Abstract
We have studied the molecular properties of in-vitro-transcribed sliced small interfering RNAs (tsli-siRNAs) as an alternative RNAi agent for chemically synthesized siRNA. We describe here a simple and cost-effective procedure for high-purity production of tsli-siRNA using bacteriophage T7 RNA polymerases. tsli-siRNAs exhibit potent gene knockdown effects, with efficacy comparable with that of chemically synthesized sli-siRNAs and classical siRNAs. Furthermore, we found that it is very easy to prepare potent tsli-siRNAs with modified bases, such as 2′-fluorine- or biotin-16-modified tsli-siRNAs. tsli-siRNAs can cause a mild innate immune response, which can be easily eliminated by alkaline phosphatase treatment. On the other hand, this feature, which can be useful as a trigger of the innate immune response, can be enhanced by polynucleotide kinase treatment. Because of the simplicity of preparation and purification, the procedure presented here could be useful for the production of RNAi or immunostimulatory reagents.
Keywords: sliced siRNA, siRNA, T7 RNA polymerase, RNAi, Ago2, miR-451, pre-miR-451 mimic
Introduction
Gene silencing by small interfering RNA (siRNA) or RNAi is a powerful technology for manipulating gene expression and can serve as a potential therapeutic strategy for treating human diseases. It is also a novel approach for controlling pest insects and for the treatment and prevention of diseases in beneficial insects for crops. Canonical siRNAs are ∼21-nt small RNAs that mimic products of Dicer-processed double-strand RNAs (dsRNA) and can be incorporated into the RNA-induced silencing complex (RISC) to trigger the degradation of mRNA targets.1 siRNAs are generally prepared by chemical synthesis, which can be rather costly if large amounts are needed, such as for clinical studies, where an average of 0.5–1 mg/kg is needed,2 or for the management of insects in agriculture, where gram or even kilogram scales are necessary.3 Endoribonuclease-prepared siRNAs (esiRNAs) can be produced both in vivo and in vitro from endoribonuclease-processed dsRNAs, with cost that can go as low as $4 per gene using esiRNA prepared in an siRNA library.4, 5, 6, 7 siRNA can also be made by Pol III promoter-driven small hairpin RNA (shRNA).8 Currently, there are two mature and effective approaches for enzyme-based low-cost siRNA production in vitro. One is E. coli endoribonuclease III-produced esiRNAs that use fully complementary dsRNA as a template, which can be adapted to large-scale production.5, 7, 9 The other approach uses highly complex endoribonuclease T1-produced siRNA pools (siPools) that use partially complementary dsRNA as the template.6 Although both approaches produce siRNAs that can be used for effective target knockdown, it is unlikely that they can be used in clinical applications because of the fact that they produce a mixture of siRNAs.5, 6, 7 Uniform production of esiRNA can be made by bacteriophage RNA polymerase (RNAP) in vitro, but the resultant siRNAs need to be in a special format, starting with a G:C base pair and ending with a C:G base pair, because of the special requirements by bacteriophage RNAPs.10, 11, 12 To solve this problem, a leader sequence can be added to the siRNA sequence to produce transcripts that can be digested by deoxyribozyme or RNase H to produce esiRNAs with the desired sequence and length.13, 14 One interesting discovery from siRNA production in vitro using bacteriophage RNAPs is that esiRNAs could trigger a type I interferon response because of having a 5′ triphosphate (5′ppp).15 Later this phenomenon was found to be mediated by RIG-I (retinoic acid-inducible gene 1),16, 17, 18 and this feature was used to produce bifunctional siRNAs that can act as reagents for both RNAi and immunostimulation.19, 20 Usually esiRNA production needs two reactions, one for sense strand and one for antisense strand, with an exception being the T7 and phi6 RNA-dependent RNAP (RdRp) combination system that uses T7 to produce sense strand and phi6 RdRp to make antisense strand.5, 7, 9 Following transcription, these two products need to be annealed, and products longer than 23 nt need to be further digested by endoribonuclease to generate 21-mer final products. The final products need to be further purified by alcohol precipitation, which results in siRNAs with low yield and low purity, or by the labor-intensive and time-consuming PAGE purification that yields products with higher purity but lower yield. Because bacteriophage RNAPs usually add 1 or more nt to the full-size product,21, 22, 23, 24 mature siRNAs may have variable lengths.6 Length variation in siRNA is troublesome because 23-mers can trigger a much stronger interferon response than 21-mers despite being different by only 2 nt.25 Some of the above problems can be overcome by esiRNA made as in-vitro-transcribed shRNA.26 This kind of esiRNA can be processed to siRNA by Dicer in vitro or in vivo, but they have not been widely adapted because Dicer processing needs a longer stem, and the processing sites require specific motifs.27, 28 Extra nucleotides added to the 3′ end of the transcripts will also affect Dicer processing sites, and prematurely terminated transcripts will affect the sequence of antisense strand.
