Abstract
The Cα—H⋅⋅⋅O hydrogen bond has been given little attention as a determinant of transmembrane helix association. Stimulated by recent calculations suggesting that such bonds can be much stronger than has been supposed, we have analyzed 11 known membrane protein structures and found that apparent carbon α hydrogen bonds cluster frequently at glycine-, serine-, and threonine-rich packing interfaces between transmembrane helices. Parallel right-handed helix–helix interactions appear to favor Cα—H⋅⋅⋅O bond formation. In particular, Cα—H⋅⋅⋅O interactions are frequent between helices having the structural motif of the glycophorin A dimer and the GxxxG pair. We suggest that Cα—H⋅⋅⋅O hydrogen bonds are important determinants of stability and, depending on packing, specificity in membrane protein folding.
The hydrogen bond is a key element in the interplay between stability and specificity in protein folding. The desolvation penalty associated with burial of polar side chains in an aqueous environment is not always fully recovered by hydrogen bond formation, so hydrogen bonds provide a small or even unfavorable net energy contribution to folding. However, the strength and directionality of hydrogen bonds make them an important factor in discriminating between correctly folded and misfolded states. Hence, polar interactions tend to contribute more to specificity than to stability in soluble proteins (1–3). Conversely, in the apolar environment of biological membranes donor and acceptor groups cannot be satisfied by the solvent, and hydrogen bonds strongly stabilize the helical conformation of membrane spanning domains (4) and can stabilize tertiary interactions as well (5–9). We are interested in the role of hydrogen bonds in the association of transmembrane helices, a stage that is pivotal in the folding of membrane proteins (4). Recently, the DeGrado group and our laboratory showed that the substitution of a single polar amino acid residue into model transmembrane helices induces homo-oligomerization (10–13); the association driven by hydrogen bonding can be strong and independent of packing details. Thus, in the apolar environment, the strength of hydrogen bonds can stabilize the association of transmembrane helices, although a lack of a need for sequence specificity could create a danger of inducing promiscuous association (10, 13).
Weaker hydrogen bonds, such as those between carbon and oxygen atoms (C—H⋅⋅⋅O), have received little attention in the membrane protein field, and their occurrence in membrane proteins has never been surveyed. The Cα is an activated carbon donor because it is bound to the electron-withdrawing amide N—H and C⩵O groups, and, in soluble proteins, hydrogen bonds between main-chain Cα—H groups and backbone or side-chain oxygen atoms are often observed (14–17). Despite its abundance, the structural contribution of the Cα—H⋅⋅⋅O hydrogen bond has been unclear and its interaction energy has been believed to be small. Recently, by using ab initio calculations, Vargas et al. (18) and Scheiner et al. (19) estimated the energy of the Cα—H⋅⋅⋅O hydrogen bond to be as much as 2.5–3.0 kcal/mol in vacuo, placing the strength of the Cα—H⋅⋅⋅O hydrogen bond at approximately one-half the energy of a “conventional” N—H⋅⋅⋅O hydrogen bond. Given the importance of electrostatic interactions in the apolar interior of a membrane lipid bilayer, this energy may be significant for providing stability in transmembrane helix–helix association, especially when several Cα—H⋅⋅⋅O interactions are coordinated at a single interface. To examine this hypothesis, we have analyzed 125 helix–helix interfaces in a database of 11 nonhomologous membrane protein structures (Table 1) in search of Cα—H⋅⋅⋅O contacts that display the geometric hallmarks of hydrogen bonds. The analysis is limited by the uncertainty in the positioning of atoms in crystal structures solved at 2- to 3-Å resolution, although even at these resolutions the position of backbone atoms and helix axes should be quite accurate. We find recurring patterns when Cα—H⋅⋅⋅O contacts cluster at interfaces between helices that display a short interaxial distance, have a preference for parallel right-handed packing, and are rich in glycine, serine, and threonine residues. Our results suggest that Cα—H⋅⋅⋅O hydrogen bonds are overlooked factors that can determine stability and, given their dependence on the packing details, also specificity in the interaction of transmembrane helices.
Table 1.
Structural database of nonhomologous helical membrane proteins
| Protein | Protein Data Base | Resolution, Å | Helix–helix interactions*† | Cα—H⋅⋅⋅O contacts‡ |
|---|---|---|---|---|
| Bacteriorhodopsin | 1c3w | 1.55 | 9 | 13 (6) |
| Calcium ATPase | 1eul | 2.60 | 15 | 11 (4) |
| Cytochrome c oxidase | 1occ | 2.80 | 48 | 53 (12) |
| Fumarate reductase | 1qla | 2.20 | 6 | 5 (2) |
| Glycerol facilitator | 1fx8 | 2.20 | 9 | 19 (11) |
| Glycophorin A | 1afo | NMR | 1 | 12 (6) |
| Light-harvesting complex II | 1lgh | 2.40 | 1 | 0 |
| Mechanosensitive channel | 1msl | 3.50 | 3 | 0 |
| Photosynthetic reaction center | 1prc | 2.30 | 13 | 19 (4) |
| Potassium channel | 1bl8 | 3.20 | 7 | 7 (3) |
| Rhodopsin | 1f88 | 2.80 | 13 | 6 (3) |
| Total | 125 | 145 (51) |
Helical segments as identified by the dssp program (21).
