17, 18. This is in part mediated by PIM2 phosphorylation of its downstream targets TSC2 (and modulation of mTORC1 activity19), the pro-apoptotic factor BAD and the protein translational inhibitor 4EBP1. PIM2 has also been implicated in the myeloma-mediated bone destruction via negative regulation of osteoblastogenesis15, 20 and more recently in the regulation of the essential DNA damage response pathway in myeloma21.
These observations suggest that therapeutically targeting PIM2 may have significant efficacy in MM. Of the PIM inhibitors5 identified so far, most are small molecule ATP-competitive kinase inhibitors that affect multiple downstream pathways22 and show more specificity to PIM1 and PIM3 than PIM223–25. In preclinical studies the PIM2 selective inhibitor LGB32122, 24 has been shown to induce MM cell death by inhibition of mTORC1 activity19, and in a phase I MM trial the pan-PIM inhibitor LGH44719, 26 had acceptable toxicity with evidence for clinical efficacy15. But other PIM inhibitors (e.g. SGI177625, AZD120815, AZD189715, 27) have shown only limited preclinical single agent activity in myeloma23. Why the different PIM inhibitors have qualitatively different efficacy in MM is not clear, although compensatory feedback mechanisms have been implicated in some studies11, 28.
We now report on a new non-ATP competitive PIM2-selective inhibitor JP11646 with potent anti-MM activity that in addition to inhibition of PIM kinase enzymatic activity has a novel mechanism of action downregulating PIM2 mRNA and protein expression. This latter effect overcomes the compensatory upregulation of PIM2 expression seen with the other PIM kinase inhibitors, and reveals previously unrecognized feedback loops regulating PIM2 expression and activity in multiple myeloma.
Materials and Methods
Cell culture
The aminopyrimidine kinase inhibitor JP11646 and other members of its family of inhibitors (Table 1), LGB321 (Novartis, Berkeley, CA) and AZD1208 (Astra Zeneca, London, UK) were synthesized and obtained from Jasco Pharmaceuticals (Woburn, MA) and were used as 10 mM stocks in DMSO. The MM cell lines MM1.S (gift from Dr. Stephen Rosen, Chicago, IL), RPMI8226, U266 and KMS11 (ATCC, Manassas, VA), were maintained in RPMI 1640 media (Mediatech Inc, Manassas, VA) containing 10% FBS (Hyclone Inc., Logan, UT), 1000 U/ml penicillin/streptomycin, 4 mM L-glutamine, 0.1 mM non-essential amino acids, 1 mM sodium pyruvate. Recombinant IL-6 was purchased from R&D Systems (Minneapolis, MN).
Table 1.
IC50 values for pan-PIM inhibitors as determined by PIM enzyme assays.
| PIM inhibitors | PIM1 IC50 (nM) | PIM2 IC50 (nM) | PIM3 IC50 (nM) | Manufacturer |
|---|---|---|---|---|
| SGI-1776 | 7 | 363 | 69 | SuperGen |
| JP11413 | 38 | 0.6 | 9 | Jasco |
| JP11416 | 36 | 1 | 6 | Jasco |
| JP11644 | 32 | 0.7 | 6 | Jasco |
| JP11646 | 24 | 0.5 | 1 | Jasco |
| JP11660 | 42 | 2 | 4 | Jasco |
| JP11662 | 50 | 9 | 4 | Jasco |
Antibodies and flow cytometry
Antibodies used for flow cytometry were obtained from Beckman Coulter (Fullerton, CA), and BD Biosciences (San Jose, CA), and those used in western immunoassays from Cell Signaling Technology (Beverly, MA). MM treatment with αCD28 mAb, neutralizing αIL6 and CTLA4Ig and flow cytometry for viability were done as described previously29.
Gene expression analysis
PIM2 expression analyses were done as described previously using datasets GSE6477 and GSE420429.
Proliferation and viability assays
Viability assays for MM cells alone or with dendritic cells (DC) were performed in 96 well flat bottom plates (1×105 cells/well in 200 µl) treated as indicated, and viability estimated by flow cytometry after 24–48 hrs as described previously29. Proliferation assays (5×104 cells/ml for RPMI8226 and KMS11, 1.25×105 cells/ml for U266 and MM1.S) were done as described previously29. LC50 and GI50 values for viability and proliferation respectively were estimated using linear regression analysis.
Western immunoassays
Western blotting and immunoassays were performed as described previously29. Blots were treated with ECL Western Blotting Substrate (Thermo Fisher Scientific, Waltham, MA) and visualized either by exposing to photographic film or acquired and analyzed using a LICOR™ Odyssey FC western blot documentation system (Lincoln, NE).
mRNA half-life analysis
MM cells in culture were incubated with transcription inhibitor actinomycin D (5 µg/ml) and cells (3×106) were collected at different time points over 12 hours, washed twice with cold PBS, RNA isolated (TRIZOL®, Invitrogen Corp, Carlsbad, CA) and quantified on a Synergy-H1 reader (BioTek Laboratories Inc, Winooski, VT), and 10–15 µg of RNA was used for northern blotting and hybridization as described previously30. RT-PCR using forward primer 5’-TCGAAGTCGCACTGCTATGG – 3’ and reverse 5’-TCGAGAGATCCACTCTGGGG – 3’ on MM1.S total RNA was used to obtain a 389 bp amplicon that was labeled using α32P –dCTP (random primer DNA labeling system, Life Technologies, Grand Island, NY) for use as probe in northern blots.
NFκB luciferase reporter assays
Cells (2×106 cells) were transfected using the Amaxa™ nucleofector system (Lonza AG, Allendale, NJ) with pGL4.32 (Promega, Madison, WI) luciferase expression vector. After 24 hours of transfection, cells were treated for a further 24–48 hrs as detailed. Samples were assayed for luciferase activity with Dual-Glo® Luciferase Assay system (Promega, Madison, WI.) as per manufacturer’s instructions.
Electromobility shift assays (EMSA)
EMSAs to detect active NFκB dimers were done as described previously31.
Gene silencing
Cells were treated with PIM2 specific siRNA from Dharmacon, Inc., using Amaxa nucleofector as described by manufacturer (Lonza Biosystems, Walkersville MD), 24 hrs prior to initiation of experiments.
In vivo murine studies
All studies were done under RPCI IACUC-approved animal use protocols. SCID/SCIDCBIgh.lblcrTac.Prkdcscid/Ros mice (Roswell Park Cancer Institute (RPCI) Laboratory Animal Resource) < 5 weeks of age were irradiated at 300 rads using a Mark II Cesium irradiator 24 hours prior to injecting them with MM1.S cells (5×106) subcutaneously under the skin on the left ventral flank. Once palpable subcutaneous tumors reached a volume of 100–200 mm3 (length × width × width/2), the mice were randomly grouped 5–6/group and drug treatments were initiated. For in vivo studies, JP11646 was prepared fresh (2.5 mg/ml, i.p) in a proprietary carrier solution of 30% modified β-cyclodextrin (Ligand Pharmaceuticals Inc, La Jolla, CA). Mice with necrotic tumors were euthanized as per IACUC guidelines. Median survival for each group was calculated using GraphPad Prism software and p values were determined using the log rank (Mantel-Cox) test. Tumor volumes were assessed twice a week over the course of the experiments. In separate studies, mice with tumors were treated with JP11646 15 µg/gm and tumor harvested 0, 1, 3 and 6 hours post injection. CD138+ MM cells were harvested from macerated tumors using αCD138 coated magnetic beads (Dynal™ beads, Invitrogen) as recommended by manufacturer, protein lysate prepared and analyzed by western immunoassays.
Statistical analysis
All experiments were performed at least twice and tests of significance was done using Student’s T test on Microsoft excel unless otherwise stated. Samples were always assayed in triplicate (n=3) unless otherwise stated.