We have previously characterized the general molecular properties of sli-siRNAs, a class of small hairpin RNAs that mimics pre-miR-451 and can be processed into potent siRNA by Ago2, not necessarily by Dicer.29 Because the sense strand of sli-siRNAs or other forms of pre-miR-451 mimics need to be cleaved by Ago2 for their RISC activation, pre-miR-451 mimics have dramatically reduced unwanted sense strand activities.29, 30, 31, 32, 33, 34 This is in contrast with the classical 21-mer siRNAs, in which both strands could be equally loaded into RISC. Consequently, strong off-target effects can result from the sense strand. Moreover, competition between the sense strand and the antisense strand to be the guide strand for 21-mer siRNA can reduce the gene silencing potency from the antisense strand. Because of their relatively longer length and being only a single strand, sli-siRNA offers the unique feature that they can be more efficiently produced in vitro and conveniently purified. Furthermore, because the Ago2 processing step will remove nucleotides after the 30th nt, longer products due to extra nucleotide added to the full-length products at RNAPs termination or shorter products generated by RNAPs premature termination (up to 3 nt) will not be a major problem for sli-siRNA.29
We find that optimized sli-siRNAs can be quickly and efficiently produced in small or large scales using T7 RNAP, and importantly, because of their relatively longer length, transcribed sliced (tsli)-siRNAs can be conveniently purified by commercially available chromatography columns. Our reporter assay and endogenous target knockdown results have established that tsli-siRNAs are highly potent in gene silencing, similar to chemically synthesized sli-siRNAs (csli-siRNAs). We also have shown that it is very easy to make sli-siRNA with modified bases, such as 2′-fluorine- (2F-tsli-siRNA) or biotin (biotin-tsli-siRNA)-modified RNA bases. Like esiRNA, we have observed a mild innate immune response by some tsli-siRNAs, but this response can be easily eliminated by alkaline phosphatase treatment. Alternatively, the immunostimulatory feature of tsli-siRNAs can be easily enhanced by polynucleotide kinase treatment to make bifunctional tsli-siRNA that is potent for both RNAi and innate immune response. Because of the simplicity of preparation and purification, and their potency in gene silencing, tsli-siRNAs will be highly desirable for both small-scale laboratory production and large-scale production using regular nucleoside triphosphates (NTPs) or modified NTPs. The method of tsli-siRNA production described here can benefit RNAi usage for both research and clinical gene therapy, as well as for manipulating viral pathogenesis and controlling insects’ infection in agriculture.
Results
Optimized Molecular Structure of sli-siRNA for Ideal Production by T7 RNAP
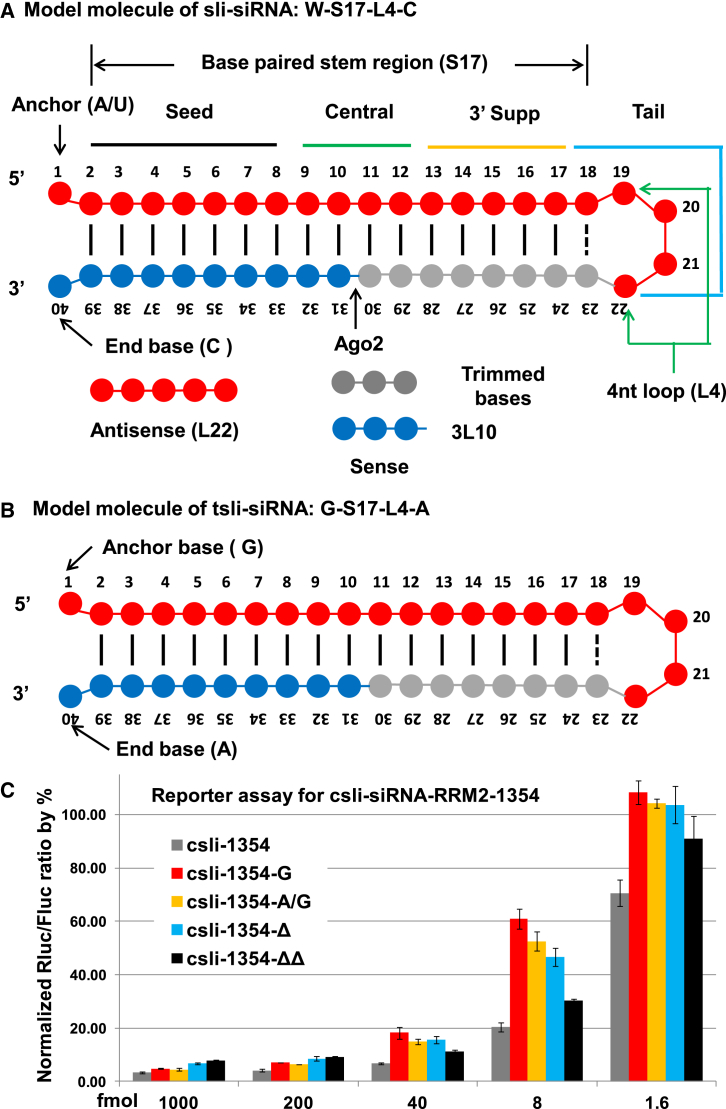
Our previously characterized canonical structure for csli-siRNA is W-S17-L4-C (Figure 1A) with the flexibility that either the 5′ end or the 3′ end can have up to 2 extra nt, or the 3′ end can be truncated by up to 3 nt without significant loss of potency. W (A or U) is the first anchor base because A and U are the preferred bases for Ago loading, S17 is the stem region with 17 base pairs, L4 is the unpaired 4-nt loop region, and the 40th nt C is the end base. We have shown that this model molecule is the most potent form among all the variants we tested.29
Figure 1.