Unique interactions in homo-oligomers.
Number of contacts with dH < 3.5 Å and ζ > 120° (number of contacts with d < 2.7 Å).
Methods
Our database of 11 nonhomologous helical membrane proteins is listed in Table 1. Hydrogen atoms were added to the crystallographic protein database coordinate files with the program REDUCE (20). Identification of the helical segments was performed with the program DSSP (21). For homo-oligomers, only a single representative of the duplicated helix–helix interaction was considered. The structural analysis of Cα—H⋅⋅⋅O contacts (distances dH and d and angles ζ, ξ, and θ, nomenclature according to Derewenda et al. (14), defined in Fig. 1A) and the calculations of interhelical distances and angles were performed with a custom-made PERL program (available from the authors on request). Interhelical angles and axial distances were calculated at the point of minimal axial distance by using the local helical axis. The local helical axes were calculated by using the coordinates of four contiguous Cα atoms according to Sugeta and Miyazawa (22) with a PERL subroutine adapted from the FORTRAN program HELANAL (23).
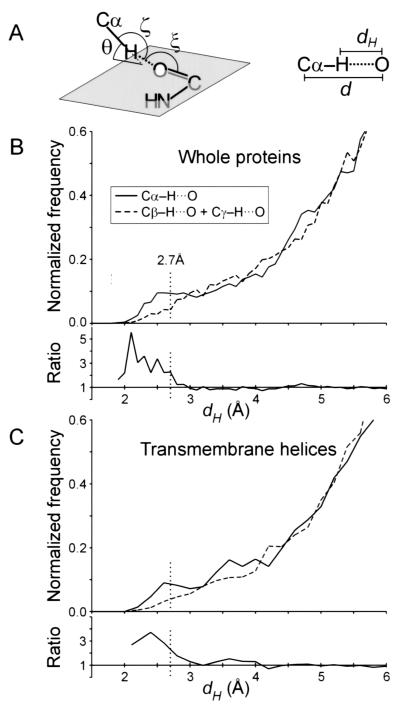
Figure 1.
(A) Definition of the geometrical parameters of the Cα—H⋅⋅⋅O hydrogen bond. Nomenclature according to Derewenda et al. (14). The ideal values (14, 18) are as follows: H—O distance, dH ≤ 2.7 Å; Cα—O distance, d ≤ 3.8 Å; Cα—H—O angle, ζ = 180°; H—O—C angle, ξ = 120°; elevation angle, θ = 0° (angle between the Cα—H vector and the amide plane). (B) Distribution of hydrogen to acceptor distances (dH) for all Cα—H donor groups (solid line), compared with that of Cβ—H + Cγ—H groups (dashed line), in the 11 membrane proteins structures. The ratio between the two curves is also shown (Cα/Cβγ). The control set is similarly sized and is composed by the Cβ—H groups of all Ile, Leu, Val, Met, Phe, and Tyr residues and the Cγ—H of Leu and Cγ1—H of Ile. Contacts below 2.7 Å have an overall frequency of occurrence that is 3 times higher for Cα donors than for the Cβγ control set. (C) Analogous distribution as in B, but limited to the subset of residues in helical transmembrane segments.
Results and Discussion
Distribution of Interhelical Cα—H⋅⋅⋅O Hydrogen Bonds in Transmembrane Helices.
The optimal geometry for a strong Cα—H⋅⋅⋅O hydrogen bond is described in Fig. 1A. The H⋅⋅⋅O distance (dH) should be smaller than 2.7 Å (the sum of the van der Waals radii), and all three atoms should be aligned (C—H—O angle ζ = 180°) (14, 17, 18). We calculated the dH distribution of all Cα hydrogen donors to backbone and side chain acceptors in our database of membrane proteins and compared it to that obtained with a similarly sized set of aliphatic Cβ and Cγ hydrogen atoms (Fig. 1B). The ratio of the two distributions shows that Cα hydrogen atoms form contacts below van der Waals separation more frequently (overall, 3 times more frequently below 2.7 Å). This is consistent with the fact that the Cα is an activated carbon donor with a higher tendency to form C—H⋅⋅⋅O hydrogen bonds. Despite the lower sample size, a similar trend is observed when the analogous distribution is calculated limited to the subset of donors and acceptor groups of transmembrane helical segments (Fig. 1C).
