Abstract
DNA vaccine can be modified to increase protein production and modulate immune response. To enhance the efficiency of a P815 mastocytoma DNA vaccine, the P1A gene sequence was optimized by substituting specific codons with synonymous ones while modulating the number of CpG motifs. The P815A murine antigen production was increased with codon-optimized plasmids. The number of CpG motifs within the P1A gene sequence modulated the immunogenicity by inducing a local increase in the cytokines involved in innate immunity. After prophylactic immunization with the optimized vaccines, tumor growth was significantly delayed and mice survival was improved. Consistently, a more pronounced intratumoral recruitment of CD8+ T cells and a memory response were observed. Therapeutic vaccination was able to delay tumor growth when the codon-optimized DNA vaccine containing the highest number of CpG motifs was used. Our data demonstrate the therapeutic potential of optimized P1A vaccine against P815 mastocytoma, and they show the dual role played by codon optimization on both protein production and innate immune activation.
Keywords: codon optimization, DNA vaccine, P815, P1A, prophylactic, therapeutic vaccine, CpG, innate immunity, cancer vaccine, tumor vaccine
Introduction
Harnessing the immune system to fight cancer has become a priority in the last few years, and it is supported by an increasing knowledge of tumor-host interactions. Among the various immunotherapy strategies that are currently being developed, DNA vaccines have many advantages, such as low cost and high stability and versatility, which allow the modulation of the encoded antigen and its intrinsic immunogenicity. Interestingly, DNA vaccines can induce not only the activation of the innate immune response but also the cellular and humoral arms of the adaptive immune system.1, 2 However, human applications of DNA vaccines have lagged, largely due to suboptimal immunogenicity compared to traditional vaccines.1, 3 Different approaches have been investigated in order to overcome this problem and enhance their efficacy.4, 5, 6, 7
Several elements appear critical for optimizing DNA vaccination. Antigen expression should be high enough to promote its presentation by antigen-presenting cells (APCs) and the activation of the adaptive immunity.8, 9 Codon optimization is an in silico technique originally based on the selection of codon triplets that have the highest tRNA frequency in the cytoplasm of the target species. Hence, codon optimization algorithms allow for an increase in the protein translation rate and mRNA stability while maintaining the typical 3D structure of the protein.10 This technique is generally used to induce greater production of a foreign protein (e.g., when a viral or a bacterial protein must be expressed in mammalian cells).11, 12 Codon optimization also allows for a modulation of the number of CpG motifs in the gene sequence. Indeed, it has been demonstrated that CpG motifs directly stimulate B cells and are recognized by Toll-like receptor 9 (TLR9) in dendritic cells (DCs), B cells, and macrophages, allowing the activation of the innate immune system.4 Hence, the addition of CpG motifs as built-in adjuvants into the plasmid sequence improves the immunogenicity of DNA vaccines.3, 13
The DNA delivery method must also be carefully selected because it not only has an influence on the magnitude of the gene expression, which depends on the delivery efficacy, but also it contributes to the immunogenicity of the DNA vaccine.14 Electroporation (EP) is a non-viral delivery method that can improve in vitro and in vivo plasmid uptake and, thereby, increase the expression level of the transgene in many cell types and tissues.15 EP utilizes electric pulses at the site of immunization that transiently destabilize the cell membrane and promote the electrophoretic movement of negatively charged DNA into the cells.16, 17 In particular, intramuscular EP promotes long-lasting gene expression and the generation of a local and systemic immune response.18, 19, 20 This technique also reduces the amount of DNA required to activate the immune system (by up to 100 times) while increasing the potency of the immune response that is generated compared to conventional DNA vaccinations.14 For these reasons, EP is being used in many clinical trials for the delivery of DNA vaccines for different pathologies.16
The P1A gene is a cancer-germline gene in mice that encodes the major tumor rejection antigen of mastocytoma P815, named P815A. This gene is activated in several tumors but is silent in normal cells, except placental trophoblasts and male germline cells. Since these cells do not bear the surface major histocompatibility complex class I (MHC class I) molecules, they are not able to present the antigen.21 Hence, immunization against this antigen does not induce autoimmune side effects. P815A shares many characteristics with human MAGE-type tumor antigens,22 suggesting that the P815 mastocytoma tumor model is relevant for future applications in human medicine.
This study aims to generate a potent immune response against P815 by optimizing the P1A antigen gene sequence. We hypothesize that optimization of the codon sequence could improve the vaccine efficacy by enhancing antigen production and inducing a stronger activation of the innate immune system. These modifications would lead to a stronger protection of mice against P815 mastocytoma. Several P1A-expressing DNA vaccines were constructed that encoded exactly the same antigenic protein but differed in terms of nucleic acid sequence. First, the impact of these modifications on gene and protein expression and on plasmid immunogenicity was studied. Then, the vaccine efficacy in DBA/2 mice challenged with P815 after a prophylactic vaccination was evaluated, analyzing mice survival and CD8+ tumor infiltration. Finally, the therapeutic efficiency of the optimized plasmid was investigated.
Results and Discussion
Codon Optimization Enhanced In Vitro Expression of P815A Antigen
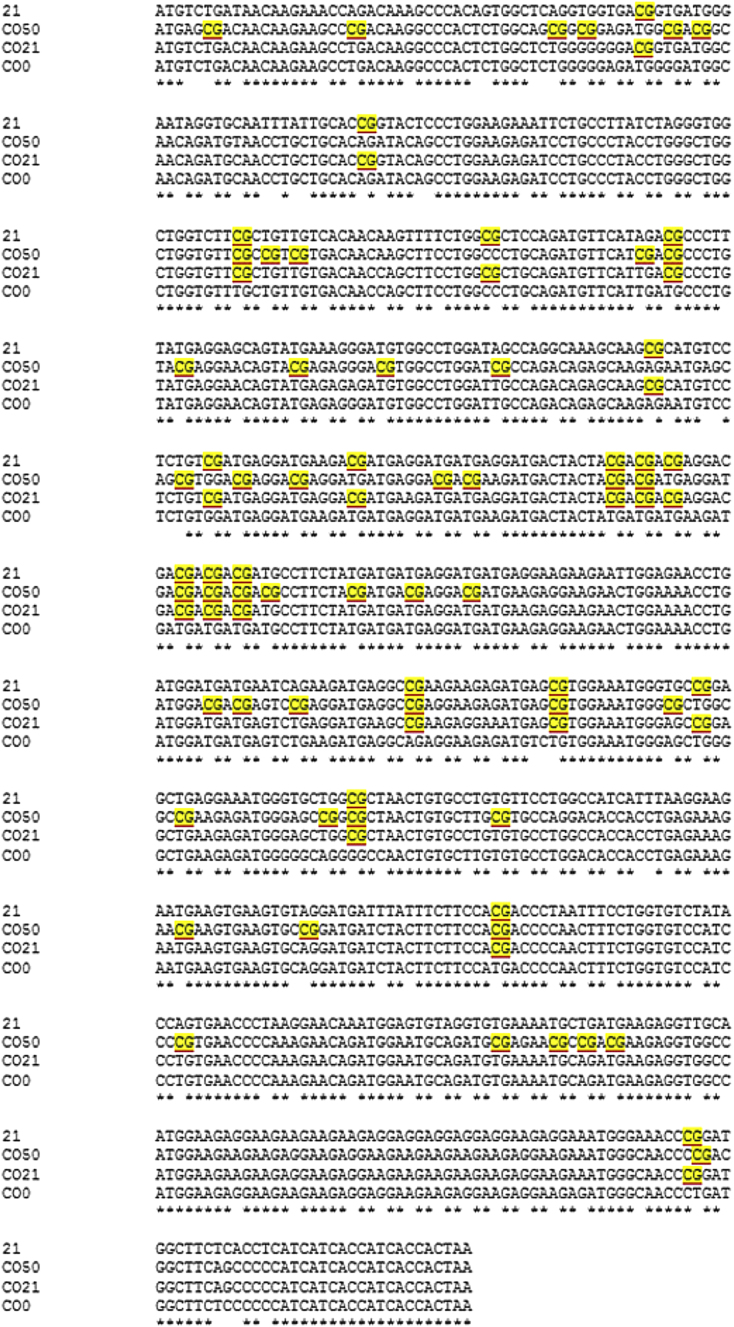
To modulate the expression of the P815A antigen and optimize a DNA vaccine against P815 mastocytoma, four different P1A-encoding plasmids were constructed. In all the constructs, a strong viral promoter (cytomegalovirus [CMV]) and a Kozak sequence were inserted upstream of the antigen sequence. Four different P1A sequences were designed. The first one contained the non-optimized wild-type P1A sequence naturally carrying 21 CpG motifs (P1A_21). The three other sequences were codon-optimized (CO) and adapted to contain zero (P1A_CO0), 21 (P1A_CO21), or 50 (P1A_CO50) CpG motifs. To estimate the production of the P815A antigen, a 6X-His-tag was added downstream of the gene sequence in the plasmids used for P1A mRNA and P815A quantification. For all the other experiments, plasmids without 6X-His-tag were used. The alignment of the four gene sequences is shown in Figure 1.
