Abstract
The use of non-viral procedures, together with CRISPR/Cas9 genome-editing technology, allows the insertion of single-copy therapeutic genes at pre-determined genomic sites, overcoming safety limitations resulting from random gene insertions of viral vectors with potential for genome damage. In this study, we demonstrate that combination of non-viral gene delivery and CRISPR/Cas9-mediated knockin via homology-directed repair can replace the use of viral vectors for the generation of genetically modified therapeutic cells. We custom-modified human adipose mesenchymal stem cells (hAMSCs), using electroporation as a transfection method and CRISPR/Cas9-mediated knockin for the introduction and stable expression of a 3 kb DNA fragment including the eGFP (selectable marker) and a variant of the herpes simplex virus 1 thymidine kinase genes (therapeutic gene), under the control of the human elongation factor 1 alpha promoter in exon 5 of the endogenous thymidine kinase 2 gene. Using a U87 glioma model in SCID mice, we show that the therapeutic capacity of the new CRISPR/Cas9-engineered hAMSCs is equivalent to that of therapeutic hAMSCs generated by introduction of the same therapeutic gene by transduction with a lentiviral vector previously published by our group. This strategy should be of general use to other applications requiring genetic modification of therapeutic cells.
Keywords: CRISPR/Cas9, cell therapy, mesenchymal stem cells, bystander suicide therapy, glioblastoma, non-invasive bioluminescence imaging, CRISPR/Cas9, CRISPR/Cas9 knockin
Introduction
Therapeutic application of cells, from multiple origins, is having a profound impact in areas of biomedical research such as regenerative medicine, inflammation, and tumor therapy.
While cell types can be selected exclusively for their intrinsic developmental potential to produce specific medical outcomes (e.g., regenerative medicine), therapeutic application can also benefit from the capacity to genetically modify cells, endowed with advantageous traits such as the potential for homing to particular therapeutic targets, to address specific medical problems.1, 2
The CRISPR/Cas9 system has revolutionized the field of genome editing since it was first described with such applicability in 2012.3 Due to its versatility, affordability, and capacity for rapid generation of custom genetically modified cells, the CRISPR/Cas9 technology is displacing other existing and available protein-guided approaches sharing the same goal, such as modified zinc-finger proteins (ZNFs) or transcription activator-like effector nucleases (TALENs).
Resulting from its capacity for editing genome sequences at precise locations, the CRISPR/Cas9 system has found application in two wide groups of gene-editing strategies. In the first class, relatively small gene disruptions, insertions, or deletions (indels) are induced at specific sites by the combined action of the RNA-guided Cas9 nuclease and the cellular DNA repair machinery related to the non-homologous end joining (NHEJ) pathway. A second class of applications involves the insertion of whole genetic elements such as promoters and genes (provided as DNA templates) via homology-directed repair (HDR).
However, the mechanisms determining the balance between these two DNA repair processes remain largely unknown, although several studies have shown that NHEJ is 3-fold more frequent than the HDR pathway.4, 5
Moreover, despite the fact that intense research is being devoted to improve the efficacy of the HDR pathway after DNA cleavage produced by Cas9 nuclease, e.g., repression by gene silencing or inhibition of proteins involved in NHEJ,6 the efficiency of this process still is low, and the preferred methodology to introduce large genetic elements for both research and clinical applications is currently the use of viral vectors due to its higher integration efficiency. An additional reason to use viral vectors as vehicles for genetic manipulation of cells has been the lack of efficient non-viral gene transfer systems, in particular for primary and stem cell lines.
Nonetheless, despite its higher technical complexity, the CRISPR/Cas9 system offers near-perfect control on integration site and number of genome integrated copies, overcoming the biosafety concerns generated by the random integration site and the number of copies resulting from viral vector procedures.
Human mesenchymal stem cells (hMSCs) are among the most promising candidates for cell therapy. These cells can differentiate under a variety of specific stimuli toward several cell lineages types including those of bone, cartilage, and fat,7 and in contrast with their embryonic counterparts, adult hMSCs have negligible tumorigenic potential.8, 9 In addition, other desirable features of these cells are their immunomodulatory capacity10 and tropism to injury and tumor sites.11
Although hMSCs traditionally have been obtained from bone marrow, other tissues, remarkably adipose tissue, have been bona fide sources of hMSCs sharing similar surface marker expression patterns and differentiation capacities.12
Previous work from this laboratory showed that human adipose mesenchymal stem cells (hAMSCs) genetically modified with lentiviral vectors for expression of a variant of the suicide gene herpes simplex virus 1 thymidine kinase (tTK) are effective vehicles to deliver cytotoxic bystander therapy against the glioblastoma tumor model. In those experiments, the therapeutic cells migrated to the tumor vasculature, differentiated to the endothelial lineage, and effectively destroyed the tumor stem cell niche, restraining tumor growth upon systemic injection of the prodrug ganciclovir (GCV) and increasing animal survival from 55 (control) to 88.5 days (treated).13, 14
The objective of the present work was to reproduce the above results using hAMSCs genetically modified by a combination of non-viral transfection and CRISPR/Cas9 genome-editing procedures in an attempt to avoid potential biohazards derived from the use of conventional lentiviral transfection procedures. We show that therapeutic hAMSCs stably expressing the tTK gene can be generated using an electroporation transfection procedure in combination with the CRISPR/Cas9 system. Moreover, we demonstrate that the resulting therapeutic cells display equivalent tumor-killing capacity to that of hAMSCs genetically modified using lentiviral vectors, thus indicating that the methodology presented in this study might be applicable to other therapy approaches involving genetic modification of therapeutic cells.