We studied the geometry of interaction of all potential Cα—H⋅⋅⋅O hydrogen bonds between transmembrane helical segments. Hydrogen bond interactions persist at a longer range than van der Waals separation and tolerate significant angular distortion (17). Operatively, we selected dH < 3.5 Å and ζ > 120° (or ζ > 90° when dH < 3.0 Å) as a comprehensive limit for recording Cα—H⋅⋅⋅O contacts as potential hydrogen bonds. We found 145 interhelical Cα—H⋅⋅⋅O contacts, 51 of which have dH < 2.7 Å in 125 helix–helix interactions between 103 helices (Table 1). About one-fourth of all helix–helix interactions contain at least two instances of interhelical Cα—H⋅⋅⋅O contacts to either backbone or side-chain oxygen acceptors. One-tenth contain at least two backbone-to-backbone Cα—H⋅⋅⋅O⩵C contacts. The distribution of these subsets of helix–helix interactions as a function of packing angle (Ω) is displayed in Fig. 2A. In the total set of interactions (blue) we found a strong bias for left-handed packing, as in an earlier analysis by Bowie (24). The bias is mainly because of the antiparallel component (57 left- vs. 25 right-handed). When one considers only those helix–helix interactions with a minimum of two Cα—H⋅⋅⋅O contacts to backbone or side-chain acceptors (red), some preference for right-handed parallel and left-handed antiparallel interactions appears. When a further requirement of at least two backbone-to-backbone Cα—H⋅⋅⋅O⩵C contacts is introduced (yellow), a dramatic selection for right-handed parallel interactions is seen (χ2 test, P < 0.005). Notably, in the −50° to −20° range of packing angles, ≈50% of all helix–helix interactions persist after the selection. Thus, parallel right-handed packing favors interhelical backbone-to-backbone contacts and Cα—H⋅⋅⋅O formation.
Figure 2.
(A) Distribution of interhelical packing angles (Ω). Interhelical packing angles were calculated as the angle between the local helical axes at the point of minimal axial distance. L-H, left-handed; R-H, right-handed; blue, all helix–helix interactions; red, helix–helix interactions with at least two Cα—H⋅⋅⋅O contacts to backbone or side-chain oxygen atoms; yellow, helix–helix interactions with at least two backbone-to-backbone Cα—H⋅⋅⋅O⩵C contacts. (B) Distribution of interhelical axial distances among the same three categories as for A.
The analysis of the occurrence of Cα—H⋅⋅⋅O contacts as a function of the interhelical axial distance is shown in Fig. 2B. All interhelical distances in the database are between 6 and 12 Å with an average of 8.9 Å. The average drops to 7.8 Å when at least two Cα—H⋅⋅⋅O contacts are present and it is further reduced to 7.0 Å when at least two backbone-to-backbone contacts are present (with no instance above 7.6 Å). Notably, below 7.0 Å all helix–helix interactions contain potential Cα—H⋅⋅⋅O⩵C backbone-to-backbone bonds.
Parallel Right-Handed Helix–Helix Interactions: Occurrence of Glycophorin A (GpA)-Like Motifs with Multiple Cα—H⋅⋅⋅O Bonds.
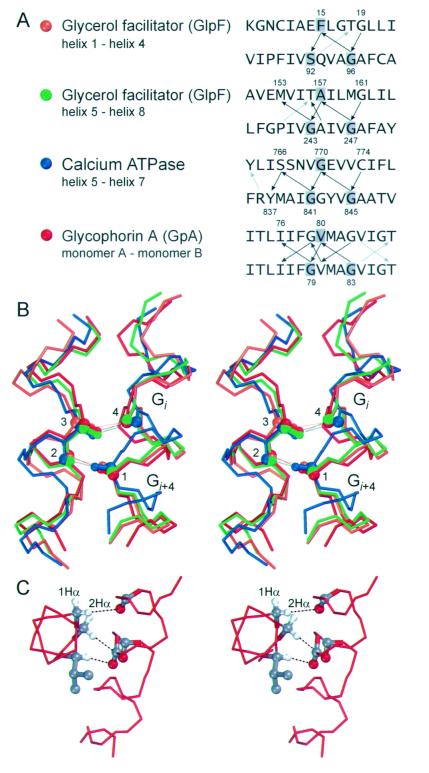
Four cases of parallel right-handed helix–helix interactions with extended Cα—H⋅⋅⋅O contacts are present in the database (Fig. 3). Two cases are found in the glycerol facilitator (GlpF) (25); a third example is observed in the calcium ATPase (26); and the fourth is found in the GpA dimer (27). The geometries of the Cα—H⋅⋅⋅O contacts are shown in Table 2 together with the ideal distance and angle values. Cα—H⋅⋅⋅O hydrogen bonds can tolerate some divergence from ideality (18), and steric factors imposed by helical packing may prevent ideal geometry. The four helix–helix interactions display very similar packing angles (−29° to −40°). The interactions also have similar interfacial residues: pairs of small residues spaced at i, i + 4 are present on all helices and, in particular, GxxxG motifs (28–30) are observed in all but one interface (a SxxxG motif is found in helix 4 of GlpF). As discussed later, the GxxxG motif is known to drive transmembrane helix association.
Figure 3.