Figure 1.
Alignment of the Four Different P1A Gene Sequences
The CpG motifs are highlighted. The stars indicate the presence of the same nucleotide in the four sequences. CO means codon optimized; 0, 21, and 50 refer to the number of unmethylated CpG motifs inside the P1A gene sequence.
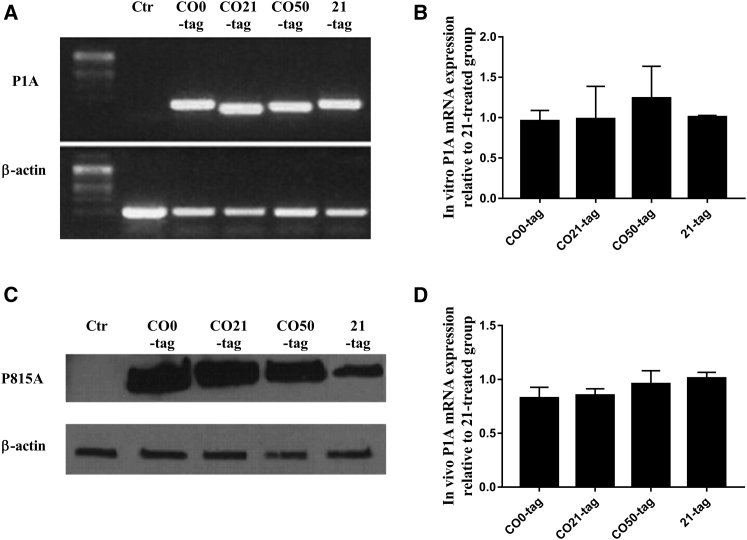
After EP of the plasmids in C2C12 murine myoblasts, the mRNA and protein expression were evaluated in vitro, using RT-PCR/qPCR and western blot, respectively. At 24 hr after EP, the P1A mRNA levels were the same for all the plasmids, without any significant differences between the optimized and non-optimized genes (Figures 2A and 2B). As expected, the western blotting analysis revealed higher levels of P815A protein for the CO genes compared to the non-optimized (Figure 2C). No difference among the three CO plasmids was noticed. P1A mRNA level was also evaluated in the electroporated muscle of DBA/2 mice 6 hr after plasmid delivery. In vivo results confirmed what was observed in vitro, showing no differences among the plasmids (Figure 2D). A similar result was obtained 48 hr after the plasmid EP (data not shown), supporting the hypothesis that the CpG motif amount did not influence the transgene mRNA expression in a short-term period.
Figure 2.
Evaluation of P1A and P815A Antigen Expression after 6X-His-Tagged Plasmid Transfection by Electroporation
(A and B) The RT-PCR (A) and qPCR (B) analysis of P1A mRNA expression 24 hr after vaccine delivery in C2C12 murine myoblasts (mean ± SEM). (C) Western blot analysis of P815A protein expression 24 hr after vaccine delivery in C2C12 murine myoblasts. (D) qPCR analysis of P1A mRNA expression 6 hr after vaccine injection in the tibial cranial muscle of DBA/2 mice (mean ± SEM). All the experiments were performed in triplicate (n = 3).
Codon optimization is often critical to enhance the production of viral or bacterial proteins in a foreign organism.11, 12 It is commonly accepted that this technique can increase the efficiency of the transgene expression, mostly acting on the codon usage.23 However, the different amount of CpG motifs inside the transgene could also influence the protein production. Controversial data are reported in the literature. Some authors demonstrated that CpG-free plasmids induced a longer protein expression.24 Interestingly, Bauer et al.25 showed a clear loss of the transgene expression following the depletion of CpGs from the coding region. Also, the effect of CpG motifs on the protein expression can vary depending on the cell type.26 Here, our results confirmed that, in the short term, the protein production was increased when optimizing a mouse gene sequence for expression in murine cells, independently of the intragenic CpG amount. This was due to improved protein translation rather than gene transcription, as the mRNA levels were not influenced by the codon modifications. The increase in antigen translation can significantly improve the immune response activation to the DNA immunogens.27, 28, 29 Hence, the use of P1A CO vaccines can be an interesting strategy to produce a strong and specific immune response in the host.
Codon Optimization Influenced Local Cytokine Expression In Vivo
In addition to an increase in the protein translation rate and protein expression, codon optimization allows for an adjustment of the number of unmethylated CpG motifs within the gene sequence. The number of CpG motifs within the P1A gene sequence was modulated. To construct the CO0 vaccine, all the CpG motifs were removed from the P1A gene sequence, while the CO50 vaccine contained 50 CpG motifs within the P1A gene. The other two vaccines, 21 and CO21, contained the wild-type number of CpG motifs. To estimate the built-in adjuvant properties of the CpG motifs and to determine the activation of the innate immune response, qPCR amplification of interleukin (IL)-1β, IL-6, IL-12, and tumor necrosis factor alpha (TNF-α) was performed. Their relative mRNA expression was assessed in the tibial cranial muscle of mice treated with the four vaccines (CO0, CO21, CO50, and 21), mice only electroporated, or untreated mice. At 6 hr after the plasmid injection, the mRNA level of all the cytokines was significantly upregulated at the vaccine injection site for all the treated groups compared to the untreated and EP control groups (Figure 3; all the treatments were significantly different [***p < 0.001] compared to untreated or EP groups, if not differently indicated).
Figure 3.
In Vivo Evaluation of Local Cytokine Production 6 hr after a Single Vaccine Injection and Electroporation in the Tibial Cranial Muscle
(A–D) The mRNA expression of (A) TNF-α, (B) IL-12, (C) IL-1β, and (D) IL-6. All the treatments are significantly different (***p < 0.001) compared to untreated or electroporation (EP) groups, unless otherwise indicated. The results are related to the untreated mice and are expressed in a logarithmic scale (n = 3; mean ± SEM).
These results clearly indicate an activation of the innate immunity in response to the double-stranded DNA (dsDNA) structure itself, due to the different DNA-sensing pathways.30, 31 Indeed, EP increases the permeability of the cell membranes, thus allowing a greater DNA uptake in the cytoplasm. This process could allow the interaction of the plasmid DNA with cytosolic DNA sensors other than TLR9, such as Zbp-1, HMGB, Dhx36, and Ifi16,32 and the production of proinflammatory cytokines and chemokines.33 These features can explain the high cytokine expression in all the treated groups and the vaccine-independent cytokine production in the case of TNF-α and IL-12 (Figures 3A and 3B). More interestingly, IL-1β and IL-6 levels increased according to the number of CpG motifs in the P1A gene. The pVAX2 backbone already contains 193 CpG motifs and is thus recognized as foreign dsDNA by TLR9 in the endosomes, as plasmid molecules could enter the cells by electroendocytosis following the electric pulse application.34, 35 However, a dramatic induction of these cytokines was observed for the vaccine containing the highest CpG number, CO50 (Figures 3C and 3D). Indeed, these cytokines were directly correlated to the presence of CpG, as it was less upregulated in the presence of CpG-free plasmid.32 Hence, even minor modifications in the number of CpG motifs might generate different innate immune responses. It has been demonstrated that the insertion of only a few CpG motifs (16–20) in an antigen-encoding plasmid augmented the production of inflammatory cytokines and allowed the activation of the adaptive immune response.36, 37 Furthermore, these motifs could have a different effect on the immune activation, depending on their adjacent nucleotides.38, 39, 40 For instance, the motif RRCGYY seems to be more immunogenic than other types of CpG.41, 42 Seventeen RRCGYY motifs were present in the pVAX2 backbone while 6, 3, and 0 motifs were found in the CO50, CO21/21, and CO0, respectively. According to Coban et al.,38 the addition of three to five strongly immunogenic CpG motifs was sufficient to increase the IL-6 and interferon (IFN)-γ levels. In a gene sequence, immunoinhibitory motifs can also be present that prevent innate immune system activation.43 The transgene in the CO50 construct did not contain some of the immunoinhibitory motifs (e.g., TTAGGG) that were present in the P1A gene sequence of the other three plasmids.
