Abstract
Long noncoding RNA (lncRNA) has been implicated in cancer, but little is known about the role of lncRNAs as regulators of tumor metastasis. In the present study, we demonstrate that lncRNA TRERNA1 acts like an enhancer of SNAI1 to promote cell invasion and migration and to contribute to metastasis of gastric cancer (GC). TRERNA1 is significantly unregulated in GCs and GC cell lines. Increased TRERNA1 is positively correlated with lymph node metastasis of GCs. RNA immunoprecipitation (RIP) and chromatin immunoprecipitation (ChIP) assays revealed that TRERNA1 functions as a scaffold to recruit EZH2 to epigenetically silence epithelial-mesenchymal transition marker CDH1 by H3K27me3 of its promoter region. TRERNA1 knockdown markedly reduced GC cell migration, invasion, tumorigenicity, and metastasis. Depletion of TRERNA1 reduced cell metastasis of GCs in vivo. Taken together, our findings indicated that TRERNA1 serves as a critical effector in GC progression by regulating CDH1 at the transcription level. It is implied that TRERNA1/CDH1 is a new potential target for GC therapy.
Keywords: TRERNA1, SNAI1, metastasis, gastric cancer, CDH1
Introduction
Gastric cancer (GC) is one of the most common human cancers and the second leading cause of cancer-related mortality worldwide despite the greater progress in the diagnosis and treatment of human malignancy.1, 2 Metastasis is a major cause of death that greatly hinders treatment success, despite rapid advances in medical technology of GC.3 Therefore, identifying pivotal genes and understanding their molecular mechanisms is a key to cancer prognosis and therapeutics for GC. Since the early 2000s, the majority of studies have focused on protein-coding genes, and a vast class of genes, including AP-1, NF-κB, SNAI1, Sp1, EGR, CREB, and ATF, is identified that was involved in the progression and metastasis of GC.4, 5, 6, 7, 8, 9, 10, 11 But so far, the molecular mechanism underlying GC development and progression remains unclear.
With the progress of DNA sequencing and analytical techniques, researchers have found that 98.5% of DNA sequences do not code for proteins and have postulated that 80% of genome sequences are transcribed into primary transcripts and have biochemical functions.12 To date, the crucial roles of these noncoding RNAs have been shown in various biological regulatory processes, especially in cancer. Long noncoding RNAs (lncRNAs) are defined as transcripts that contain more than 200 nucleotides that do not display the typical properties of well-characterized protein-coding RNAs that act as decoys and guides to facilitate both proximal and distal macromolecular interactions,13, 14, 15, 16 and accumulating evidence also suggests that lncRNAs play roles in regulating a wide range of cellular processes, including apoptosis, proliferation, cell migration, and invasion through affecting various aspects of protein, DNA, and RNA expression and interactions.17, 18, 19, 20, 21 Recent evidence has demonstrated that the aberrant upregulated expression of lncRNAs, such as TGU1, SNHG5, H19, HOXA11-AS, and UCA1, is involved in cell invasion and metastasis of GC.22, 23, 24, 25 Therefore, it is very important for clinical diagnosis and therapy of GC to explore the biological function of more lncRNAs.
lncRNA TRERNA1 was identified and located close to SNAI1, which is an epithelial-mesenchymal transition (EMT) master regulator transcription factor. A retrospective analysis shows that TRERNA1 is overexpressed in lymph node metastatic human breast cancer samples and suggests that TRERNA1 plays an important role in cell invasion and metastasis.26, 27 In the present study, we found the expression of lncRNA TRERNA1 was significantly upregulated in tumor tissues compared with that in normal tissues in GC. Enhanced expression of TRERNA1 was correlated with lymph node metastasis in GC. Our data showed that TRERNA1 contributed to GC cell invasion and metastasis both in vitro and in vivo. Further studies have shown that TRERNA1 not just inhibits the expression of CDH1 by enhancing SNAI1 expression, but also epigenetically silenced CDH1 by recruiting EZH2 to carry out histone methylation of its promoter region. These studies will help to clarify the role of TRERNA1 in GC progression and as a potential therapeutic target.
Results
Increased Expression Level of lncRNA TRERNA1 Is Correlated with Lymph Node Metastasis in GCs
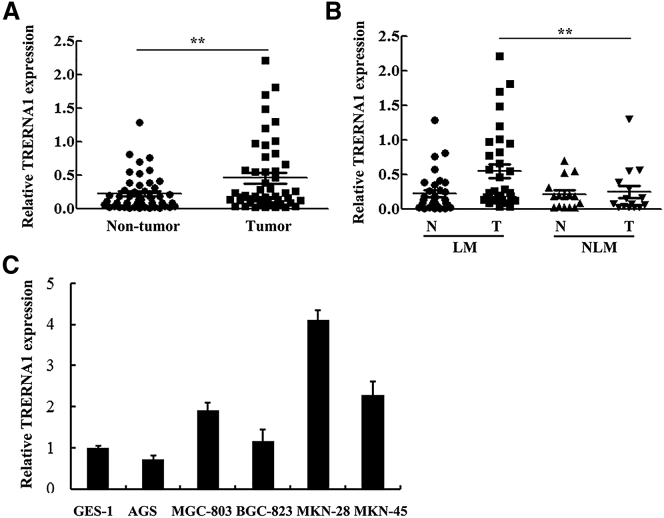
To explore the potential relationship between the expression pattern of TRERNA1 with GC carcinogenesis and progression, we examined the expression levels of lncRNA in 48 GC tissues with paired non-tumor tissue as control. As revealed by qRT-PCR analysis, the expression level of lncRNA TRERNA1 was remarkably higher in tumor tissues compared with adjacent non-tumor tissues (Figure 1A). After evaluating the correlation between TRERNA1 and the clinicopathological characteristics, we found a significant relationship between TRERNA1 expression level and lymph node metastasis in GC specimens (Figure 1B), although no significant correlation was observed among TRERNA1 to GC patients’ age, gender, TNM staging, and the differentiation degree of tumors (Table 1). In six GC cell lines, compared with the immortalized normal gastric epithelial cell line GES-1, there is upregulated expression of TRERNA1 in four cell lines, including KMN-28, MKN-45, BGC-823, and MGC-803, and downregulated expression of TRERNA1 in the AGS cell (Figure 1C). These results indicated the potential role of TRERNA1 during GC carcinogenesis, particularly in the metastasis process.