Parallel right-handed helix–helix interactions with extended networks of Cα—H⋅⋅⋅O contacts (GpA-like motifs). (A) Schematic representation of the structure of the glycerol facilitator (GlpF, Protein Database ID 1fx8), the calcium ATPase (1eul), and GpA (model 19 in 1afo). The page is parallel to the plane of the membrane. The color coding corresponds to the interactions shown in B. The interaction between helices 2 and 6 in GlpF (yellow) is antiparallel and is shown in Fig. 4A. Some extramembranous regions of the calcium ATPase were removed for clarity. (B) Apparent networks of Cα—H⋅⋅⋅O hydrogen bonds at the interface of four right-handed helix–helix interactions. Some side-chain atoms have been removed for clarity. In the GpA homodimer (red) each interaction occurs symmetrically on both sides but only one set of interactions is shown for clarity. Apparent hydrogen bonds are denoted with dots (⋅⋅⋅) and the distance (dH) in Å is indicated. The interhelical axial distance (a.d.) and packing angle (Ω) of the helix–helix interactions are also indicated.
Table 2.
Geometry* of Cα—H⋅⋅⋅O contacts in GpA-like motifs
| Donor | Acceptor | dH, Å | d, Å | ζ, deg | ξ, deg | θ, deg |
|---|---|---|---|---|---|---|
| Ideal values | ||||||
| C—H | O⩵C | ≤ 2.7 | ≤ 3.8 | 180 | 120 | 0 |
| Glycophorin A (1afo): model 19† (average of 20 models‡) | ||||||
| G79—2Hα | I76—O | 2.6 (3.5) | 3.6 (4.5) | 156 | 119 | 37 |
| V80—Hα | G79—O | 2.1 (2.5) | 2.9 (3.3) | 129 | 102 | 71 |
| G83—2Hα | V80—O | 2.6 (2.8) | 3.7 (3.9) | 170 | 112 | 40 |
| V84—Hα | T87—Oγ | 2.6 (2.7) | 3.5 (3.6) | 140 | 133 | — |
| Glycerol facilitator (1fx8): helix 1–helix 4 | ||||||
| F15—Hα | S92—O | 2.5 | 3.5 | 149 | 118 | 59 |
| G19—1Hα | G96—O | 2.6 | 3.3 | 122 | 103 | 45 |
| Q93—Hα | E14—O | 3.2 | 4.3 | 156 | 114 | 84 |
| Q93—Hα | T18—Oγ | 2.7 | 3.6 | 142 | 94 | — |
| G96—2Hα | F15—O | 2.4 | 3.4 | 145 | 123 | 45 |
| Glycerol facilitator (1fx8): helix 5–helix 8 | ||||||
| P240—Hα | T156—Oγ | 2.5 | 3.4 | 147 | 89 | — |
| G243—2Hα | M153—O | 2.8 | 3.5 | 124 | 113 | 47 |
| A244—Hα | T156—O | 3.1 | 4.1 | 148 | 104 | 82 |
| A157—Hα | G243—O | 2.2 | 3.0 | 123 | 127 | 50 |
| G247—2Hα | A157—O | 2.4 | 3.3 | 134 | 107 | 57 |
| G161—1Hα | G247—O | 3.3 | 3.9 | 116 | 105 | 34 |
| Calcium ATPase (1eul): helix 5–helix 7 | ||||||
| A762—Hα | T837—Oη | 3.3 | 4.0 | 127 | 67 | 33 |
| S776—Hα | T837—O | 3.4 | 4.2 | 130 | 93 | 6 |
| G770—1Hα | G841—O | 3.1 | 3.8 | 116 | 93 | 13 |
| C774—Hα | G845—O | 2.6 | 3.2 | 117 | 120 | 5 |
| G841—1Hα | S766—O | 2.9 | 3.4 | 111 | 137 | 6 |
| G845—1Hα | G770—O | 2.5 | 3.3 | 129 | 91 | 10 |
Although the four right-handed structures have similar packing angles and interfacial residues, they, most strikingly, also share a pattern of Cα—H⋅⋅⋅O connectivity. In Fig. 4A, the backbone-to-backbone Cα—H⋅⋅⋅O⩵C contacts are represented by black arrows, and the backbone-to-side chain Cα—H⋅⋅⋅O contacts by gray arrows. All backbone-to-backbone contacts occur with the same periodicity, between one residue and two residues spaced at i, i + 4 on the opposite helix. The i, i + 4 connectivity extends up to three helical turns. The GxxxG interaction motifs are central to this connectivity, as highlighted in the backbone superimposition of the four structures in Fig. 4B. The four connecting atoms used to align the structures are displayed in ball representation. On the right are shown the helices containing the GxxxG (SxxxG) motifs with the carbonyl oxygen of the Gly at i (4) and the Cα of the Gly at i + 4 (1); on the left, the Cα (3) and the oxygen (2) of the residue that interacts with the two Gly residues. The interhelical packing angles of −29° to −40° appear to favor the i, i + 4 connectivity. The relative rotation that has to be applied to two opposing helices to align the vector joining the Cα and the carbonyl oxygen of a residue on one helix (vector 2-3), with the vector joining a carbonyl oxygen with the Cα at i − 4 on the opposing helix (vector 1-4), is approximately −35°.