Model Molecules of sli-siRNA and 5′ End Variants
(A) Model molecule of csli-siRNA. (B) Model molecule of tsli-siRNA. (C) Reporter assay of 5′ end variants of csli-RRM2-1354-G: adding a G to the 5′ end of csli-RRM2-1354 (addition). -A/G, replacing the anchor nucleotide A with a G (replacement); -Δ, removing the anchor nucleotide A; -ΔΔ, removing the first A and second A from the 5′ end (truncation). Error bars indicate SD.
Based on the T7 promoter (T7pro) sequences used in nature, the T7pro used in in vitro transcription was optimized as T7pro-17 (5′-TAA TAC GAC TCA CTA TA-3′). Efficient T7 transcription also needs the conserved sequence 5′-GGG AGA-3′ (+1 to +6) immediately downstream of T7pro-17.35 Any base change at −1 to −10 results in at least a 50% decrease of transcription activity (Figure S1A). The G at +1 is important because the best transcription efficiency is only 33% when +1 is an H (A, C, T) instead of a G. But when G at +2 was replaced with an H base, at least 50% T7 RNAP strength could be maintained. All replacement of G by an H base or an A base by a B (C, G, T) base at +3 to +6 had a much weaker effect on T7 RNAP strength (Figure S1B).35 Based on the above results, we conclude that the G-S17-L4-A form of tsli-siRNA will give the best yield (Figure 1B).
G-Anchored csli-siRNAs and 5′ End or 3′ End Nucleotide Addition or Truncation Variants of csli-siRNAs Are Highly Active
In our previous publication, we have shown that although a W as the anchor base is preferred by sli-siRNA/siRNA, replacing them with an S base (G or C) has only about a 10% decrease on the potency.29 Therefore, the G-S17-L4-A form of tsli-siRNA could be a compromise model molecule that has both high yield and high potency. The other concern for in vitro production of RNA by bacterial phage RNAPs is that they usually produce heterogeneous RNA transcripts with difference size because of non-template addition of up to 2 nt at either or both 5′ and 3′ ends. Therefore, the same template can produce up to a total of nine different transcripts, with the canonical form as the dominate form, and up to eight of the 5′ and/or 3′ end nucleotide addition forms that can comprise up to 30% of the RNA population from some templates.22, 23, 24, 36, 37 Our previous results showed up to a 2-nt addition in both 5′ and 3′ ends (csli-siRNA-887 target RRM2 gene) has a marginal effect on the potency, especially the 3′ end addition can be tailed up to 5 nt because it will be cut off during sli-siRNA maturation.29 In the current study, to further examine the potency of G-anchored csli-siRNA, we tested several variants of csli-siRNA-1354 (target RRM2 gene): G-W-S17-L4-C, in which a G is appended to the 5′ end of a csli-siRNA (csli-siRNA-g); G-S17-L4-C, a swapping variant in which the anchor W is swapped with a G; and Δ-S17-L4-C and ΔΔ-S16-L4-C, two truncation variants in which 1 or 2 nt is truncated from the 5′end of a csli-siRNA, respectively. Reporter assay results showed there is less than a 5-fold decrease of potency caused by these modifications when lower doses were used, and the difference is marginal when higher doses were used (Figure 1C). These results demonstrate that both appending and swapping variants are good options for converting csli-siRNA to tsli-siRNA, and suggest that tsli-siRNAs produced by transcription starting at +2 or +3 position can maintain high potency.