Figure 1.
LncRNA TRERNA1 Is Upregulated in GC Tissues and Cells
(A) Relative expression of lncRNA TRERNA1 in GC tissues compared with the corresponding non-tumor tissues (n = 48). (B) Patients with LM showed high levels of lncRNA TRERNA1 compared with NLM patients. (C) qRT-PCR analysis of relative expression of lncRNA TRERNA1 in GC cells. lncRNA TRERNA1 expression levels were normalized to β-actin. Data are means ± SD. **p < 0.01. LM, lymph node metastasis; N, non-tumor tissues; NLM, no lymph node metastasis; T, tumor tissues.
Table 1.
Clinicopathological Correlation of TRERNA1 Expression in GC Cases
| Feature | Expression of TRERNA1 |
p Value | |
|---|---|---|---|
| T > N | T ≤ N | ||
| Gender | 0.809 | ||
| Male | 21 | 12 | |
| Female | 9 | 6 | |
| Age | 0.582 | ||
| ≥60 | 23 | 15 | |
| <60 | 7 | 3 | |
| Histologic Grade | 0.825 | ||
| Poor | 13 | 7 | |
| Moderate | 6 | 5 | |
| High | 11 | 6 | |
| Lymph Node Metastasis | 0.034a | ||
| Yes | 21 | 7 | |
| No | 9 | 11 | |
| TNM Staging | 0.878 | ||
| Stage I/II | 19 | 11 | |
| Stage III/IV | 11 | 7 | |
| Lauren’s Classification | 0.414 | ||
| Diffuse type | 18 | 9 | |
| Intestinal type | 12 | 9 | |
N, non-tumor tissues; T, tumor tissues.
χ2 test, *p < 0.05.
TRERNA1 Promotes GC Cell Migration, Invasion, and Metastasis In Vitro
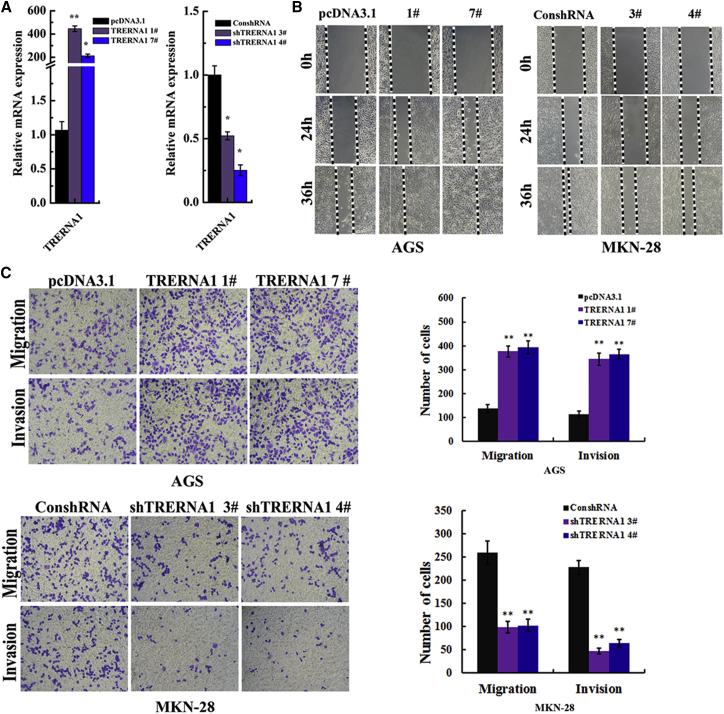
To assess the potential role of TRERNA1 in GC carcinogenesis, we obtained stable monoclonal cells to test and compare the cell phenotype after TRERNA1 knockdown and overexpression in MKN-28 and AGS cells (Figure 2A). Wound healing assay showed that AGS cells that stably enforced TRERNA1 displayed a notably faster recovery compared with control cells; conversely, MKN-28 cells that knocked down TRERNA1 showed a slower recovery compared with control cells (Figure 2B). Similarly, in transwell migration assay and matrigel invasion assay, enforced TRERNA1 expression level increased the ability of cell migration and invasion in AGS cells (p < 0.05; Figure 2C); however, suppressed TRERNA1 expression level decreased the ability of cell migration and invasion in MKN-28 cells (p < 0.05; Figure 2C). These results implied an important role of TRERNA1 in cell migration and invasion in GCs.