The delivery method also influenced the cytokine production, as EP alone significantly increased the IL-1β level compared to the untreated group. Roos et al.44 also demonstrated a dramatic upregulation of several cytokines, among others IL-1β, and chemokines involved in defense responses, immune responses, inflammatory responses, chemotaxis, and MHC class I receptor activity when DNA was delivered by EP.44 The upregulation of these cytokines was correlated with the potentiation of the immune response induced by the delivered DNA.44, 45
Oligodeoxynucleotides (ODNs) containing unmethylated CpG motifs (CpG-ODNs) have been widely used alone or as vaccine adjuvants that are co-delivered with the antigen-encoding vaccine in order to accelerate the induction, increase the maximum level, and extend the duration of the induced immune response.46, 47, 48 Indeed, CpG-ODN can activate signaling by TLR9 on cells of the innate and adaptive immune system, leading to the production of several pro-inflammatory cytokines, such as IL-1β, IL-6, TNF-α, and IL-12.48, 49, 50, 51, 52 In particular, IL-6 and IL-1β play an important role in activation of the innate immune system. Indeed, IL-6 helps naive CD8+ T cells to proliferate and acquire lytic capability in the absence of stimulation by specific T cell receptors (TCRs). It has also been demonstrated that IL-6, in synergy with IL-1, can augment IL-2 responsiveness of CD8+ T cells and prime them for subsequent stimulation via the TCR.53, 54
Codon Optimization of Plasmids and Modulation of Their CpG Motif Content Significantly Influenced P815 Tumor Growth and Mouse Survival after Prophylactic Vaccination
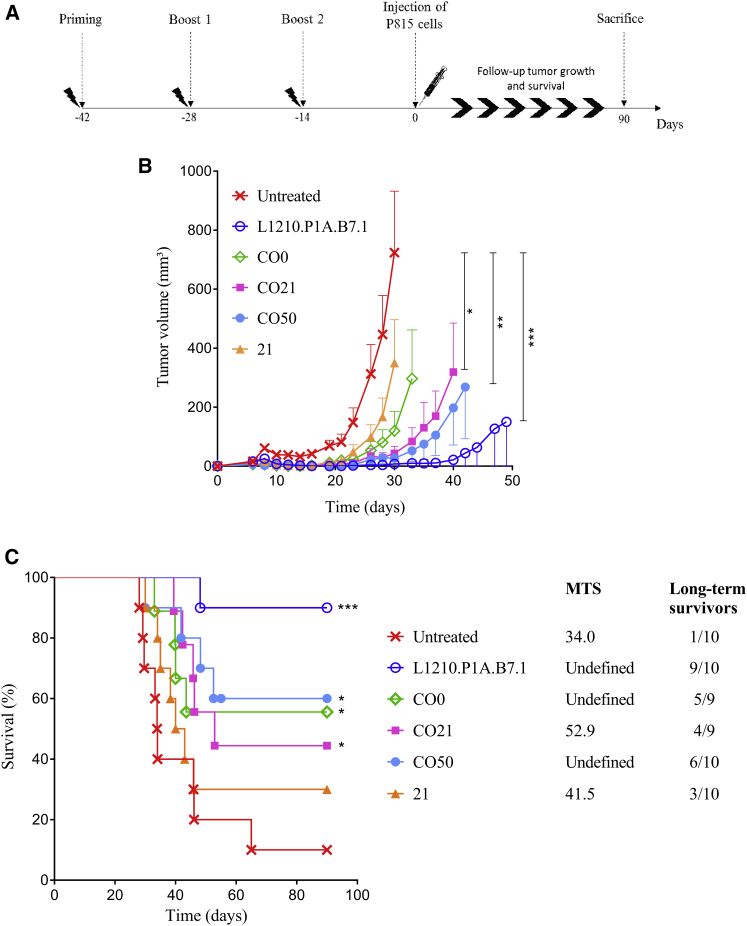
To evaluate the impact of plasmid optimization on mice survival and tumor growth, mice were vaccinated and then challenged with P815 mastocytoma tumor cells (protocol shown in Figure 4A). The initial evolution of the tumor growth was similar for all the groups (Figure 4B). At 3 days after the challenge, all the mice had palpable tumors, demonstrating the aggressive nature of the tumors. Tumor volume reached a size of 50–100 mm3 around days 5–7. Between days 10 and 20, the tumors of the untreated mice reached a plateau and then they started to re-grow faster than the others; at day 32, 60% of untreated mice were already dead (Figure 4C). However, the tumors of all treated mice were found to be impalpable between days 10 and 20. The tumors that did not start to re-grow between days 20 and 35 were considered to be definitively rejected. Interestingly, 40%–60% of mice treated with a CO vaccine completely rejected the tumor 10–15 days after the injection. The most significant tumor growth delay was observed in mice treated with the CO50 vaccine (Figure 4B). Furthermore, the mice treated with the CO sequences survived longer than the mice in the untreated group or the group treated with the non-optimized vaccine (21). Again, the greatest survival among the vaccine-treated mice was reached in mice treated with the CO50 vaccine (Figure 4C), as 60% of the mice survived the challenge. In the L1210.P1A.B7.1-treated group, only one mouse did not survive the challenge, confirming the efficiency of this treatment for prophylactic vaccination.55 As previously studied, immunization with L1210 cells expressing P1A and B7.1 efficiently protects mice against a P815 challenge,55 by the induction of a very strong P1A-specific cytotoxic activity. These cells were used as positive controls in a few studies.55, 56, 57
Figure 4.
P815 Challenge after Immunization with Intramuscular Electroporation of Four Different pVAX2-P1A Vaccines
Mice were treated with CO0, CO21 (n = 9), CO50, 21, or by intraperitoneal injection of L1210.P1A.B7.1 cells (n = 10). (A) Prophylactic vaccination protocol. (B) Evolution of tumor volume (mm3) after P815 challenge as a function of time (days) (mean ± SEM). Statistical analysis is referred to the untreated group: two-way ANOVA, column factor (***p < 0.001, **p < 0.01, and *p < 0.05). (C) Survival curve representing the percentage of mice alive (%) as a function of time (days). Statistical analysis, log rank (Mantel-Cox) test (***p < 0.001 and *p < 0.05). In the legend, the median survival time (MST, days) of P815-challenged DBA/2 mice after prophylactic vaccination and the number of long-term survivors are shown.
It also has been demonstrated that the administration of the wild-type P1A gene (the same as the 21 vaccine in this study) could delay tumor growth and increase mouse survival, due to an augmented cytotoxic T lymphocyte (CTL) activation.56 Here, better results were obtained using CO vaccines. Codon optimization induced an increased production of antigen and a modulation of the CpG motifs that further strengthened the in vivo effect of the vaccine and generated a stronger immunity.
To evaluate the presence of memory T cells, long-term survivors were re-challenged 4 weeks after the end of the experiment in the other flank with 106 P815 mastocytoma cells. All the mice rejected the tumor 8–10 days after the re-injection (data not shown). This result is representative of the generation of long-lasting immunity, a critical feature for successful vaccinations.
CD8+ Lymphocyte Infiltration in Tumor Was Higher Using CO Plasmids with Higher CpG Content
To evaluate the potency of the anti-tumor immune response in mice that received a prophylactic vaccination but did not survive to the P815 challenge, tumors were collected and analyzed by immunohistochemistry and the CD8+ lymphocyte infiltration was evaluated. Figure 5 depicts a series of representative sections for all the groups of animals (n = 3 for all the groups, except L1210.P1A.B7.1, as only one mouse developed the tumor in this group). CD3 (green) and CD8 (red) staining were merged (yellow) in order to exclude natural killer (NK) cells from the CD8+ T lymphocyte evaluation. In the groups treated with CO plasmids containing 21 or 50 CpG motifs, more CD8+ cells were observed than in the untreated group or the group vaccinated with the non-optimized plasmid or with the CO0. Furthermore, in the CO21- and CO50-treated groups, the formation of several CD3+/CD8+ aggregates all over the tumor was detected, especially in the tumor stroma and in the invasion front of the tumor. Again, the highest CD3+/CD8+ tumor infiltration was reached using the CO50 vaccine. Conversely, in the untreated and 21 groups, the presence of CD3+/CD8+ cells was sporadic. An elevated presence of CD8+ T lymphocytes in the tumor stroma of the CO21 and CO50 groups, but not in the untreated, CO0, or 21 groups, was confirmed by H&E staining with 3,3'-diaminobenzidine (DAB) directed against the anti-CD8+ antibody (data not shown). The images from the only mouse treated with L1210.P1A.B7.1 cells that developed the tumor are also shown in Figure 5, and the result was similar to the CO50 group.