Results
Vectors for HDR-Based Gene Editing by CRISPR/Cas9 Knockin
In human cells, thymidine kinase 1 and 2 genes (TK1 and TK2) encode thymidine kinase activities that are expressed in cell-cycle-dependent and constitutive manners, respectively. The constitutively expressed TK2 gene sequence was subjected to analysis for CRISPR/Cas9-mediated knockin via HDR. Particularly, a single-guide RNA (sgRNA) for Cas9 nuclease (target sequence) located at exon 5 of TK2 gene was chosen for insertion because of both its predicted presence in all the splice variants of the gene and its infrequent off-target effects.
Replacement of the TK2 gene by the viral tTK counterpart was considered an adequate strategy because it would preserve a constitutive thymidine kinase activity during normal cell function, while reducing the level of competing endogenous thymidine triphosphate when cytotoxic GCV-triphosphate was required for therapy.
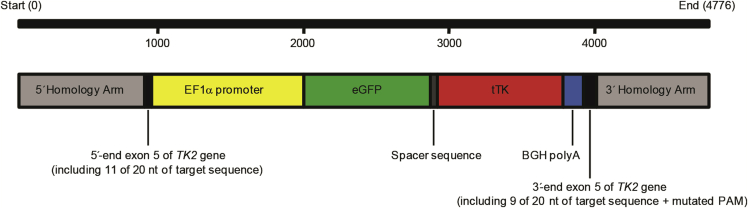
Previous to editing, the integrity of the selected target sequence in hAMSCs was assessed by PCR and DNA sequencing of the purified amplicon (Figure S1). The donor construct for CRISPR/Cas9-mediated integration at exon 5 of the hAMSCs TK2 gene was designed to comprise the human elongation factor 1 alpha (EF1α) promoter, eGFP and tTK genes, and the bovine polyadenylation signal (BGH pA). This construct was flanked 5′ and 3′ by homology arms to direct CRISPR/Cas9-mediated knockin (Figure 1; Materials and Methods). The resulting 4,776 bp fragment was inserted into a pMA vector to generate pMA-DONR-EF1α-EGFP-tTK. The selected target sequence was cloned into a different vector including Cas9 nuclease (pCRISPR-Cas9/CD4-TK2).
Figure 1.
Donor Sequence Design for HDR Induced by CRISPR/Cas9 Activity
PAM, protospacer adjacent motif.
CRISPR/Cas9-Engineered hAMSCs Generation and Validation
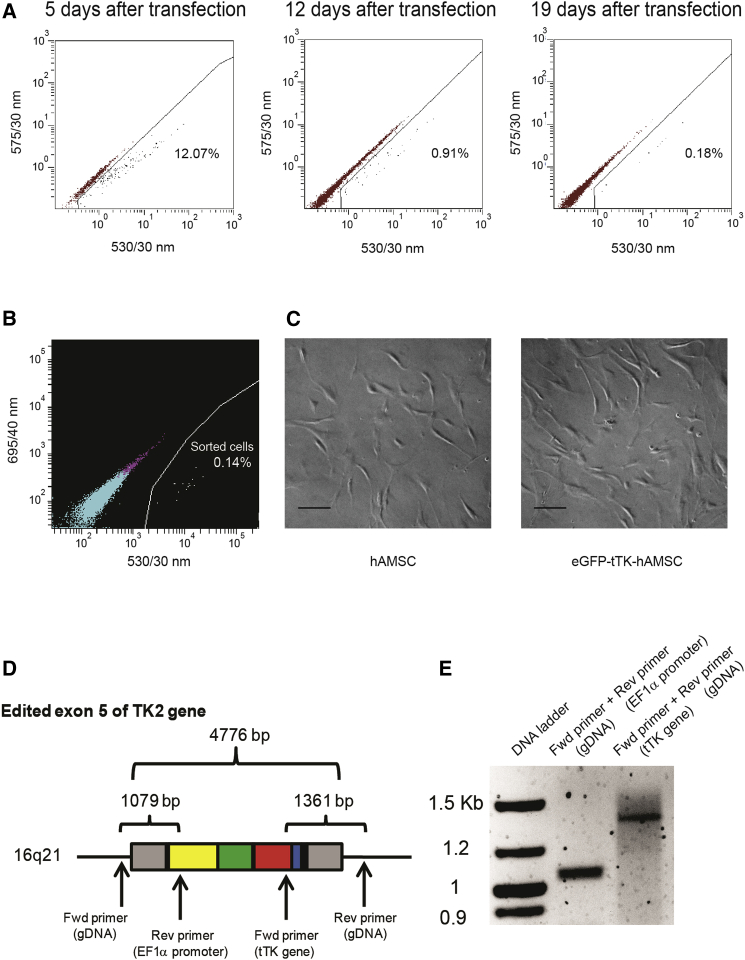
Constructs pMA-DONR-EF1α-EGFP-tTK and pCRISPR-Cas9/CD4-TK2 were co-transfected in hAMSCs using an electroporation procedure that yielded 12% EGFP-positive cells on day 5 post-transfection (Figure 2A). However, a fraction of the EGFP-positive cells is expected to only transiently express the reporter or to not be co-transfected with both constructs. In order to eliminate these unwanted cells, the reduction in eGFP expression was monitored until it reached the lowest stable expression level (Figure 2A), at which point remaining EGFP-positive cells were sorted (Figure 2B) and expanded. The resulting putative CRISPR/Cas9-modified cells (namely eGFP-tTK-hAMSCs) were morphologically indistinguishable from unmodified hAMSCs (Figure 2C).
