Abstract
Antisense oligonucleotide (ASO) gapmers downregulate gene expression by inducing enzyme-dependent degradation of targeted RNA and represent a promising therapeutic platform for addressing previously undruggable genes. Unfortunately, their therapeutic application, particularly that of the more potent chemistries (e.g., locked-nucleic-acid-containing gapmers), has been hampered by their frequent hepatoxicity, which could be driven by hybridization-mediated interactions. An early de-risking of this liability is a crucial component of developing safe, ASO-based drugs. To rank ASOs based on their effect on the liver, we have developed an acute screen in the mouse that can be applied early in the drug development cycle. A single-dose (3-day) screen with streamlined endpoints (i.e., plasma transaminase levels and liver weights) was observed to be predictive of ASO hepatotoxicity ranking established based on a repeat-dose (15 day) study. Furthermore, to study the underlying mechanisms of liver toxicity, we applied transcriptome profiling and pathway analyses and show that adverse in vivo liver phenotypes correlate with the number of potent, hybridization-mediated off-target effects (OTEs). We propose that a combination of in silico OTE predictions, streamlined in vivo hepatotoxicity screening, and a transcriptome-wide selectivity screen is a valid approach to identifying and progressing safer compounds.
Keywords: antisense oligonucleotides, ASOs, off-target effects, OTEs, hepatotoxicity, locked nucleic acids, LNA, gene silencing, RNArcher, selectivity
Introduction
Antisense oligonucleotide (ASO) gapmers are short, synthetic oligonucleotides that localize to the cytoplasm and nucleus and downregulate gene expression by inducing RNase-H1-dependent degradation of their RNA target. They represent a promising therapy to address previously undruggable genes, as they have the potential to selectively target the entire transcriptome, with relatively short design and optimization cycle times. However, their application has been hampered by frequent hepatoxicity, which is often observed following systemic administration of phosphorothioated (PS) ASOs containing various sugar modifications.1, 2 For the first- and second-generation chemistries, it is generally recognized that the hepatic damage is a downstream effect of the immunostimulatory nature of the ASOs, the extent of which can vary from sequence to sequence. In rodents, ASO accumulation in the liver is associated with lymphohistiocytic inflammatory infiltrates.3 Depending on the degree of oligonucleotide-induced inflammation, varying levels of hepatotoxicity have been observed and are accompanied by correlative increases in plasma transaminases (aspartate transaminase [AST] and alanine transaminase [ALT]). ASOs that are both profoundly immunostimulatory and hepatotoxic in rodents have also been identified, although the latter is a direct result of the former.4
Locked nucleic acid (LNA) gapmers, which are chemically modified RNA analogs and classified as third-generation chemistries, are generally viewed as being less immunostimulatory relative to previous chemistries.5, 6 Literature does, however, indicate that, compared to other modifications, LNAs are associated with a higher frequency of hepatotoxicity.1, 7, 8, 9 Early de-risking of hepatoxicity is an important enabler to identify safe ASO drugs, as is identification of selective sequences. To address this issue, the Oligonucleotide Safety Working Group (OSWG)10 clearly recommended assessment of off-target effects (OTEs) for ASOs during drug discovery and development, both computationally and experimentally. Conventionally, ASO-associated liver and renal toxicities, as well as immunotoxicity, are assessed mainly using rodent in vivo tests, which involve multiple drug applications and microscopic tissue examinations.6 Although animal toxicology studies do not guarantee that all potential safety issues will be identified for humans, these studies remain the best and most well-characterized way to predict the presence of all toxicities.10 However, such screens are lengthy and require large numbers of animals, which poses a significant challenge for ASO development from an ethical, resource, and time point of view.
In addition, for developing safe and effective oligonucleotide drugs, it is important to understand the underlying mechanism behind their hepatotoxic potential. However, such processes are poorly understood and are likely to be caused by a combination of (a) hybridization-dependent off-target toxicity due to hybridization of the respective ASO to non-intended RNA targets because of sequence similarity1, 11 and (b) hybridization-independent toxicity caused by binding of the ASO to intracellular proteins, thereby interfering with their function and, ultimately, leading to toxicity.12, 13 We have previously shown that a more exhaustive OTE assessment, utilizing both in silico predictions and in vitro confirmation in relevant cell lines, is a crucial aspect of ASO drug optimization.11 Importantly, with RNase H1 activity residing predominantly within the nucleus, ASOs are as likely to interact with intronic sequences as with exonic regions, leading to a considerable number of interactions with primary RNAs.
In this study, we tested the hepatotoxic potential of LNA gapmers designed against two human targets and demonstrate that the adverse effect of ASOs on mouse liver can be effectively ranked based on a single-dose (3-day) screening protocol. Using a streamlined readout of plasma transaminases alongside liver weights, we obtained a hepatotoxicity ranking equivalent to that based on a conventional 15-day repeat-dose study with detailed histopathological evaluation (GSK2910557A ≥ GSK2910632A > GSK2910584A ≫ GSK2910613A). Furthermore, we utilized RNA sequencing (RNA-seq) analysis to study the relationship between observed hepatotoxicity and unintended hybridization-mediated events. We found a significant correlation between the grade of adverse liver phenotype and the number of potent OTEs. Our observations suggest that judicious computational analyses should be routinely performed during antisense drug design to minimize the chances of encountering interactions with unintended genes. Prioritizing ASOs with high selectivity (against both exonic and intronic sequences) and utilizing the streamlined mouse screen will shorten the development cycle and ensure that the most promising designs are progressed.
Results
Comparison between Single- and Repeat-Dose Screening Studies
The primary objective of this part of the study was to determine whether a single-dose (acute) screen, with a streamlined readout of plasma ALT/AST levels and liver weights (and no histopathological assessment), was predictive of hepatotoxicity ranking based on a 15-day repeat-dose study. The rationale behind the design was that such a screen can be applied at an earlier stage of ASO optimization, as it requires less compound and fewer resources to run. Four different human BACH1 (BTB and CNC Homology 1, Basic Leucine Zipper Transcription Factor 1)-targeting LNA gapmers were selected for this investigation, based on the high target potency that had been demonstrated in primary in vitro pharmacology screening.11
Estimating Hepatotoxicity of BACH1-Targeting ASOs Using a Full Histopathological Evaluation
Male CD1 mice were dosed on five separate occasions, twice weekly over a period of 15 days. A dose of 30 mg/kg was chosen based on a literate review of two key publications available at the time the work was conducted,9, 14 which showed that such a dose should be sufficient to characterize the profile of liver toxicity for the chemistry. No treatment-related clinical signs or changes in body weight were noted with any of the four human BACH1 ASOs administrated on five occasions over the 15-day period (data not shown), indicating that the treatments were well tolerated relative to the vehicle (i.e., PBS) control. Significant increases in plasma ALT/AST levels were noted on day 7 in groups treated with GSK2910557A (ratio of group mean to vehicle controls = 4.1×/2.9×) and GSK2910584A (1.9×/2.1×), with no changes observed with the remaining two ASOs (GSK2910613A and GSK2910632A). On day 16, significant increases in plasma transaminases were noted with the majority of the ASO treatments relative to the vehicle control group, the exception being GSK2910613A. GSK2910632A induced the greatest effect, with 42×/25× increases in ALT/AST. GSK2910557A caused moderate increases (12×/4.1×), with modest increases noted for GSK2910584A (3.4×/2.8×). In general, there was agreement between the measured increases in plasma transaminases and the type and/or severity of morphological changes noted in the animals (summarized in Table 1). Frequent treatment-related effects based on review of the H&E sections of the livers included hepatocellular single-cell necrosis and increased mitotic rate, centrilobular hepatocellular cytoplasmic alteration and inflammatory cell infiltration, and rare instances of centrilobular fibrosis and hepatocellular vacuolation. Interestingly, no treatment-related liver pathology was seen in animals given GSK2910613A, which reflected the absence of ALT/AST increases seen in this group (hematology data are shown in Table S1). Based on these analyses, we gave the LNAs the following hepatotoxic potential ranking: GSK2910557A ≥ GSK2910632A > GSK2910584A ≫ GSK2910613A.