Figure 4.
(A) Connectivity of the networks of apparent Cα—H⋅⋅⋅O hydrogen bonds in the four parallel right-handed GpA-like motifs. The arrows show the interactions in the donor-to-acceptor direction. Black arrows: backbone-to-backbone Cα—H⋅⋅⋅O bonds; gray arrows: backbone-to-side chain bonds. Backbone-to-backbone Cα—H⋅⋅⋅O⩵C contacts occur between an amino acid residue on one helix and two residues spaced at i, i + 4 on the opposite helix in all structures. The three residues involved in the alignment in B are shaded in the sequences. (B) Superimposition of the four structures aligned using the carbonyl oxygen (4) at i and the Cα (1) at i + 4 of the GxxxG (SxxxG) motifs on the helix on the right; and the Cα (3) and carbonyl oxygen (2) of the residue that interacts with the two Gly residues with apparent Cα—H⋅⋅⋅O bonds. The overall rms deviation of the superimposition calculated on the backbone atoms of 13 residues on each helix is 1.6 Å. (C) The 2Hα of glycine residues is oriented roughly in the same direction of the Hα of the residue at i + 1 and i − 3. When a GxxxG motif is present a potential interaction interface arises. The example shows the interaction of Gly-79, Val-80, and Gly-83 of GpA that donate to carbonyl oxygen atoms spaced at i, i + 3 and i + 4 on the opposite monomer.
Among the four structures, the GpA dimer is the only one with GxxxG pairs present on both interacting helix surfaces (Fig. 4A). GxxxG was identified by Russ and Engelman (29) as a major motif driving oligomerization in a screen of randomized interaction interfaces. Furthermore, small residues spaced at i, i + 4 have increased occurrence in transmembrane helices; in particular, GxxxG is the most biased pair in transmembrane helices, being strongly over-represented in both single-span and multispan membrane proteins (28). Finally, several mutagenesis studies support the involvement of the GxxxG motif in transmembrane helix oligomerization and interaction (30–35). In addition to favoring interhelical backbone contacts because of the lack of a side chain, Gly residues also increase the opportunities for Cα—H⋅⋅⋅O formation with two α-hydrogen atoms that can act as donors (denoted as 1Hα and 2Hα). In a helical conformation, the 2Hα of Gly (the hydrogen that is stereochemically in the position of the side chain in other amino acids) points in the same direction as the Hα of the residues at i + 1 and i − 3. Thus, in a GXxxG motif three Hα atoms (specifically, the 2Hα atoms of each Gly residue, and the Hα of the residue X at i + 1 with respect to the first Gly) are oriented roughly parallel and a potential interaction surface arises. This is illustrated in Fig. 4C, where the Cα—H of Gly-79, Val-80, and Gly-83 of GpA are donors to carbonyl oxygen atoms on the opposite side spaced at i, i + 3 and i + 4. A similar pattern is also found in GlpF (donors Gly-243, Ala-244, and Gly-247). Hence, the present data suggest that the GxxxG motif drives transmembrane helix association in part by favoring Cα—H⋅⋅⋅ O⩵C interactions. We have found GxxxG pairs only in parallel right-handed “GpA-like” interaction motifs, although numerous interfacial Gly residues occur in all identified interhelical networks with backbone-to-backbone contacts. It will be interesting to see whether the expectation of finding GxxxG pairs frequently—and perhaps mainly—occurring in GpA-like motifs will be met once more structural data become available.
Other Networks of Interhelical Cα—H⋅⋅⋅O Bonds.
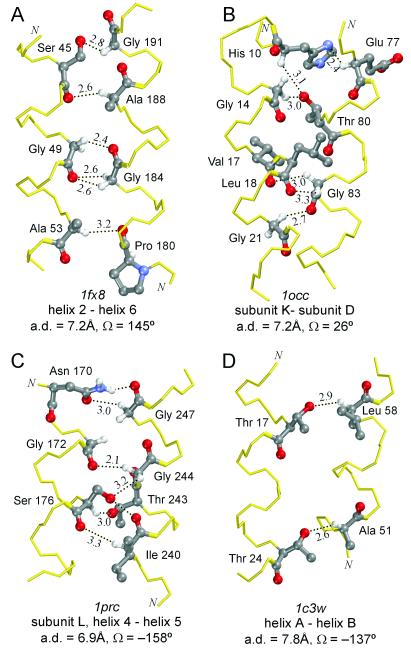
Fig. 5 shows four more examples of helix–helix interfaces with apparent Cα—H⋅⋅⋅O bonds: three very extensive networks (A–C) containing several backbone-to-backbone Cα—H⋅⋅⋅O contacts occur in an antiparallel right-handed interaction (A, GlpF), a parallel left-handed interaction (B, cytochrome c oxidase), and an antiparallel left-handed interaction (C, photosynthetic reaction center). The interaction of Fig. 5A is the third case found in GlpF. As with the other two interactions, it is right-handed and characterized by small interfacial amino acid residues spaced at i, i + 4 engaged in Cα—H⋅⋅⋅O contacts (AxxxGxxxS on helix 2 and GxxxA on helix 6), but it is antiparallel. An example from bacteriorhodopsin (bR) is shown in Fig. 5D. The structure of bR has been solved at 1.55 Å and is currently the highest-resolution structure for a membrane protein. bR has interhelical distances in the range 7.8–10.6 Å, above the 7.6 Å that appears to be the approximate limit for backbone-to-backbone Cα—H⋅⋅⋅O⩵C formation. However, backbone-to-side-chain Cα—H⋅⋅⋅O bonds are observed.