Figure 2.
Generation and Validation Strategy for CRISPR/Cas9-Mediated Knockin in hAMSCs
(A) FACS analysis of EGFP expression level following co-transfection by electroporation of pCRISPR-Cas9/CD4-TK2 and pMA-DONR-EF1α-EGFP-tTK. EGFP expression in hAMSCs decreased progressively until stabilization. (B) EGFP-expressing hAMSCs (0.14% from the total population) were sorted at day 22 post-transfection and expanded. (C) Bright-field images showing control and sorted hAMSCs. Scale bars, 20 μm. (D) Schematic representation of the TK2 exon 5 after integration and predicted amplicons for validation of integration; arrows indicate the hybridization site for forward and reverse primers flanking 5′ and 3′ insert junctions at exon 5. (E) Agarose gel showing the size of predicted PCR amplicons from amplification of genomic DNA extracted from sorted and expanded hAMSCs after CRISPR/Cas9 modification.
Evaluation of donor construct integration in the pre-designed hAMSCs genomic location was performed by PCR amplification of the 5′ and 3′ junctions of the construct insertion site, using primers complementary to 5′ and 3′ genomic regions of the TK2 gene, together with primers hybridizing in the integrated EF1α promoter and tTK gene (Figures 2D and 2E). In addition, predicted amplicons corresponding to the integration region were extracted from agarose gels and sequenced (Figure S2). No amplification in untransfected hAMSCs was detected using these pairs of primers (data not shown).
CRISPR/Cas9-Engineered hAMSCs Mediate Bystander Tumor Cell Killing In Vitro
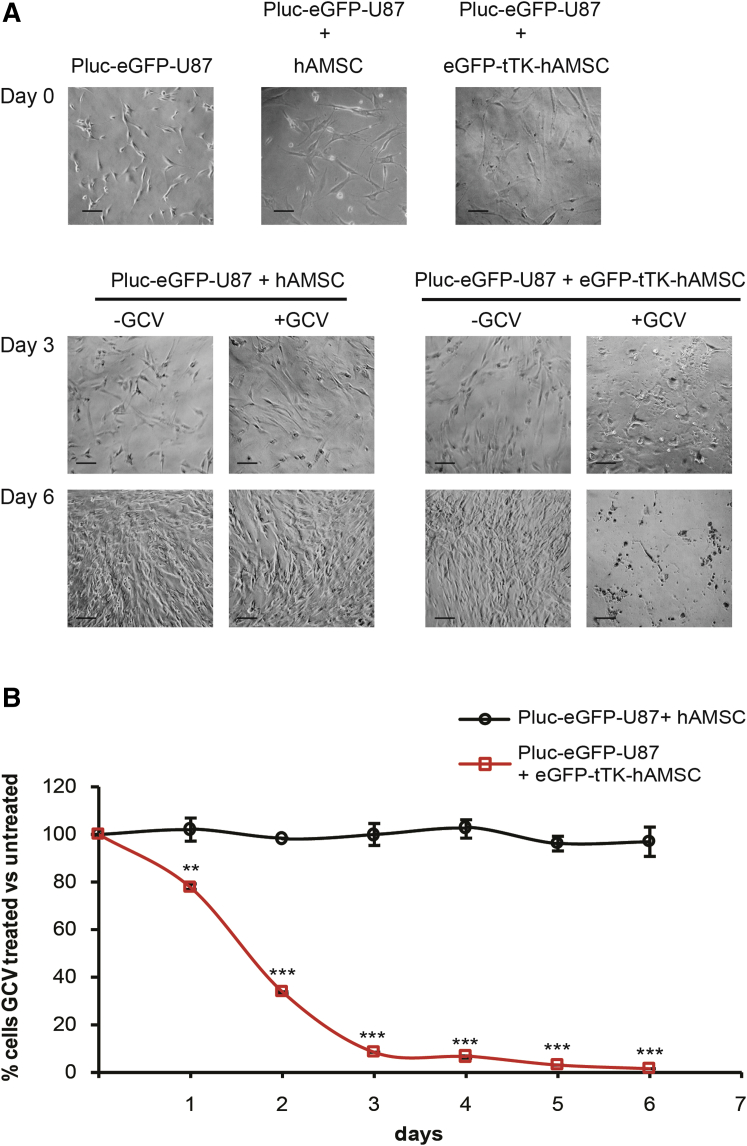
To evaluate the capacity of CRISPR/Cas9-modified hAMSCs to exert bystander killing in vitro, co-cultures of luciferase-expressing U87 tumor cells (referred as Pluc-EGFP-U87) with either unmodified (control) or EGFP-tTK-hAMSCs were seeded in a 1:4 ratio, as indicated for optimal results by previous studies in our laboratory,13 and treated with GCV. The results showed a potent bystander effect mediated by CRISPR/Cas9-modified hAMSCs, with only 8% surviving tumor cells relative to the initial number seeded after 6 days of GCV treatment (Figure 3). Conversely, there was no significant effect on the survival of tumor cells co-cultured with control hAMSCs plus GCV treatment, also suggesting negligible cytotoxicity of GCV prodrug in agreement with previous observations.13, 15
Figure 3.
Evaluation of the Bystander Killing Effect of CRISPR/Cas9-Modified hAMSCs In Vitro
(A) Representative bright-field images of single Pluc-EGFP-U87, and co-cultures with either control hAMSCs or EGFP-tTK-hAMSCs at days 0, 3, and 6 of GCV treatment. Scale bars, 20 μm. (B) Graph showing the fraction of Pluc-EGFP-U87 cells surviving GCV treatment. Tumor cells were mixed at ratio 1:4 with control hAMSCs or with EGFP-tTK-hAMSCs and treated with GCV (4 μg/mL). **p < 0.01; ***p < 0.001 compared with the control group. Error bars represent mean ± SD.