Table 1.
Summary of Histopathological Observations Made in the 15-Day Repeat Dose Study with BACH1-Targeting ASOs
| Finding | (No. of Animals with) Grade of Finding |
||||
|---|---|---|---|---|---|
| PBS | GSK2910613A | GSK2910584A | GSK2910632A | GSK2910557A | |
| Degeneration/necrosis; hepatocyte; localizeda | (1) low | ||||
| Single-cell necrosis; hepatocyteb | (1) low | (6) low | |||
| Cytoplasmic alteration; centrilobular; hepatocytebc | (1) low | (5) low; (1) high | (5) low; (2) high | ||
| Microvacuolation; hepatocyte; centrilobularb | (1) high | ||||
| Mitotic increase; hepatocyteb | (3) low | (2) low; (1) high | (3) low; (4) high | ||
| Inflammatory cell infiltrate; mixed cella | (2) low | (5) low | (7) low | (7) low | (1) low |
| Inflammatory cell infiltrate; centrilobular; mixed cellb | (1) low | (1) low | (6) low | ||
| Plasma ALT day 7 | 1 | 1 | 1.93** | 1.53 | 4.12** |
| Plasma ALT day 16 | 1 | 1.22 | 3.37 | 41.68** | 12.39** |
| Plasma AST day 7 | 1 | 0.97 | 2.13** | 1.5* | 2.94** |
| Plasma AST day 16 | 1 | 1.06 | 2.76 | 24.85** | 4.07 |
The number in parentheses reflects the number of animals for which the given phenotype was observed (out of eight animals for treated groups and seven animals for the PBS group). The text annotation shows the grade of finding, which is classified as “low” (i.e., minimal or low) or “high” (i.e., moderate or marked). The group mean ratios of plasma ALT/plasma AST were all normalized to the vehicle PBS control group mean. 0.05 > *p > 0.01; **p < 0.01.
Considered to be a background finding and not related to BACH1 ASO treatment.
Findings were observed within the same areas of the liver and are considered to be a continuum of the same pathology.
Possibly hypertrophy, but no liver weights were available for confirmation.
Hepatotoxicity Evaluation from a Streamlined Single-Dose 3-Day Screen Shows a Similar Ranking to that from the Repeat Dose 15-Day Study
To conduct a single-dose screen, we used the same four 16-mer human BACH1-targeting ASOs that had been assessed in the repeat-dose study, along with a 14-mer LNA gapmer (“Sequence 1”) that served as a positive control. Sequence 1, which targets murine glucocorticoid receptor (GR), was identified in the literature as being profoundly hepatotoxic.15 Male CD1 mice were intravenously dosed with 100 mg/kg of each of the ASOs on day 1, while organs were harvested and final measurements were collected on day 4 (72 hr after dosing). No treatment-related clinical signs or changes in body weights were noted with any of the four human BACH1 ASOs administered at 100 mg/kg (data not shown), indicating that the treatments were well tolerated relative to vehicle control. No clinical signs were noted with Sequence 1 (dosed at 30 mg/kg) until day 3. On day 4, one of four mice was found dead in the cage, and two more were extremely subdued, reluctant to move, cold to the touch, and had hunched posture. Another animal displayed similar signs, although of a lower severity. All three mice were euthanized early on day 4, due to their clinical signs, and blood samples were obtained from only two of them. Relative to day 2, Sequence-1-treated animals also exhibited decreases in day-4 body weights of between 0.86× and 0.92×.
Relative to the PBS control group, treatment with 100 mg/kg GSK2910613A and GSK2910584A did not result in a significant effect on plasma transaminase levels or body-weight-corrected liver weights (Table 2). Treatment with GSK2910557A and GSK2910632A—BACH1 ASOs that were the most hepatotoxic in the 15-day repeat-dose study (Table 1)—was associated with significant increases in liver weights (1.22× and 1.23×, respectively). The increases in liver weights were accompanied by slight increases in the groups’ mean ALT/AST levels (relative to the PBS control group), which were significant in the case of GSK2910557A. Based on the measurements collected from three mice, which survived dosing with 30 mg/kg of Sequence 1, the mean liver weights were 1.27× higher than that of PBS control (SD ± 0.07). The mean plasma ALT (757.5 U/L) and AST (429 U/L) levels were also markedly higher than those seen in the PBS control group (ALT = 31.5 U/L, SD ± 6.5; AST = 40.8 U/L, SD ± 10.8), confirming that Sequence 1 is profoundly hepatotoxic in the mouse. Animals treated with Sequence 1 had a significant, 1.81× increase in group mean spleen weights and an ∼2-fold decrease in group mean total white blood cell (WBC) counts, which can be attributed to an ∼4-fold decrease in monocytes. With Sequence 1 previously reported as highly toxic after repeat dosing in the mouse,15 the high degree of hepatotoxicity observed in the acute screen increases the confidence in the study design. No other ASOs caused any significant effect on spleen weights or change in WBCs (Table S2). Based on the findings in this study, the LNA gapmers can be ranked as Sequence 1 ≫ GSK2910557A ≥ GSK2910632A > GSK2910584A > GSK2910613A, which is similar to the ranking established in the 15-day repeat-dose study results.
Table 2.
Summary of Group Mean Plasma ALT/AST Levels and Liver and Spleen Weights, All Expressed as Group Mean Ratios versus the PBS Control Group Mean, for Treatments Used in Acute Study 1: BACH1
| Treatment | Hepatotox | Plasma ALT | Plasma AST | Liver Weight | Spleen Weight |
|---|---|---|---|---|---|
| PBS | 1 | 1 | 1 | 1 | |
| GSK2910613A | − | 1.06 | 0.87 | 1.02 | 1.01 |
| GSK2910584A | − | 1.01 | 1.12 | 1.13 | 1.16 |
| GSK2910632A | + | 1.56 | 1.34 | 1.23** | 0.98 |
| GSK2910557A | + | 1.75* | 1.45* | 1.22** | 1.23 |
| Sequence 1 | ++ | 24.05** | 10.53** | 1.27** | 1.81** |
The ASOs are coded to represent an arbitrary threshold for high (++), low (+), and no (−) evidence of hepatotoxicity. Liver-weight ratios are based on body-weight-corrected values. *p > 0.01; **p < 0.01. The values were all normalized to vehicle PBS control and show a statistical significance for each observation (using ANOVA and post-hoc Dunnett’s test).
Ranking Adverse Liver Phenotype Potential for an Independent Set of 14-mer PAR-2-Targeting ASOs
As a part of an independent drug discovery program, we also evaluated ASOs that target a different gene, Protease-Activated Receptor 2 (PAR-2). The second study utilized the established acute, 3-day screen to assess the hepatotoxicity of seven 14-mer ASOs, targeting the human PAR-2 gene, and a single murine surrogate ASO (GSK3025301A). No consistent treatment-related clinical signs were noted with any of the PAR-2 ASOs or the rodent surrogate ASO during the course of the study, although some body weight changes were observed. In general, these comprised modest decreases or increases in body weight gain relative to the vehicle control group and were not considered adverse. In three treatment groups, the increases in body weights were influenced by single animals per group gaining a relatively large amount of weight (3.9 g, 4.3 g, and 4.0 g for GSK3025422A, GSK3025566A, and GSK3025301A, respectively). Overall, the eight tested ASO gapmers were well tolerated, and body weight changes did not appear to clearly correlate with increases in markers reflective of hepatotoxicity (plasma ALT and AST, as well as liver weight), which are summarized in Table 3.
Table 3.