Figure 2.
TRERNA1 Regulates Gastric Cancer Cell Migration and Invasion In Vitro
(A) Overexpression and knockdown efficiency of TRERNA1 after transfection of the overexpression and shRNA construct in AGS and MKN-28 cells. (B) Wound healing assay showed that the enforced expression of TRERNA1 displayed a notably faster recovery compared with control cells in AGS cells (left); conversely, there was a slower recovery in TRERNA1 knockdown clones compared with control in MKN-28 cells (right). (C) TRERNA1 modulated GC cell migration and invasion in vitro by transwell migration and invasion assays in AGS and MKN-28 cells. *p < 0.05; **p < 0.01.
Depletion of TRERNA1 Reduced Cell Metastasis of GCs In Vivo
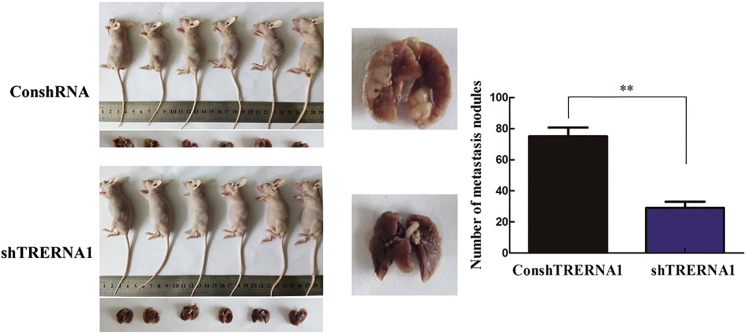
To validate the important role of TRERNA1 in cell metastasis of GCs in vivo, MKN-28 cells that are transfected with TRERNA1-short hairpin RNA (shRNA) or negative control (NC)-shRNA were inoculated into nude mice by tail vein injection. After injection for 45 days, the treated nude mice were sacrificed to dissect lungs for analysis. As shown in Figure 3, inhibition of TRERNA1 by shRNA effectively suppressed tumor cells metastasis compared with the control. These data further demonstrated the important role of TRERNA1 in GC tumor cell metastasis both in vitro and in vivo.
Figure 3.
TRERNA1 Stable Knockdown in MKN-28 Cells Significantly Reduced the Number of Visible Nodules on the Lung Surface after Tail Vein Injection in Nude Mice
Lungs from each group are shown (left). The middle panel is representative of pulmonary nodules, and the right panel is a statistical analysis of the number of nodules from two groups. Bar graph represents the number of visible nodules on the lung surface. Error bars represent mean ± SD. p < 0.05, **p < 0.01.
TRERNA1 Regulated Nearby Gene SNAI1 Expression by Acting Like an Enhancer
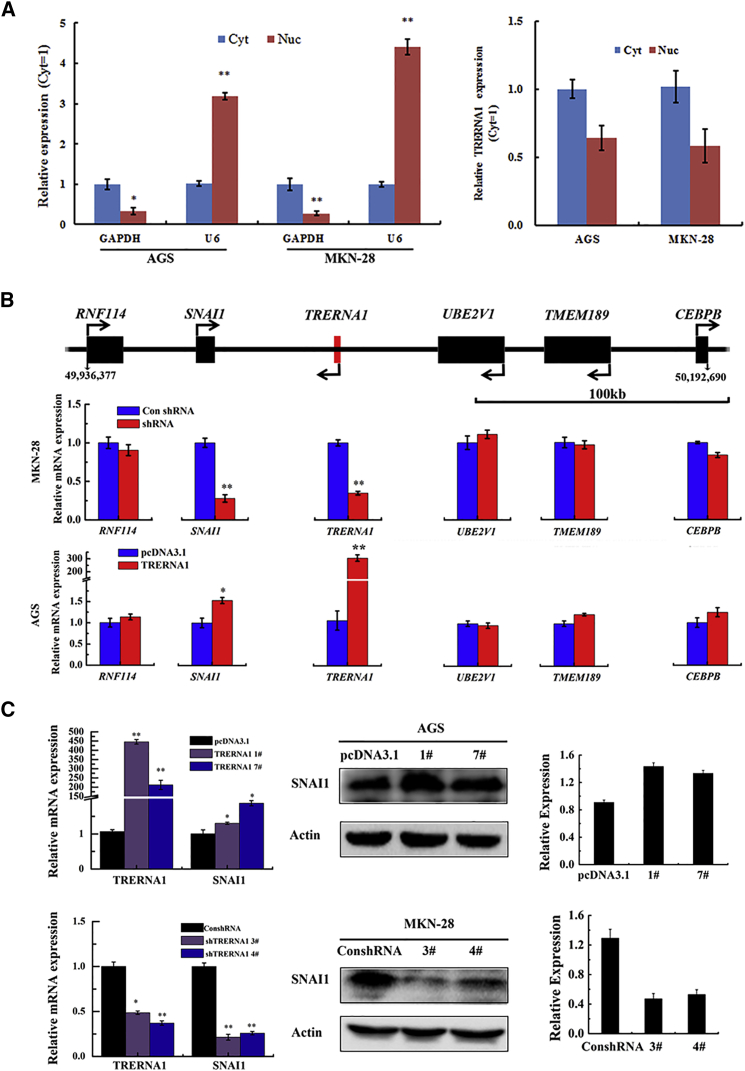
Accumulating evidence shows that lncRNA participates in multiple cell processes by regulating gene expression in different levels. In order to explore the function and mechanism of TRERNA1 on cell migration and invasion, we investigated the subcellular location of TRERNA1 in GC cells. We fractionated GC cells into cytoplasmic and nuclear fractions, and separation was confirmed by the presence of GAPDH mRNA in the cytoplasm fraction and nuclear U6 mRNA predominantly in the nuclear fraction (Figure 4A, right). qRT-PCR results showed that TRERNA1 distributes in both the cytoplasm (AGS: 58.7% ± 3.78%, MKN-28: 65.6% ± 4.65%) and the nucleus of GC cells (AGS: 41.3% ± 2.54%, MKN-28: 36.4% ± 3.725%) (Figure 4A, left). The data indicate that TRERNA1 may play a regulatory role in both the cytoplasm and nucleus in GCs.