CRISPR/Cas9-Engineered hAMSCs Mediate Bystander Tumor Cell Killing In Vivo
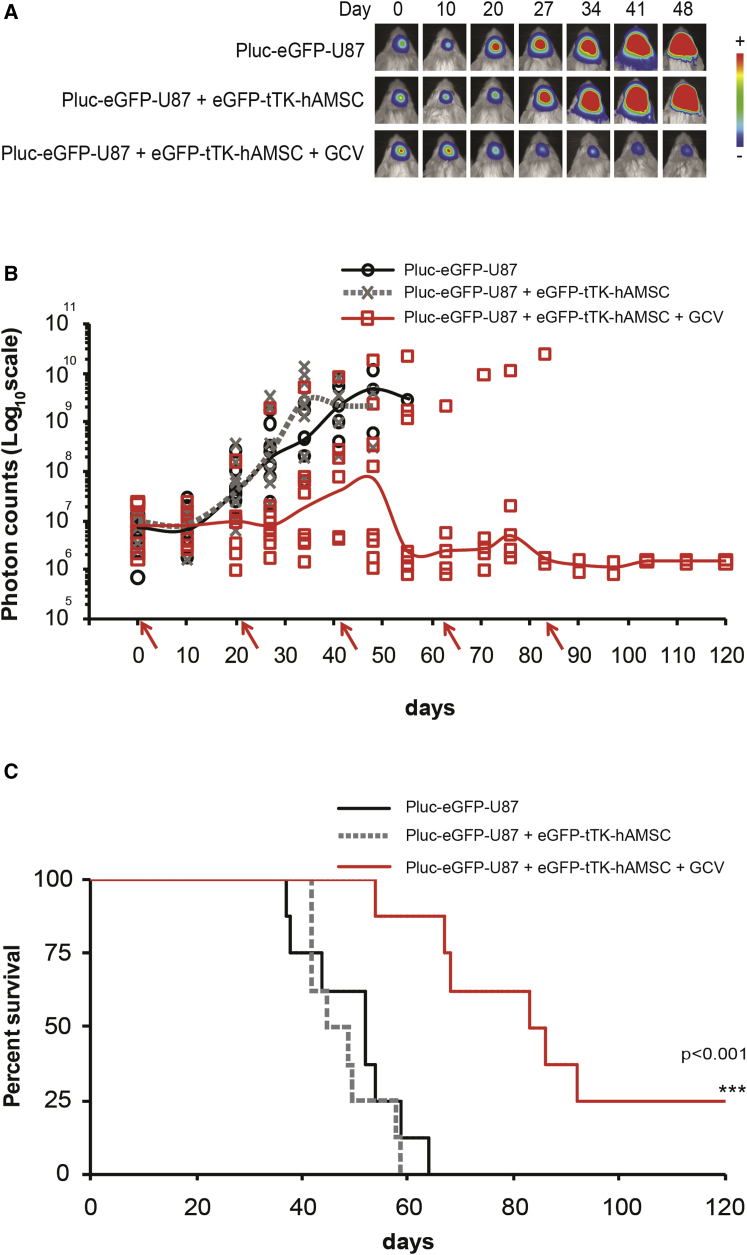
To assess the therapeutic effectiveness of CRISPR/Cas9-edited hAMSCs in vivo, we stereotactically inoculated a group of 24 six-week-old SCID mice with 6 × 104 Pluc-EGFP-U87 at specific brain coordinates, as established in previous glioblastoma therapy models, and allowed them to grow for 5 days. At this point, tumor growth was evaluated by non-invasive bioluminescence imaging (BLI) and the animals were separated into three groups homogenized by tumor size (Figure S3A). Following a 48-hr period for recovery from anesthesia, two of the three groups of animals were subjected to intracranial injection at the same coordinates of tumor implantation, with 2.4 × 105 CRISPR/Cas9-modified hAMSCs. Mice in one of the groups bearing tumor cells and modified hAMSCs were treated by intraperitoneal (i.p.) injection of GCV (5 days per week) beginning 72 hr after cell implantation. Non-invasive BLI was used to monitor tumor development weekly during the experiment and every 3 weeks, mice from the two groups initially treated with therapeutic hAMSCs were re-injected at the same location with the same number of CRISPR/Cas9-modified cells.
As shown in Figure 4, administration of EGFP-tTK-hAMSCs in combination with GCV delayed, and in some cases even abrogated, tumor growth (Figures 4A, 4B, and S3B), resulting in stabilization of the disease with no apparent behavioral changes in surviving animals. A Kaplan-Meier graph (Figure 4C) shows a significant (p < 0.001) increase in the survival of animals treated with hAMSCs and GCV (84.5 days), relative to the control group receiving hAMSCs, but not GCV, or the control group bearing only tumor cells (median survivals of 52 and 47 days, respectively). No significant difference in survival was detected between the two control groups.
Figure 4.
CRISPR/Cas9-Modified hAMSCs Are Effective against U87 Tumors in SCID Mice
(A) Overlay of bioluminescence and bright-field images of representative animals from each experimental group showing tumor growth development through time (arbitrary color palette ranging from blue = low intensity to red = high-intensity light emission). (B) Graph showing the median (solid line) of light emission intensity recorded from brain tumor areas and from individual animals (symbols) (n = 8). Arrows in the x axis indicate re-inoculation of therapeutic hAMSCs. (C) Kaplan-Meier graph illustrating survival curves for each experimental group. ***p < 0.001, EGFP-tTK-hAMSCs + GCV-treated group versus each control group.