To examine the potency of tsli-siRNAs against endogenous targets, we designed tsli-siRNAs for the Stat3 gene, converting csli-siRNA-RRM2-1354 or -Plk1 (target Plk1 gene) to a tsli- version by appending a G at the 5′ end (G addition variants), or converting csli-siRNA-ARX1 and -ARX3 (both target the same ARX gene) to a tsli- version by replacing the anchor base A or U with a G, respectively (anchor nucleotide/G replacement variants). We compared these tsli-siRNAs for silencing potency side by side with corresponding csli-siRNAs and their base-modified tsli-siRNA versions containing 2′-F-cytidine triphosphate (CTP) and 2′-F-uridine triphosphate (UTP) (2F-tsli-siRNAs) or biotin-16-UTP (biotin-tsli-siRNA). The 2′-fluoro-incorporated 2F-tsli-siRNAs are supposed to have better stability than tsli-siRNA because RNA transcripts produced by the same Dura-transcripts kit have shown that 2′-fluoro-incorporated RNA is resistant to RNase A and DNase.38, 39 The biotin-16-UTP-incorporated biotin-tsli-siRNA could provide an alternative way to biotinylate tsli-siRNA for potential RNAi applications, such as identifying the Ago2-formed RISC complex.40
The Requirement of G as the Anchor Nucleotide for High Yield of T7 Transcription
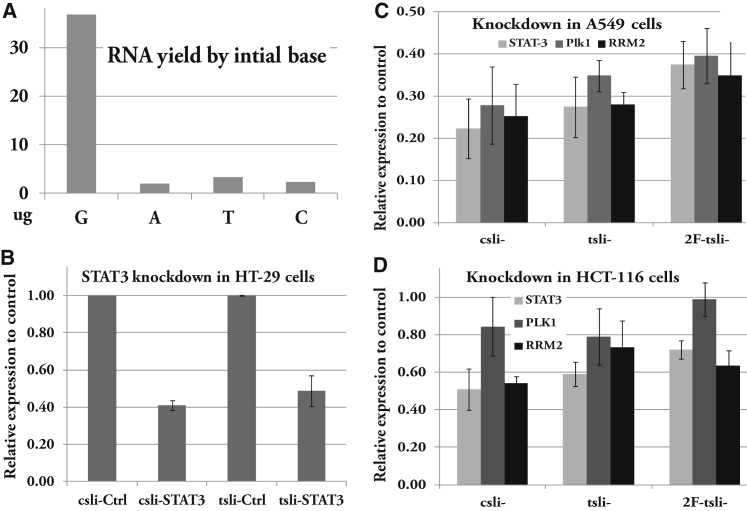
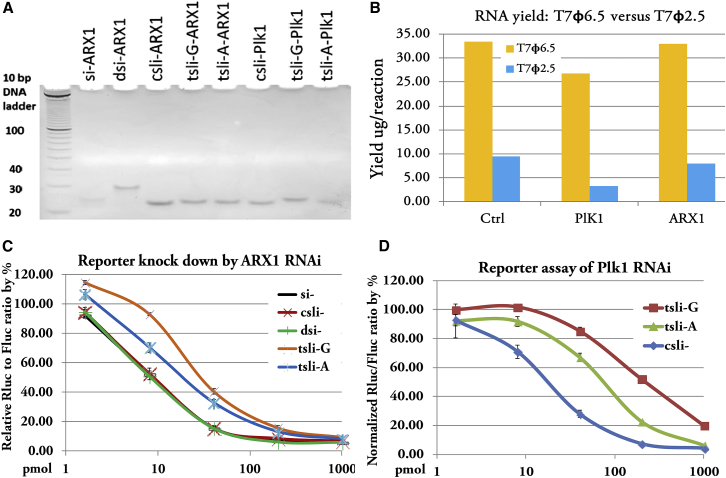
We first validated the requirement of a G at the +1 position for efficient T7 transcription. Results using an N (G, A, T, or C) base as the starting base clearly showed that G at +1 position gives the highest transcription yield, whereas all H bases at +1 position gave a very poor yield (Figure 2A), indicating that a G base at +1 position is absolutely required for high-yield production of tsli-siRNAs. Whereas both appending and swapping variants of csli-siRNAs resulted in transcription products with both high yield and uniformity in size, an A base as the starting base gave poor yields and the products appeared as a smear on PAGE gel (Figure S2).
Figure 2.
Knocking Down Stat3, RRM2, and Plk1 by csli-, tsli-, and 2F-tsli-siRNAs
Final concentration of 10 nM for each RNAi reagent was used in transfection for qPCR assay. (A) Testing tsli-stat3 production using any N as T7 transcription bases. (B) qPCR assay of knockdown Stat3 in HT-29 cells by csli- and tsli-siRNAs. (C) qPCR assay of knockdown Stat3, RRM2, and Plk1 in A549 cells by csli-, tsli-, and 2F-tsli-siRNAs. (D) qPCR assay of knockdown Stat3, RRM2, and Plk1 in HCT-116 cells by csli-, tsli-, and 2F-tsli-siRNAs. Details of qPCR procedure and results calculation were provided in the Materials and Methods. Error bars indicate SD.
We also tested the effect of the amount of template used on the yield of transcription. There was no big difference in the yield when a 100 ng to 1 μg template was used per reaction. The yield nearly reached the plateau when a 200–300 ng template was used per reaction (Figure S3).