Figure 4.
TRERNA1 Regulates SNIA1 Function Like an Enhancer
(A) The relative distribution of TRERNA1 in the cytoplasm and nucleus as identified using qRT-PCR; GAPDH was used as a cytosolic marker, and U6 was used as a nuclear marker in AGS and MKN-28 cells. (B) TRERNA1 is a regulator of SNAI1 expression. The relative position of the adjacent gene to the TRERNA1 (upper) and the effects on mRNA levels for the surrounding genes after knockdown or enforced expression of TRERNA1 in MKN-28 and AGS cells as determined by qRT-PCR (middle and lower panels) are shown. Scale bar, 100 kb. (C) Western blot assays detected the expression of SNAI1 after the transfection of the overexpression and shRNA TRERNA1 plasmid in GC cell lines. *p < 0.05, **p < 0.01. Cyt, cytoplasm; Nuc, nucleus.
TRERNA1 is located at chromosome 20q13.13. In 150 kb neighbor regions to TRERNA1 there are five protein-coding genes: SNAI1, UBE2V1, TMEM189, CEBPB, and RNF114 (Figure 4B, upper). We assay these gene expression levels in knockdown or overexpression of TRERNA1 by qRT-PCR (primers are listed in Table S1). Depletion of TRERNA1 led to decreased expression of their neighboring protein-coding gene SNAI1, but did not affect the other protein-coding genes surrounding TRERNA1 in mRNA level (Figure 4B, middle). Conversely, overexpression of TERNA1 increased the expression of SNAI1 (Figure 4B, lower), but did not affect the other protein-coding genes surrounding TRERNA1. The same results observed in western blot analysis demonstrated that TRERNA1 was able to regulate the expression of SNAI1 protein levels in TRERNA1 knockdown and overexpression of GC cell clones (Figure 4C). These results implied that TRERNA1 functions as an enhancer in regulating the protein-coding gene SNAI1 expression in cis.
TRERNA1 Enhances SNAI1 and Modulates CDH1 Expression in GC Cells
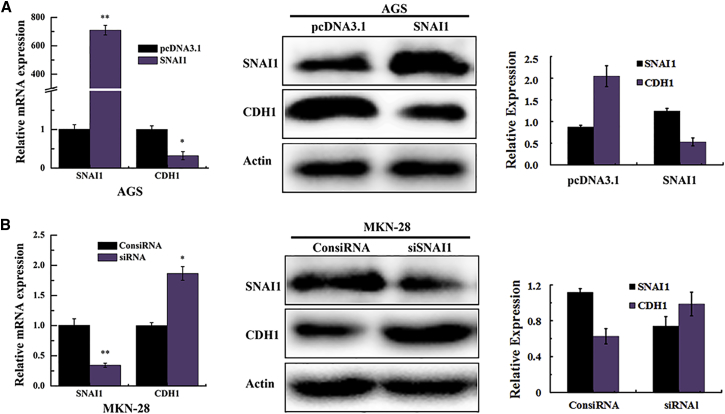
SNAI1 is a member of the snail zinc-finger family, which has been shown to function as the regulators of cell migration, adhesion, and EMT in cancer progression.28, 29, 30 Previous studies have shown that SNAI1 binds to E-boxes of CDH1 promoter to repress its transcription.31 Downregulated CDH1 is one of the hallmarks of EMT in tumor. To investigate whether SNAI1 is a repressor of CDH1 in GC cells, we detected the SNAI1 expression level either in RNA interference or in enforced expression. The results showed that SNAI1 overexpression significantly reduced the expression of CDH1 in AGS cell lines (Figure 5A); on the contrary, downregulation of SNAI1 contributes to increased expression of the CDH1 in MKN-28 cell lines (Figure 5B).
Figure 5.
Transcription Factor SNAI1 Represses the Expression of CDH1
(A and B) The expression of CDH1 after transfection of the overexpression construct (A) and siRNA (B) of SNAI1 in AGS and MKN-28 cells detected by qRT-PCR and western blot assays. *p < 0.05; **p < 0.01.
Then, we evaluated the effects of TRERNA1 on gene transcripts and protein levels of EMT-related genes, which were suspected to initiate the invasion process of many tumor cells by qRT-PCR and western blot in GC cells. Enforced expression of TRERNA1 in AGS cells significantly suppressed expression of CDH1; conversely, knockdown of TRERNA1 in MKN-28 cells that increased the expression of CDH1 was determined by qRT-PCR (Figure 6A). Similar results were observed that TRERNA1 modulates the expression of CDH1 proteins after knockdown or overexpression of TRERNA1 in GC cells (Figure 6B). These data clearly provide insights into the mechanisms that TRERNA1 upregulated SNAI1 to suppress CDH1 expression and contributed to GC cell invasion and metastasis.