Summary of Group Mean Plasma ALT/AST Levels and Liver and Spleen Weights, All Expressed as Group Mean Ratios versus the PBS Control Group Mean, for Treatments Used in Acute Study 2: PAR-2
| Treatment | Hepatotox | Plasma ALT | Plasma AST | Liver Weight | Spleen Weight |
|---|---|---|---|---|---|
| PBS | 1 | 1 | 1 | 1 | |
| GSK3025617A | − | 1.1 | 1.02 | 1.07 | 0.97 |
| GSK3025508A | − | 1.11 | 0.93 | 1.07 | 0.94 |
| GSK3025301A | − | 1.42 | 1.12 | 1.2** | 1.09 |
| GSK3025456A | + | 1.61 | 1.36 | 1.3** | 0.87 |
| GSK3025566A | + | 2.91 | 1.3 | 1.24** | 1.08 |
| GSK3025466A | ++ | 9.17* | 3.54* | 1.21** | 1.06 |
| GSK3025638A | ++ | 9.37* | 2.66* | 1.22** | 1.1 |
| GSK3025422A | ++ | 38.39** | 11.59** | 1.45** | 0.97 |
The ASOs are coded to represent an arbitrary score for high (++), low (+), and no (−) evidence of hepatotoxicity. Liver-weight ratios are based on body-weight-corrected values. *p > 0.01; **p < 0.01. The values were all normalized to vehicle PBS control and show a statistical significance for each observation (using ANOVA and post-hoc Dunnett’s test).
Based on the plasma transaminase levels measured in both acute studies, the 14-mer PAR-2 ASOs were found to have a higher proportion of sequences that caused high-grade hepatotoxicity when compared to the 16-mer BACH1 ASOs. While ASO length and sequence/chemical design appear to influence the mismatch (MM) and gap tolerance, in general, decreasing oligonucleotide length increases the number of potential transcriptome binding sites, with similarity high enough for interaction to occur. As expected, the group mean increases in plasma ALT and AST were closely correlated (Pearson’s r = 0.997, p = 6.33E-09). Treatment with GSK3025422A, GSK3025638A, and GSK3025466A caused a high degree of hepatotoxicity (applying a threshold of ≥5-fold increases in ALT). A lower level of toxicity was evident following treatment with GSK3025566A and GSK3025456A (increases in group mean ALT between 1.5- and 4.9-fold, with larger increases in individual animals), while the remaining LNA gapmers showed no significant increase in the ALT levels. Out of the eight sequences evaluated in the screen, GSK3025422A stood out as causing, by far, the greatest increase in ALT and AST levels (∼38× and ∼12×, respectively), accompanied by the largest increase in mean liver weight (1.4× increase, adjusted to body weight) relative to PBS control. Corresponding clinical chemistry and hematology data are shown in Table S3.
14-mer ASOs Show Lower Tolerability in a 4-Week Repeat-Dose Study in Rat Compared to 16-mer ASOs
We have also evaluated a number of LNA gapmers for hepatotoxic potential in rat 4-week repeat-dose studies (Table S4). Two 16-mers (GSK29105456A and GSK2910613A) caused minimal effects in the rat at a dose of 40 mg/kg given twice weekly for 4 weeks. The three 14-mers evaluated were generally more hepatotoxic (a trend we also observed in mouse), with only one (GSK3025647A) being tolerated for 4 weeks’ dosing. The rats dosed with the other two 14-mer compounds had to be euthanized humanely before the end of the study. Overall, we have found that the shorter 14-mer designs are more likely to be associated with hepatic effects compared to the longer 16-mer designs. This is in agreement with the findings in the mouse model indicated in Tables 2 and 3.
Translatability of Mouse In Vivo Model to Human
One of the premises of our work is that ASOs can be initially selected based on the number of putative human gene hybridization-dependent off-target interactions that can be predicted in silico, using sequence alignment algorithms. We previously showed that the OTE predictions are verified in vitro in cells known to express both the intended target and off-target mRNAs using standard qPCR. However, the toxicity assays used in this study are performed in rodents. Although ASOs designed against human genes are used in a different species, the degree and frequency of OTEs do appear to translate between species for a given ASO (although the precise off-target effects will be species-specific). To test for this, we repeated the OTE predictions with our in-house-developed software (RNArcher) using mouse transcriptome. Next, we performed correlation analysis between the numbers of predicted OTEs in mouse and human for all oligonucleotides used in the study (Figure S1). A high concordance of data was found (r = 0.93, p = 4.89E-06, and remained high even after removing the top two data points: r = 0.70, p = 0.016) for mouse and human, respectively, indicating that the non-selective designs in human appear to be non-selective in the mouse.
High Confirmation Rate of In Silico Predicted OTEs in Mouse Liver
We subsequently applied transcriptome profiling and pathway analyses on both PAR-2- and BACH1-targeting ASOs to test whether the in vivo animal liver toxicities correlate with downregulation of off-target genes (OTEs). Hybridization-mediated OTEs were predicted for all tested LNA gapmers using in-house-developed software (RNArcher)11 and overlapped with the results of differential expression (DE) analysis derived from the liver tissue of mice treated with the corresponding ASO. The computational methodology presented in Kamola et al.11 was further refined to increase the sensitivity of the alignment detection. Genes with very low baseline expression (fewer than ten reads overlapping a gene and those identified as low abundance by independent filtering in DESeq2) were removed, as this is best practice. OTEs were defined as sequences having up to one or two MMs and/or gaps for 14- and 16-mer ASOs, respectively. The level was established based on previous experience with regard to MMs and gap tolerance and the results shown in Kamola et al.11 The length of oligonucleotides significantly alters the probability of encountering hits with high similarity; the shorter the sequence, the harder it is to design a “clean” ASO (given a similar chemical design), especially so given the length of intronic regions that can be targeted. To illustrate this, the average number of putative one-MM hits for 16-mer BACH1 ASOs in the study is 16, while all of the 14-mer PAR-2 ASOs have over 140 putative one-MM hits. Furthermore, the number of potential hits at lower levels is far too high for accurate analysis: due to algorithm limitations, it becomes increasingly difficult to find such alignments and to distinguish between true OTEs and downstream effects. “Background genes” were defined as those genes that were not predicted to have any interaction site with the ASO (no target site with fewer than three MMs and/or gaps).