Comparing the Potency of tsli-siRNA with csli-siRNA in Gene Silencing
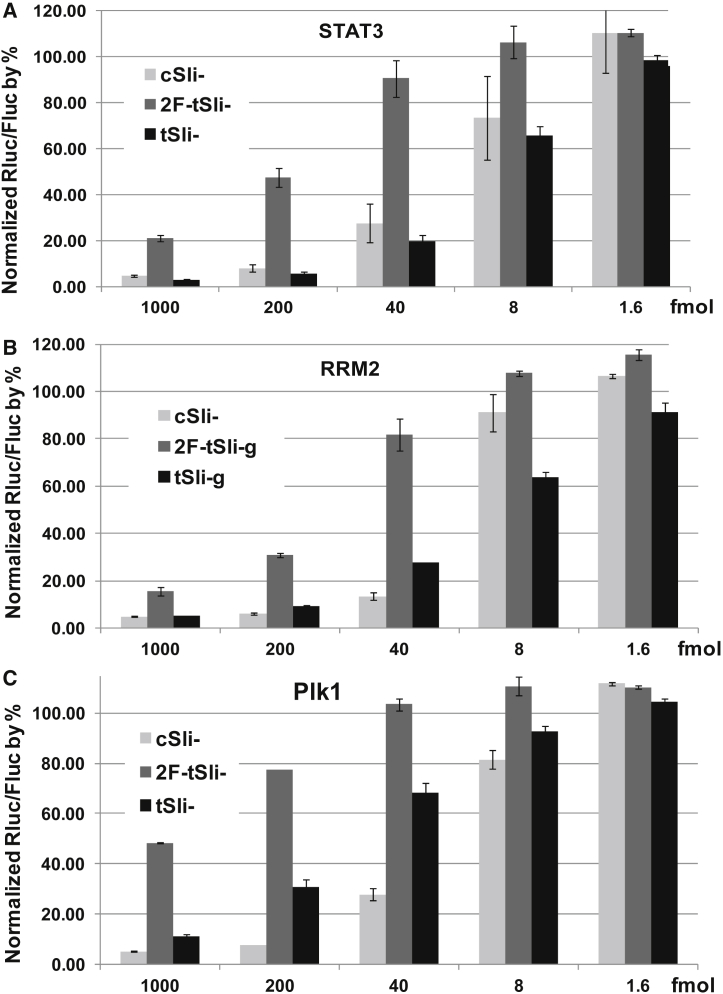
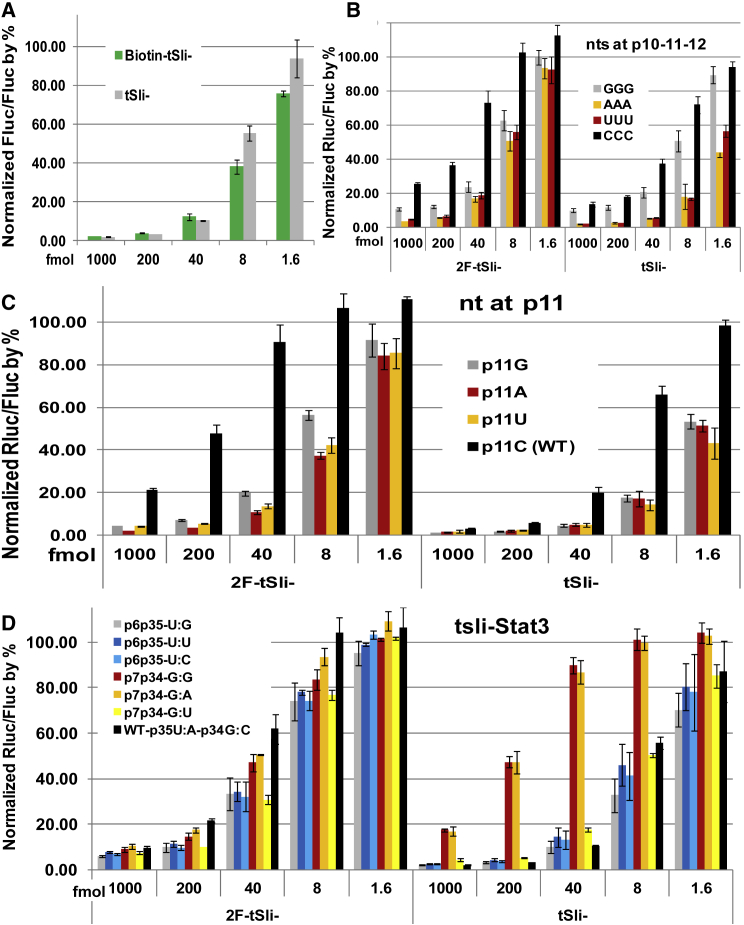
Using qPCR, we observed similar potency in gene silencing for csli- and tsli-siRNA by measuring target knockdown. However, 2F-tsli-siRNAs are observably less potent than csli- or tsli-siRNAs in general (Figures 2B–2D). We further quantified their potency using a reporter assay, and the results agreed well with our qPCR data. Again, we observed similar potency for csli- and tsli-, but less potency for 2F-tsli- (Figure 3). Using the reporter assay, we also tested tsli-siRNA replacement variants: tsli-siRNA-ARX1 and -ARX3, and found that they were highly active, similar to csli-siRNAs (Figure S4). In contrast with the less potent 2F-tsli-, biotin-tsli-Stat3 is as potent as tsli-Stat3 in the reporter assay (Figure 4A). We found that 2F-tsli-siRNAs are not as homogeneous in size as tsli-siRNAs or biotin-tsli-siRNAs when they were visualized by PAGE gel (Figure S5).
Figure 3.
Reporter Assay to Compare Potency of csli-, tsli-, and 2F-tsli-siRNAs Targeting Stat3, RRM2, and Plk1
(A) Knockdown Stat3 reporter in HEK293 cells by csli-, tsli-, and 2F-tsli-siRNAs. (B) Knockdown RRM2 reporter in HEK293 cells by csli-, tsli-, and 2F-tsli-siRNAs. (C) Knockdown Plk1 reporter in HEK293 cells by csli-, tsli-, and 2F-tsli-siRNAs. Detailed reporter assay procedure and results calculation were provided in the Materials and Methods. Error bars indicate SD.
Figure 4.
Effects on Potency by Different Bases at Position p10-11-12, Slicing Nucleotide, and Different Base Pairing at p6:p25 and p7:p34
(A) Reporter assay to compare potency of tsli-Stat3 with biotin-tsli-Stat3. (B) Reporter assay to compare effects on potency by triple nucleotides at position p10-11-12. (C) Reporter assay to compare effects on potency by different slicing nucleotide. (D) Reporter assay to check effects on potency by different base pairing at p6:p25 and p7:p34. Detailed reporter assay procedure and results calculation were provided in the Materials and Methods. Error bars indicate SD.