Figure 6.
TRERNA1 Regulates EMT Marker Expression in GC Cells
(A) qRT-PCR was used to measure expression levels of EMT-related markers (CDH1, N-cadherin, Vimentin, and β-catenin) in AGS and MKN-28 cells, which transfected with construct (pcDNA3.1+TRERNA1) or shRNA-TRERNA and normalized to β-actin expression. (B) TRERNA1 affects the expression of EMT-related genes in GC cells as determined by western blot analysis. *p < 0.05; **p < 0.01.
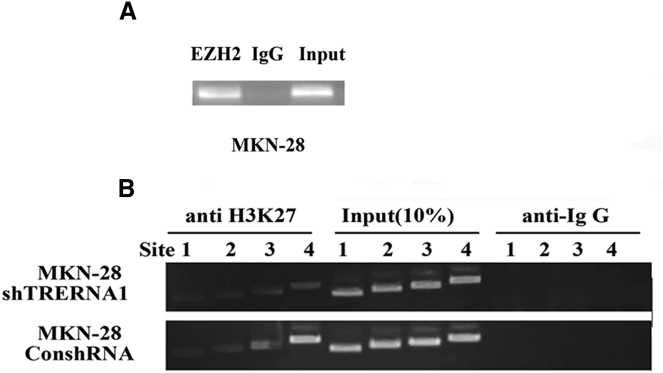
lncRNA TRERNA1 Altered the H3K27me3 Level of CDH1 Gene by Binding to EZH2
It has been found that the expression of CDH1 is not only regulated by the transcription factor SNAI1, but also by epigenetic modification, such as microRNA (miRNA), DNA methylation, and histone modification.32, 33, 34, 35, 36 It is well known that lncRNA-mediated epigenetic gene silencing by PRC2 appears to be an evolutionarily conserved mechanism between plants and animals,37 we first detected whether TRERNA1 combined with EZH2 using RNA binding protein immunoprecipitation technology (RIP), and we observed a strong binding of TRERNA1 to EZH2 (Figure 7A). Next, we conducted chromatin immunoprecipitation (ChIP) assays to map histone modifications in the proximal region of the CDH1 promoter in MKN-28 cells. We found that knockdown of TRERNA1 decreased the H3K27me3 levels of CDH1 promoter compared with the control (Figure 7B). These results demonstrated that TRERNA1 epigenetically silenced CDH1 expression by physical association with EZH2 in GC cells.
Figure 7.
TRERNA1 Represses Transcription of CDH1 by Recruiting PRC2 Subunits EZH2 to the CDH1 Promoter
(A) The TRERNA1 directly interacts with PRC2 subunits EZH2 by RIP assay (RIP experiments using anti-EZH2 antibody and IgG as a negative control were performed in MKN-28 cells, and the coprecipitated RNA was subjected to PCR for TRERNA1). (B) ChIP-PCR of H3K27me3 enrichment of the promoter region of the CDH1 after knockdown TRERNA1 in MKN-28 cells.
Discussion
GC is one of the leading causes of cancer death worldwide, especially in China.38, 39 Despite the fact that strategies in the treatment of advanced and metastatic cancer have improved dramatically in recent years, metastasis leads to most cancer deaths and remains one of the most enigmatic aspects of GC.2, 40, 41 Therefore, understanding the dynamic interactions of these pathways will help us to identify promising molecular targets for cancer therapy and key obstacles to their clinical treatment.42
lncRNAs have been shown to be involved in cellular processes, including apoptosis, proliferation, cell migration, and invasion.19, 21, 43 Evidence has also indicated that lncRNAs act as crucial determinants of GC metastasis.22, 44, 45 Identification of lncRNAs involved in cancer progression will further improve our understanding in cancer. lncRNAs can exert their regulatory function through a variety of mechanisms, including chromatin remodeling, RNA processing, localization, translation, and mRNA stability and even as a competing endogenous RNA.46, 47, 48
Despite the fact that growing evidence shows that aberrant lncRNA expression plays a key role in carcinogenesis and progression of cancer,49 the underlying biological and molecular mechanisms of lncRNAs in diverse tumors are not yet fully elucidated.
TRERNA1 was first reported by Ørom et al.,26 who indicated that TRERNA1 plays an important role in the activation of critical regulators of development and differentiation. Gumireddy et al.27 showed that TRERNA1 is upregulated in paired clinical breast cancer primary and lymph node metastasis samples, and that its expression stimulates tumor invasion in vitro and metastasis in vivo. Our results from investigation of GC showed that TRERNA1 was significantly upregulated in GC tissues and correlated with lymph node metastasis in GC. In vitro and in vivo, the data showed that TRERNA1 exerts an oncogenic function in GC cell invasion and metastasis. Further molecular mechanism study indicated that TRERNA1 modulates CDH1 expression by enhancing SNAI1 expression. To our surprise, we found that TRERNA1 epigenetically regulates the expression of the CDH1 gene by recruiting to EZH2. These results suggested that lncRNA TRERNA1 regulates CDH1 expression and contributes to EMT by various molecular mechanisms. Our studies not only help to clarify the role of TRERNA1 in GC progression and metastasis, but also as a potential therapeutic target. Importantly, our data implied that TRERNA1 can regulate tumor-related gene expression by multiple mechanisms in diverse cancers.