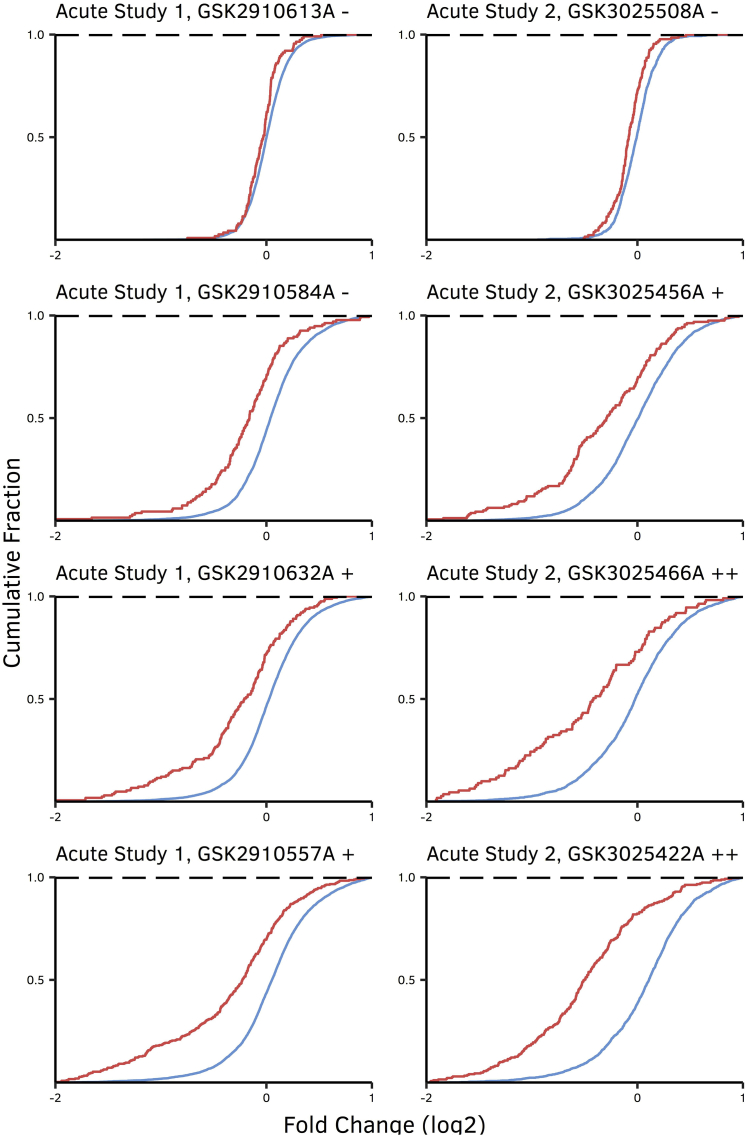
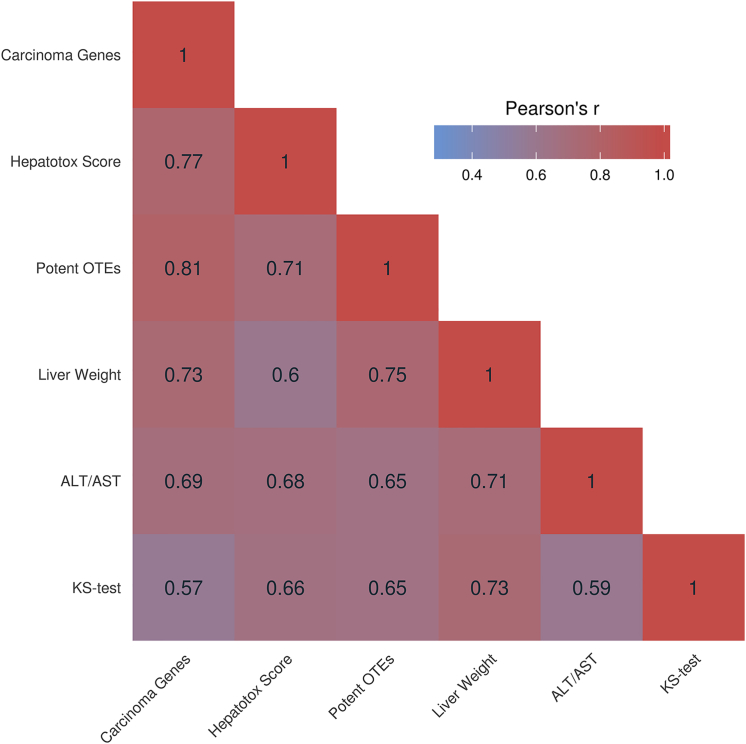
There were considerable differences in expression profiles between the OTE and background gene groups (Figure 1), with predicted OTEs being more downregulated. This observation is in agreement with the hypothesis that the OTE group is enriched for true ASO hits where gene downregulation is expected, while the background gene group is enriched for downstream pathway changes where equal numbers of up- and downregulated gene changes are expected. Critically, the difference in fold change profiles between OTEs and background genes not only confirmed that hybridization-mediated exonic and intronic OTEs translate in vivo but also highlighted the abundance and magnitude of such interactions. All the interactions were found to be statistically significant based on the Kolmogorov-Smirnov test (KS test) (Table 4). The LNA gapmers in Figure 1 were ordered from those with the lowest potential toxicity at the top to those that showed highest toxicity at the bottom. Most importantly, the magnitude of difference between the groups (KS-test values) correlates with the hepatotoxicity score of the corresponding ASO (r = 0.66, p = 0.019). Subsequently, the number of confirmed/potent OTEs was established by extracting putative hits (zero to one and zero to two MMs for 14- and 16-mers, respectively) that were found to have a fold change ⩽ −0.5 with an associated p value ⩽ 0.01. A high degree of correlation was observed between the number of potent OTEs and the average of ALT/AST levels (r = 0.65, p = 0.022) and the hepatotoxicity score (r = 0.71, p = 0.0095) for the corresponding treatment group. A correlation matrix that shows the dependence between KS-test values, average ALT/AST levels, liver weights, number of potent OTEs and hepatotoxicity scores is shown in Figure 2. Sequence 1 was excluded from all aforementioned analyses, as its high toxicity translated into a profound downregulation of the majority of the expressed genes, making it challenging to differentiate between OTEs and downstream changes in expression. Overall, our data demonstrate that animal liver toxicities and in vivo off-target gene downregulation for a set of tested ASOs are significantly correlated and that ASOs that are more hepatotoxic are also less selective.
Figure 1.
Expression Profiles of OTEs, Indicated in Red, Compared to Transcripts Not Predicted to Interact with the ASO, Indicated in Blue
OTEs were defined as genes with zero- to one-MM and zero- to two-MM alignments relative to a particular oligonucleotide, for 14- and 16-mer ASOs, respectively. The difference in profiles and their significance were calculated using the KS test and are shown in Table 4. The ASOs are coded to represent an arbitrary score for high (++), low (+), and no (-) evidence of hepatotoxicity.
Table 4.
Table Summarizing Observed OTEs and Pathway Analyses Performed for ASOs Used in Acute Studies 1 and 2
| ASO IDa | Hepatotoxb | KS Testc | p Value | Potent OTEs | OTE Level | Liver Hyperplasiad | Hepatocellular Carcinomad |
|---|---|---|---|---|---|---|---|
| GSK2910613A | − | 0.18 | 1.01E-03 | 0 | 0–2 MM | 7 | 6 |
| GSK2910584A | − | 0.3 | 2.97E-11 | 18 | 200 | 194 | |
| GSK2910632A | + | 0.32 | 3.11E-15 | 39 | 270 | 259 | |
| GSK2910557A | + | 0.35 | <2.2E-16 | 123 | 563 | 544 | |
| GSK3025617A | − | 0.29 | 5.52E-08 | 14 | 0–1 MM | 17 | 17 |
| GSK3025508A | − | 0.22 | 2.63E-05 | 0 | 0 | 0 | |
| GSK3025301A | − | 0.34 | 6.33E-15 | 41 | 219 | 211 | |
| GSK3025456A | + | 0.31 | 3.96E-14 | 50 | 448 | 431 | |
| GSK3025566A | + | 0.54 | <2.2E-16 | 43 | 224 | 221 | |
| GSK3025466A | ++ | 0.37 | 1.07E-14 | 55 | 795 | 771 | |
| GSK3025638A | ++ | 0.39 | <2.2E-16 | 67 | 214 | 209 | |
| GSK3025422A | ++ | 0.51 | <2.2E-16 | 124 | 831 | 807 |
Sequence 1 was excluded from the table, as the treatment resulted in downregulation of the majority of expressed genes, making it challenging to verify OTE confirmation or to perform pathway analysis.
The ASOs are coded to represent an arbitrary score for high (++), low (+), and no (−) evidence of hepatotoxicity.
The KS test was completed to calculate the difference (i.e., distance between the empirical distribution function) in expression profiles of genes annotated as OTEs and remaining (“background”) genes.
Number of activated genes connected to liver hyperplasia or hepatocellular carcinoma.
Figure 2.
Correlation Matrix Showing Dependence between Hepatotoxicity Descriptors
Pearson’s correlation coefficients are presented for KS-test values, average ALT/AST levels, liver weights, number of potent OTEs, hepatotoxicity scores, and number of activated genes annotated as involved in hepatocellular carcinoma for all ASOs used in acute studies 1 and 2 (excluding Sequence 1).