Optimization of the tsli-siRNA Molecular Structure
Next, we explored the optimization of the tsli-siRNA molecular structure to facilitate its production and enhance its potency. There are many possibilities to optimize tsli-siRNA molecular structure because the potency of siRNA is guide sequence and target context dependent. We decided to compare the potential effect of slicing bases on the potency of tsli-siRNA because sli-siRNA depends on slicing its passenger strand to activate sli-RISC for subsequent target silencing, and this potential effect has not been addressed in previous publications for pre-miR-451 mimics. We first tested all four kinds of triple Ns at the central region of siRNA targeting (position p10-11-12), finding that AAA or TTT is much better than GGG or CCC, with CCC being the worst (Figure 4B); clearly a W base is a much better choice at these positions. 2F-CTP also led to less potency at these positions. Then, we checked how nucleotide changes at p11, the slicing nucleotide, would affect potency and yield of transcription. The results showed a W base at p11 was much better than a G or a C base, and again a C base was worse than any D (G, A, T) base in both potency and yield of transcription (Figure 4C; Figures S6A and S6B). Therefore, G-S17-p11W-L4-A represents the optimized tsli-siRNA form.
We also evaluated the effect of different base pairing at p6:p25 and p7:p34 on the potency of gene silencing. It remains a puzzle why most base pairing on pre-miR-451 from all species is conserved except that p6:p35 base pairing is almost equally used as G:U, G:C, and G:G in all species. Furthermore, a G:U base pair is almost equally potent to G:C base pair for target cleavage and repression, whereas a G:G mismatch shows marginally weaker potency among all three types of base pairing.29 Whereas p6:p35 pairs using wobble pairing or mismatch did not have much effect on the potency in gene silencing, mismatches for p7:p34 dramatically reduced the potency for both tsli-siRNA and 2F-tsli-siRNA (Figure 4D). The yield for tsli-siRNA variants was also affected. One benefit using p6:p35 wobble pair or mismatches is that the yield for both tsli- and 2F-tsli-siRNA is increased by 10%–20% compared with the wild-type, indicating that wobble pair or mismatches introduced into the templates may help T7 RNAP to open the hairpin structure and result in higher yield (Figure S6).
Innate Immunostimulatory Effect by tsli-siRNAs
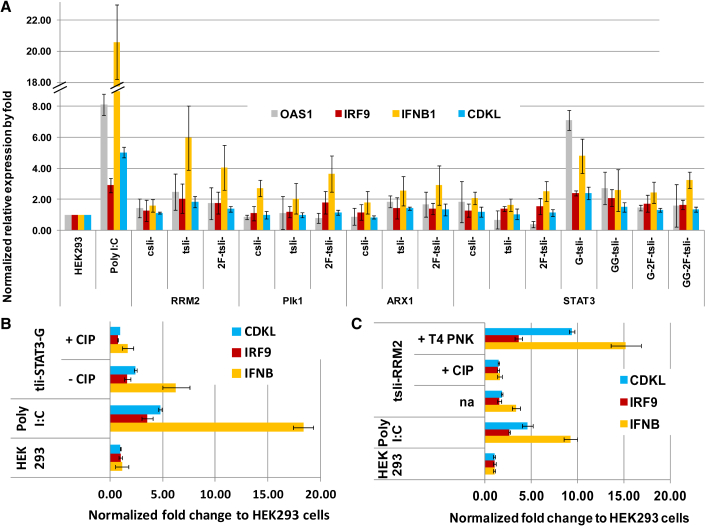
Some bacteriophage RNAP transcripts have 5′ppp added during in vitro transcriptions, and 5′ppp can induce an innate immune response, such as induction of interferon α and β,15, 16, 18 so it is expected that tsli-siRNAs may have a similar immunostimulatory effect. We also produced G- and GG-tsli-siRNA versions because Gn- (n ≥ 2) appending to T7 RNAP transcripts was shown to reduce type I interferon induction.26
We transfected tsli-siRNAs to HEK293 cells and measured expression of several interferon-related genes, and compared the expression level of these genes with polyinosinic:polycytidylic acid [poly(I:C)]-induced interferon response. We noticed that some tsli-siRNAs did cause a mild interferon response, and GG- appending reduced this effect. Our data also showed the innate immunostimulatory feature of tsli-siRNAs is most likely sequence dependent. For example, tsli-RRM2-1354, -ARX1 exhibited much higher innate immune response than tsli-Stat3, -Plk1, or -ARX3. We also observed that G-tsli-Stat3 exhibited an elevated innate immune response (Figure 5A), which implies that G-tsli-siRNA may have enhanced immunostimulatory potency.
Figure 5.