Results
PIM2 is overexpressed in multiple myeloma
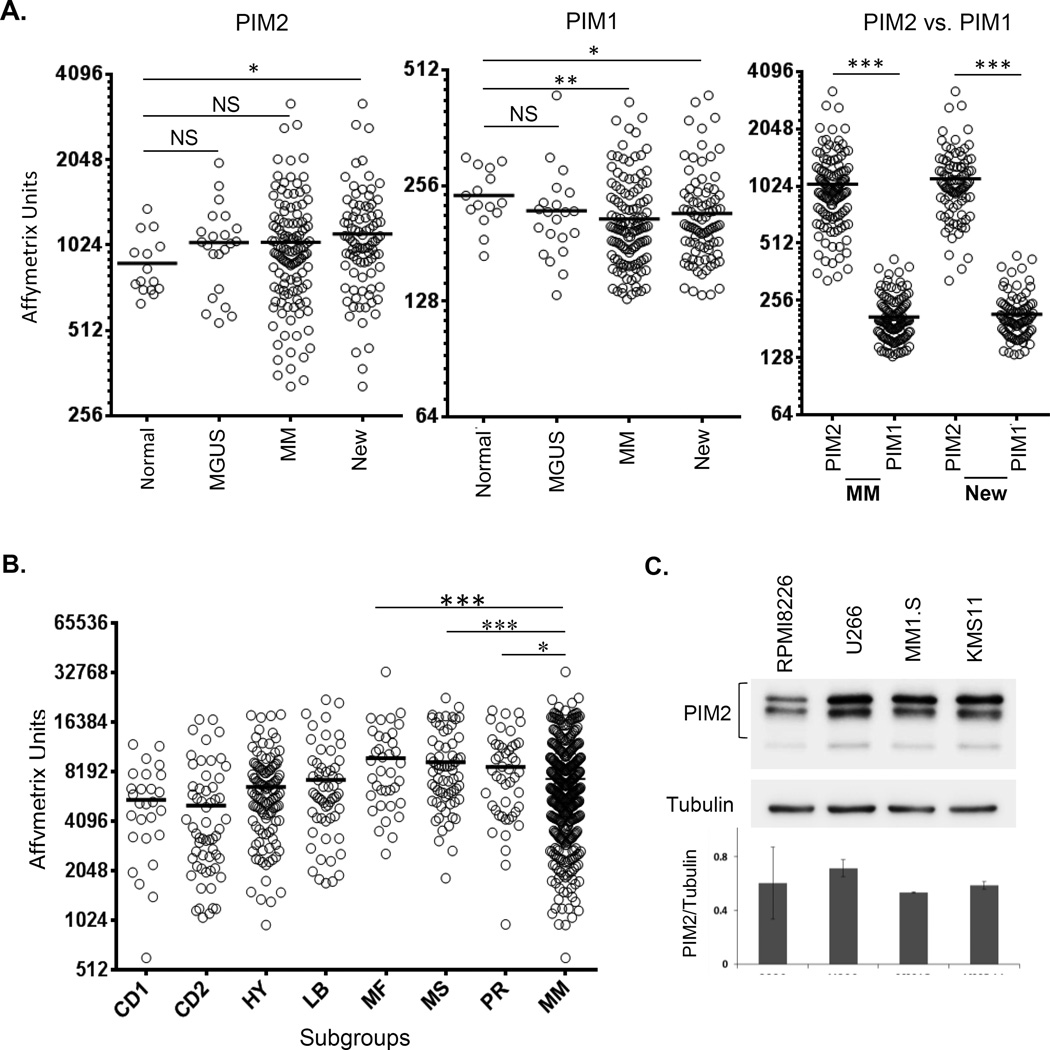
Our analysis of publicly available MM patient data sets shows increasing expression of PIM2 with disease progression from normal PC to myeloma (MM), with significantly greater expression in newly diagnosed MM patients (New) vs. normal PC (Fig. 1A, left). Re-analysis did not show any significant difference in PIM2 expression if the relapsed patients were removed from the total MM patient population (not shown). The increased PIM2 expression in MM versus normal controls was further verified by performing a similar analysis of PIM2 expression on another dataset GSE3975432 (sFig. 1). This was specific to PIM2, as PIM1 expression significantly decreased from normal PC to MM (Fig. 1A, center). PIM3 expression values being statistically insignificant were excluded. PIM1 expression in the whole MM group or newly diagnosed patients was significantly lower (~5 fold, p<0.0001) than PIM2 (Fig. 1A, right), consistent with previous studies demonstrating the predominance of PIM2 overexpression in myeloma15. Within the myeloma patient subgroups33, PIM2 expression was significantly higher in the poor prognostic MF subgroup compared to the whole MM group (Fig. 1B). PIM2 expression in subgroup MS and PR also show significantly higher PIM2 expression from the whole MM population. Western immunoassays revealed similar levels of protein expression of all three isoforms of PIM2 (34, 38 and 40 kDa) in the MM cell lines RPMI8226, MM1.S, U266 and KMS11 (Fig. 1C).
Figure 1.
Analysis of expression of PIM1 and PIM2 in myeloma patients and cell lines. A) Gene expression datasets from CD138+ plasma cells from normal controls, MGUS and myeloma patients (total and newly diagnosed (New)) showing PIM2 (left) and PIM1 (center) expression, and absolute expression of PIM2 vs. PIM1 within total MM population and in the newly diagnosed subset (right). B) PIM2 levels in different genetic signature subgroups within MM group. Horizontal bars show mean expression. NS=Not significant, *p<0.05, **p<0.01, ***p<0.001. C) Western blot analysis of PIM2 expression (representative of 3 experiments) in 4 MM cell lines. Densitometric analysis for PIM2 expression relative to tubulin is shown on the bottom panel.
JP11646 is selective PIM2 inhibitor
PIM2 overexpression in MM (in particular in the MF signature subgroup) suggests it may be a therapeutic target for small molecule kinase inhibition. In cell-free enzyme assays with PIM kinases, a novel series of PIM kinase inhibitors (JP11413-11662, US patents US 8,563,539 B2, US 8,901,145 B2, US 8,927,525 B2, US 9,073,903 B2, & US 9,157,077 B2) (Table 1) demonstrated low nanomolar IC50 values and showed greater inhibition of PIM2 than PIM1 or PIM3, with JP11646 being the most potent (IC50 values of 24, 0.5 and 1 nM for PIM1, PIM2 and PIM3 respectively) (sTable 1). JP11646 inhibitory effect on other kinases as assessed using a kinase selectivity panel was considerably less (sTable 1).
The low Km of PIM2 for ATP22 necessitates high potency of the ATP-competitive PIM2 inhibitors to block PIM2 enzymatic activity. Pharmacokinetic studies of JP11646 matched that of a freely reversible inhibitor standard (sFig. 2A), with slopes of 67 and 63 respectively for test and standard. EC50 concentrations show only a 1.67-fold increase across a 100-fold ATP change (0.15 – 0.25 nM, sFig. 2B) and the replot of EC50 values against ATP concentrations (sFig 2C) that match a non-competitive (NC) inhibitor are consistent with the non-competitive nature of JP11646 binding to PIM2 with respect to ATP. Using the Cheng and Prusoff algorithm34, a Ki of 0.165 nM was determined for JP11646. Thus, JP11646 differs from other reported PIM2 inhibitors by being freely reversible and ATP non-competitive. Since inhibiting PIM kinases is known to affect myeloma survival13, 24, 25, and most of the current PIM inhibitors preferentially inhibit PIM1 or PIM3 over PIM2, we next examined the effect of selective PIM2 inhibition by JP11646 treatment on downstream PIM2 targets in myeloma cells.
Effect of JP11646-mediated inhibition on downstream molecular targets of PIM2
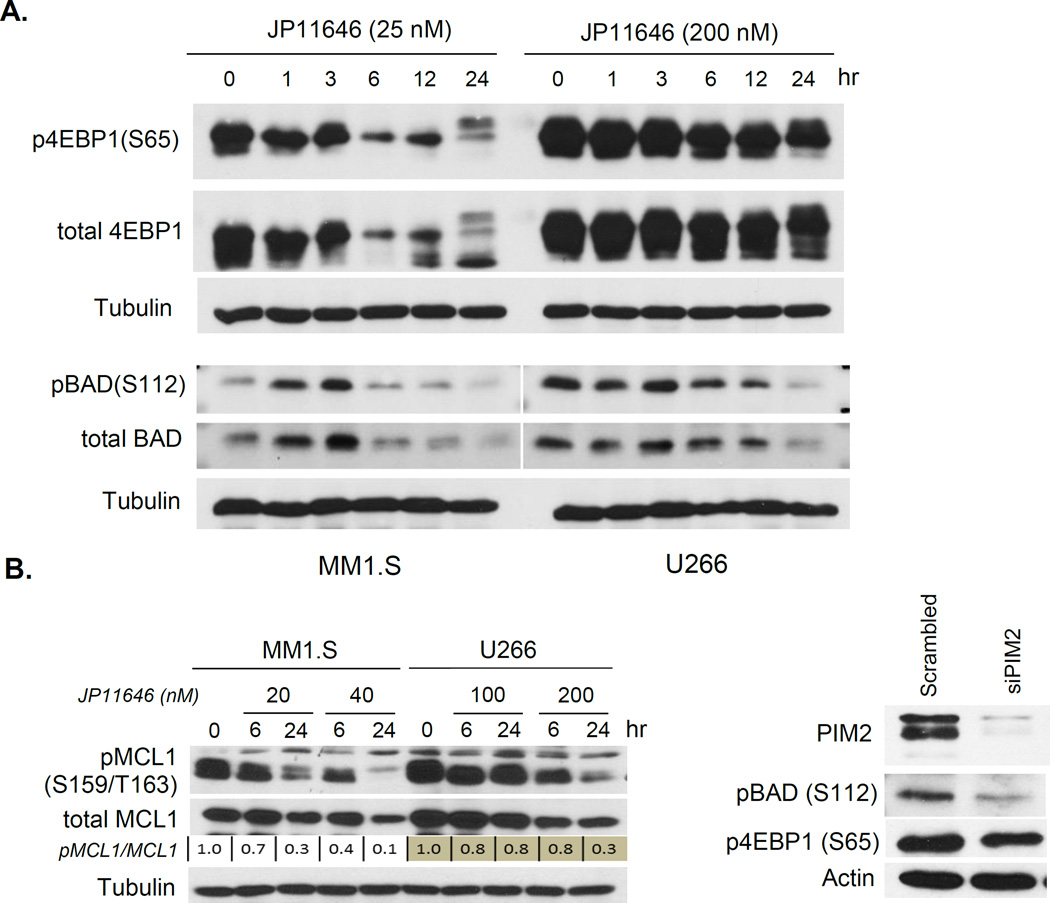
PIM2 mediates its pro-survival role by phosphorylating a number of downstream targets including 4EBP135, BAD22, 36, 37 and MCL138, which all play central roles in MM cell proliferation and survival. PIM2 phosphorylation and inhibition of the negative regulator of translation 4 E-BP1 is important for enhancing the Unfolded Protein Response (UPR)39, and phosphorylation of the pro-apoptotic factor BAD blocks BAD activation-induced cell death in myeloma3. MM1.S and U266 treated with JP11646 show a progressive decrease in p4EBP1 (S65) and pBAD (S112) over time, as well as decreases in the total levels of these proteins (which may be in part due to positive feedback from the phosphorylated forms on total expression7)(Fig. 2A). In addition, there was a dose-dependent decrease in the more potent phosphorylated form of the anti-apoptotic factor MCL1 (Ser159/Thr163) to 10% (MM1.S) or 30% (U266) of total MCL1 compared to untreated cells (Fig. 2B Left). Total MCL1 levels also demonstrate a less marked dose-dependent decrease over time, likely due to the role of active pMCL1 in sustaining total MCL1 levels38. Silencing PIM2 with siRNA (Fig. 2B Right) produce similar decreases in pBAD and p4EBP1 levels, suggesting that the effect of JP11646 is due to its inhibition of PIM2. Pathway analysis of whole transcriptome analysis of untreated vs. JP11646 treated MM1.S and U266 cells (sTable 2) demonstrated in both cell lines a disproportionate effect on components of nucleotide and amino acid metabolism pathways as well as the UPR response pathway, (sTable 3).