On the basis of the data from our study, we propose the following model for clarifying the molecular mechanism of TRERNA1 involved in GC progression and metastasis. TRERNA1 fraction in the nucleus acts as an enhancer-like function to suppress CDH1 expression by enhancing transcription of the SNAI1 gene and epigenetically silencing the expression of CDH1 by binding to PRC2.
Materials and Methods
Collection of Tissue Specimens
A total of 48 fresh GC tissues and paired adjacent non-tumor tissues were obtained from patients who had undergone surgical resection between 2010 and 2014 at the Third Affiliated Hospital of Harbin Medical University, China. Histopathological diagnosis was confirmed by an experienced pathologist. All of the tissue samples were washed with sterile PBS before being snap frozen in liquid nitrogen until total RNA was extracted. No patients had been treated with radiotherapy or chemotherapy before surgery. This study was approved by the Ethics Committee of Southeast University, and informed consent was obtained from each patient involved in the study.
Cell Culture
The GC cell lines (MKN-28, MKN-45, BGC-823, AGS, and MGC-803) and the human gastric epithelial cell line (GES-1) were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences. All cell lines were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum (Wisent), 100 U/mL penicillin, and 100 mg/mL streptomycin (Invitrogen) in an incubator humidified with 5% CO2 at 37°C.
RNA Isolation and Quantitative Real-Time PCR
Total RNA was extracted from tissues or cultured cells using TRIzol reagent (Invitrogen). For qRT-PCR, RNA was reverse transcribed to cDNA by using a Reverse Transcription Kit (Takara). qRT-PCR analyses were performed using SYBR Green (Takara). The results were normalized to the expression of β-actin. The primers were listed in Table S1. All experiments were performed using the 2−ΔΔCt method, and each experiment was performed in triplicate.
Plasmid Construction and Cell Transfection
The cDNA of lncRNA TRERNA1 (NR_051976.1) were synthesized by GENEWIZ and then cloned into pcDNA3.1. DNA segments of TRERNA1-shRNA and small interfering RNA (siRNA) for SNAI1 were synthesized by GENEWIZ and ligated into the BglII/Hind III sites of pSUPER-EGFP vector after annealing.50 The cDNA of SNAI1 was isolated by RT-PCR and then cloned into the Hind III/EcoR I sites of pcDNA3.1. The primers sequences for PCR amplification are as follows: forward 5′-CCCCAAGCTTATGCCGCGCTCTTTCCTCGTCAG-3′ and reverse 5′-GGTGGAATTCTCAGCGGGGACATCCTGAGCAGC-3′. Transfections were performed using Lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen). The stable cell clones were selected with G418.
Wound Healing, Migration, and Invasion Assays
Wound healing, migration, and invasion assays were performed as described previously.33 Cells were cultured in standard conditions until 80%–90% confluence during a scratch wound was generated using a 200 μL pipette tip, and the spread of the wound closure was observed under the microscope. For transwell migration assay, cells (4 × 105) were plated within the top chamber (Millipore) with the non-coated membrane. For invasion assays, matrigel (BD Biosciences) was polymerized in transwell inserts for 30 min at 37°C. In both assays, cells were plated within the top chamber in medium without serum; the lower chamber filled with 10% FBS was used as a chemoattractant. After 36 hr incubation, the cells that did not migrate or invade through the pores were gently removed with a cotton swab. All of the cells were stained with crystal violet and counted in five fields by inverted microscope. All experiments were repeated independently three times.
Xenograft Mouse Model
Four-week-old male nude mice, weighing 20 g, were housed in an air-conditioned room and maintained under pathogen-free (SPF) conditions. Ten nude mice were randomly divided into two groups, and MKN-28 cells stably transfected with sh-TRERNA1 or empty vector were injected into the tail veins of 10 mice (1 × 106 cells/mouse). After 45 days, the nude mice were sacrificed to dissect lungs for quantitative analysis of lung metastases. All animal studies were approved by the Animal Welfare and Research Ethics Committee at Southeast University, and all protocols were conducted strictly in accordance with the Guide for the Care and Use of Laboratory Animals.
Western Blot Assay and Antibodies
Cells protein lysates were separated by 10% SDS-PAGE, transferred to 0.22 μm NC membranes (Sigma), and incubated with specific antibodies. Anti-CDH1 and SNAI1 (Bioworld Technology), anti-β-catenin, and N-cadherin (4061) (Cell Signaling Technology), Vimentin (sc-6260) (Santa Cruz Biotechnology), and anti-β-actin (Sigma-Aldrich) were used as controls. Protein detection was performed with Super Signal Chemiluminescence Substrate (Pierce).
Subcellular Fractionation Location
Separation of the nuclear and cytosolic fractions was performed using the PARIS Kit (Life Technologies) according to the manufacturer’s instructions, and RNA was used for subsequent reverse transcription reaction and real-time PCRs (SYBR Premix Ex Taq; TaKaRa). The primer sequences are shown in Table S1.
Chromatin Immunoprecipitation Assays
ChIP assays were performed using the Chromatin Immunoprecipitation Kit according to the manufacturer’s instructions (Millipore). Immunoprecipitations were performed using anti-H3K27me3 (Millipore) and/or normal mouse IgG used as the negative control. PCR was performed with the primers designed from the sequence of the CDH1 promoter listed in Table S2, covering upstream of the transcription start site of the CDH1 gene.