The flow diagram in Figure S4 summarizes the steps and time used in this approach.
Discussion
In the current work we use a non-viral approach, combining electroporation and the CRISPR/Cas9 system, to generate therapeutic hAMSCs genetically modified by insertion of one copy of the cytotoxic tTK and the selectable marker eGFP in a pre-defined site of the hAMSCs TK2 gene. Moreover, we also demonstrate that the therapeutic cells, thus generated, are very effective for killing a glioblastoma tumor model in SCID mice. Thus, we now show a significant increase in animal survival of 35 days between control and treated animals, which is in accordance with the 33-day survival increase obtained previously using lentivirus-modified hAMSCs.13 This level of survival was associated with stabilization of the disease state in surviving animals.
We have previously shown that hAMSCs modified using lentiviral vectors for expressing the tTK gene home to the vascular system of tumors, differentiate to the endothelial lineage in the proximity of tumor stem cells, and in the presence of the prodrug GCV effectively destroy the tumor.
There are reports indicating that MSCs may help and also hinder tumor growth.16 However, when used as delivery vehicles of cytotoxic tTK, their potential tumor-helping capacity is overshadowed by their cytotoxic effect in the presence of GCV.15 Moreover, it is clear from the control groups included in our in vivo experiment (tumors treated with CRISPR/Cas9-modified hAMSCs in which no GCV was administered and tumors not bearing cells) that both develop at the same rate. Thus, in this instance, hAMSCs by themselves had no positive or negative effects on tumor growth.
However, although very efficient, the use of lentiviral vectors for the generation of genetically modified therapeutic cells has important drawbacks resulting from the insertion of an unpredictable number of foreign DNA copies at random sites of the genome, with the potential for impairing important gene functions and cell transformation.
Human cells express cell-cycle-dependent (TK1) and constitutive (TK2) thymidine kinase activities. In the current work, the TK2 locus was selected for insertion of the therapeutic tTK gene in order to ensure the capacity to generate the cytotoxic agent after CGV administration even in non-proliferating therapeutic cells.13 A specific DNA sequence within exon 5 of the gene was the chosen target for insertion because this particular exon is predicted to be present in all the splice variants of the endogenous gene.
The transgene insert was designed to comprise a chimeric DNA sequence formed by joining the coding sequences for the therapeutic tTK and the selectable marker eGFP genes, both regulated by the constitutively active human EF1α promoter. Additionally, 5′ and 3′ homology arms complementary to the target insertion site were included to direct insertion via the HDR pathway after DNA cleavage exerted by the CRISPR/Cas9 nuclease.
Different transfection procedures, including the use of three different standard types of lipoplexes, polyplex-SPION combinations, and a magnetofection procedure, were explored to co-transfect pCRISPR-Cas9/CD4-TK2 and pMA-DONR-EF1α-EGFP-tTK plasmids into hAMSCs. However, none of the indicated procedures yielded more than 3% of EGFP-positive hAMSC transfectants (data not shown).
Constructs for donor transgenes and CRISPR/Cas9 nuclease were most effectively co-transfected by electroporation into hAMSCs (12% EGFP transfectants), as assessed by flow cytometry and EGFP fluorescence, a value still low compared with the standard 50%–90% efficiency of viral transfections.
Nevertheless, the yield of bona fide inserts was, as expected, considerably lower (0.14% of the total initial cells) because a large fraction of the EGFP-positive transfectants only transiently expressed the reporter and disappeared during cell culture without undergoing insertion via HDR.
Cells permanently expressing the EGFP reporter were selected by fluorescence-activated cell sorting (FACS) and shown by two PCR-based procedures to have been effectively modified and to have contained the transgene insert at the expected exon 5 site of the TK2 gene.
Although the CRISPR/Cas9 system is effective for generating small targeted indels, large insertions, as in the current case, are known to occur with considerably lower efficiency.17, 18 Low frequency of transgene insertion, together with the need for culture expansion to produce large numbers of cells for therapeutic applications, may be considered a handicap for this strategy at this stage. However, gene-editing procedures, such as the current one, are in their infancy, and considerable progress in efficiency should be expected in the near future. Because the goal is the generation of safe therapeutic cells for human clinical use, continued efforts to improve in this technical aspect should be a priority.
The new CRISPR/Cas9-modified hAMSCs were effective, in the presence of GCV, for bystander killing of tumor cells in culture and in a live glioblastoma tumor model.
For this purpose, we used our standard non-invasive BLI procedure to monitor the capacity of the new CRISPR/Cas9-engineered therapeutic hAMSCs to kill luciferase-expressing U87 tumors in SCID mice. In these experiments, we were able to show that the new therapeutic hAMSCs had a remarkable capacity to destroy tumors and significantly prolong the survival of mice from 47 (untreated controls) to 84.5 days (CRISPR/Cas9-modfied hAMSCs + GCV), in fact a degree of therapeutic effectiveness very similar to that published previously,13 where untreated animals survived for 55 days and those treated with lentivirus-modified hAMSCs survived 88.5 days.
Even though solid evidence is not provided for the number of transgene copies per cell in these experiments, CRISPR/Cas9-mediated knockin is expected to result in the insertion of a single copy per genome.17 Nevertheless, Cas9 nuclease activity may produce off-target strand excision/repair events at predictable sites similar to the target sequence.18 Off-target effects may result in excisions that are mis-repaired, generating mutations with potential for affecting gene function. However, this information is available before starting the genetic modification of therapeutic cells in comparison with the potential and unpredictable damage of lentivirus random insertion.