Figure 5.
Additional helix–helix interactions with multiple Cα—H⋅⋅⋅O hydrogen bonds at various interhelical packing angles. (A) Antiparallel right-handed interaction from the GlpF. (B) Parallel left-handed interaction from bovine heart cytochrome c oxidase. (C) Antiparallel left-handed interaction from the photosynthetic reaction center. (D) Antiparallel left-handed interaction with apparent backbone-to-side-chain Cα—H⋅⋅⋅O bonds from bacteriorhodopsin.
It should be noted that the examples of Fig. 5 do not have the thematic packing angles and connectivity seen in the right-handed parallel interactions. Instead, the central element characterizing all observed cases with backbone-to-backbone Cα—H⋅⋅⋅O⩵C networks is the presence of numerous interfacial Gly, Ser, and Thr residues.
The Versatility of Glycine in Interhelical Cα—H⋅⋅⋅O Bond Networks.
Gly residues are frequent in transmembrane helices, constituting 8% of the amino acid composition (28). Gly residues permit short interhelical separation, and it has previously been suggested that they might participate in Cα—H⋅⋅⋅O bonds (36). In our analysis, 23% of all helical Gly residues appear to be involved as donors and 10% as acceptors in Cα—H⋅⋅⋅O contacts. As previously discussed, Gly residues increase the opportunity for Cα—H⋅⋅⋅O bond formation because the second Hα of glycine points roughly in the same direction as the Hα of the residues at i + 1 and i − 3 (Fig. 4C). In addition, both the Hα atoms of a Gly residue can simultaneously form Cα—H⋅⋅⋅O bonds to either different acceptors (Gly-83 in Fig. 5B; Gly-244 in Fig. 5C) or the same acceptor oxygen (bidentate interactions, Gly-184 in Fig. 5A). Hence, the versatility of Gly residues appears to favor Cα—H⋅⋅⋅O network formation.
Serine and Threonine Allow Cα—H⋅⋅⋅Oγ Contacts at Longer Interhelical Distances.
Ser and Thr residues are frequently involved in interhelical Cα—H⋅⋅⋅O bonds. They each constitute 5% of the amino acid composition of transmembrane helices (28), where they are well tolerated because the donor potential of their polar side chains can be satisfied by forming O—H⋅⋅⋅O hydrogen bonds to the carbonyl at i − 4 or i − 3 on the same helix (37, 38). For this reason they have a weak tendency to form interhelical O—H⋅⋅⋅O hydrogen bonds (11, 13). The Oγ, however, is available as an acceptor and is displaced from the helix axis. In our database 24% of all Ser and 20% of all Thr residues appear to be involved in Cα—H⋅⋅⋅Oγ bonds. In particular, in GpA, Thr-87 allows the formation of an additional Cα—H⋅⋅⋅O when the interhelical distance is too large for the Cα—H of Val-84 to reach the backbone oxygen on the opposite helix (Fig. 3B). Consistently, the isosteric mutation of Thr-87 to Val results in partial destabilization of the GpA dimer (32). Moreover, the GxxxGxxxT motif was found by Russ and Engelman to be among the strongly associating helices in their selection of randomized interfaces (29) and the triplet is also strongly over-represented in transmembrane sequences (28). Thus, the frequent occurrence of Ser and Thr residues in transmembrane helices could be in part linked to their ability to engage in Cα—H⋅⋅⋅O hydrogen bonds.
Concluding Remarks.
By using known structures of helical membrane proteins, we have found common features in a number of helix–helix interfaces: (i) networks of apparent Cα—H⋅⋅⋅O bonds; (ii) abundant interfacial Gly, Ser, and Thr residues; and (iii) short interhelical axial distances. In particular, Gly residues permit backbone-to-backbone Cα—H⋅⋅⋅O⩵C formation by allowing short interhelical axial distances; the two α-hydrogen atoms of Gly also increase the opportunities for Cα—H⋅⋅⋅O formation. Ser and Thr side-chain hydroxyl groups allow Cα—H⋅⋅⋅O interaction at longer interhelical distances. In addition, Cα—H⋅⋅⋅O contacts appear more frequently in parallel right-handed helix–helix interactions. Finally, right-handed parallel GpA-like motifs, in which GxxxG pairs promote extended Cα—H⋅⋅⋅O network formation, are a recurrent theme.