RIP
RIP experiments were performed using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) according to the manufacturer’s instructions. Antibody for RIP assays of EZH2 was from Abcam (ab3748). The co-precipitated RNAs were detected by conventional RT-PCR.
Statistical Analysis
All statistical analyses were performed using SPSS 20.0 software. The significance of differences between groups was estimated by Student’s t tests and χ2 tests as appropriate. Pearson’s correlation coefficient was calculated using Prism5 software (GraphPad). A p value < 0.05 was considered statistically significant.
Author Contributions
H.F. designed the experiments and modified the manuscript. H.W. conducted the experiments and wrote the paper. W.S., Y.H., Z.C., and P.G. performed the animal experiment and data collection. Statistical analysis was performed by by X.L. and X.S. Y.Q. analyzed the data from qRT-PCR. K.Z. and M.Z. collected the clinical sample and analyzed the clinicopathological characteristics.
Conflicts of Interest
We declare that we have no conflicts of interest.
Acknowledgments
This work was supported by the grants from National Natural Science Foundation of China (81672414 and 81472548). The project was supported by the Jiangsu Provincial Natural Science Foundation—Youth Foundation (BK20160667). This study was also supported by the Foundational Research Funds for the Central University (grant 2242016k41034).
Footnotes
Supplemental Information includes two tables and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.06.021.
Supplemental Information
References
- 1.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Bass A.J., Thorsson V., Shmulevich I., Reynolds S.M., Miller M., Bernard B., Hinoue T., Laird P.W., Curtis C., Shen H., Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta G.P., Massagué J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Redlak M.J., Power J.J., Miller T.A. Prevention of deoxycholate-induced gastric apoptosis by aspirin: roles of NF-kappaB and PKC signaling. J. Surg. Res. 2008;145:66–73. doi: 10.1016/j.jss.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 5.Safe S., Abdelrahim M. Sp transcription factor family and its role in cancer. Eur. J. Cancer. 2005;41:2438–2448. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Schewe D.M., Leupold J.H., Boyd D.D., Lengyel E.R., Wang H., Gruetzner K.U., Schildberg F.W., Jauch K.W., Allgayer H. Tumor-specific transcription factor binding to an activator protein-2/Sp1 element of the urokinase-type plasminogen activator receptor promoter in a first large series of resected gastrointestinal cancers. Clin. Cancer Res. 2003;9:2267–2276. [PubMed] [Google Scholar]
- 7.Szalad A., Katakowski M., Zheng X., Jiang F., Chopp M. Transcription factor Sp1 induces ADAM17 and contributes to tumor cell invasiveness under hypoxia. J. Exp. Clin. Cancer Res. 2009;28:129. doi: 10.1186/1756-9966-28-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitsuno Y., Yoshida H., Maeda S., Ogura K., Hirata Y., Kawabe T., Shiratori Y., Omata M. Helicobacter pylori induced transactivation of SRE and AP-1 through the ERK signalling pathway in gastric cancer cells. Gut. 2001;49:18–22. doi: 10.1136/gut.49.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdel-Latif M.M., Windle H.J., Davies A., Volkov Y., Kelleher D. A new mechanism of gastric epithelial injury induced by acid exposure: the role of Egr-1 and ERK signaling pathways. J. Cell. Biochem. 2009;108:249–260. doi: 10.1002/jcb.22247. [DOI] [PubMed] [Google Scholar]
- 10.Pradeep A., Sharma C., Sathyanarayana P., Albanese C., Fleming J.V., Wang T.C., Wolfe M.M., Baker K.M., Pestell R.G., Rana B. Gastrin-mediated activation of cyclin D1 transcription involves beta-catenin and CREB pathways in gastric cancer cells. Oncogene. 2004;23:3689–3699. doi: 10.1038/sj.onc.1207454. [DOI] [PubMed] [Google Scholar]
- 11.Chai J.Y., Jones M.K., Tarnawski A.S. Serum response factor is a critical requirement for VEGF signaling in gastric microvascular endothelial cells and VEGF-induced angiogenesis: insight into the mechanisms. Gastroenterology. 2004;126:A557. doi: 10.1096/fj.03-1232fje. [DOI] [PubMed] [Google Scholar]
- 12.Niu D.K., Jiang L. Can ENCODE tell us how much junk DNA we carry in our genome? Biochem. Biophys. Res. Commun. 2013;430:1340–1343. doi: 10.1016/j.bbrc.2012.12.074. [DOI] [PubMed] [Google Scholar]
- 13.Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., FANTOM Consortium. RIKEN Genome Exploration Research Group Phase I & II Team Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 14.Ulitsky I., Bartel D.P. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J., Jung C., Xu J., Wang H., Deng S., Bernad L., Arenas-Huertero C., Chua N.H. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell. 2012;24:4333–4345. doi: 10.1105/tpc.112.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang F., Zhang L., Zhang C. Long noncoding RNAs and tumorigenesis: genetic associations, molecular mechanisms, and therapeutic strategies. Tumour Biol. 2016;37:163–175. doi: 10.1007/s13277-015-4445-4. [DOI] [PubMed] [Google Scholar]
- 17.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 18.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilusz J.E., Sunwoo H., Spector D.L. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G.Y., Zhu Y.Y., Zhang Y.Q. The functional role of long non-coding RNA in digestive system carcinomas. Bull. Cancer. 2014;101:E27–E31. doi: 10.1684/bdc.2014.2023. [DOI] [PubMed] [Google Scholar]