Manipulation of 5′ppp-Triggered Interferon Response
HEK293 cells were transfected with poly(I:C) or several tsli-siRNAs. The final concentration of 10 nM for each RNAi reagent was used in transfection for qPCR assay. Gene expression level changes in OAS1, IRF9, CDKL, and IFNB relative to GAPDH were measured by qPCR. (A) Mild interferon response was observed from all four tsli-siRNAs, with tsli-RRM2 having the strongest response among them. G-tsli-Stat3 exhibited a much stronger response than tsli-Stat3, and GG-tsli-Stat3 reversed this effect to some extent. (B) CIP treatment minimized the strong interferon response by G-tsli-Stat3. (C) CIP treatment minimized and T4 PNK treatment elevated the interferon response by tsli-RRM2. Fold changes in gene expression were normalized to untreated HEK293 cells. Details of qPCR procedure and results calculation were provided in the Materials and Methods. Error bars indicate SD.
We modified the procedure for tsli-siRNA production by adding calf intestinal alkaline phosphatase (CIP) and DNase together at the template DNA-removing step to remove 5′ppp and DNA simultaneously. The results showed that the immune response trigger by 5′ppp was successfully minimized (Figures 5B and 5C). Although the immune response from siRNA should be avoided for most target knockdown RNAi applications, an immune response can be beneficial in some applications. For example, bifunctional siRNAs that can trigger both innate immune response and RNAi can be used to overcome immune resistance from cancers and immune evasion by viruses, and also knockdown oncogenes or viral pathogenesis genes.41, 42, 43, 44, 45, 46, 47 To increase the amount of tsli-siRNA molecules with 5′ppp, we added T4 polynucleotide kinase (PNK) and DNase together at the template DNA-removing step. This addition resulted in enhanced interferon responses, indicating that the population of tsli-siRNAs with 5′ppp was increased by PNK treatment (Figure 5C). Interestingly, tsli-siRNA-triggered immune response is also sequence dependent, because tsli-Stat3, a tsli-siRNA that has the weakest immune response among all the tested ones, was not significantly affected by T4 PNK treatment (Figure S7).
An Alternative Approach to Produce tsli-siRNAs with Higher Potency
The above optimized tsli-siRNA form, G-S17-p11W-L4-A, is derived from the conserved T7 (5′-TAA TAC GAC TCA CTA TA-3′) promoter that exists in nature. It is also called class III ϕ6.5 T7pro.48 The G requirement (tsli-siRNA-G, G as the first nucleotide) is a compromised approach for producing sli-siRNAs with both higher yield and higher potency. It was previously reported that an alternative T7pro, the class II ϕ2.5 promoter (5′-TAA TAC GAC TCA CTA TT-3′), which only differs from the ϕ6.5 T7pro used above by the last base, prefers A as the initiation base. The ϕ2.5 T7pro was also shown to produce RNA with superior 5′ homogeneity over the ϕ6.5 T7pro and with comparable total RNA yields.48 Therefore, we explored the possibility of producing tsli-siRNA with A as the first nucleotide (tsli-siRNA-A), which may increase potency because the mid domain of argonautes prefers binding to A or U during RISC loading.49, 50, 51 The tsli-siRNA-A will have the same sequence as the A-S17-L4-C form of csli-siRNA or differ by the first nucleotide from the U-S17-L4-C form of csli-siRNA. Because the difference in silencing from both A and U forms of csli-siRNA is minimal,29 we expected that tsli-siRNA-A will be comparable with both A and U forms of csli-siRNAs. We produced both tsli-siRNA-A and tsli-siRNA-G forms and compared them in both potency and yield. Although PAGE gel showed both -G and -A forms of tsli-siRNAs were as clean as their csli-siRNA form (Figure 6A), the yield of the ϕ2.5 promoter is about 70% less than the ϕ6.5 promoter’s yield in all three sequences (-control, -ARX1, and -Plk1) being tested (Figure 6B). Our data also showed both tsli-siRNA-A-ARX1 and tsli-siRNA-A-PlK1 had higher potency than their corresponding tsli-siRNA-G-ARX1 and tsli-siRNA-G-PlK1 (Figures 6C and 6D). Therefore, when higher potency is desired, the tsli-siRNA-A form can be produced using the same procedure as the tsli-siRNA-G form, albeit at a lower yield.
Figure 6.
tsli-siRNA Production by ϕ6.5 T7 Promoter versus ϕ2.5 T7 Promoter
(A) Visualized classical siRNA, dicer substrate siRNA (dsiRNA), chemically synthesized sli-siRNA (csli-siRNA), ϕ6.5 T7 promoter-produced tsli-siRNA-G, and ϕ2.5 T7 promoter-produced tsli-siRNA-A on 12% native PAGE gel. (B) In vitro transcription RNA yield by T7 promoter variants (100 ng of template DNA was used per reaction). G and A forms of tsli-siRNA-ctrl, -Plk1, and -ARX1 were produced by ϕ6.5 and ϕ2.5 T7 promoter, respectively. (C) Reporter assay to compare potency of siRNA-ARX1 (si-), csli-siRNA-ARX1 (csli-), dsiRNA-ARX1 (dsi-), tsli-siRNA-G-ARX1 (tsli-G), and tsli-siRNA-A-ARX1 (tsli-A). (D) Reporter assay to compare potency of tsli-siRNA-G-Plk1 (tsli-G), tsli-siRNA-A-Plk1 (tsli-A), and csli-siRNA-Plk1 (csli-). Detailed reporter assay procedure and results calculation were provided in the Materials and Methods. Error bars indicate SD.