Cα and carbonyl O atoms are typically shielded by side chains in an α-helix, and so tend to be inaccessible. Derewenda et al. (14) defined C—H⋅⋅⋅O contacts between helices as “esoteric” in soluble proteins and, in fact, they did not report a single instance of helical Cα—H⋅⋅⋅O contacts with dH < 2.7 Å in 13 high-resolution structures (1–2 Å). Conversely, we have found 51 such contacts in 11 membrane proteins. Such a discrepancy is not due simply to the lower resolution of our database; instead, it depends on the frequency of Gly residues, which are rare in helical segments of soluble proteins but prevalent in transmembrane helices. It seems highly probable that Cα—H⋅⋅⋅O hydrogen bonds exist in helix–helix interfaces having numerous Gly residues, very short interaxial distances, and backbone contacts. Although Cα—H⋅⋅⋅O hydrogen bonding could be merely incidental to the optimal packing achieved with close-range interhelical contacts, the key is then the relative energy contribution of the Cα—H⋅⋅⋅O bond. If the estimate of 2.5–3.0 kcal/mol of interaction energy for the Cα—H⋅⋅⋅O hydrogen bond is even approximately correct, several coordinated interactions would approximate or even exceed the energy of the N—H⋅⋅⋅O hydrogen bond, which is known to drive nonspecific transmembrane helix association (10–13). However, packing would still be central in helix–helix interactions promoted by coordinated Cα—H⋅⋅⋅O hydrogen bonds because main-chain atoms come in contact only between helices that fit together well. Moreover, because Cα—H⋅⋅⋅O hydrogen bonds are weaker than main-chain N—H⋅⋅⋅O⩵C, helix distortion would represent a substantial cost for their optimization. Thus, even if the energy contribution of backbone-to-backbone Cα—H⋅⋅⋅O⩵C interactions turned out to be overwhelming with respect to the dispersion component of packing, their formation would be much more dependent on the specifics of local geometry than that of O—H⋅⋅⋅O or N—H⋅⋅⋅O bonds between ends of flexible side chains, thus reducing the dangers of nonspecific association. The Cα—H⋅⋅⋅O could then be a more controllable and cooperative alternative to O—H⋅⋅⋅O or N—H⋅⋅⋅O bonds for exploiting the strength and directionality of hydrogen bonds in the hydrophobic environment and achieving, simultaneously, stability and specificity in transmembrane helix–helix interactions.
Acknowledgments
We thank J. Cabral, M. Cocco, R. Curran, J. Dawson, K. Ho, A. Lee, H. Li, M. Lemmon, N. Luscombe, K. MacKenzie, J.-L. Popot, J. Qian, B. Russ, and F. Zhou for helpful discussions and critical reading of the manuscript. This work was supported by grants from the National Institutes of Health and the National Science Foundation.
Abbreviations
- GpA
glycophorin A
- GlpF
glycerol facilitator
References
- 1.Hendsch Z S, Tidor B. Protein Sci. 1994;3:211–226. doi: 10.1002/pro.5560030206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hendsch Z S, Tidor B. Protein Sci. 1999;8:1381–1392. doi: 10.1110/ps.8.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheinerman F B, Norel R, Honig B. Curr Opin Struct Biol. 2000;10:153–159. doi: 10.1016/s0959-440x(00)00065-8. [DOI] [PubMed] [Google Scholar]
- 4.Popot J L, Engelman D M. Biochemistry. 1990;29:4031–4037. doi: 10.1021/bi00469a001. [DOI] [PubMed] [Google Scholar]
- 5.Deisenhofer J, Epp O, Sinning I, Michel H. J Mol Biol. 1995;246:429–457. doi: 10.1006/jmbi.1994.0097. [DOI] [PubMed] [Google Scholar]
- 6.Palczewski K, Kumasaka T, Hori T, Behnke C A, Motoshima H, Fox B A, Le Trong I, Teller D C, Okada T, Stenkamp R E, et al. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 7.Popot J L, Engelman D M. Annu Rev Biochem. 2000;69:881–922. doi: 10.1146/annurev.biochem.69.1.881. [DOI] [PubMed] [Google Scholar]
- 8.Ubarretxena-Belandia I, Engelman D M. Curr Opin Struct Biol. 2001;11:370–376. doi: 10.1016/s0959-440x(00)00217-7. [DOI] [PubMed] [Google Scholar]
- 9.Snijder H J, Ubarretxena-Belandia I, Blaauw M, Kalk K H, Verheij H M, Egmond M R, Dekker N, Dijkstra B W. Nature (London) 1999;401:717–721. doi: 10.1038/44890. [DOI] [PubMed] [Google Scholar]