Activation of Mouse Pathways Related to Toxic Response
There were no common pathways found across all ASOs, suggesting that diverse mechanisms are responsible for the toxicological phenotype observed with each ASO. As expected, the most common theme reported in the pathway analysis was liver damage/necrosis; however, this is mainly observed for the most hepatotoxic PAR-2-targeting ASOs. The evaluation did reveal a significant correlation between number of potent OTEs and genes and interactions annotated as being involved in hepatotoxicity. Specifically, the “hepatocellular carcinoma” and “liver hyperplasia/hyperproliferation” groups were closely correlated (in both cases, for all ASOs excluding Sequence 1: r = 0.81, p = 0.0015; Figure 2; Table 4). It is likely that the high toxicity caused by potent OTEs is causal of hepatocyte death (e.g., via apoptosis or necrosis) (hepatocellular carcinoma), which, in turn, activates the regenerative functions of the liver (liver hyperplasia/hyperproliferation). This concurs with the histopathology observations of the 15-day repeat-dose study, where toxicity reflected “mitotic increase” (Table 1). A detailed pathway analysis, based on all differentially expressed genes, is shown in Table 5 for ASOs with low and high hepatotoxicity.
Table 5.
Summary of Top Pathways and Regulator Effect Networks Performed in IPA for ASOs Classified as Having Low or High Evidence of Hepatotoxicity
| ASO ID | Top Regulator Effect Networks/Toxicology-Related Pathway | Consistency Score/Overlap (p Value) |
|---|---|---|
| GSK2910632A (+) | hypoplasia of lymphatic system, osteosarcoma (+4 more) | 35.988 |
| cell viability of tumor cell lines (+4 more) | 5.485 | |
| apoptosis, viral infection | 4.523 | |
| NRF2-mediated oxidative stress response | 8.5% (3.08E-04) | |
| GSK2910557A (+) | apoptosis of hepatocytes (+3 more) | 24.167 |
| apoptosis, infection of mammalia, viral infection | 22.045 | |
| infection of mammalia, leukemia, metabolism of DNA (+3 more) | 14.878 | |
| benign connective or soft-tissue neoplasm (+4 more) | 13.294 | |
| renal necrosis/cell death | 12.1% (9.19E-05) | |
| Sequence 1 (++) | liver necrosis/cell death | 57.8% (2.83E-22) |
| production of nitric oxide and reactive oxygen species in macrophages | 55.0% (1.58E-12) | |
| mitochondrial dysfunction | 55.0% (5.96E-12) | |
| acute-phase response signaling | 53.3% (1.62E-10) | |
| GSK3025456A (+) | development of malignant tumor (+3 more) | 9 |
| NRF2-mediated oxidative stress response | 21.8% (2.43E-16) | |
| GSK3025566A (+) | apoptosis of cancer cells (+4 more) | 12.882 |
| apoptosis of fibrosarcoma cell lines (+1 more) | 10.733 | |
| apoptosis of fibrosarcoma cell lines | 9.192 | |
| apoptosis of cancer cells (+4 more) | 7.034 | |
| apoptosis of cancer cells (+4 more) | 5.004 | |
| NRF2-mediated oxidative stress response | 11.7% (4.82E-07) | |
| GSK3025466A (++) | inflammatory response | 4.249 |
| TREM1 signaling | 40.0% (1.30E-11) | |
| Increases liver damage | 34.9% (7.18E-12) | |
| role of pattern recognition receptors in recognition of bacteria and viruses | 29.9% (6.60E-10) | |
| IL-8 signaling | 26.6% (2.30E-10) | |
| hepatic fibrosis/hepatic stellate cell activation | 25.7% (1.95E-09) | |
| liver necrosis/cell death | 25.3% (5.39E-13) | |
| GSK3025638A (++) | antiviral response of cells, apoptosis of microglia (+8 more) | 154.477 |
| antiviral response of cells, apoptosis of microglia (+9 more) | 145.831 | |
| antiviral response of cells, apoptosis of microglia (+7 more) | 79.024 | |
| antiviral response of cells, apoptosis of microglia (+13 more) | 52.343 | |
| activation of cells, infection by RNA virus (+7 more) | 46.445 | |
| interferon signaling | 32.4% (1.77E-09) | |
| activation of IRF by cytosolic pattern recognition receptors | 21.9% (3.30E-09) | |
| role of RIG1-like receptors in antiviral innate immunity | 15.6% (2.93E-04) | |
| increases glomerular injury | 13.2% (8.16E-04) | |
| role of pattern recognition receptors in recognition of bacteria and viruses | 9.4% (3.34E-04) | |
| renal necrosis/cell death | 5.4% (1.44E-03) | |
| GSK3025422A (++) | injury of renal tubule | 16.1 |
| gliomatosis,hepatic steatosis, viral infection | 13.72 | |
| apoptosis of mesangial cells | 9.5 | |
| acute renal failure panel (rat) | 33.9% (8.01E-07) |
ASOs were classified as having low (+) or high (++) evidence of hepatotoxicity. IRF, interferon regulatory factor.
There was a variable degree of immune activation observed in most ASO treatments. However, the effect generally appears to be weak, and it is difficult to judge whether it is caused by the immunostimulatory potential of ASOs or is reflective of inflammatory cell infiltrates around areas of hepatocellular necrosis (as was observed based on H&E histological assessment of the livers from the 15-day repeat-dose screen; Table 1). The pathway changes annotated for GSK3025638A appear to indicate a higher than average level of immunostimulation (evidence of Toll-like receptor and other pathogen associate molecular pattern receptor activation is observed in the IPA results). Based on WBC changes measured in the mouse screen, circulating neutrophils were increased by approximately 2-fold (relative to the PBS control group) after treatment with GSK3025638A (Table S3). The pathway results also suggest that the high toxicity of Sequence 1 might be related to damage caused to mitochondria, leading to liver necrosis. Of particular interest is that Sequence 1, being the most toxic ASO based on clinical observations and transaminase changes, does not seem to cause any immune effects at the pathway analysis level. Similarly, GSK3025422A was not associated with a marked level of immunostimulation (the increase in monocytes shown in Table S3 can be attributed to liver toxicity) that would explain its extreme effects on the liver (note that the results do suggest cellular injury). The difference between OTEs and background genes for GSK3025422A was the highest among all tested ASOs (based on KS test), and the additive/synergistic effects of silencing a large number of genes is the most likely driver of the adverse liver phenotype. Unsurprisingly, as the OTEs causing the toxic effect are sequence dependent, the initiating events of the toxicity will be different between each ASO. The high number of confirmed OTEs also makes it very challenging to pinpoint the hits whose knockdown may be responsible for the adverse phenotype in the liver.
Discussion
Given its sensitivity to LNA-gapmer-mediated hepatotoxicity, and the relatively low amounts of compound needed for dosing, the mouse makes an effective early stage model for screening antisense therapeutics. For this reason, we undertook a series of studies to establish single-dose screens in the mouse to rank ASOs based on liver response phenotype. Such an approach allows de-selection of sequences that generated unacceptable toxicity and progression of the “cleanest” ASOs at an earlier stage of the drug optimization (as it requires less compound and fewer resources to run). Based on the findings in the acute mouse screen, we deselected a number of PAR-2 ASOs (GSK3025456A, GSK3025566A, GSK3025466A, GSK3025638A, and GSK3025422A) as having undesirable levels of hepatotoxicity. Having subsequently assessed two BACH1 and three PAR-2 ASOs in 28 repeat-dose studies in rat (Table S4), we have found the acute mouse screen to be broadly predictive of the overall ranking of hepatotoxicity in this second rodent species (albeit for a smaller set of compounds). While the screen does not measure systemic exposure (i.e., toxicokinetics) or ASO levels in the tissue, it is widely known that there is a high level of ASO accumulation in the liver, which is only second to that observed in proximal tubule cells of the kidney.16, 17
We have conducted limited studies in murine primary hepatocytes, using the same four BACH1 and Sequence 1 gapmers detailed in this paper, to explore whether an in vitro screen could be used. The ASOs were incubated for 72 hr at concentrations ranging from 0.1 μM to 1 mM, and lactate degydrogenase (LDH) leakage was used as a measure of cytotoxicity (unpublished data). In our hands, we found the liver perfusions to be technically challenging, with unreliable hepatocyte yields and viability. In addition, we noted that high background/media control LDH leakage, which made ranking of the ASO gapmers that had subtle differences in hepatotoxicity in vivo difficult. We were only able to differentiate the grossly hepatotoxic ASOs (such as Sequence 1) from non-hepatotoxic ASOs (such as GSK2910613A). Due to the technical challenges, we moved away from this approach in favor of the acute mouse screen. Recently, Sewing and colleagues18 published a much more rigorous evaluation of in vitro screening, with primary human and murine hepatocytes for ASOs. This clearly demonstrates that it is possible to overcome the technical difficulties of working with mouse hepatocytes and that such an in vitro screening approach has utility in predicting the hepatotoxicity observed in repeat-dose mouse screens. We envisage that both strategies are equally suited for screening and prioritization of a relatively large number of ASOs. However, once the best one to two ASOs have been selected, it is advisable to take them through the more thorough conventional toxicological analyses. This should include a literature-based review of the known (and theoretical) health consequences of inhibiting any confirmed off-target genes, as well as extensive in vivo non-clinical safety studies.