In addition, evaluation of off-target effects was not explored because for efficient generation of large numbers of therapeutic cells, it was necessary to genetically modify cell populations rather than single clones. Attempts to evaluate the overall off-target modification frequency in DNA from such heterogeneous cell populations would be non-informative, while using DNA from a single-cell clone would produce information not representative of the whole. Most relevant to this work is that the new CRISPR/Cas9-engineered hAMSCs with a putative single copy of the therapeutic tTK gene per genome are as effective for tumor killing as those produced by viral transduction.
As we have also observed in previous work, some animals were refractory to therapy probably due to a variety of factors. It is possible, but unlikely, that variations in the response to therapy result from differences in the engineered hAMSCs. Populations of hAMSCs as extracted from donors were used for practical purposes. Besides, generation of enough cells for therapy starting from single-cell clones would have required extensive cell expansion, far beyond the proliferative capacity adscribed to mesenchymal stem cells before engaging the senescence program.19
In fact, the CRISPR/Cas9-modified hAMSCs are expected to be, due to the nature of the modification, more uniform in terms of therapeutic gene content than virus-modified cells, and the variations we see in the animals treated with GCV are well within the variations inherent to the use of animal models. The most plausible explanation is that the observed variations result from the difficulty of inoculating therapeutic cells exactly at the same tumor site in each re-inoculation.
Significantly, in most cases, tumor growth was delayed or kept at check during the duration of the experiment while therapeutic cells and GCV were administered. This result indicates that this approach has the potential to lead to the stabilization of the disease state.
The clinical cell therapy strategy we describe here should become a feasible alternative to current ones for therapeutic cell generation, improving biosafety, while preserving therapeutic capacity.
Materials and Methods
Cell Lines and Culture
hAMSCs were isolated from an 18-year-old white female (BMI > 25), with adipose tissue derived from cosmetic subdermal liposuction of the lateral hip, as described elsewhere,15 after written informed consent. Work with human samples was approved by the Bioethical Subcommittee of Superior Council of Scientific Research (CSIC). In brief, lipoaspirate was suspended in 1× collagenase type I (Invitrogen) solution, incubated at 37°C, and inactivated by addition of DMEM containing 10% heat-inactivated fetal bovine serum (FBS). hAMSCs were isolated by plastic adherence technique and grown in DMEM-high glucose (hg) with 10% FBS (Sigma), 2 mM l-glutamine (Sigma), and 50 U/mL penicillin/streptomycin (Sigma).
The human glioblastoma cell line U87 MG (HTB-14; ATCC) was grown in nutrient mixture DMEM-Ham’s F12 containing 10% heat-inactivated FBS, 2 mM l-glutamine, and 50 U/mL penicillin/streptomycin. The U87 MG cell line was transduced with lentiviral vector pRRL-Pluc-IRES-EGFP to generate the Pluc-EGFP-U87 cell line as described previously.13
Cas9/sgRNA and Donor Constructs Design
GeneArt CRISPR Search and Design Tool (Thermo Fisher Scientific) and CRISPR Design tool developed by Dr. Zhang’s laboratory (http://crispr.mit.edu/) were used to design sgRNA for Cas9 nuclease targeting the TK2 gene. A particular target sequence in exon 5 was selected following analysis for its common presence in all predicted TK2 splicing variants. The target sequence of 20 nt (5′-TGTACCACGATGCCTCTCGC-3′), including the protospacer adjacent motif (PAM; 5′-TGG-3′), was selected for its predicted high score and reduced off-target effects.
The mentioned TK2 sgRNA was synthesized and subcloned into GeneArt CRISPR Nuclease (CD4) Vector (GeneArt Gene Synthesis; Thermo Fisher) to generate pCRISPR-Cas9/CD4-TK2 (9.8 kb). In parallel, the donor sequence was designed in silico to include (5′ to 3′) 813 nt comprising 795 nt of the 5′ flanking intron and 18 nt of exon 5 of the TK2 gene including 11 of 20 nt of the Cas9 target sequence (namely 5′ homology arm), followed by the EF1α promoter, a chimeric coding sequence comprising the eGFP gene and the sr39tk truncated version of the herpes simplex virus 1 thymidine kinase gene20 ended by the BGH polyA signal, plus 807 nt comprising the rest of the TK2 exon 5 (72 nt), including 9 of 20 nt of the target sequence and 735 nt of the 3′ flanking intron (namely 3′ homology arm) (Data S1). This 4,776 bp fragment was then synthesized de novo and cloned into pMA plasmid (GeneArt Gene Synthesis; Thermo Fisher) to generate pMA-DONR-EF1α-EGFP-tTK (7.1 kb).
Transfection Procedure
hAMSCs were co-transfected with pCRISPR-Cas9/CD4-TK2 and pMA-DONR-EF1α-EGFP-tTK by electroporation with an NEPA21 electroporator (NEPA GENE) following manufacturer’s instructions and applying 175 V and 5 ms length poring pulse.
Flow Cytometry
EGFP expression was monitored with a Gallios cytometer (Becton Dickinson), and data were analyzed using the FlowJo software (Tree Star). Positive EGFP-expressing cells were sorted using a FACSAria Fusion II cell sorter (Becton Dickinson). Analysis of data was performed using the BD FACSDiva 8.0.1 software. In both types of determinations, single cells were identified on the basis of forward scatter area (FSC-A), FSC width (FSC-W), and side scatter area (SSC-A). EGFP fluorescence was graphed using a bidimensional dot plot of 488 nm excitation and emission detected at 530/30 versus 575/30 nm or 530/30 versus 695/40 nm, respectively.