Figure 2.
Modulation of PIM2 associated cellular activity by JP11646. A) Protein lysates from MM cells treated with JP11646 was immunoassayed for phosphorylated forms of the PIM2 substrates 4EBP1 and BAD. Tubulin is used as loading control. B, Left) Western assay of MCL1 and its active form p-MCL1 on proteins lysates from MM cells treated with two concentrations of JP11646 across 2 time points. Densitometric quantitation normalized (to tubulin) expression of pMCL1/MCl1 relative to respective untreated 0 hr control is shown as numbers below the figures. B, Right) MM1.S were transfected with scrambled siRNA control or PIM2 siRNA, and after 48 hours cell lysates were analyzed for PIM2, pBAD, p4EBP1 and actin expression by western blot. C) MM1.S treated with melphalan (Mel) (2 µM) or bortezomib (BTZ) (1.5 nM) or dexamethasone (Dex) (5 µM) with or without sublethal concentration of JP11646 (10 nM) and cell viability determined by flow cytometry after 48 hrs (Left). Viability of U266 cells treated with Mel (4 µM), BTZ (2.5 nM) with or without JP11646 (100 nM) (Right). *p<0.05, **p<0.01. All data are representative from at least 2 independent replicate experiments.
Effect of JP11646-mediated PIM2 inhibition on MM proliferation and viability
We next examined the effect of PIM2 inhibition by JP11646 on MM proliferation and survival. We found that the anti-proliferative effect of JP11646 treatment was greatest in the MF-signature cell line MM1.S (Growth Inhibition (GI50) of 5 nM), although the sensitivity against the other MF-signature cell line RPMI8226 (GI50 of 36 nM) was similar to the non-MF cell lines (15 nM for MS signature KMS11, and 37 nM for CD1 signature U266) (Table 2, sFig. 3A). However, JP11646’s inhibition of cell viability was more pronounced for the MF-signature cell lines RPMI8226 and MM1.S (LC50 values of 46 and 16 nM respectively), in comparison to 67 and 222 nM for KMS11 and U266 respectively (Table 2, sFig. 3B).
TABLE 2.
GI50 and LC50 values for JP11646 and other pan-PIM inhibitors determined by cell based assays.
| PIM inhibitors | GI50 (nM) (Growth/proliferation) | |||
| RPMI8226 | U266 | MM1S | KMS11 | |
| JP11646 | 36 | 37 | 5 | 15 |
| AZD1208 | 2,626 | 4,272 | 711 | 4,666 |
| LGB321 | 156 | 342 | 22 | 123 |
| JP12461 | 30,797 | 14,053 | 624 | 9,814 |
| LC50 (nM) (Viability) | ||||
| JP11646 | 46 | 222 | 16 | 67 |
| AZD1208 | 30,013 | 56,561 | 23,678 | 31,320 |
| LGB321 | 8,024 | 8,471 | 501 | 25,284 |
| JP12461 | 91,112 | 90,665 | 38,273 | 58,787 |
When compared to the ATP-competitive PIM inhibitors, JP11646 show >80 fold and >4 fold greater inhibition of proliferation (GI50 of 5–37 nM) versus AZD1208 (0.7 – 4.7 µM) or LGB321 (22 – 343 nM) respectively (Table 2, sFig. 3C–F). For viability, JP11646 showed >760 fold and >230 fold inhibition (LC50 of 16–222 nM) compared to AZD1208 (23 – 56 µM) and LGB321 (0.5 – 25 µM) respectively (Table 2, sFig. 3G–J). Like JP116464, both AZD1208 and LGB321 were more effective inducing cell death in the MF-signature cell lines.
In addition to single agent activity, the ability to sensitize MM cells in combination with other anti-myeloma chemotherapies was tested using sub lethal concentrations of JP11646 (10 nM for MM1.S (viability of 87%) and 100 nM for U266 (viability of 81%)). In MM1.S, cell viability with melphalan decreased from 36% (alone) to 15% in combination with JP11646; with dexamethasone, from 55% (alone) to 8% in combination with JP11646; and with bortezomib from 18% (alone) to 2% in combination with JP11646 (sFig. 3K, left). In the more resistant cell line U266, viability in melphalan alone (58%) or bortezomib alone (85%) was significantly reduced to 39% or 53% respectively in combination with JP11646 (sFig. 3K, Right). Consistent with the single agent findings, there was greater efficacy of the bortezomib + JP11646 combination in MF-signature cell line MM1.S than with CD1-signature cell line U266.
These data demonstrate that PIM inhibition with JP11646 causes greater inhibition of proliferation and cell viability than seen with the other ATP-competitive inhibitors, despite the equivalent ability to inhibit PIM kinase activity in cell-free assays15, 40. One on-target explanation for this disparity is that inhibition with JP11646 is affecting PIM2 kinase activity through mechanisms in addition to blocking enzymatic activity.
JP11646 treatment inhibits PIM2 expression in myeloma cells
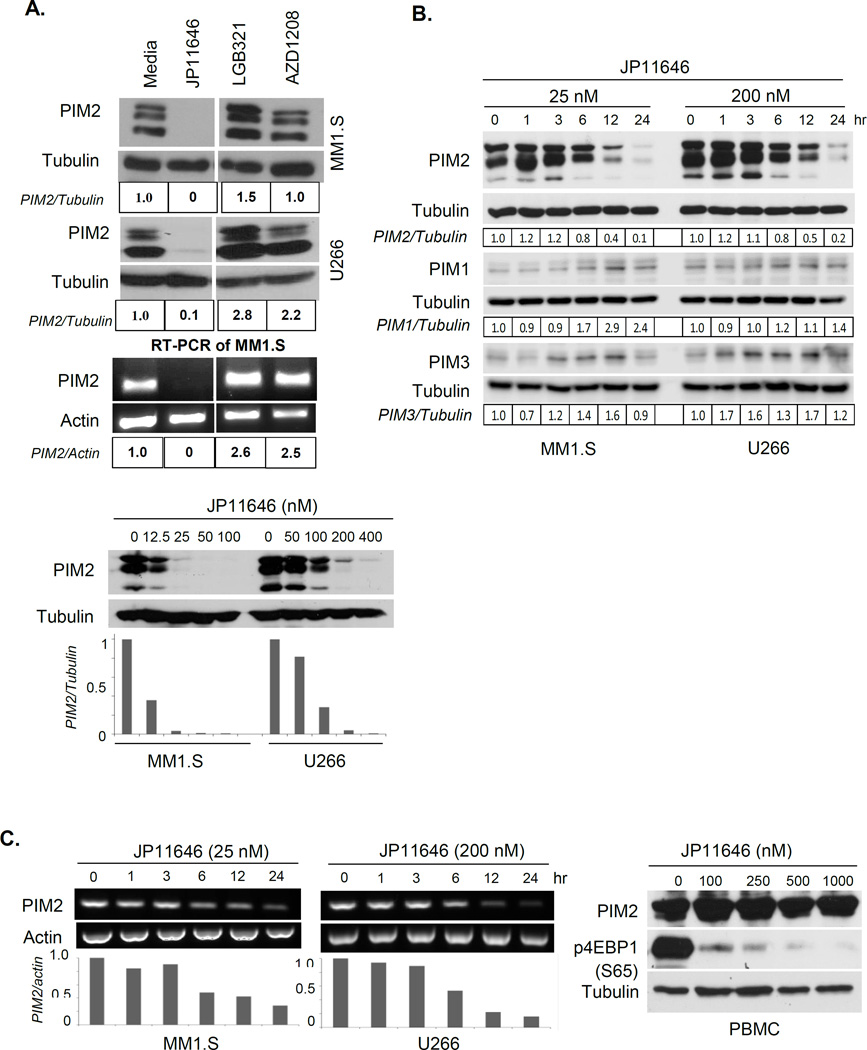
Since the PIMs are constitutively active, total activity is regulated by PIM expression levels. We found that in contrast to the unchanged-to-upregulated expression of PIM2 following treatment with LC50 doses of AZD1208 or LGB321, JP11646 treatment decreased PIM2 protein and mRNA levels to undetectable levels (Fig. 3A, top). These findings suggest that inhibition of PIM2 kinase activity can induce compensatory upregulation of PIM2 expression, and the ability of JP11646 to also inhibit PIM2 expression may account for its increased potency. The concentration and time dependency of JP11646 effect on PIM2 expression was further characterized by treating MM cells with increasing doses over 24 hrs, or treating with LC50 doses over a time course. In both MM1.S and U266, decreases in PIM2 expression were observed at concentrations as low as 12.5 and 50 nM, with 90% loss at 100 and 400 nM respectively (Fig. 3A, bottom). Over time, a slight increase at 1 hour was followed by a progressive decrease between 3 and 6 hrs (86% and 83% respectively). This was specific to PIM2, as in the same experiment there was a concomitant increase in PIM1 and PIM3 expression (Fig. 3B). Similar decreases in PIM2 mRNA levels were also observed (30% and 16% of control respectively, Fig. 3C, Left). Surprisingly, in normal primary human peripheral blood mononuclear cells (PBMC) there was no effect of JP11646 on PIM2 expression, even though the substantial decreases in p4EBP1 indicated inhibition of PIM2 kinase activity (Fig. 3C, Right). Why the effect on PIM2 expression in normal PBMC vs. MM cells is different is not clear, although one possibility is that normal PBMC are largely quiescent.