Based on the data shown in Figures 1 and 2 and Table 4, we can conclude that ASOs that are more hepatotoxic are also less selective. Our data show that liver toxicity of LNA-containing gapmer ASOs is associated with their ability to hybridize to off-target transcripts and downregulate their expression. This concurs with recent in vivo studies, which demonstrate that hepatotoxicity is attenuated by RNase H1-mediated knockdown.19, 20 Whether a given interaction will result in cleavage depends on many factors, such as ASO tolerance for MMs (which appears to be driven by sequence and chemical design), position of MMs and/or gaps, target-ASO affinity, target site accessibility, competition for the RNase H1 enzyme with other binding regions, number of interaction sites within a gene, transcript abundance and turnover, and more. As current understanding of those aspects is not sufficient for computational modeling and estimation (especially affinity, which we previously showed to be a highly predictive parameter), our data are solely relying on identifying interactions based on sequence complementarity. This presents the most straightforward and reliable estimator of hepatotoxic potential at the ASO design stage, although it will inevitably produce false-positive hits. While a single OTE could potentially translate into profound toxicity, the adverse phenotype in the mouse study seems to correlate with the number of potent of interactions. Higher numbers of putative hits increase the probability of encountering a potent interaction or several weaker hits that could lead additive or synergistic effects. The MM tolerance of LNA oligonucleotides could also explain why 14-mer ASOs were broadly more hepatotoxic than the 16-mer designs, as it is easier to avoid putative OTEs with a longer sequence (based on in-house experience with ASO design; data not shown). This observation increases confidence in the approach proposed in our previous study,11 where we recommend designing and/or selecting ASOs with the highest predicted selectivity (i.e., lowest number of putative intronic and exonic off-target hits identified in a database of coding and non-coding genes). We also envisage that further improvements to an oligonucleotide safety profile will likely come from refinement in non-sequence-related design components such as length, chemical modifications, and gap size.
While the high numbers of confirmed OTEs make it impossible to pinpoint the precise transcriptional source(s) of toxicity, it appears that the number of potent, unintended interactions is a key concern. The high correlation between potent OTEs and hepatotoxicity scores (r = 0.71, p = 0.0095) suggests that the limited immunostimulation observed with the tested LNA gapmers may occur alongside hepatotoxicity, and it is unlikely that the latter is dependent on the former. This is highlighted by the two most hepatotoxic ASOs we studied (Sequence 1 and GSK3025422A), which show little to no immune activation (based on Ingenuity Pathway Analysis; IPA). The high number of potent OTEs confirmed for both of those ASOs is, thus, the most likely driver of observed phenotype. Further investigation will help determine the precise route to cell death. It has to be noted that increases in spleen weights—an endpoint that tends to correlate with potent immunostimulatory ASOs and that was observed with most first- and, frequently, with second-generation chemistries16—were generally not observed in these studies.
While it is currently not possible to quantify the confirmatory rate of OTEs for a given oligonucleotide, the possibility of encountering potent interactions will certainly be minimized by progressing ASOs with the lowest number of predicted interactions. Considering selectivity at the ASO design stage is applicable, regardless of oligonucleotide length and design, its affinity to hits with partial sequence complementarity (which is very labor intensive and challenging to estimate), or target tissue. Our data show that a subsequent in vivo and/or in vitro hepatotoxicity screen should also be performed before investing resources in more robust repeat-dose tolerability and toxicology studies. The presented acute mouse screen was found to be predictive of findings in the more established 15-day mouse study and is more amenable to evaluating larger numbers of compound in a shorter time and with fewer resources. We have utilized this approach to deselect PAR-2 leads and have found it to be broadly predictive of observations in a 28-day, repeat-dose rat study (in terms of hepatotoxicity). Therefore, until further refinements in ASO chemistries, our data suggest that a combination of in silico OTE predictions, streamlined in vivo hepatotoxicity screening, and transcriptome-wide selectivity screen, is a valid approach to identifying and progressing safe compounds.
Materials and Methods
Oligonucleotide Sequences
The BACH1- and PAR-2-targeting ASO gapmers were designed and supplied by Exiqon A/S as >85% pure. The sequences were fully phosphorothioated and contained six LNAs and between four and six LNAs for 16- and 14-mer designs, respectively. Sequence 1 was synthesized by Exiqon based on the sequence and spiking pattern detailed in a study by Stanton et al.15
Animal Treatment and Ethical Standards
All animal studies detailed within this paper were ethically reviewed and carried out in accordance with the Animals (Scientific Procedures) Act 1986 and the GSK Policy on the Care, Welfare and Treatment of Animals. The CD1 mouse strain was chosen, as there is considerable knowledge of this strain’s general pathology and its response to a wide variety of drugs within GSK. For ASOs, doses of between 3 mg/kg and 5 mg/kg are considered to be at the upper end of the typical dose range assessed in clinical trials.21, 22, 23 Using a formula to calculate human equivalent doses (HEDs) based on allometric scaling (body-surface-area-adjusted dose), the 30 mg/kg and 100 mg/kg doses used in the mouse studies translate into HEDs of 2.4 mg/kg and 8.1 mg/kg, respectively.24 Male Crl:CD1 (ICR) mice (4–5 weeks old at the start of dosing) were obtained from Charles River UK. All animals were acclimated for 1 week prior to being randomized to study groups. They were group housed in temperature- and humidity-controlled conditions with 12-hr/12-hr light/dark cycling. Water and diet (Purina Mills International 5CR4 rodent diet) were supplied ad libitum. Pre-treatment clinical observations were made on day −3, and body weights were recorded on days −3 and −1. These observations were then made daily throughout the treatment and off-treatment periods and just prior to termination. Immediately prior to termination, mice were anesthetized with isofluorane and exsanguinated via the abdominal vena cava.
Repeat-Dose, 15-Day Study
Up to eight male CD1 mice per group were dosed on days 1, 4, 8, 11, and 15 (5 mL/kg) with the ASOs formulated in 1× PBS (pH 7.4) at 6 mg/mL (Table S5). Clinical observations were made 3 days before the study, three times on the treatment days, and once daily on off-dose days, with body weight being measured 3 days before the study and before each treatment. On day 7 (72 hr after the second dose) and day 16 (24 hr after the fifth and last dose), 0.3 mL blood was taken into lithium heparin tubes and plasma prepared, or into potassium EDTA tubes for hematology analysis. Animals were euthanized on day 16, 24 hr after the fifth dose, and terminal body weights were recorded (liver and spleen weights were omitted in error for this study). Liver samples were obtained post-mortem (fixed in 10% formalin for 24 hr, processed to wax blocks, sectioned, and stained with H&E) for histopathological assessment. A qualified regulatory veterinary pathologist read the liver sections from the study and made the histological diagnosis (summarized in Table 1).