Figure 5.
Representative Images of Immunostaining of CD3 and CD8 Cells in Tumors
Mice were injected and electroporated in the tibial muscle with the different P1A vaccines (CO0, CO21, CO50, and 21) or injected intraperitoneally with L1210.P1A.B7.1 cells (positive control) and compared to the untreated group. When the mice were sacrificed, tumors were harvested and fluorescent immunohistochemistry was performed for CD3+ (FITC, green) and CD8+ (APC, red) T cells. Sections were counterstained with DAPI. The final pictures represent merged images. The objective used was 10× and the scale bar represents 100 μm; n = 3 for all the groups, except L1210.P1A.B7.1 (n = 1), as only one mouse developed the tumor.
Recently, Sobottka et al.58 showed that a higher number of CD8+ tumor-infiltrating lymphocytes, especially infiltrative-margin lymphocytes (i.e., lymphocytes resident in the peripheral areas of the invasive tumor cells representing the invasion front of the tumor), was associated with increased disease-free survival in human breast cancer.58 Hence, even in mice that eventually developed the tumor, the elevated lymphocyte infiltration obtained in mice treated with the CO vaccines containing the higher number of CpG motifs could be responsible for the slower tumor growth and the prolonged survival time.
Therapeutic Vaccination Using the CO P1A-Encoding Vaccine with the Highest Number of CpG Motifs Decreased Tumor Growth Rate and Prolonged Mouse Survival
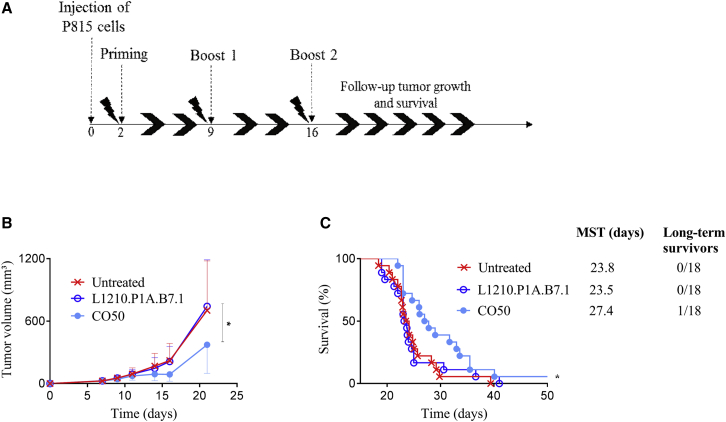
To assess the therapeutic potential of P1A vaccines, the best plasmid from the prophylactic vaccination, pVAX2-P1A_CO50, was evaluated. P815 tumor cells were injected 2 days before the priming with the vaccine, followed by two boosts administered weekly (Figure 6A). At 2 days after the tumor cell injection, all the mice developed a tumor and no difference was seen until day 10, when the tumor growth rate decreased for the CO50-treated group, reaching a plateau between days 10 and 16 (Figure 6B). At day 16, the tumor volume in the CO50-treated mice was approximately half of the tumor volume in the untreated mice. Surprisingly, the L1210.P1A.B7.1 treatment was ineffective in delaying the tumor growth despite its efficacy in the prophylactic approach. Starting from day 16, all the tumors grew approximately at the same rate, leaving a gap between the CO50-treated group and the others. Furthermore, treatment with the CO50 vaccine led to an increase in mouse survival (Figure 6C). However, only 6% of mice completely rejected the tumor, suggesting the necessity of a combined therapy to counteract the tumor-immunosuppressing microenvironment.
Figure 6.
P815 Injection followed by Immunization with Intramuscular Electroporation of CO50 or Intraperitoneal Injection of L1210.P1A.B7.1 Cells
(A) Therapeutic vaccination protocol. (B) Evolution of tumor volume (mm3) after P815 challenge as a function of time (days) (mean ± SEM). Statistical analysis is compared to the untreated group: two-way ANOVA, column factor (*p < 0.05). (C) Survival curve representing the percentage of mice alive (%) as a function of time (days). Statistical analysis, log rank (Mantel-Cox) test (*p < 0.05); n = 18 for all the groups.
Although several DNA vaccines directed against P815 have been already tested in prophylactic cancer vaccination,56, 59, 60 only one previous study has obtained a promising result after a therapeutic vaccination. In 2004, Ni et al.61 tested P1A replicon DNA as a therapeutic vaccine, and they found a significant increase in mouse survival. However, the dose of P815 cells injected was lower (5 × 104 cells) than the dose used in this study (106 cells). Other studies have demonstrated the potential of P1A vaccines for a therapeutic approach if used in combination with adjuvants or other therapies that can slow tumor development.62, 63 Here the administration by EP of an optimized P1A vaccine alone effectively increased the survival of the tumor-bearing DBA/2 mice after P815 tumor injection.
Conclusions
Despite promising results in preclinical models, the low immunogenicity of DNA vaccines currently limits their development as cancer vaccines for human applications. This study aimed to assess whether the optimization of the antigen gene sequence could improve the immune response generated by a P1A DNA cancer vaccine against the P815 tumor, a model for vaccines directed against human MAGE-type tumor antigens.55
Four P1A sequences were constructed and delivered in vivo by intramuscular EP. Three of them were CO and contained various numbers of CpG motifs. All the CO sequences were able to significantly delay the tumor growth and increase the survival of the mice following a prophylactic vaccination (Figure 4). Among the different P1A constructs, the most successful vaccine was the one that contained the highest number of CpG motifs. Indeed, this vaccine had the combined effect of enhanced antigen production coupled with the induction of IL-1β and IL-6 cytokine expression at the site of administration as well as greater CD8+ lymphocyte infiltration in the tumors of the vaccinated mice. The same vaccine was also able to delay the tumor growth and increase the survival of the mice under therapeutic vaccination conditions. For the therapeutic vaccination, the CO vaccine containing 50 CpG motifs in the P1A sequence was more efficient than the L1210.P1A.B7.1 positive control,57 which was completely ineffective in that context (Figure 6). However, vaccine therapy still needs to address the immunosuppressive tumor microenvironment, which prevents the establishment of a strong immunity in an established tumor. Hence, a combination of DNA vaccination with other treatment modalities that simultaneously aim to reduce tumor growth or inhibit the immunosuppressive tumor environment may be the best strategy to eradicate cancer and improve the efficacy of DNA cancer vaccines.
Materials and Methods
Plasmid Optimization and Production
Three CO P1A sequences with different numbers of CpG motifs were designed in silico, using the GeneOptimizer algorithm from GeneArt Gene Synthesis (Thermo Fisher Scientific), to enable codon optimization in mice.64 Then the genes were synthesized (Gene Art, Thermo Fisher Scientific) and subcloned into a pVAX2 vector containing the CMV promoter, as previously described.56 The plasmids were named pVAX2-P1A_CO0 (CO0), pVAX2-P1A_CO21 (CO21), and pVAX2-P1A_CO50 (CO50), where CO means that the P1A sequence has been CO and 0, 21, and 50 refer to the number of CpG motifs in the P1A sequence. A fourth non-optimized plasmid, pVAX2-P1A_21 (21), containing the wild-type P1A sequence was also used in this study. Upstream of the gene sequence, a Kozak sequence was inserted in order to improve the translation efficiency.65 For the analysis of in vitro expression, a 6X-His-tag motif was added downstream of the P1A gene sequence to facilitate protein quantification. To avoid any bias, no tag was included for the immunization experiments. All the sequences encoding the tumor rejection antigen P1A with the related modifications are shown in Figure 1. All plasmids were sequenced to ensure the correct nucleotide sequence (Beckman Coulter Genomics), and the vaccines were amplified and purified using an EndoFree Plasmid Giga Kit (QIAGEN), according to the manufacturer’s protocol. Optical density at 260 nm was used to determine DNA concentration. Plasmids were diluted in PBS and stored at −20°C before use.