Figure 3.
Inhibition by JP11646 downregulates PIM2 protein and mRNA expression. A, top) MM1.S cells were treated with LC50 concentrations of JP11646 (20 nM), LGB321 (500 nM) and AZD1208 (25 µM) over 24 hours before analyzing protein lysates by western blot (top). Similarly treated U266 cells are shown below. RT-PCR analysis on total mRNA from the MM1.S samples is shown in the bottom panel. Numbers below each panel indicate PIM2 expression first normalized to loading control (tubulin or actin) relative to untreated media control (tubulin or actin). A, bottom) MM cells (MM1.S or U266) treated with a dosing-range of JP11646 around the respective LC50 for each cell line. Relative expression of normalized PIM2 values shown below. B) MM cells (MM1.S or U266) treated with JP11646 and samples collected at different time points and westerns for PIM2, PIM1 and PIM3 are shown. Densitometric analysis of PIM expression (normalized to tubulin) relative to untreated control is shown below each panel. C) RT-PCR analysis of PIM2 mRNA expression for the same samples, with actin as loading control. Relative PIM2 expression to 0 hr control is shown below (left). Normal PBMC was treated with a dose-range of JP11646 (0–1000 nM) for 24 hours and then analyzed by Western blot for PIM2, p4EBP1 and tubulin expression (Right). D) MM cells were treated with JP11646 at 20 nM for MM1.S and 200 nM for U266 over 24 hours, after which cells were washed 3 times to remove JP11646 and incubated in fresh media without JP11646 (0 hr) for up to 48 hrs post washing. An untreated control (Pre) was also included. Protein lysates were western immunoassayed for PIM2 and tubulin (Top) while total RNA was analyzed by RT-PCR analysis for PIM2 expression with actin and GAPDH as loading controls (Bottom). A negative (neg, no template) control was also included. All data are representative from at least 2 independent replicate experiments.
Since JP11646 is a reversible inhibitor, we examined whether sustained treatment was necessary for the inhibition of PIM2 expression. MM1.S and U266 treated with JP11646 for 24 hrs, then washed and cultured in media without JP11646, showed continued inhibition of both PIM2 protein (Fig. 3D, Upper) and mRNA expression (Fig. 3D, Lower) over 12 (MM1.S) to 24 hours (U266) – and also continued to die at similar rates as cells continuously cultured in the presence of JP11646 (sFig. 4).
Loss of PIM2 mRNA expression suggests that either gene transcription or mRNA stability was being affected, as PIM mRNA are known to have destabilizing AUUUA sequences in their 3’ regions41. The half-life (t1/2) of PIM2 mRNA in untreated MM1.S and U266 cells following actinomycin D treatment (3.2 hr and 5.6 hr respectively (sFig 5)), was similar to what we observed with JP11646 treatment (Fig. 3D, 6 hrs and 5.9 hrs respectively), suggesting that PIM2 mRNA stability was not being affected, and instead points to an effect at the level of PIM2 transcription.
PIM2 inhibition overcomes growth factor and pro-survival signaling in myeloma
The preceding data suggests that JP11646 represents a new class of PIM2 inhibitors with a novel mechanism of action that simultaneously inhibits PIM2 kinase activity and gene expression – and this affords greater potency in part by abrogating compensatory upregulation of PIM2 expression. The pathways that regulate PIM2 expression are not well characterized5, but current studies13, 42–45 suggest interlocking loops where growth factor signals regulate PIM2 expression and that PIM2 is part of the downstream signaling pathway of the receptors for these growth factors.
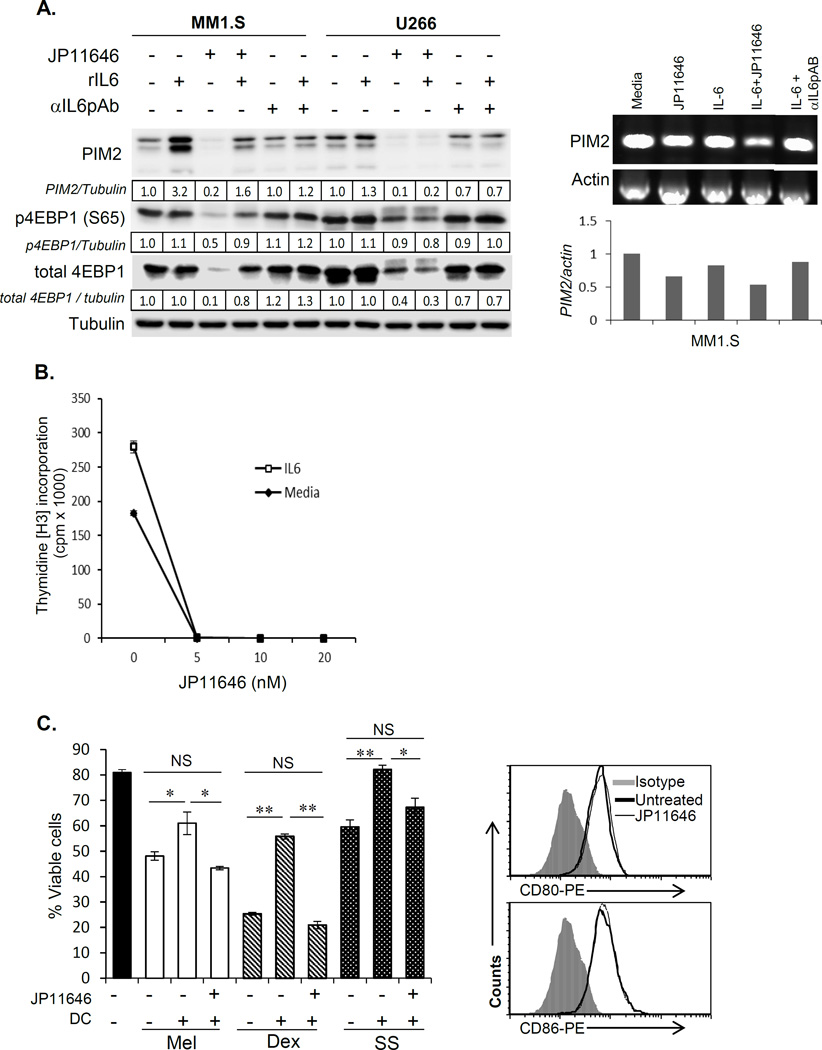
To assess this in the context of MM, we first examined the effect of the important stromally-derived MM survival factor IL-646, 47 on PIM2 expression, and the effect of PIM2 inhibition on IL-6 mediated cellular responses. In MM1.S and U266, recombinant IL-6 (rIL-6) induced upregulation of PIM2 (3.2 and 1.3 fold respectively), which was reversed by neutralizing αIL6 antibody (Fig. 4A, Left). JP11646-mediated inhibition of PIM2 expression (>80% in MM1.S and >90% in U266) was partially reversed by rIL-6 in MM1.S, along with partial restoration of phospho- and total 4EBP1 levels. But IL-6 had no effect in U266, possibly because an autocrine IL-6 growth loop makes U266 less responsive to exogenous IL-648. Interestingly, IL-6 had no effect on PIM2 mRNA expression in either setting (Fig. 4A, Right), suggesting IL-6 may be regulating PIM2 expression post-transcriptionally.
Figure 4.