Single/Acute-Dose, 3-Day Study
For both acute studies, male CD1 mice were intravenously dosed with 100 mg/kg of each of the ASOs on day 1 (Table S6). The exception was Sequence 1, which was only administrated at 30 mg/kg due to concerns over its tolerability at a higher dose. A dose of 100 mg/kg was selected, as it was below the total cumulative dose that was administrated in the 15-day repeat-dose study but was still anticipated to be sufficient for the more hepatotoxic ASOs to cause ALT and AST increases. Four to five mice per group were dosed on day 1 (5 mL/kg), with ASOs formulated in 1× PBS (pH 7.4) at 20 mg/mL and 6 mg/mL, for 100 mg/kg and 30 mg/kg doses, respectively. Clinical observations and body weight measurements were taken as described previously for the repeat-dose study. On day 4 (72 hr after dosing), 0.3 mL blood was taken into lithium heparin tubes, and plasma was prepared, or taken into potassium EDTA tubes for hematology analysis. Animals were euthanized 72 hr after dosing, and terminal body weight and liver and spleen weights were recorded. Approximately 100 mg of the caudate lobe from each mouse was also snap frozen in liquid nitrogen and stored at −70°C for future investigative work. Histopathology was not assessed in either acute study.
Transaminase and Hematology Analysis
Plasma ALT/AST levels were determined using the Advia Chemistry System (Siemens), loading 150 μL plasma. Total leukocyte and leukocyte differential cell counts (neutrophils, lymphocytes, monocytes, eosinophils, basophils, and large unstained cells) were assessed using the Advia 2120 system (Siemens), loading 178 μL whole blood. Numbers of animals used are detailed in the table legends. Statistical significance between treatment groups was calculated using ANOVA with Dunnett’s post hoc test.
RNA-Seq Profiling
Four animals were selected per treatment group from the acute-dose screens (all available animals from Acute Study 1 were used, and we randomly selected four out of five animals per group from Acute Study 2), except for the group dosed with Sequence 1 (in Acute Study 1), where only three animals were available. Total RNA was extracted from ∼50 mg snap-frozen liver using the RNeasy Plus Mini Kit (QIAGEN) (starting with 800 μL Buffer RLT Plus and extracting the RNA from 350 μL lysate) and MagNA Lyser Green Beads (Roche) (tissues were shredded for 33 s at 6,200 rpm). Samples were then quantified, and their integrity was assessed using the Qubit RNA BR (Broad Range) Assay Kit (Thermo Fisher Scientific) and an Agilent 2100 Bioanalyzer RNA 6000 Nano Kit (Agilent Technologies), respectively. The extracted RNA had an average RNA integrity number (RIN) score of 8.9, and no sample with a RIN score <8.1 was used for sequencing. RNA-seq libraries were prepared with a TruSeq Stranded Total RNA LT Sample Prep Kit with Ribo-Zero Human/Mouse/Rat Set A (Illumina). The library preparation followed the manufacturer’s manual, except for two modifications made to the thermal cycler parameters. First, the time of the “Elution 2-Frag-Prime” program was reduced from 8 min to 6 min to increase the length of the RNA fragments. Second, the number of cycles (11 instead of 15) and the time of the initial step in each cycle (30 s instead of 10 s at the first 98°C step) were both modified to minimize PCR-based amplification bias (PCR program). Three rounds of qPCR quantification of the final libraries were performed with the KAPA Library Quantification Kit (KAPA Biosystems) on a QuantStudio 12K Flex Real-Time PCR System (Thermo Fisher Scientific). Between six and seven samples were multiplexed per lane, after which the libraries were clustered using a HiSeq SBS Kit v4 and a HiSeq PE Cluster Kit v4 cBot (Illumina). Paired-end (PE) sequencing (2 × 76) was then performed on two separate HiSeq 1500 (Illumina) runs.
Data Analysis and OTE Predictions
RNA-seq data are available in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress), with the accession number ArrayExpress: E-MTAB-5180. Sequence reads were mapped to a primary assembly of the mouse genome (GRCm38) with STAR v2.4.2a,25 using the default parameters. Read summarization was completed using featureCounts v1.4.6-p426 based on gene annotation provided by the GENCODE M6 release.27 DE was established at the gene level with the DESeq2 v3.128 library using standard parameters. Putative hybridization-mediated off-target effects were identified and annotated using RNArcher.11 Pathway analysis was completed using IPA using a log2 of fold-change threshold of 0.6 with an associated p value threshold of 0.01 (Ingenuity System: build version, 346717M; content version, 24390178). Data processing, analysis, and visualization were performed in the R software environment and Python.
Author Contributions
J.D.P. conceived and designed the acute screen assay. T.M. study-directed the BACH1 ASO 15-day mouse and 28-day rat screens. K.R. study-directed the acute mouse screens. P.C. study-directed the PAR-2 28-day rat screen. J.R. and J.D.P. were the nonclincal safety project representatives for the BACH1 ASO program, and J.R. was a representative for the PAR-2 ASO programs. T.McK. and A.R. were the study pathologists for the BACH1 and PAR-2 programs, respectively. P.E. carried out the clinical chemistry and hematology analyses. A.F., S.M., and K.C. carried out the study post-mortems, tissue processing, and H&E slide staining. M.R.E., S.A.H., K.C., and K. Moores were members of Therapeutic Oligonucleotide DPU at GSK, who developed and progressed the ASOs used in the study. P.J.K. and K. Maratou extracted, processed, and sequenced the mouse liver RNA. P.J.K. performed RNA-seq analysis, OTE predictions, bioinformatics investigation, and data visualizations. P.A.W. provided pathway information based on IPA software and uploaded the data to ArrayExpress. P.J.K., J.D.P., K. Maratou, and P.A.W. wrote and revised the manuscript. J.D.P., P.A.W., K. Maratou, N.J.G., and T.W.G. supervised the work. All authors read and approved the final manuscript.
Acknowledgments
The project was supported by a EPSRC Industrial CASE Award (Imperial College London, GlaxoSmithKline and Public Health England; EP/J502017/1) to P.J.K. Analysis and manuscript preparation were finalized at the Laboratory for Medical Science Mathematics, RIKEN Center for Integrative Medical Sciences in Yokohama, Japan, under T. Tsunoda and supported by a JSPS postdoctoral fellowship (15F15776) and a JST CREST grant (JPMJCR1412). Funding for the open access charge was provided by GlaxoSmithKline. The authors would like to thank the anonymous reviewers who provided very helpful suggestions and comments.
Footnotes
Supplemental Information includes one figure and six tables and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.07.003.