- 22.Wiestler B., Capper D., Hovestadt V., Sill M., Jones D.T., Hartmann C., Felsberg J., Platten M., Feiden W., Keyvani K. Assessing CpG island methylator phenotype, 1p/19q codeletion, and MGMT promoter methylation from epigenome-wide data in the biomarker cohort of the NOA-04 trial. Neuro-oncol. 2014;16:1630–1638. doi: 10.1093/neuonc/nou138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang E., He X., Yin D., Han L., Qiu M., Xu T., Xia R., Xu L., Yin R., De W. Increased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57. Cell Death Dis. 2016;7:e2109. doi: 10.1038/cddis.2015.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao L., Han T., Li Y., Sun J., Zhang S., Liu Y., Shan B., Zheng D., Shi J. The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4. FASEB J. 2016;31:893–903. doi: 10.1096/fj.201600994R. [DOI] [PubMed] [Google Scholar]
- 25.Sun M., Nie F., Wang Y., Zhang Z., Hou J., He D., Xie M., Xu L., De W., Wang Z., Wang J. LncRNA HOXA11-AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299–6310. doi: 10.1158/0008-5472.CAN-16-0356. [DOI] [PubMed] [Google Scholar]
- 26.Ørom U.A., Derrien T., Beringer M., Gumireddy K., Gardini A., Bussotti G., Lai F., Zytnicki M., Notredame C., Huang Q. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gumireddy K., Li A., Yan J., Setoyama T., Johannes G.J., Orom U.A., Tchou J., Liu Q., Zhang L., Speicher D.W. Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. EMBO J. 2013;32:2672–2684. doi: 10.1038/emboj.2013.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrallo-Gimeno A., Nieto M.A. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- 29.Nieto M.A. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 30.Peinado H., Olmeda D., Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 31.Batlle E., Sancho E., Francí C., Domínguez D., Monfar M., Baulida J., García De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 32.Grady W.M., Willis J., Guilford P.J., Dunbier A.K., Toro T.T., Lynch H., Wiesner G., Ferguson K., Eng C., Park J.G. Methylation of the CDH1 promoter as the second genetic hit in hereditary diffuse gastric cancer. Nat. Genet. 2000;26:16–17. doi: 10.1038/79120. [DOI] [PubMed] [Google Scholar]
- 33.Cui H., Wang L., Gong P., Zhao C., Zhang S., Zhang K., Zhou R., Zhao Z., Fan H. Deregulation between miR-29b/c and DNMT3A is associated with epigenetic silencing of the CDH1 gene, affecting cell migration and invasion in gastric cancer. PLoS ONE. 2015;10:e0123926. doi: 10.1371/journal.pone.0123926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simon J.A., Lange C.A. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat. Res. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 35.Herranz N., Pasini D., Díaz V.M., Francí C., Gutierrez A., Dave N., Escrivà M., Hernandez-Muñoz I., Di Croce L., Helin K. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol. Cell. Biol. 2008;28:4772–4781. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao Q., Yu J., Dhanasekaran S.M., Kim J.H., Mani R.S., Tomlins S.A., Mehra R., Laxman B., Cao X., Yu J. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27:7274–7284. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heo J.B., Lee Y.S., Sung S. Epigenetic regulation by long noncoding RNAs in plants. Chromosome Res. 2013;21:685–693. doi: 10.1007/s10577-013-9392-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 39.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 40.Chaffer C.L., Weinberg R.A. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 41.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 42.Steeg P.S. Tumor metastasis: mechanistic insights and clinical challenges. Nat. Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 43.Guttman M., Donaghey J., Carey B.W., Garber M., Grenier J.K., Munson G., Young G., Lucas A.B., Ach R., Bruhn L. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Z., Wang R., Zhang T., Dong X. Hypoxia/lncRNA-AK123072/EGFR pathway induced metastasis and invasion in gastric cancer. Int. J. Clin. Exp. Med. 2015;8:19954–19968. [PMC free article] [PubMed] [Google Scholar]
- 45.Lai J., Nie W., Zhang W., Wang Y., Xie R., Wang Y., Gu J., Xu J., Song W., Yang F. Transcriptional regulation of the p73 gene by Nrf-2 and promoter CpG methylation in human breast cancer. Oncotarget. 2014;5:6909–6922. doi: 10.18632/oncotarget.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xing Z., Lin A., Li C., Liang K., Wang S., Liu Y., Park P.K., Qin L., Wei Y., Hawke D.H. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159:1110–1125. doi: 10.1016/j.cell.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heo J.B., Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 49.Fang X.Y., Pan H.F., Leng R.X., Ye D.Q. Long noncoding RNAs: novel insights into gastric cancer. Cancer Lett. 2015;356(2 Pt B):357–366. doi: 10.1016/j.canlet.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 50.Jordà M., Olmeda D., Vinyals A., Valero E., Cubillo E., Llorens A., Cano A., Fabra A. Upregulation of MMP-9 in MDCK epithelial cell line in response to expression of the Snail transcription factor. J. Cell Sci. 2005;118:3371–3385. doi: 10.1242/jcs.02465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.