PIM2 inhibition overcomes the proliferative and pro-survival effects of IL-6 and CD28 signaling. A, Left) MM cells were treated with IL-6 (25 ng/ml), JP11646 (20 nM for MM1.S and 200 nM for U266) and western assayed for PIM2 and its substrate 4EBP1. Sometimes neutralizing rabbit anti-hIL-6 was also included (1 µg/ml) to specifically examine the role of IL-6 in any effect observed. Tubulin was used as loading controls (Left). A, Right) Total RNA from MM cells cultured with IL-6 with or without JP11646 or neutralizing rabbit anti-IL6 antibodies were tested for PIM2 mRNA levels by semi-quantitative RT-PCR. PCR was performed over 22 cycles. Densitometric quantitation showing PIM2 mRNA levels relative to untreated control is shown below the figure. B) Proliferation assays (3H-thymidine incorporation) were performed on MM1.S cells cultured with recombinant IL6 (25 ng/ml) and treated with a dose range of JP11646 (0–20 nM). C, Left) MM1.S cells were cultured alone or with DC +/− sublethal concentration JP11646 (20 nM). Cell death was induced with melphalan (Mel, 10 µM), dexamethasone (Dex, 5 µM) or by serum starvation (SS) and viability assessed after 48 hrs. C, Right) DC were cultured alone +/− JP11646 (20 nM) for 48 hrs and analyzed by FACS for surface expression of CD28 ligands CD80 or CD86. F) Viability assays on MM1.S cells cultured under serum starvation to induce cell death with or without CD28 activation by CD28.2 mAb (25 µg/ml) +/− JP11646 (10 and 20 nM) over 48 hrs. Viability was quantitated by flow cytometry. D) MM1.S cells were cultured under low serum conditions (0.5% FBS) +/− CTLA4Ig (100 µg/ml) +/− JP11646 (20 nM) for 48 hr. Western blot analysis of PIM2 expression in MM cells cultured with or without CTLA4Ig for 48 hrs is shown in the inset. *p<0.05, **p<0.01, ***p<0.001, NS-Not significant. All data are representative from at least 2 independent replicate experiments.
In addition to IL-6 regulation of PIM2 expression, we also found that PIM2 inhibition by sub lethal doses of JP11646 completely blocked rIL-6-induced MM cell proliferation (Fig. 4B). Altogether these findings indicate that PIM2 is both a target of IL-6 signaling as well as a component of the biological response pathways to IL-6 signaling.
We had previously shown that the T cell costimulatory molecule CD28, also overexpressed in poor prognostic MF-signature MM patient subgroup29, when activated directly (agonistic antibodies) or indirectly via coculture with CD80+/CD86+ dendritic cells (DC) confers resistance to chemotherapy-induced cell death29, 31, 49, 50. We first assessed whether PIM2 was a component of the CD28-mediated pro-survival effect in MM-DC cocultures. We found that DC-mediated protection of MM1.S cells against death caused by melphalan, dexamethasone, or serum starvation was completely reversed by PIM2 inhibition by sublethal concentrations of JP11646 (Fig. 4C, Left) – with no effect on DC expression of CD28 ligands CD80/CD86 (Fig. 4C, Right) or DC viability (not shown). Similarly, the pro-survival effect of directly activating CD28 on MM cells alone with agonistic mAb was also completely reversed by JP11646 treatment (Fig. 4D, Left). Furthermore, CD28 activation had no effect on JP11646-induced reduction of PIM2 expression or its phosphorylation of its targets (sFig. 6). Conversely, blocking CD28 activation with the inhibitor CTLA4-Ig (MM cells express both CD28 and CD86, which transduces a cis pro-survival signal between MM cells themselves29, 51) significantly enhanced JP11646 cytotoxicity in MM1.S (Fig. 4D, Right). These findings suggest there is previously unrecognized crosstalk between the CD28 and PIM2 signaling pathways, and this is further supported by the observation that CTLA4-Ig treatment also decreased PIM2 expression (Fig. 4D right, inset), suggesting that constitutive CD28 signaling may also be involved in regulation of PIM2 expression.
Inhibition of PIM2 inhibits CD28-induced NFκB signaling in myeloma
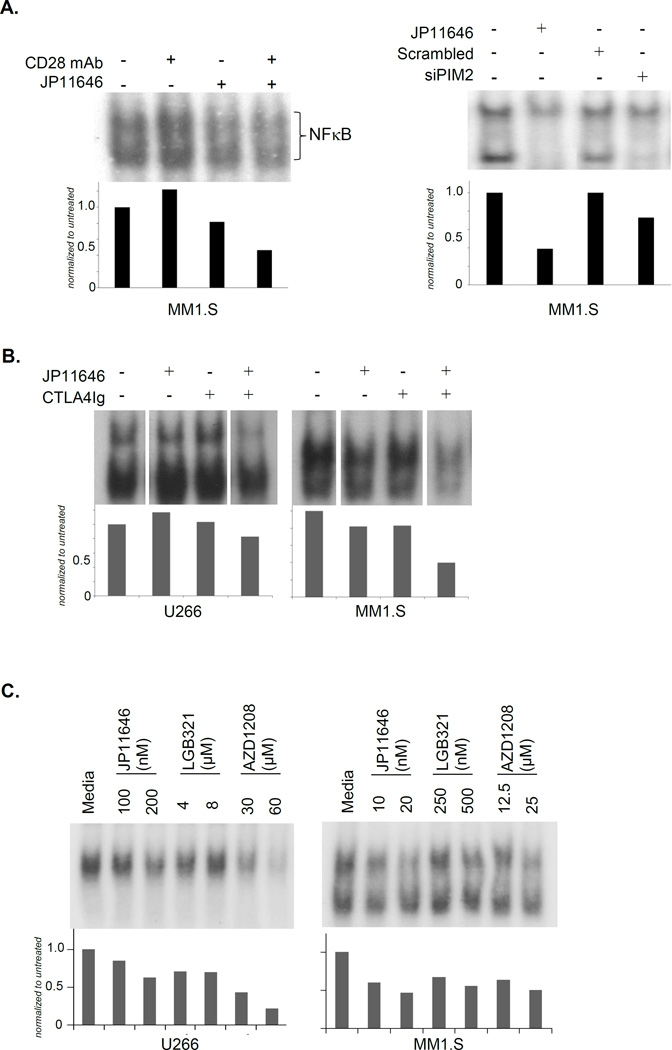
A major survival signal induced downstream of CD28 activation in MM is NFκB31, but a role for PIM2 in the CD28-->NFκB pathway has not been previously identified. We found that while PIM2 inhibition with JP11646 alone had little effect on basal NFκB signaling, it blocked NFκB activation induced by anti-CD28 mAb (Fig. 5A, Left). This was also seen when PIM2 inhibition was accomplished with siRNA knockdown instead of JP11646 (Fig. 5A, Right), suggesting that PIM2 is in the CD28-->NFκB signaling pathway. In addition, consistent with sensitization of MM cells to JP11646 by CTLA4-Ig, we found that CTLA4-Ig + sublethal doses of JP11646 inhibited NFκB signaling to substantially lower levels than with either alone (Fig. 5B). Similar effects were also observed with AZD1208 and LGB321 (Fig. 5C). The loss of NFκB signaling was supported by NFκB luciferase reporter assays, where there was a dose-dependent decrease in luciferase activity - with RPMI8226 showing a plateau in response (Fig. 5D, Left). When NFκB activity was assessed at a fixed concentration of JP11646 over time, we observed a brief transient increase at 1 hour followed by a progressive decrease in activity over 12 hours (Fig. 5D, Right). These findings suggest the previously unrecognized possibility that PIM2 is involved in NFκB signaling downstream of CD28 and other pathways.
Figure 5.
PIM2 inhibition abrogates CD28-induced NFκB signaling in MM cells. A) EMSA on protein lysates from MM1.S cells treated with CD28 mAb ± JP11646 (20 nM) (Left) or transfected with scrambled or PIM2 specific siRNA and treated after 48 hrs with JP11646 (30 nM) for a further 24 hours before being lysed and analyzed by EMSA (Right). NFκB bands were quantified and shown relative to untreated controls below each figure. B) MM1.S cells treated with CTLA4Ig (100 µg/ml) with or without JP11646 (20 nM for MM1.S or 200 nM for U266) for 24 hours were lysed and analyzed for NFκB activity by EMSA. C) EMSA was done similarly on MM cells treated with LC50 or half LC50 concentrations of JP11646, AZD1208 or LGB321 for 24 hours. Densitometry relative to untreated is shown below. D) MM1.S cells were transfected with an NFκB luciferase reporter construct and treated 24 hours later with a dose-range of JP11646. Cell lysates were assayed for luciferase activity and plotted against JP11646 concentration (Left). MM1.S cells similarly transfected with NFκB luciferase reporter constructs were treated with JP11646 (20 nM) and then collected over time and assayed for luciferase activity (Right). **p<0.01, ***p<0.001 and NS-not significant. All data are representative from at least 2 independent replicate experiments.