Validation of CRISPR-Mediated Knockin in hAMSCs
The target sequence for the Cas9 nuclease in the TK2 locus of hAMSCs was first validated by PCR amplification of genomic DNA using flanking primers (forward primer 5′-CTCCCTTCCTCCCCACTCTA-3′ and reverse primer 5′-AATTCAGCAGGTCTGGGATG-3′) (Thermo Fisher), yielding a 1.3 kb length amplicon (Figure S1). Following electrophoresis in 1% agarose gels, the PCR-amplified amplicon products were extracted, purified (QIAquick gel extraction kit; QIAGEN), and subjected to capillary sequencing using the BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems) and an ABI 3730 DNA analyzer (Applied Biosystems) based on the Sanger method.
Donor construct integration in the pre-defined hAMSCs genomic location was performed by PCR amplification of the junction sequences 5′ and 3′ flanking the insertion site within exon 5 of the TK2 gene, using primers hybridizing to genomic DNA (5′-CTGTGCCTGGCTGGTATTTT-3′ and 5′-TAGGCCTAAGCAGCCATGTT-3′, respectively), together with primers hybridizing in integrated EF1α promoter and tTK gene (5′-TAAGTGCAGTAGTGCCCGTG-3′ and 5′-CACGGCGACTACTGCACTTA-3′, respectively). PCR products were run in an agarose gel, and predicted amplicons were extracted, purified, and sequenced as described above (Figure S2).
Cell Proliferation Analysis
Pluc-EGFP-U87 cells (3 × 103) were mixed in 1:4 ratio with control or EGFP-tTK-hAMSCs on 24-well plates (Corning) in DMEM supplemented with 10% FBS. Following 24 hr of culture, 4 μg/mL GCV (Cymevene; Roche) was added to the corresponding wells. The number of Pluc-EGFP-U87 cells was monitored during the following 6 days by BLI after luciferin administration (Regis Technologies) using an ImagEM X2 C9100-23BEM-CCD imaging system (Hamamatsu Photonics) cooled at −80°C. Photon emission was recorded in charge-coupled device (ccd) mode during 60 s at binning 1 × 1, and image quantification was done using the Hokawo 2.7 image analysis software (Hamamatsu Photonics).
Each condition was assayed in six replicates. Additionally, bright-field images were taken every day during the experiment.
Animal Experiments
Adult 6-week-old SCID mice were purchased (Charles River) and kept under pathogen-free conditions in laminar flow cages and maintained on a regular 12 hr light/dark cycle (8:00–20:00 light period) at a constant 25°C. Animal maintenance and experiments were performed in accordance with established guidelines of the Catalan Government and a protocol (DAAM 8856) approved by Direcció General del Medi Natural, Generalitat de Catalunya. Experimental groups (n = 8 × 3) comprised equal numbers of male and female animals. Food and water were available ad libitum.
For cell implantation, animals were anesthetized by i.p. injections of xylazine (3.3 mg/kg Rompun; Bayer) and ketamine (1.39 mg/kg, Imalgene 100; Merial Laboratories). Subsequently, mice were restrained in a stereotactic frame (Stoelting); heads were secured using a nose clamp and two ear bars, and a skin flap was surgically lifted to expose the skull surface. For stereotactic cell implantations, a cell suspension was injected at a 0.6 μL/min rate using a Hamilton syringe series 700 (Sigma) at 0.6 mm posterior, 2 mm lateral, and 2.75 mm depth with respect to the Bregma. The scalp was closed by suture, and the animals were placed in individual recovery cages and supplied with buprenorphine (Buprecare; Divasa-Farmavic). For brain tumor generation, 6 × 104 Pluc-EGFP-U87 cells stably expressing firefly luciferase were injected and allowed to grow for 5 days. On day 5 after tumor implantation, experimental animals were subjected to non-invasive BLI and then separated into experimental groups, producing equivalent quantities of light (Figure S3A). On day 7 after tumor cell implantation, 2.4 × 105 EGFP-tTK-hAMSCs per animal were inoculated, as described above, at tumor implantation site in two of the experimental groups. On day 10 after tumor cell implantation, animals in one of the groups bearing therapeutic hAMSCs were subject to a weekly protocol of i.p. injection of GCV (50 mg/kg) for 5 consecutive days followed by 2 resting days without the prodrug. Therapeutic cell implantation was repeated every 3 weeks, with previous GCV withdrawal (48 hr before cell implantation), and repeated up to five times in surviving animals.
In Vivo Non-invasive BLI
To monitor Pluc-EGFP-U87 tumor growth, we anesthetized mice as described above and then injected them i.p. with 150 μL of luciferin (16.7 mg/mL in PBS). Animals were placed individually in the detection chamber of an ImagEM X2 C9100-23BEM-CCD imaging system (Hamamatsu Photonics) cooled at −80°C. Images were acquired from the dorsal direction, and photon emission was recorded in ccd mode during 60 s at binning 1 × 1. A second set of images of the animals was obtained using a white-light source inside the detection chamber, in order to register the position of the luminescence signal. Mice were imaged weekly during experiments. Quantification and analysis of photons recorded in images was done using the Hokawo 2.7 image analysis software (Hamamatsu Photonics). Light measurements were expressed as median of photon counts (PHC) after subtraction of background.