Cell Lines
C2C12 murine myoblasts were kindly provided by Professor Marc Francaux (Université Catholique de Louvain). They were cultivated without reaching more than 70% confluence to avoid their differentiation into myotubes. P815B mastocytoma cells and L1210.P1A.B7.1 leukemia cells were obtained from Dr. Catherine Uyttenhove (Ludwig Institute for Cancer Research). L1210.P1A.B7.1 cells derived from a DBA/2 mouse were stably transfected to express the P1A antigen and the B7-1 costimulatory molecule.53 All the cells were cultured at 37°C in 5% CO2 in DMEM supplemented with 10% fetal calf serum and 1% penicillin/streptomycin. Specifically, L1210.P1A.B.7.1 cells were cultured adding 1.5 μg/mL puromycin.
Animals
DBA/2 female mice were obtained from Janvier. Mice were between 5 and 6 weeks old at the beginning of the experiments. Water and food were provided ad libitum. All experimental protocols using mice were approved by the Ethical Committee for Animal Care and Use of the Medical Sector of the Université Catholique de Louvain (UCL/MD/2011/007 and UCL/MD/2016/001).
In Vitro Plasmid Transfection
The 10 μL at the concentration of 1 μg/μL of the tagged plasmids was electroporated into 106 C2C12 cells suspended in 100 μL pulsing buffer (10 mM phosphate, 1 mM MgCl2, and 250 mM sucrose [pH 7.4]), using the following protocol: 230 V, 4 ms, and one pulse. EP was performed in a 2-mm gap cuvette in a BTX Gemini X2 Electroporation System (Thermo Fisher Scientific). After pulsing, 25 μL of fetal bovine serum (FBS) was added and the cuvette was incubated for 5 min at 37°C. Cells were suspended in media and seeded in a 96-well plate (106 cells/well).
Evaluation of Antigen Expression
qPCR
At 24 hr after transfection, cells were lysed and total RNA was isolated using TRIzol reagent (Thermo Fisher Scientific). The quality and quantity of RNA were evaluated using a nanospectrophotometer (NanoDrop 2000, Thermo Fisher Scientific). RNA (1 μg) was reverse transcribed using a first standard synthesis system (SuperScript, Thermo Fisher Scientific) and oligo(dT) primers (Eurogentec), according to the supplier’s protocol. The resulting cDNA was used as a template for 30 cycles of semiquantitative PCR in a T100 thermocycler (Bio-Rad). Primers for β-actin (housekeeping gene) and the four different P1A genes were used to amplify respective cDNA by PCR. The PCR products were subjected to electrophoresis on a SYBR Safe (Thermo Fisher Scientific) -stained 1.5% agarose gel.
SYBR green real-time qPCR (GoTaq qPCR MasterMix kit, Promega) was conducted on a StepOne Plus Real-Time PCR System (Thermo Fisher Scientific) in order to detect the expression of P1A in vitro 24 hr after vaccination and in vivo 6 hr after the vaccine injection in the tibial muscle of DBA/2 mice. Analysis of the melting curves was performed to ensure the purity of PCR products. The results were analyzed with the StepOne Software V2.1. The P1A mRNA expression was calculated relative to the expression of corresponding β-actin, according to the delta-delta Ct method, and all the results were normalized to non-transfected cells. Primers for P1A were designed using Primer Blast software based on the consensus of sequences from GenBank.
Western Blotting
P815A protein production was evaluated in C2C12 cells 24 hr after plasmid transfection. Cells were lysed and proteins isolated using radioimmunoprecipitation assay (RIPA) buffer (Thermo Fisher Scientific), according to manufacturer’s protocol. Proteins were quantified using a micro bicinchoninic acid (BCA) test (Thermo Fisher Scientific). Purified proteins (10 μg) were loaded in a Mini-PROTEAN TGX Precast Gel (Bio-Rad) and separated at 300 V for 10 min. Proteins were transferred onto a 0.2-μm nitrocellulose membrane (Bio-Rad), blocked with 5% milk in 0.05% Tween 20-PBS for 1 hr at room temperature, and washed with a solution of 0.05% Tween 20-TBS. Monoclonal antibody (Mab) against 6X-His-tag (Abcam) diluted 1:1,000 in 0.2% milk was added in order to detect P815A and incubated for 2 hr at room temperature with shaking. Mab anti-actin was used as the control (Abcam). After washing with Tween 20-TBS, the membrane was incubated with a solution of streptavidine-horseradish peroxidase (HRP) (R&D Systems, dilution 1:200) for 20 min at room temperature and then washed with 0.05% Tween 20-TBS. Membranes were visualized using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific) and X-ray Film for Western Blot Detection (Thermo Fisher Scientific). Dot values were semi-quantitated with GelQuantNET software and the actin dots were used for normalization. Non-transfected C2C12 cell lysate was used as the negative control (Ctr).
In Vivo Delivery of DNA Vaccine
Mice were anesthetized with 140–150 μL solution of 10 mg/mL ketamine (Ketalar, Pfizer) and 1 mg/mL xylazine (Sigma). The left paw was shaved using a rodent shaver (Aesculap Exacta shaver, AgnTho’s). DBA/2 mice were vaccinated with P1A-encoding plasmids: 50 μg plasmid diluted in 30 μL PBS was injected in the left tibial cranial muscle and electroporated (200 V/cm, eight pulses, 20 ms with 500-ms pause between pulses), as described in Vandermeulen et al.56 Briefly, after the vaccine injection, a conductive gel was placed on the left paw to ensure electrical contact with the skin (Aquasonic 100, ultrasound transmission gel, Parker Labs). The paw was then placed between 4-mm plate BTX caliper electrodes (VWR International). The pulses were delivered by a BTX Gemini Electroporation System (VWR International).
Cytokine Expression
The mice were sacrificed 6 hr after a single vaccine injection. The tibial cranial muscle was withdrawn and stored in RNA later solution (Thermo Fisher Scientific) at −20°C before RNA extraction, according to the previous protocol (see qPCR above). The muscle was used to detect the local expression of inflammatory cytokines by qPCR. Primers for TNF-α and IL-6 were designed using Primer Blast software based on the consensus of sequences from GenBank. The other primers can be found elsewhere.66, 67 A complete list of the primers used is shown in Table 1. All experiments were performed in triplicate.
Table 1.
Primer Sequences for PCR
| Oligo Name | Primer Sequence (5′ → 3′) |
|---|---|
| CO0-tag forward | AGA-TGG-GGA-TGG-CAA-CAG-ATG |
| CO0-tag reverse | GGC-CAC-ATC-CCT-CTC-ATA-CT |
| CO21-tag forward | GGA-AGA-GAT-CCT-GCC-CTA-CC |
| CO21-tag reverse | CTG-TTC-CTC-ATA-CAG-GGC-GT |
| CO50-tag forward | GCG-ACG-GCA-ACA-GAT-GTA-AC |
| CO50-tag reverse | GTA-CAG-GGC-GTC-GAT-GAA-CA |
| 21-tag forward | CCA-CGA-CCC-TAA-TTT-CCT-GGT |
| 21-tag reverse | GTG-GTG-ATG-GTG-ATG-ATG-AGG-T |
| β-actin forward | TAC-AAT-GAG-CTG-CGT-GTG-GCC-C |
| β-actin reverse | AGG-ATG-GCG-TGA-GGG-AGA-GCA-T |
| IL-6 forward | CCG-GAG-AGG-AGA-CTT-CAC-AG |
| IL-6 reverse | TCC-ACG-ATT-TCC-CAG-AGA-AC |
| TNF-α forward | CAT-CTT-CTC-AAA-ATT-CGA-GTG-ACA-A |
| TNF-α reverse | TGG-GAG-TAG-ACA-AGG-TAC-AAC-CC |
| IL-1β forward | AAC-TGT-TCC-TGA-ACT-CAA-CTG-T |
| IL-1β reverse | GAG-ATT-TGA-AGC-TGG-ATG-CTC-T |
| IL-12 forward | GGA-AGC-ACG-GCA-GCA-GAA-TA |
| IL-12 reverse | AAC-TTG-AGG-GAG-AAG-TAG-GAA-TGG |
Prophylactic and Therapeutic Vaccinations
Each mouse received three vaccine administrations (one priming and two boosts). Vaccine injections were performed biweekly before the tumor challenge (n = 9–10) or weekly after the tumor challenge (n = 18) for the prophylactic and therapeutic DNA immunization studies, respectively (Figures 4A and 6A, respectively). The therapeutic study is a combination of two separate experiments; the total number of mice is 18. As a positive control, mice were immunized with two intra-peritoneal injections of L1210.P1A.B.7.1 cells (106 living cells) in 100 μL PBS at an interval of 2 weeks or 1 week for prophylactic56, 57 and therapeutic vaccination, respectively. L1210.P1A.B7.1 positive control is a leukemia cell line derived from DBA/2 mice. These cells were stably transfected with the cosmid C1A.3.1 and the cDNA of B7, as described in Gajewski et al.53 Non-immunized mice were used as a negative control.