In vivo efficacy of PIM2 inhibition
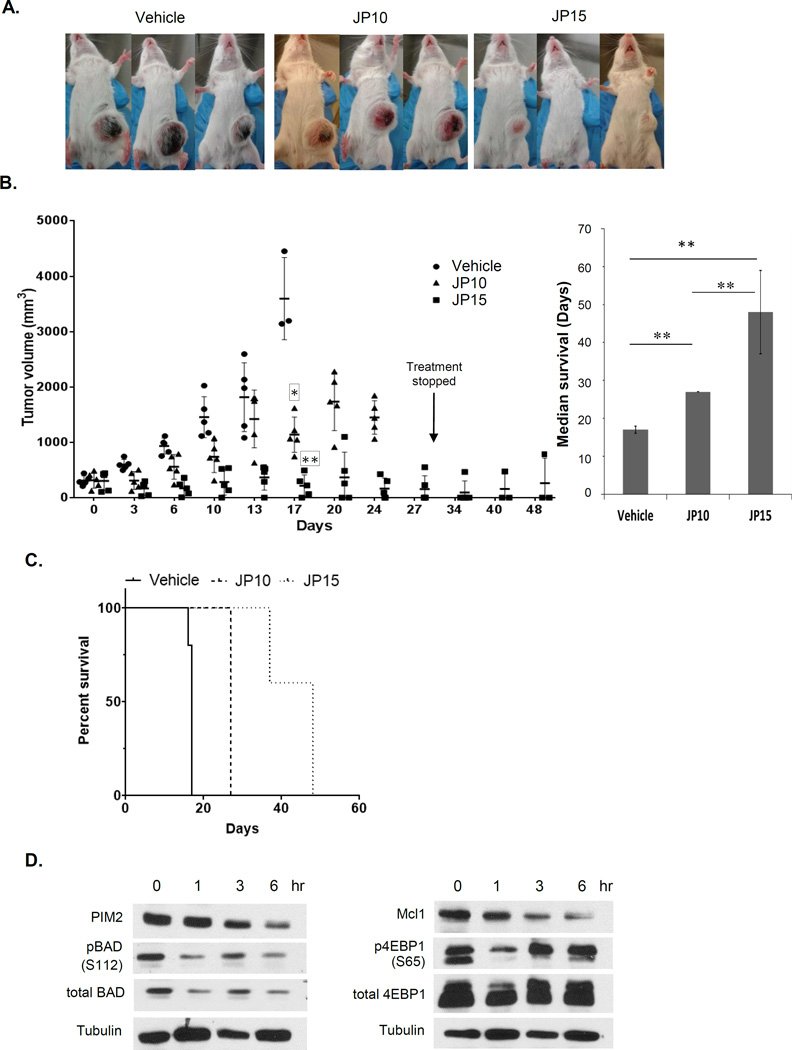
To assess whether PIM2 inhibition by JP11646 had in vivo anti-MM efficacy, scid/scid mice were implanted with MM1.S, and after tumor reached 100 mm3, these mice were treated with JP11646 at 10 (JP10) or 15 µg/gm (JP15) via i.p. injection given consecutively for 2 or 3 times a week. The JP15 dosage was found to have superior anti-tumor efficacy when compared to vehicle alone or JP10 (Fig. 6A) with toxicity limited to weight loss that was readily managed with dietary supplementation. At week 2, the relative tumor volumes were reduced to 49% (JP10 group) or 89% (JP15) of the untreated controls. At the end of week 4, JP15 group showed >91% decrease in tumor compared to control at week 2 (control was euthanized at week 2 due to necrotic tumors). Mice in the JP10 group continued to see a growth in tumor volume, though not at the same rate as in the vehicle control group (Fig. 6B, Left). Median survival in vehicle control group was 17 days, 27 days for JP10 and 48 days for JP15 group (Fig 6B, right) as calculated by Kaplan-Meier analysis (Fig. 6C). After treatment was stopped at day 27, two of the three surviving mice in the JP15 group did not show any regrowth of tumors over the remaining 2 weeks of observation.
Figure 6.
PIM2 inhibition by JP11646 has anti-myeloma activity in murine models. A) Pictures show mice with subcutaneous tumors of MM1.S at day 14 of treatment from groups vehicle (30% modified β-cyclodextrin), JP10 (JP11646 i.p. 10 mg/kg given consecutively 3 days /wk) or JP15 (JP11646 i.p. 15 mg/kg given 2 days/wk). B, Left) Plot of disease burden in mice followed over the entire experimental period of 48 days. For the sake of clarity, groups are staggered while plotting at each time point to avoid overlap. B, Right) Median survival (days) for the different groups with range (error bars) are shown. p-values were determined using the log rank (Mantel-Cox) test. C) Kaplan-Meier plots for survival was plotted using Graphpad Prism. D) Mice with 100–200 mm3 tumors were treated with a single dose of JP11646 at 15 mg/kg, and then tumors collected immediately or after 1,3 or 6 hrs, lysed and immunoassayed for PIM2, pBAD (S112), BAD, MCL1, pEBP1 (S65), total 4EBP1 or Tubulin. * p<0.05, ** p<0.01, ***p<0.001, NS-Not significant.
To examine whether decrease in substrate phosphorylation by JP11646 in vitro happened in vivo, SCID/SCID mice were implanted subcutaneously with MM1.S and when tumors reached 100 mm3 were given a single dose of JP11646 at 15 µg/gm, and tumors harvested at different time points post injection. Western assays show a similar time dependent decrease in PIM2 levels with subsequent drop in both pBAD, p4E-BP1 and total MCL1 as we have observed in vitro (Fig. 6C).
Discussion
The pleotropic roles of the PIM kinases in proliferation and survival in cancer, the overexpression of PIM2 in multiple myeloma (especially in the poor prognosis MF-signature group) and reduced potential toxicity as compared to pan-PIM inhibition underscores the rationale to target PIM252. While some PIM2 inhibitors (LGB32119, 22, 24 and LGH44719, 26) show promising anti-MM efficacy15, others do not23 – and the basis for these differences remains largely unclear.
We now report on a novel non-ATP competitive PIM2 selective inhibitor JP11646 that not only inhibits kinase activity and downstream phosphorylation of PIM2 targets 4EBP1, BAD and MCL1, but also has the unique ability to simultaneously inhibit expression of PIM2 at the protein and mRNA levels, both in vitro and in vivo. This likely accounts for its great potency inhibiting MM cell proliferation and viability compared to the ATP-competitive PIM inhibitors (AZD1208, LGB321) which do not inhibit PIM2 expression – and in fact, induce a compensatory increase in PIM2 expression that likely blunts their anti-proliferative/pro-apoptotic effect. The effect of JP11646 inhibition on both PIM2 kinase activity and expression suggest the existence of additional previously unrecognized feedback loops, as what controls PIM expression in general is not well characterized. To begin to understand this better, we looked at potential off-target effects of JP11646 by screening it against a panel of 150 kinases using Millipore’s KinaseProfiler™ (Billerica, MA). While PIM2 gave the lowest IC50 of 0.5 nM, the only other kinase that showed comparable inhibition was the PIM family member PIM3 with an IC50 of >1 nM (>2 fold of PIM2), while 7 more kinases gave IC50 values 36 – 197 fold greater than PIM2 (sTable 1). The rest of the kinases gave IC50 values >200 fold that of PIM2 (not shown). Gene expression analysis of MM cells treated with JP11646 over 24 hr show several common genes that are most upregulated or downregulated (sTable 2). Pathway analysis also show dysregulation of genes that are part of essential nucleotide/amino acid metabolism and the UPR response pathways (sTable 3). However, given that PIM2 appears to be involved in multiple signaling pathways in myeloma (e.g. IL-6 and CD28 in our studies), it is difficult to definitively define from the analysis an “off target” effect of JP11646. We also found that unlike PIM2, JP11646 treatment induced either no change (U266) or a 1.8-fold increase (MM1.S) in PIM1 gene expression. While it is certainly possible that JP11646 is affecting some pathway other than PIM2 that regulates PIM expression, the observations that the effect of JP11646 inhibition is mirrored by PIM2 knockdown (suggesting PIM2 specificity) while not affecting other PIM members (PIM1/3) at least in U266 point to a particular role for PIM2 signaling in regulating its own expression.
But it remains to be investigated whether JP11646 represents a new class of dual-action PIM2 inhibitors that alters PIM2 signaling in a qualitatively different manner than other ATP-competitive PIM inhibitors to inhibit PIM2 expression, or by interdicting other components of the feedback loops.
We have also found that JP11646 downregulates PIM2 expression in acute myeloid leukemia and a wide range of solid tumor cell lines (KP and GF, manuscript in preparation), indicating that this effect is not myeloma-specific. Why JP11646 inhibits PIM2 kinase activity but not PIM2 expression in normal human PBMC is not clear, but raises the possibility that this effect in malignant cells is due to proliferation and/or aberrant growth signals – and that JP11646 is inhibiting some component (PIM2?) in the downstream signaling cascades (STAT3, STAT5, NFκB) that also drive PIM2 transcription7, 9, 53, 54. However, this does not explain why IL-6 (which induces STAT3 and STAT5 signaling) has no effect on constitutive PIM2 mRNA expression, and cannot rescue JP11646-induced downregulation in MM cells. Nor does it explain why AZD1208 or LGB321, which we have shown can inhibit NFκB signaling to a similar extent as JP11646, do not affect PIM2 expression. Alternatively, JP11646 may be directly affecting the transcriptional regulation of PIM2, as many transcription factors and associated machinery are regulated by phosphorylation. For example, the FoxP3 transcription factor regulates PIM2 gene expression in Tregs55. Further studies are underway to define the underlying mechanisms.