Statistical Analysis
Unpaired two-tailed Student’s t test was used for statistical analysis of in vitro cell killing assays. Gehan-Breslow-Wilcoxon test was performed with GraphPad Prism 5 software (GraphPad Software) and applied to determine statistical significance in animal survival experiments: *p < 0.05; **p < 0.01; ***p < 0.001.
Author Contributions
O.M.-C., J.B., N.R., C.G., and S.B. conceived and designed the experiments. O.M.-C. and M.G.-R. performed the experiments. O.M.-C. analyzed the data. S.B. contributed with reagents/materials. O.M.-C. and J.B. wrote the paper.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by MINECO/FEDER (grants SAF2015-64927-C2-1-R and SAF2015-64927-C2-2-R) and the Instituto de Salud Carlos III (Red Temática de Investigación Cooperativa en Terapia Celular-TERCEL). We would like to thank M.A. Pastor and C. Bestard (flow cytometry) for their excellent technical assistance.
Footnotes
Supplemental Information includes four figures and data and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.07.012.
Supplemental Information
References
- 1.Dodson B.P., Levine A.D. Challenges in the translation and commercialization of cell therapies. BMC Biotechnol. 2015;15:70. doi: 10.1186/s12896-015-0190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson H.J., Rafiq S., Brentjens R.J. Driving CAR T-cells forward. Nat. Rev. Clin. Oncol. 2016;13:370–383. doi: 10.1038/nrclinonc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mao Z., Bozzella M., Seluanov A., Gorbunova V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst.) 2008;7:1765–1771. doi: 10.1016/j.dnarep.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kühn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 7.Pittenger M.F., Mackay A.M., Beck S.C., Jaiswal R.K., Douglas R., Mosca J.D., Moorman M.A., Simonetti D.W., Craig S., Marshak D.R. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 8.Herberts C.A., Kwa M.S.G., Hermsen H.P.H. Risk factors in the development of stem cell therapy. J. Transl. Med. 2011;9:29. doi: 10.1186/1479-5876-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vilalta M., Dégano I.R., Bagó J., Gould D., Santos M., García-Arranz M., Ayats R., Fuster C., Chernajovsky Y., García-Olmo D. Biodistribution, long-term survival, and safety of human adipose tissue-derived mesenchymal stem cells transplanted in nude mice by high sensitivity non-invasive bioluminescence imaging. Stem Cells Dev. 2008;17:993–1003. doi: 10.1089/scd.2007.0201. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs S.A., Roobrouck V.D., Verfaillie C.M., Van Gool S.W. Immunological characteristics of human mesenchymal stem cells and multipotent adult progenitor cells. Immunol. Cell Biol. 2013;91:32–39. doi: 10.1038/icb.2012.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pendleton C., Li Q., Chesler D.A., Yuan K., Guerrero-Cazares H., Quinones-Hinojosa A. Mesenchymal stem cells derived from adipose tissue vs bone marrow: in vitro comparison of their tropism towards gliomas. PLoS ONE. 2013;8:e58198. doi: 10.1371/journal.pone.0058198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Ugarte D.A., Alfonso Z., Zuk P.A., Elbarbary A., Zhu M., Ashjian P., Benhaim P., Hedrick M.H., Fraser J.K. Differential expression of stem cell mobilization-associated molecules on multi-lineage cells from adipose tissue and bone marrow. Immunol. Lett. 2003;89:267–270. doi: 10.1016/s0165-2478(03)00108-1. [DOI] [PubMed] [Google Scholar]
- 13.Alieva M., Bagó J.R., Aguilar E., Soler-Botija C., Vila O.F., Molet J., Gambhir S.S., Rubio N., Blanco J. Glioblastoma therapy with cytotoxic mesenchymal stromal cells optimized by bioluminescence imaging of tumor and therapeutic cell response. PLoS ONE. 2012;7:e35148. doi: 10.1371/journal.pone.0035148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bagó J.R., Alieva M., Soler C., Rubio N., Blanco J. Endothelial differentiation of adipose tissue-derived mesenchymal stromal cells in glioma tumors: implications for cell-based therapy. Mol. Ther. 2013;21:1758–1766. doi: 10.1038/mt.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vilalta M., Dégano I.R., Bagó J., Aguilar E., Gambhir S.S., Rubio N., Blanco J. Human adipose tissue-derived mesenchymal stromal cells as vehicles for tumor bystander effect: a model based on bioluminescence imaging. Gene Ther. 2009;16:547–557. doi: 10.1038/gt.2008.176. [DOI] [PubMed] [Google Scholar]
- 16.Klopp A.H., Gupta A., Spaeth E., Andreeff M., Marini F., 3rd Concise review: dissecting a discrepancy in the literature: do mesenchymal stem cells support or suppress tumor growth? Stem Cells. 2011;29:11–19. doi: 10.1002/stem.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He X., Tan C., Wang F., Wang Y., Zhou R., Cui D., You W., Zhao H., Ren J., Feng B. Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Res. 2016;44:e85. doi: 10.1093/nar/gkw064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner W., Horn P., Castoldi M., Diehlmann A., Bork S., Saffrich R., Benes V., Blake J., Pfister S., Eckstein V., Ho A.D. Replicative senescence of mesenchymal stem cells: a continuous and organized process. PLoS ONE. 2008;3:e2213. doi: 10.1371/journal.pone.0002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ray P., De A., Min J.J., Tsien R.Y., Gambhir S.S. Imaging tri-fusion multimodality reporter gene expression in living subjects. Cancer Res. 2004;64:1323–1330. doi: 10.1158/0008-5472.can-03-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.