Tumor Implantation and Tumor Growth Measurement
At 2 weeks after the second boost (prophylactic) or 2 days before the priming (therapeutic), 106 P1A-expressing P815B cells diluted in 100 μL PBS were injected into the right flank of mice. An electronic digital caliper was used to measure the tumor length, width, and height three times a week. Tumor volume was calculated as length × width × height (in mm3). Mice were sacrificed when the tumor volume was larger than 1,500 mm3 or when they were in poor condition and expected to die shortly.
Immediately after sacrifice, tumors from mice that received a prophylactic vaccination were withdrawn and immediately fixed overnight in 4% paraformaldehyde (PFA) and then cryopreserved in 30% sucrose before Tissue-Tek OCT embedding (Sakura Finetek). At 90 days after the first challenge, surviving mice were re-challenged on the left flank using the same protocol as the first challenge.
Immunohistochemistry
To determine the tumor CD8+ T lymphocyte infiltration, tumors embedded in OCT were sectioned at 10 μm using a cryostat (Leica Microsystems), and the sections were stained with antibodies directed against the murine CD3 and CD8. After permeabilization with 0.2% (v/v) Triton X-100 (Sigma) in PBS and blocking in 10% (w/v) goat serum, 5% rat serum, and 2% BSA in PBS for 1 hr at room temperature, the primary antibodies (rat CD8a-allophycocyanin [APC] 1:250 [clone 53-6.7, BD Biosciences] and CD3e-fluorescein isothiocyanate (FITC) 1:1,000 [clone 145-2C11, BD Biosciences]) were applied to the slides for 1 hr at room temperature. After being washed with PBS, the sections were mounted using Vectashield Mounting Medium (Vector Laboratories) containing DAPI to visualize the cell nuclei. The slides were imaged using a structured illumination AxioImager microscope (Zeiss, 10× objective).
Statistical Analysis
Statistical analyses were performed using the software GraphPad Prism 5 for Windows. Survival curves were compared using a Mantel-Cox (log rank) test. *p < 0.05, **p < 0.01, and ***p < 0.001 were indicative of statistically significant differences.
Author Contributions
A.L., G.V., and V.P. designed the study. A.L., G.V., and K.V. collected the data. A.L. and G.V. analyzed and interpreted the data. A.L., G.V., and V.P. wrote the manuscript that was validated by all authors.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
The authors thank Caroline Bouzin and Nicolas Dauguet for their interesting suggestions. G.V. is supported by a FIRST spin-off grant 1610437 from the Walloon Region.
References
- 1.Tiptiri-Kourpeti A., Spyridopoulou K., Pappa A., Chlichlia K. DNA vaccines to attack cancer: Strategies for improving immunogenicity and efficacy. Pharmacol. Ther. 2016;165:32–49. doi: 10.1016/j.pharmthera.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Herrada A.A., Rojas-Colonelli N., González-Figueroa P., Roco J., Oyarce C., Ligtenberg M.A., Lladser A. Harnessing DNA-induced immune responses for improving cancer vaccines. Hum. Vaccin. Immunother. 2012;8:1682–1693. doi: 10.4161/hv.22345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rocha J., Cicéron F., de Sanctis D., Lelimousin M., Chazalet V., Lerouxel O., Breton C. Structure of Arabidopsis thaliana FUT1 Reveals a Variant of the GT-B Class Fold and Provides Insight into Xyloglucan Fucosylation. Plant Cell. 2016;28:2352–2364. doi: 10.1105/tpc.16.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kobiyama K., Jounai N., Aoshi T., Tozuka M., Takeshita F., Coban C., Ishii K.J. Innate Immune Signaling by, and Genetic Adjuvants for DNA Vaccination. Vaccines (Basel) 2013;1:278–292. doi: 10.3390/vaccines1030278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li L.L., Wang H.R., Zhou Z.Y., Luo J., Xiao X.Q., Wang X.L., Li J.T., Zhou Y.B., Zeng Y. One-prime multi-boost strategy immunization with recombinant DNA, adenovirus, and MVA vector vaccines expressing HPV16 L1 induces potent, sustained, and specific immune response in mice. Antiviral Res. 2016;128:20–27. doi: 10.1016/j.antiviral.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 6.Seo S.H., Jin H.T., Park S.H., Youn J.I., Sung Y.C. Optimal induction of HPV DNA vaccine-induced CD8+ T cell responses and therapeutic antitumor effect by antigen engineering and electroporation. Vaccine. 2009;27:5906–5912. doi: 10.1016/j.vaccine.2009.07.033. [DOI] [PubMed] [Google Scholar]
- 7.Saade F., Petrovsky N. Technologies for enhanced efficacy of DNA vaccines. Expert Rev. Vaccines. 2012;11:189–209. doi: 10.1586/erv.11.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benteyn D., Anguille S., Van Lint S., Heirman C., Van Nuffel A.M., Corthals J., Ochsenreither S., Waelput W., Van Beneden K., Breckpot K. Design of an Optimized Wilms’ Tumor 1 (WT1) mRNA Construct for Enhanced WT1 Expression and Improved Immunogenicity In Vitro and In Vivo. Mol. Ther. Nucleic Acids. 2013;2:e134. doi: 10.1038/mtna.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang S., Hackett A., Jia N., Zhang C., Zhang L., Parker C., Zhou A., Li J., Cao W.C., Huang Z. Polyvalent DNA vaccines expressing HA antigens of H5N1 influenza viruses with an optimized leader sequence elicit cross-protective antibody responses. PLoS ONE. 2011;6:e28757. doi: 10.1371/journal.pone.0028757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stachyra A., Redkiewicz P., Kosson P., Protasiuk A., Góra-Sochacka A., Kudla G., Sirko A. Codon optimization of antigen coding sequences improves the immune potential of DNA vaccines against avian influenza virus H5N1 in mice and chickens. Virol. J. 2016;13:143. doi: 10.1186/s12985-016-0599-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamdan F.F., Mousa A., Ribeiro P. Codon optimization improves heterologous expression of a Schistosoma mansoni cDNA in HEK293 cells. Parasitol. Res. 2002;88:583–586. doi: 10.1007/s00436-001-0585-0. [DOI] [PubMed] [Google Scholar]
- 12.Robinson F., Jackson R.J., Smith C.W.J. Expression of human nPTB is limited by extreme suboptimal codon content. PLoS ONE. 2008;3:e1801. doi: 10.1371/journal.pone.0001801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kojima Y., Xin K.Q., Ooki T., Hamajima K., Oikawa T., Shinoda K., Ozaki T., Hoshino Y., Jounai N., Nakazawa M. Adjuvant effect of multi-CpG motifs on an HIV-1 DNA vaccine. Vaccine. 2002;20:2857–2865. doi: 10.1016/s0264-410x(02)00238-4. [DOI] [PubMed] [Google Scholar]
- 14.Jorritsma S.H., Gowans E.J., Grubor-Bauk B., Wijesundara D.K. Delivery methods to increase cellular uptake and immunogenicity of DNA vaccines. Vaccine. 2016;34:5488–5494. doi: 10.1016/j.vaccine.2016.09.062. [DOI] [PubMed] [Google Scholar]
- 15.Kusumanto Y.H., Mulder N.H., Dam W.A., Losen M., De Baets M.H., Meijer C., Hospers G.A. Improvement of in vivo transfer of plasmid DNA in muscle: comparison of electroporation versus ultrasound. Drug Deliv. 2007;14:273–277. doi: 10.1080/10717540601098807. [DOI] [PubMed] [Google Scholar]
- 16.Lambricht L., Lopes A., Kos S., Sersa G., Préat V., Vandermeulen G. Clinical potential of electroporation for gene therapy and DNA vaccine delivery. Expert Opin. Drug Deliv. 2016;13:295–310. doi: 10.1517/17425247.2016.1121990. [DOI] [PubMed] [Google Scholar]