The ability of JP11646 to abrogate key pro-myeloma effects of stromal IL-6 and DC-mediated CD28 activation is of particular clinical significance, given the central role the microenvironment plays in the survival and therapy-resistance of MM cells. The additional complexity that PIM2 may be both a component of the upstream signal transduction pathway (of both IL-6 and CD28) and a downstream gene target of these signaling pathways further suggests interlocking feedback loops where PIM2 function affects PIM2 expression. Our findings that PIM2 is involved in activating the key NFκB pro-survival pathway (also reported in pro-B cells56 and hepatocellular carcinoma57) suggests the molecular mechanism by which the PIM2 inhibitors kill MM cells. These multiple anti-MM effects of JP11646 inhibition are reflected by in vivo efficacy, with complete eradication of tumors in 40% of treated mice. Additionally, recent studies using small molecule inhibitors of PIM2 or siRNA show that PIM2 inhibition can reverse bone loss and tumor progression in multiple myeloma20, suggesting that in addition to a direct anti-myeloma effect that JP11646 may ameliorate MM-associated bony disease.
Altogether our results suggest the existence of previously unrecognized feedback loop(s) where PIM2 kinase activity regulates PIM2 gene expression in multiple myeloma and other malignant cells, and that JP11646 represents a novel class of PIM2 inhibitors that interdicts this feedback. This abrogates the upregulation of PIM2 expression following PIM kinase inhibition, resulting in significantly greater inhibition of MM proliferation and survival. Our findings that similar anti-tumor activity and mechanisms of action are seen in other hematologic malignancies and solid tumors suggest that this “dual inhibition” of PIM2 may have applicability to a broad range of cancers.
Supplementary Material
Acknowledgments
CB and JC are scientific co-founders and the president and vice president respectively of Jasco Pharmaceuticals. This work was funded by Jasco Pharmaceuticals, R01 CA121044 and R01 AI100157.
JRN, KP, JC, TH and CB performed the experiments and derived the data shown. JRN wrote the paper. JC, CB, GF and KPL supervised the work performed, and reviewed and edited the manuscript.
Footnotes
Conflict of interest: The other authors declare no conflicts of interest.
Supplementary information is available at Leukemia’s website.
References
- 1.Cuypers HT, Selten G, Quint W, Zijlstra M, Maandag ER, Boelens W, et al. Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell. 1984 May;37(1):141–150. doi: 10.1016/0092-8674(84)90309-x. [DOI] [PubMed] [Google Scholar]
- 2.Nawijn MC, Alendar A, Berns A. For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat Rev Cancer. 2011 Jan;11(1):23–34. doi: 10.1038/nrc2986. [DOI] [PubMed] [Google Scholar]
- 3.Yan B, Zemskova M, Holder S, Chin V, Kraft A, Koskinen PJ, et al. The PIM-2 kinase phosphorylates BAD on serine 112 and reverses BAD-induced cell death. J Biol Chem. 2003 Nov 14;278(46):45358–45367. doi: 10.1074/jbc.M307933200. [DOI] [PubMed] [Google Scholar]
- 4.Siu A, Virtanen C, Jongstra J. PIM kinase isoform specific regulation of MIG6 expression and EGFR signaling in prostate cancer cells. Oncotarget. 2011 Dec;2(12):1134–1144. doi: 10.18632/oncotarget.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narlik-Grassow M, Blanco-Aparicio C, Carnero A. The PIM family of serine/threonine kinases in cancer. Medicinal research reviews. 2014 Jan;34(1):136–159. doi: 10.1002/med.21284. [DOI] [PubMed] [Google Scholar]
- 6.Gong J, Wang J, Ren K, Liu C, Li B, Shi Y. Serine/threonine kinase Pim-2 promotes liver tumorigenesis induction through mediating survival and preventing apoptosis of liver cell. The Journal of surgical research. 2009 May 1;153(1):17–22. doi: 10.1016/j.jss.2008.03.033. [DOI] [PubMed] [Google Scholar]
- 7.Fox CJ, Hammerman PS, Cinalli RM, Master SR, Chodosh LA, Thompson CB. The serine/threonine kinase Pim-2 is a transcriptionally regulated apoptotic inhibitor. Genes & development. 2003 Aug 1;17(15):1841–1854. doi: 10.1101/gad.1105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bachmann M, Moroy T. The serine/threonine kinase Pim-1. Int J Biochem Cell Biol. 2005 Apr;37(4):726–730. doi: 10.1016/j.biocel.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Adam K, Lambert M, Lestang E, Champenois G, Dusanter-Fourt I, Tamburini J, et al. Control of Pim2 kinase stability and expression in transformed human haematopoietic cells. Bioscience reports. 2015;35(6) doi: 10.1042/BSR20150217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hallermann C, Niermann C, Fischer RJ, Schulze HJ. New prognostic relevant factors in primary cutaneous diffuse large B-cell lymphomas. J Am Acad Dermatol. 2007 Apr;56(4):588–597. doi: 10.1016/j.jaad.2006.12.026. [DOI] [PubMed] [Google Scholar]
- 11.Brault L, Gasser C, Bracher F, Huber K, Knapp S, Schwaller J. PIM serine/threonine kinases in the pathogenesis and therapy of hematologic malignancies and solid cancers. Haematologica. 2010 Jun;95(6):1004–1015. doi: 10.3324/haematol.2009.017079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim KT, Baird K, Ahn JY, Meltzer P, Lilly M, Levis M, et al. Pim-1 is up-regulated by constitutively activated FLT3 and plays a role in FLT3-mediated cell survival. Blood. 2005 Feb 15;105(4):1759–1767. doi: 10.1182/blood-2004-05-2006. [DOI] [PubMed] [Google Scholar]
- 13.Asano J, Nakano A, Oda A, Amou H, Hiasa M, Takeuchi K, et al. The serine/threonine kinase Pim-2 is a novel anti-apoptotic mediator in myeloma cells. Leukemia. 2011 Jul;25(7):1182–1188. doi: 10.1038/leu.2011.60. [DOI] [PubMed] [Google Scholar]
- 14.Claudio JO, Masih-Khan E, Tang H, Goncalves J, Voralia M, Li ZH, et al. A molecular compendium of genes expressed in multiple myeloma. Blood. 2002 Sep 15;100(6):2175–2186. doi: 10.1182/blood-2002-01-0008. [DOI] [PubMed] [Google Scholar]
- 15.Keane NA, Reidy M, Natoni A, Raab MS, O'Dwyer M. Targeting the Pim kinases in multiple myeloma. Blood cancer journal. 2015;5:e325. doi: 10.1038/bcj.2015.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nachbaur DM, Herold M, Maneschg A, Huber H. Serum levels of interleukin-6 in multiple myeloma and other hematological disorders: correlation with disease activity and other prognostic parameters. Ann Hematol. 1991 Feb-Mar;62(2–3):54–58. doi: 10.1007/BF01714900. [DOI] [PubMed] [Google Scholar]
- 17.Puthier D, Derenne S, Barille S, Moreau P, Harousseau JL, Bataille R, et al. Mcl-1 and Bcl-xL are co-regulated by IL-6 in human myeloma cells. Br J Haematol. 1999 Nov;107(2):392–395. doi: 10.1046/j.1365-2141.1999.01705.x. [DOI] [PubMed] [Google Scholar]
- 18.Tu Y, Renner S, Xu F, Fleishman A, Taylor J, Weisz J, et al. BCL-X expression in multiple myeloma: possible indicator of chemoresistance. Cancer research. 1998 Jan 15;58(2):256–262. [PubMed] [Google Scholar]
- 19.Lu J, Zavorotinskaya T, Dai Y, Niu XH, Castillo J, Sim J, et al. Pim2 is required for maintaining multiple myeloma cell growth through modulating TSC2 phosphorylation. Blood. 2013 Aug 29;122(9):1610–1620. doi: 10.1182/blood-2013-01-481457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiasa M, Teramachi J, Oda A, Amachi R, Harada T, Nakamura S, et al. Pim-2 kinase is an important target of treatment for tumor progression and bone loss in myeloma. Leukemia. 2015 Jan;29(1):207–217. doi: 10.1038/leu.2014.147. [DOI] [PubMed] [Google Scholar]
- 21.Ramachandran J, Santo L, Siu KT, Panaroni C, Raje N. Pim2 is important for regulating DNA damage response in multiple myeloma cells. Blood cancer journal. 2016 Aug 26;6(8):e462. doi: 10.1038/bcj.2016.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia PD, Langowski JL, Wang Y, Chen M, Castillo J, Fanton C, et al. Pan-PIM kinase inhibition provides a novel therapy for treating hematologic cancers. Clin Cancer Res. 2014 Apr 1;20(7):1834–1845. doi: 10.1158/1078-0432.CCR-13-2062. [DOI] [PubMed] [Google Scholar]