Supplemental Information
References
- 1.Burdick A.D., Sciabola S., Mantena S.R., Hollingshead B.D., Stanton R., Warneke J.A., Zeng M., Martsen E., Medvedev A., Makarov S.S. Sequence motifs associated with hepatotoxicity of locked nucleic acid--modified antisense oligonucleotides. Nucleic Acids Res. 2014;42:4882–4891. doi: 10.1093/nar/gku142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crooke S.T. Second Edition. CRC Press; 2007. Antisense Drug Technology: Principles, Strategies, and Applications. [Google Scholar]
- 3.Zanardi T.A., Han S.-C., Jeong E.J., Rime S., Yu R.Z., Chakravarty K., Henry S.P. Pharmacodynamics and subchronic toxicity in mice and monkeys of ISIS 388626, a second-generation antisense oligonucleotide that targets human sodium glucose cotransporter 2. J. Pharmacol. Exp. Ther. 2012;343:489–496. doi: 10.1124/jpet.112.197426. [DOI] [PubMed] [Google Scholar]
- 4.Burel S.A., Machemer T., Ragone F.L., Kato H., Cauntay P., Greenlee S., Salim A., Gaarde W.A., Hung G., Peralta R. Unique O-methoxyethyl ribose-DNA chimeric oligonucleotide induces an atypical melanoma differentiation-associated gene 5-dependent induction of type I interferon response. J. Pharmacol. Exp. Ther. 2012;342:150–162. doi: 10.1124/jpet.112.193789. [DOI] [PubMed] [Google Scholar]
- 5.Vollmer J., Jepsen J.S., Uhlmann E., Schetter C., Jurk M., Wader T., Wüllner M., Krieg A.M. Modulation of CpG oligodeoxynucleotide-mediated immune stimulation by locked nucleic acid (LNA) Oligonucleotides. 2004;14:23–31. doi: 10.1089/154545704322988021. [DOI] [PubMed] [Google Scholar]
- 6.Frazier K.S. Antisense oligonucleotide therapies: the promise and the challenges from a toxicologic pathologist’s perspective. Toxicol. Pathol. 2015;43:78–89. doi: 10.1177/0192623314551840. [DOI] [PubMed] [Google Scholar]
- 7.Kakiuchi-Kiyota S., Koza-Taylor P.H., Mantena S.R., Nelms L.F., Enayetallah A.E., Hollingshead B.D., Burdick A.D., Reed L.A., Warneke J.A., Whiteley L.O. Comparison of hepatic transcription profiles of locked ribonucleic acid antisense oligonucleotides: evidence of distinct pathways contributing to non-target mediated toxicity in mice. Toxicol. Sci. 2014;138:234–248. doi: 10.1093/toxsci/kft278. [DOI] [PubMed] [Google Scholar]
- 8.Hagedorn P.H., Yakimov V., Ottosen S., Kammler S., Nielsen N.F., Høg A.M., Hedtjärn M., Meldgaard M., Møller M.R., Orum H. Hepatotoxic potential of therapeutic oligonucleotides can be predicted from their sequence and modification pattern. Nucleic Acid Ther. 2013;23:302–310. doi: 10.1089/nat.2013.0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swayze E.E., Siwkowski A.M., Wancewicz E.V., Migawa M.T., Wyrzykiewicz T.K., Hung G., Monia B.P., Bennett C.F. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 2007;35:687–700. doi: 10.1093/nar/gkl1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindow M., Vornlocher H.-P., Riley D., Kornbrust D.J., Burchard J., Whiteley L.O., Kamens J., Thompson J.D., Nochur S., Younis H. Assessing unintended hybridization-induced biological effects of oligonucleotides. Nat. Biotechnol. 2012;30:920–923. doi: 10.1038/nbt.2376. [DOI] [PubMed] [Google Scholar]
- 11.Kamola P.J., Kitson J.D.A., Turner G., Maratou K., Eriksson S., Panjwani A., Warnock L.C., Douillard Guilloux G.A., Moores K., Koppe E.L. In silico and in vitro evaluation of exonic and intronic off-target effects form a critical element of therapeutic ASO gapmer optimization. Nucleic Acids Res. 2015;43:8638–8650. doi: 10.1093/nar/gkv857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang X.-H., Shen W., Sun H., Kinberger G.A., Prakash T.P., Nichols J.G., Crooke S.T. Hsp90 protein interacts with phosphorothioate oligonucleotides containing hydrophobic 2′-modifications and enhances antisense activity. Nucleic Acids Res. 2016;44:3892–3907. doi: 10.1093/nar/gkw144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kakiuchi-Kiyota S., Whiteley L.O., Ryan A.M., Mathialagan N. Development of a method for profiling protein interactions with LNA-modified antisense oligonucleotides using protein microarrays. Nucleic Acid Ther. 2016;26:93–101. doi: 10.1089/nat.2015.0576. [DOI] [PubMed] [Google Scholar]
- 14.Koch T., Ørum H. Locked nucleic acid. In: Crooke S.T., editor. Antisense Drug Technology: Principles, Strategies, and Applications. Second Edition. CRC Press; 2008. pp. 519–562. [Google Scholar]
- 15.Stanton R., Sciabola S., Salatto C., Weng Y., Moshinsky D., Little J., Walters E., Kreeger J., DiMattia D., Chen T. Chemical modification study of antisense gapmers. Nucleic Acid Ther. 2012;22:344–359. doi: 10.1089/nat.2012.0366. [DOI] [PubMed] [Google Scholar]
- 16.Henry S.P., Kim T.-W., Kramer-Stickland K., Zanardi T.A., Fey R.A., Levin A.A. Toxicologic properties of 2′ O-methoxyethyl chimeric antisense inhibitors in animals and man. In: Crooke S.T., editor. Antisense Drug Technology: Principles, Strategies, and Applications. Second Edition. CRC Press; 2007. pp. 327–364. [Google Scholar]
- 17.Lendvai G., Velikyan I., Bergström M., Estrada S., Laryea D., Välilä M., Salomäki S., Långström B., Roivainen A. Biodistribution of 68Ga-labelled phosphodiester, phosphorothioate, and 2′-O-methyl phosphodiester oligonucleotides in normal rats. Eur. J. Pharm. Sci. 2005;26:26–38. doi: 10.1016/j.ejps.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 18.Sewing S., Boess F., Moisan A., Bertinetti-Lapatki C., Minz T., Hedtjaern M., Tessier Y., Schuler F., Singer T., Roth A.B. Establishment of a predictive in vitro assay for assessment of the hepatotoxic potential of oligonucleotide drugs. PLoS ONE. 2016;11:e0159431. doi: 10.1371/journal.pone.0159431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burel S.A., Hart C.E., Cauntay P., Hsiao J., Machemer T., Katz M., Watt A., Bui H.H., Younis H., Sabripour M. Hepatotoxicity of high affinity gapmer antisense oligonucleotides is mediated by RNase H1 dependent promiscuous reduction of very long pre-mRNA transcripts. Nucleic Acids Res. 2015;80:436–445. doi: 10.1093/nar/gkv1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasuya T., Hori S., Watanabe A., Nakajima M., Gahara Y., Rokushima M., Yanagimoto T., Kugimiya A. Ribonuclease H1-dependent hepatotoxicity caused by locked nucleic acid-modified gapmer antisense oligonucleotides. Sci. Rep. 2016;6:30377. doi: 10.1038/srep30377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Poelgeest E.P., Hodges M.R., Moerland M., Tessier Y., Levin A.A., Persson R., Lindholm M.W., Dumong Erichsen K., Ørum H., Cohen A.F., Burggraaf J. Antisense-mediated reduction of proprotein convertase subtilisin/kexin type 9 (PCSK9): a first-in-human randomized, placebo-controlled trial. Br. J. Clin. Pharmacol. 2015;80:1350–1361. doi: 10.1111/bcp.12738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rabinovich-Guilatt L., Elgart A., Erisson L., Willsie S.K., Tessler S., Barnett-Griness O., Pande A., Spiegelstein O. Impact of dosing regimen of custirsen, an antisense oligonucleotide, on safety, tolerability and cardiac repolarization in healthy subjects. Br. J. Clin. Pharmacol. 2015;80:436–445. doi: 10.1111/bcp.12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsimikas S., Viney N.J., Hughes S.G., Singleton W., Graham M.J., Baker B.F., Burkey J.L., Yang Q., Marcovina S.M., Geary R.S. Antisense therapy targeting apolipoprotein(a): a randomised, double-blind, placebo-controlled phase 1 study. Lancet. 2015;386:1472–1483. doi: 10.1016/S0140-6736(15)61252-1. [DOI] [PubMed] [Google Scholar]
- 24.Reagan-Shaw S., Nihal M., Ahmad N. Dose translation from animal to human studies revisited. FASEB J. 2008;22:659–661. doi: 10.1096/fj.07-9574LSF. [DOI] [PubMed] [Google Scholar]
- 25.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 27.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.