- 17.Yarmush M.L., Golberg A., Serša G., Kotnik T., Miklavčič D. Electroporation-based technologies for medicine: principles, applications, and challenges. Annu. Rev. Biomed. Eng. 2014;16:295–320. doi: 10.1146/annurev-bioeng-071813-104622. [DOI] [PubMed] [Google Scholar]
- 18.Kang T.H., Monie A., Wu L.S., Pang X., Hung C.F., Wu T.C. Enhancement of protein vaccine potency by in vivo electroporation mediated intramuscular injection. Vaccine. 2011;29:1082–1089. doi: 10.1016/j.vaccine.2010.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel V., Valentin A., Kulkarni V., Rosati M., Bergamaschi C., Jalah R., Alicea C., Minang J.T., Trivett M.T., Ohlen C. Long-lasting humoral and cellular immune responses and mucosal dissemination after intramuscular DNA immunization. Vaccine. 2010;28:4827–4836. doi: 10.1016/j.vaccine.2010.04.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Best S.R., Peng S., Juang C.-M., Hung C.-F., Hannaman D., Saunders J.R., Wu T.C., Pai S.I. Administration of HPV DNA vaccine via electroporation elicits the strongest CD8+ T cell immune responses compared to intramuscular injection and intradermal gene gun delivery. Vaccine. 2009;27:5450–5459. doi: 10.1016/j.vaccine.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uyttenhove C., Godfraind C., Lethé B., Amar-Costesec A., Renauld J.-C., Gajewski T.F., Duffour M.T., Warnier G., Boon T., Van den Eynde B.J. The expression of mouse gene P1A in testis does not prevent safe induction of cytolytic T cells against a P1A-encoded tumor antigen. Int. J. Cancer. 1997;70:349–356. doi: 10.1002/(sici)1097-0215(19970127)70:3<349::aid-ijc17>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 22.Rosato A., Milan G., Collavo D., Zanovello P. DNA-based vaccination against tumors expressing the P1A antigen. Methods. 1999;19:187–190. doi: 10.1006/meth.1999.0844. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Z., Dang Y., Zhou M., Li L., Yu C.H., Fu J., Chen S., Liu Y. Codon usage is an important determinant of gene expression levels largely through its effects on transcription. Proc. Natl. Acad. Sci. USA. 2016;113:E6117–E6125. doi: 10.1073/pnas.1606724113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hyde S.C., Pringle I.A., Abdullah S., Lawton A.E., Davies L.A., Varathalingam A., Nunez-Alonso G., Green A.M., Bazzani R.P., Sumner-Jones S.G. CpG-free plasmids confer reduced inflammation and sustained pulmonary gene expression. Nat. Biotechnol. 2008;26:549–551. doi: 10.1038/nbt1399. [DOI] [PubMed] [Google Scholar]
- 25.Bauer A.P., Leikam D., Krinner S., Notka F., Ludwig C., Längst G., Wagner R. The impact of intragenic CpG content on gene expression. Nucleic Acids Res. 2010;38:3891–3908. doi: 10.1093/nar/gkq115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Low P.T., Lai M.I., Ngai S.C., Abdullah S. Transgene expression from CpG-reduced lentiviral gene delivery vectors in vitro. Gene. 2014;533:451–455. doi: 10.1016/j.gene.2013.09.075. [DOI] [PubMed] [Google Scholar]
- 27.Siegismund C.S., Hohn O., Kurth R., Norley S. Enhanced T- and B-cell responses to simian immunodeficiency virus (SIV)agm, SIVmac and human immunodeficiency virus type 1 Gag DNA immunization and identification of novel T-cell epitopes in mice via codon optimization. J. Gen. Virol. 2009;90:2513–2518. doi: 10.1099/vir.0.013730-0. [DOI] [PubMed] [Google Scholar]
- 28.Deml L., Bojak A., Steck S., Graf M., Wild J., Schirmbeck R., Wolf H., Wagner R. Multiple effects of codon usage optimization on expression and immunogenicity of DNA candidate vaccines encoding the human immunodeficiency virus type 1 Gag protein. J. Virol. 2001;75:10991–11001. doi: 10.1128/JVI.75.22.10991-11001.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ko H.-J., Ko S.-Y., Kim Y.-J., Lee E.-G., Cho S.-N., Kang C.-Y. Optimization of codon usage enhances the immunogenicity of a DNA vaccine encoding mycobacterial antigen Ag85B. Infect. Immun. 2005;73:5666–5674. doi: 10.1128/IAI.73.9.5666-5674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barber G.N. Cytoplasmic DNA innate immune pathways. Immunol. Rev. 2011;243:99–108. doi: 10.1111/j.1600-065X.2011.01051.x. [DOI] [PubMed] [Google Scholar]
- 31.Suschak J.J., Wang S., Fitzgerald K.A., Lu S. A cGAS-Independent STING/IRF7 Pathway Mediates the Immunogenicity of DNA Vaccines. J. Immunol. 2016;196:310–316. doi: 10.4049/jimmunol.1501836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mann C.J., Anguela X.M., Montané J., Obach M., Roca C., Ruzo A., Otaegui P., Mir L.M., Bosch F. Molecular signature of the immune and tissue response to non-coding plasmid DNA in skeletal muscle after electrotransfer. Gene Ther. 2012;19:1177–1186. doi: 10.1038/gt.2011.198. [DOI] [PubMed] [Google Scholar]
- 33.Znidar K., Bosnjak M., Cemazar M., Heller L.C. Cytosolic DNA Sensor Upregulation Accompanies DNA Electrotransfer in B16.F10 Melanoma Cells. Mol. Ther. Nucleic Acids. 2016;5:e322. doi: 10.1038/mtna.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Markelc B., Skvarca E., Dolinsek T., Kloboves V.P., Coer A., Sersa G., Cemazar M. Inhibitor of endocytosis impairs gene electrotransfer to mouse muscle in vivo. Bioelectrochemistry. 2015;103:111–119. doi: 10.1016/j.bioelechem.2014.08.020. [DOI] [PubMed] [Google Scholar]
- 35.Rosazza C., Phez E., Escoffre J.M., Cézanne L., Zumbusch A., Rols M.P. Cholesterol implications in plasmid DNA electrotransfer: Evidence for the involvement of endocytotic pathways. Int. J. Pharm. 2012;423:134–143. doi: 10.1016/j.ijpharm.2011.05.024. [DOI] [PubMed] [Google Scholar]
- 36.Šimčíková M., Prather K.L.J., Prazeres D.M.F., Monteiro G.A. Towards effective non-viral gene delivery vector. Biotechnol. Genet. Eng. Rev. 2015;31:82–107. doi: 10.1080/02648725.2016.1178011. [DOI] [PubMed] [Google Scholar]
- 37.Mitsui M., Nishikawa M., Zang L., Ando M., Hattori K., Takahashi Y., Watanabe Y., Takakura Y. Effect of the content of unmethylated CpG dinucleotides in plasmid DNA on the sustainability of transgene expression. J. Gene Med. 2009;11:435–443. doi: 10.1002/jgm.1317. [DOI] [PubMed] [Google Scholar]
- 38.Coban C., Ishii K.J., Gursel M., Klinman D.M., Kumar N. Effect of plasmid backbone modification by different human CpG motifs on the immunogenicity of DNA vaccine vectors. J. Leukoc. Biol. 2005;78:647–655. doi: 10.1189/jlb.1104627. [DOI] [PubMed] [Google Scholar]
- 39.Yew N.S., Wang K.X., Przybylska M., Bagley R.G., Stedman M., Marshall J., Scheule R.K., Cheng S.H. Contribution of plasmid DNA to inflammation in the lung after administration of cationic lipid:pDNA complexes. Hum. Gene Ther. 1999;10:223–234. doi: 10.1089/10430349950019011. [DOI] [PubMed] [Google Scholar]
- 40.Krieg A.M. CpG motifs in bacterial DNA and their immune effects. Annu. Rev. Immunol. 2002;20:709–760. doi: 10.1146/annurev.immunol.20.100301.064842. [DOI] [PubMed] [Google Scholar]
- 41.Kant R., de Vos W.M., Palva A., Satokari R. Immunostimulatory CpG motifs in the genomes of gut bacteria and their role in human health and disease. J. Med. Microbiol. 2014;63:293–308. doi: 10.1099/jmm.0.064220-0. [DOI] [PubMed] [Google Scholar]
- 42.Kwon H.-J., Lee K.-W., Yu S.H., Han J.H., Kim D.-S. NF-kappaB-dependent regulation of tumor necrosis factor-α gene expression by CpG-oligodeoxynucleotides. Biochem. Biophys. Res. Commun. 2003;311:129–138. doi: 10.1016/j.bbrc.2003.09.168. [DOI] [PubMed] [Google Scholar]