- 23.Cervantes-Gomez F, Chen LS, Orlowski RZ, Gandhi V. Biological effects of the Pim kinase inhibitor, SGI-1776, in multiple myeloma. Clin Lymphoma Myeloma Leuk. 2013 Sep;13(Suppl 2):S317–329. doi: 10.1016/j.clml.2013.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keeton EK, McEachern K, Dillman KS, Palakurthi S, Cao Y, Grondine MR, et al. AZD1208, a potent and selective pan-Pim kinase inhibitor, demonstrates efficacy in preclinical models of acute myeloid leukemia. Blood. 2014 Feb 6;123(6):905–913. doi: 10.1182/blood-2013-04-495366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foulks JM, Carpenter KJ, Luo B, Xu Y, Senina A, Nix R, et al. A small-molecule inhibitor of PIM kinases as a potential treatment for urothelial carcinomas. Neoplasia. 2014 May;16(5):403–412. doi: 10.1016/j.neo.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chesi M, Matthews GM, Garbitt VM, Palmer SE, Shortt J, Lefebure M, et al. Drug response in a genetically engineered mouse model of multiple myeloma is predictive of clinical efficacy. Blood. 2012 Jul 12;120(2):376–385. doi: 10.1182/blood-2012-02-412783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meja K, Stengel C, Sellar R, Huszar D, Davies BR, Gale RE, et al. PIM and AKT kinase inhibitors show synergistic cytotoxicity in acute myeloid leukaemia that is associated with convergence on mTOR and MCL1 pathways. Br J Haematol. 2014 Oct;167(1):69–79. doi: 10.1111/bjh.13013. [DOI] [PubMed] [Google Scholar]
- 28.Mikkers H, Nawijn M, Allen J, Brouwers C, Verhoeven E, Jonkers J, et al. Mice deficient for all PIM kinases display reduced body size and impaired responses to hematopoietic growth factors. Molecular and cellular biology. 2004 Jul;24(13):6104–6115. doi: 10.1128/MCB.24.13.6104-6115.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nair JR, Carlson LM, Koorella C, Rozanski CH, Byrne GE, Bergsagel PL, et al. CD28 expressed on malignant plasma cells induces a prosurvival and immunosuppressive microenvironment. J Immunol. 2011 Aug 1;187(3):1243–1253. doi: 10.4049/jimmunol.1100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ausubel FM. Current protocols in molecular biology. John Wiley & Sons, Inc; 1987. Analysis of RNA by Northern and Slot Blot hybridization. [DOI] [PubMed] [Google Scholar]
- 31.Bahlis NJ, King AM, Kolonias D, Carlson LM, Liu HY, Hussein MA, et al. CD28-mediated regulation of multiple myeloma cell proliferation and survival. Blood. 2007 Jun 1;109(11):5002–5010. doi: 10.1182/blood-2006-03-012542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chauhan D, Tian Z, Nicholson B, Kumar KG, Zhou B, Carrasco R, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer cell. 2012 Sep 11;22(3):345–358. doi: 10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, et al. The molecular classification of multiple myeloma. Blood. 2006 Sep 15;108(6):2020–2028. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973 Dec 1;22(23):3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 35.Gingras AC, Kennedy SG, O'Leary MA, Sonenberg N, Hay N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes & development. 1998 Feb 15;12(4):502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hideshima T, Nakamura N, Chauhan D, Anderson KC. Biologic sequelae of interleukin-6 induced PI3-K/Akt signaling in multiple myeloma. Oncogene. 2001 Sep 20;20(42):5991–6000. doi: 10.1038/sj.onc.1204833. [DOI] [PubMed] [Google Scholar]
- 37.Baumann P, Schneider L, Mandl-Weber S, Oduncu F, Schmidmaier R. Simultaneous targeting of PI3K and mTOR with NVP-BGT226 is highly effective in multiple myeloma. Anticancer Drugs. 2012 Jan;23(1):131–138. doi: 10.1097/CAD.0b013e32834c8683. [DOI] [PubMed] [Google Scholar]
- 38.Le Gouill S, Podar K, Harousseau JL, Anderson KC. Mcl-1 regulation and its role in multiple myeloma. Cell Cycle. 2004 Oct;3(10):1259–1262. doi: 10.4161/cc.3.10.1196. [DOI] [PubMed] [Google Scholar]
- 39.Matsuo J, Tsukumo Y, Sakurai J, Tsukahara S, Park HR, Shin-ya K, et al. Preventing the unfolded protein response via aberrant activation of 4E-binding protein 1 by versipelostatin. Cancer Sci. 2009 Feb;100(2):327–333. doi: 10.1111/j.1349-7006.2008.01036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morwick T. Pim kinase inhibitors: a survey of the patent literature. Expert Opin Ther Pat. 2010 Feb;20(2):193–212. doi: 10.1517/13543770903496442. [DOI] [PubMed] [Google Scholar]
- 41.Wingett D, Reeves R, Magnuson NS. Stability changes in pim-1 proto-oncogene mRNA after mitogen stimulation of normal lymphocytes. J Immunol. 1991 Nov 15;147(10):3653–3659. [PubMed] [Google Scholar]
- 42.Uddin N, Kim RK, Yoo KC, Kim YH, Cui YH, Kim IG, et al. Persistent activation of STAT3 by PIM2-driven positive feedback loop for epithelial-mesenchymal transition in breast cancer. Cancer Sci. 2015 Jun;106(6):718–725. doi: 10.1111/cas.12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bansal K, Kapoor N, Narayana Y, Puzo G, Gilleron M, Balaji KN. PIM2 Induced COX-2 and MMP-9 expression in macrophages requires PI3K and Notch1 signaling. PLoS One. 2009;4(3):e4911. doi: 10.1371/journal.pone.0004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu Z, Zhao X, Ge Y, Zhang T, Huang L, Zhou X, et al. A regulatory feedback loop between HIF-1alpha and PIM2 in HepG2 cells. PLoS One. 2014;9(2):e88301. doi: 10.1371/journal.pone.0088301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narayana Y, Bansal K, Sinha AY, Kapoor N, Puzo G, Gilleron M, et al. SOCS3 expression induced by PIM2 requires PKC and PI3K signaling. Mol Immunol. 2009 Sep;46(15):2947–2954. doi: 10.1016/j.molimm.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 46.Kawano M, Hirano T, Matsuda T, Taga T, Horii Y, Iwato K, et al. Autocrine generation and requirement of BSF-2/IL-6 for human multiple myelomas. Nature. 1988 Mar 3;332(6159):83–85. doi: 10.1038/332083a0. [DOI] [PubMed] [Google Scholar]
- 47.Hardin J, MacLeod S, Grigorieva I, Chang R, Barlogie B, Xiao H, et al. Interleukin-6 prevents dexamethasone-induced myeloma cell death. Blood. 1994 Nov 1;84(9):3063–3070. [PubMed] [Google Scholar]
- 48.Schwab G, Siegall CB, Aarden LA, Neckers LM, Nordan RP. Characterization of an interleukin-6-mediated autocrine growth loop in the human multiple myeloma cell line, U266. Blood. 1991 Feb 1;77(3):587–593. [PubMed] [Google Scholar]
- 49.Nair JR, Rozanski C, Lee KP. CD28: old dog, new tricks. CD28 in plasma cell/multiple myeloma biology. Adv Exp Med Biol. 2009;633:55–69. doi: 10.1007/978-0-387-79311-5_6. [DOI] [PubMed] [Google Scholar]
- 50.Rozanski CH, Utley A, Carlson LM, Farren MR, Murray M, Russell LM, et al. CD28 Promotes Plasma Cell Survival, Sustained Antibody Responses, and BLIMP-1 Upregulation through Its Distal PYAP Proline Motif. J Immunol. 2015 May 15;194(10):4717–4728. doi: 10.4049/jimmunol.1402260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murray ME, Gavile CM, Nair JR, Koorella C, Carlson LM, Buac D, et al. CD28-mediated pro-survival signaling induces chemotherapeutic resistance in multiple myeloma. Blood. 2014 Jun 12;123(24):3770–3779. doi: 10.1182/blood-2013-10-530964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muraski JA, Rota M, Misao Y, Fransioli J, Cottage C, Gude N, et al. Pim-1 regulates cardiomyocyte survival downstream of Akt. Nat Med. 2007 Dec;13(12):1467–1475. doi: 10.1038/nm1671. [DOI] [PubMed] [Google Scholar]
- 53.Shirogane T, Fukada T, Muller JM, Shima DT, Hibi M, Hirano T. Synergistic roles for Pim-1 and c-Myc in STAT3-mediated cell cycle progression and antiapoptosis. Immunity. 1999 Dec;11(6):709–719. doi: 10.1016/s1074-7613(00)80145-4. [DOI] [PubMed] [Google Scholar]
- 54.Li J, Peet GW, Balzarano D, Li X, Massa P, Barton RW, et al. Novel NEMO/IkappaB kinase and NF-kappa B target genes at the pre-B to immature B cell transition. J Biol Chem. 2001 May 25;276(21):18579–18590. doi: 10.1074/jbc.M100846200. [DOI] [PubMed] [Google Scholar]
- 55.Basu S, Golovina T, Mikheeva T, June CH, Riley JL. Cutting edge: Foxp3-mediated induction of pim 2 allows human T regulatory cells to preferentially expand in rapamycin. J Immunol. 2008 May 1;180(9):5794–5798. doi: 10.4049/jimmunol.180.9.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hammerman PS, Fox CJ, Cinalli RM, Xu A, Wagner JD, Lindsten T, et al. Lymphocyte transformation by Pim-2 is dependent on nuclear factor-kappaB activation. Cancer research. 2004 Nov 15;64(22):8341–8348. doi: 10.1158/0008-5472.CAN-04-2284. [DOI] [PubMed] [Google Scholar]
- 57.Ren K, Zhang W, Shi Y, Gong J. Pim-2 activates API-5 to inhibit the apoptosis of hepatocellular carcinoma cells through NF-kappaB pathway. Pathol Oncol Res. 2010 Jun;16(2):229–237. doi: 10.1007/s12253-009-9215-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.