- 10.Zhou F X, Cocco M J, Russ W P, Brunger A T, Engelman D M. Nat Struct Biol. 2000;7:154–160. doi: 10.1038/72430. [DOI] [PubMed] [Google Scholar]
- 11.Zhou F X, Merianos H J, Brunger A T, Engelman D M. Proc Natl Acad Sci USA. 2001;98:2250–2255. doi: 10.1073/pnas.041593698. . (First Published February 13, 2001; 10.1073/pnas.041593698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choma C, Gratkowski H, Lear J D, DeGrado W F. Nat Struct Biol. 2000;7:161–166. doi: 10.1038/72440. [DOI] [PubMed] [Google Scholar]
- 13.Gratkowski H, Lear J D, DeGrado W F. Proc Natl Acad Sci USA. 2001;98:880–885. doi: 10.1073/pnas.98.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derewenda Z S, Lee L, Derewenda U. J Mol Biol. 1995;252:248–262. doi: 10.1006/jmbi.1995.0492. [DOI] [PubMed] [Google Scholar]
- 15.Bella J, Berman H M. J Mol Biol. 1996;264:734–742. doi: 10.1006/jmbi.1996.0673. [DOI] [PubMed] [Google Scholar]
- 16.Fabiola G F, Krishnaswamy S, Nagarajan V, Pattabhi V. Acta Crystallogr D. 1997;53:316–320. doi: 10.1107/S0907444997000383. [DOI] [PubMed] [Google Scholar]
- 17.Desiraju G R, Steiner T. The Weak Hydrogen Bond in Structural Chemistry and Biology. Oxford: Oxford Univ. Press; 1999. [Google Scholar]
- 18.Vargas R, Garza J, Dixon D A, Hay B P. J Am Chem Soc. 2000;122:4750–4755. [Google Scholar]
- 19.Scheiner S, Kar T, Gu Y. J Biol Chem. 2001;276:9832–9837. doi: 10.1074/jbc.M010770200. [DOI] [PubMed] [Google Scholar]
- 20.Word J M, Lovell S C, Richardson J S, Richardson D C. J Mol Biol. 1999;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 21.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 22.Sugeta H, Miyazawa T. Biopolymers. 1967;5:673–679. [Google Scholar]
- 23.Bansal M, Kumar S, Velavan R. J Biomol Struct Dyn. 2000;17:811–819. doi: 10.1080/07391102.2000.10506570. [DOI] [PubMed] [Google Scholar]
- 24.Bowie J U. J Mol Biol. 1997;272:780–789. doi: 10.1006/jmbi.1997.1279. [DOI] [PubMed] [Google Scholar]
- 25.Fu D, Libson A, Miercke L J, Weitzman C, Nollert P, Krucinski J, Stroud R M. Science. 2000;290:481–486. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- 26.Toyoshima C, Nakasako M, Nomura H, Ogawa H. Nature (London) 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 27.MacKenzie K R, Prestegard J H, Engelman D M. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 28.Senes A, Gerstein M, Engelman D M. J Mol Biol. 2000;296:921–936. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- 29.Russ W P, Engelman D M. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 30.Brosig B, Langosch D. Protein Sci. 1998;7:1052–1056. doi: 10.1002/pro.5560070423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Font M, Feliubadalo L, Estivill X, Nunes V V, Golomb E, Kreiss Y, Pras E, Bisceglia L, d'Adamo A P, Zelante L, et al. Hum Mol Genet. 2001;10:305–316. doi: 10.1093/hmg/10.4.305. [DOI] [PubMed] [Google Scholar]
- 32.Lemmon M A, Treutlein H R, Adams P D, Brunger A T, Engelman D M. Nat Struct Biol. 1994;1:157–163. doi: 10.1038/nsb0394-157. [DOI] [PubMed] [Google Scholar]
- 33.Op De Beeck A, Montserret R, Duvet S, Cocquerel L, Cacan R, Barberot B, Le Maire M, Penin F, Dubuisson J. J Biol Chem. 2000;275:31428–31437. doi: 10.1074/jbc.M003003200. [DOI] [PubMed] [Google Scholar]
- 34.McClain M S, Cao P, Cover T L. Infect Immun. 2001;69:1181–1184. doi: 10.1128/IAI.69.2.1181-1184.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang C, Deber C M. J Biol Chem. 2000;275:16155–16159. doi: 10.1074/jbc.M000723200. [DOI] [PubMed] [Google Scholar]
- 36.Javadpour M M, Eilers M, Groesbeek M, Smith S O. Biophys J. 1999;77:1609–1618. doi: 10.1016/S0006-3495(99)77009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gray T M, Matthews B W. J Mol Biol. 1984;175:75–81. doi: 10.1016/0022-2836(84)90446-7. [DOI] [PubMed] [Google Scholar]
- 38.Ballesteros J A, Deupi X, Olivella M, Haaksma E E, Pardo L. Biophys J. 2000;79:2754–2760. doi: 10.1016/S0006-3495(00)76514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]