- 43.Kaminski J.J., Schattgen S.A., Tzeng T.C., Bode C., Klinman D.M., Fitzgerald K.A. Synthetic oligodeoxynucleotides containing suppressive TTAGGG motifs inhibit AIM2 inflammasome activation. J. Immunol. 2013;191:3876–3883. doi: 10.4049/jimmunol.1300530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roos A.-K., Eriksson F., Timmons J.A., Gerhardt J., Nyman U., Gudmundsdotter L., Bråve A., Wahren B., Pisa P. Skin electroporation: effects on transgene expression, DNA persistence and local tissue environment. PLoS ONE. 2009;4:e7226. doi: 10.1371/journal.pone.0007226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lambricht L., Vanvarenberg K., De Beuckelaer A., Van Hoecke L., Grooten J., Ucakar B., Lipnik P., Sanders N.N., Lienenklaus S., Préat V., Vandermeulen G. Coadministration of a Plasmid Encoding HIV-1 Gag Enhances the Efficacy of Cancer DNA Vaccines. Mol. Ther. 2016;24:1686–1696. doi: 10.1038/mt.2016.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klinman D.M., Klaschik S., Sato T., Tross D. CpG oligonucleotides as adjuvants for vaccines targeting infectious diseases. Adv. Drug Deliv. Rev. 2009;61:248–255. doi: 10.1016/j.addr.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 47.Sato T., Shimosato T., Ueda A., Ishigatsubo Y., Klinman D.M. Intrapulmonary Delivery of CpG Microparticles Eliminates Lung Tumors. Mol. Cancer Ther. 2015;14:2198–2205. doi: 10.1158/1535-7163.MCT-15-0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shirota H., Tross D., Klinman D.M. CpG Oligonucleotides as Cancer Vaccine Adjuvants. Vaccines (Basel) 2015;3:390–407. doi: 10.3390/vaccines3020390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Cesare M., Sfondrini L., Pennati M., De Marco C., Motta V., Tagliabue E., Deraco M., Balsari A., Zaffaroni N. CpG-oligodeoxynucleotides exert remarkable antitumor activity against diffuse malignant peritoneal mesothelioma orthotopic xenografts. J. Transl. Med. 2016;14:25. doi: 10.1186/s12967-016-0781-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho H.C., Kim B.H., Kim K., Park J.Y., Chang J.H., Kim S.K. Cancer immunotherapeutic effects of novel CpG ODN in murine tumor model. Int. Immunopharmacol. 2008;8:1401–1407. doi: 10.1016/j.intimp.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 51.Murad Y.M., Clay T.M. CpG oligodeoxynucleotides as TLR9 agonists: therapeutic applications in cancer. BioDrugs. 2009;23:361–375. doi: 10.2165/11316930-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 52.Morecki S., Slavin S. Immunoregulation of GVHD by triggering the innate immune system with CpG. Expert Rev. Hematol. 2009;2:443–453. doi: 10.1586/ehm.09.29. [DOI] [PubMed] [Google Scholar]
- 53.Gajewski T.F., Renauld J.C., Van Pel A., Boon T. Costimulation with B7-1, IL-6, and IL-12 is sufficient for primary generation of murine antitumor cytolytic T lymphocytes in vitro. J. Immunol. 1995;154:5637–5648. [PubMed] [Google Scholar]
- 54.Ramanathan S., Gagnon J., Ilangumaran S. Antigen-nonspecific activation of CD8+ T lymphocytes by cytokines: relevance to immunity, autoimmunity, and cancer. Arch. Immunol. Ther. Exp. (Warsz.) 2008;56:311–323. doi: 10.1007/s00005-008-0033-2. [DOI] [PubMed] [Google Scholar]
- 55.Brändle D., Bilsborough J., Rülicke T., Uyttenhove C., Boon T., Van den Eynde B.J. The shared tumor-specific antigen encoded by mouse gene P1A is a target not only for cytolytic T lymphocytes but also for tumor rejection. Eur. J. Immunol. 1998;28:4010–4019. doi: 10.1002/(SICI)1521-4141(199812)28:12<4010::AID-IMMU4010>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 56.Vandermeulen G., Uyttenhove C., De Plaen E., Van den Eynde B.J., Préat V. Intramuscular electroporation of a P1A-encoding plasmid vaccine delays P815 mastocytoma growth. Bioelectrochemistry. 2014;100:112–118. doi: 10.1016/j.bioelechem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 57.Näslund T.I., Uyttenhove C., Nordström E.K.L., Colau D., Warnier G., Jondal M., Van den Eynde B.J., Liljeström P. Comparative prime-boost vaccinations using Semliki Forest virus, adenovirus, and ALVAC vectors demonstrate differences in the generation of a protective central memory CTL response against the P815 tumor. J. Immunol. 2007;178:6761–6769. doi: 10.4049/jimmunol.178.11.6761. [DOI] [PubMed] [Google Scholar]
- 58.Sobottka B., Pestalozzi B., Fink D., Moch H., Varga Z. Similar lymphocytic infiltration pattern in primary breast cancer and their corresponding distant metastases. OncoImmunology. 2016;5:e1153208. doi: 10.1080/2162402X.2016.1153208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malm M., Sikut R., Krohn K., Blazevic V. GTU-MultiHIV DNA vaccine results in protection in a novel P815 tumor challenge model. Vaccine. 2007;25:3293–3301. doi: 10.1016/j.vaccine.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 60.Wu Y., Wan Y., Bian J., Zhao J., Jia Z., Zhou L., Zhou W., Tan Y. Phage display particles expressing tumor-specific antigens induce preventive and therapeutic anti-tumor immunity in murine p815 model. Int. J. Cancer. 2002;98:748–753. doi: 10.1002/ijc.10260. [DOI] [PubMed] [Google Scholar]
- 61.Ni B., Lin Z., Zhou L., Wang L., Jia Z., Zhou W., Diciommo D.P., Zhao J., Bremner R., Wu Y. Induction of P815 tumor immunity by DNA-based recombinant Semliki Forest virus or replicon DNA expressing the P1A gene. Cancer Detect. Prev. 2004;28:418–425. doi: 10.1016/j.cdp.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 62.Liu H.-L., Wu Y.-Z., Zhao J.-P., Ni B., Jia Z.-C., Zhou W., Zou L.Y. Effective elicitation of anti-tumor immunity by collocation of antigen with encoding gene in the same vaccine. Immunol. Lett. 2003;89:167–173. doi: 10.1016/s0165-2478(03)00132-9. [DOI] [PubMed] [Google Scholar]
- 63.Kano Y., Iguchi T., Matsui H., Adachi K., Sakoda Y., Miyakawa T., Doi S., Hazama S., Nagano H., Ueyama Y., Tamada K. Combined adjuvants of poly(I:C) plus LAG-3-Ig improve antitumor effects of tumor-specific T cells, preventing their exhaustion. Cancer Sci. 2016;107:398–406. doi: 10.1111/cas.12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raab D., Graf M., Notka F., Schödl T., Wagner R. The GeneOptimizer Algorithm: using a sliding window approach to cope with the vast sequence space in multiparameter DNA sequence optimization. Syst. Synth. Biol. 2010;4:215–225. doi: 10.1007/s11693-010-9062-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Olafsdóttir G., Svansson V., Ingvarsson S., Marti E., Torsteinsdóttir S. In vitro analysis of expression vectors for DNA vaccination of horses: the effect of a Kozak sequence. Acta Vet. Scand. 2008;50:44. doi: 10.1186/1751-0147-50-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Z., Wu X., Zhang Y., Zhou L., Li L., Yu Y., Wang L. Discrepant roles of CpG ODN on acute alcohol-induced liver injury in mice. Int. Immunopharmacol. 2012;12:526–533. doi: 10.1016/j.intimp.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 67.Giulietti A., Overbergh L., Valckx D., Decallonne B., Bouillon R., Mathieu C. An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods. 2001;25:386–401. doi: 10.1006/meth.2001.1261. [DOI] [PubMed] [Google Scholar]