Discussion
Today, RNAi is commonly used in research, and there is increasing use in clinical trials and agriculture. However, the high cost for chemical synthesis of RNAi reagents at scales necessary for these applications is a significant and potentially limiting factor.2, 3 RNAi reagents also need to be optimized by extensive screening to obtain high potency in target gene silencing.
Many potent siRNAs already have been identified, and siRNAs have been shown to be producible in mammalian cells or E. coli, which makes large-scale production or RNAi expression libraries possible at low cost.4, 9, 52, 53, 54 But these procedures are complicated and, due to the fact that in-vitro-produced siRNAs are not uniform, it will be difficult for them to be approved as drugs.6 Here, we present a very simple procedure to produce small hairpin RNA as a special form of miR-451 mimics that can act as potent RNAi reagents as well as the innate immune response triggers.
We optimized the csli-siRNA molecule to facilitate production using T7 RNAP. Our data showed T7 RNAP can use regular NTPs or 2F- or biotin-modified NTPs to produce tsli-, 2F-tsli-, or biotin-tsli-siRNAs as potent as csli-siRNAs. Our results also indicate that using a W base at the slicing position p11 and introducing wobble pair or mismatches for the p6:p35 base pair could further enhance potency or yield.
Although tsli-siRNAs produced by ϕ6.5 T7pro must use G as its anchor nucleotide, which may reduce their potency by as much as 5-fold compared with csli-siRNAs (Figure 1C) or canonical siRNAs when lower doses were used (Figure 6), the gene silencing potency for siRNAs also highly depends on their sequence and target accessibility, and they are usually used at a saturating dose. 5′-G-containing small RNAs do ubiquitously exist in nature; about 14% endogenous miRNAs use G as their first nucleotide in human (miRBase release 21), and 22G siRNA that start with a G are abundant in C. elegans.55, 56
If high potency is desired, the ϕ2.5 T7pro can be used to produce tsli-siRNA-A, which has higher potency than tsli-siRNA-G (Figure 6). If a high amount is desired, the ϕ6.5 T7pro should be used to produce tsli-siRNA-G, and it is very easy to convert a W base start siRNA/csli-siRNA to a tsli-siRNA by just appending a G or replacing the anchor W base with a G. If chemical synthesis of an RNAi reagent is preferred for some particular applications, any potent tsli-siRNA can be easily converted to csli-siRNA or siRNA by an anchor base G to W replacement.
However, our data showed tsli-siRNAs, both the G and A forms, showed less potency than canonical siRNA, csli-siRNA, or dicer substrate siRNA (dsi-siRNA) (Figures 2 and 6). One cause of this is probably the heterogeneity of RNAs produced by bacteriophage RNAPs. The other reason is, to keep the procedure simple, we used spin column to purify in vitro transcription-produced RNAs. The spin-column method of purification may result in lower purity for the tsli-siRNAs when compared with chemically synthesized siRNA, csli-siRNA, or dsiRNA.
Although tsli-siRNA can trigger the interferon response, this potential problem can be easily minimized by alkaline phosphatase treatment. On the other hand, this immunostimulatory property can be beneficial in some applications, in which tsli-siRNA may be used as triggers for both interferon response and RNAi to achieve synergistic effects, such as applications to treat virus infection or cancers.42, 46, 57 Interestingly, this immunostimulatory feature can be easily enhanced by PNK treatment, and we believe it could be further enhanced by adding immunostimulatory motifs to the tsli-siRNA sequence.
We estimate that the cost for tsli-siRNA-G is much less than other methods (only about $10/reaction and $5–$10/nmol calculated based on 1 nmol siRNA equals about 12 μg of siRNA), and the whole procedure can be finished within 2 hr. The cost per reaction for tsli-siRNA-A is the same as tsli-siRNA-G, but the yield is reduced by about 70%. The same RNA oligo (csli-siRNA) costs about $120 each and about $130 for a 21-mer siRNA duplex (based on the minimum order for the same RNA oligo from Integrated DNA Technologies [IDT], which is 100 nmol).
In summary, we have developed a simple method that can be used to produce sli-siRNAs or other forms of pre-miR-451 mimics with relatively high quality and high yield in a cost-effective and rapid manner. We believe that tsli-siRNAs will be a novel RNAi reagent and could be used to screen potent siRNAs with greatly reduced cost. The fact that tsli-siRNA can be easily adapted to small-scale laboratory production or scaled up for industrial production led us to believe that the tsli-siRNA will be a very attractive RNAi reagent for siRNA applications in research and siRNA drug development for cancers, viruses, and insects. Furthermore, tsli-siRNA with both immunostimulatory and RNAi features will be a very useful reagent to confront the sudden emergence of a drug-resistant pandemic virus strain that lacks effective antibody or when the antibody cannot be manufactured in a short time.
Materials and Methods
Cell Lines and Cell Culture
HEK293, A549, HCT-116, and HT-29 cells were maintained in high glucose (4.5 g/L) DMEM supplemented with 2 mM glutamine, 10% FBS, and 2 mM penicillin/streptomycin. Cells were incubated at 37°C, 5% CO2.
Transfection
For reporter assays, RNAi triggers and reporter constructs were co-transfected into cells by using Lipofectamine 2000 (Thermo Fisher Scientific) as previously reported.29 For each experiment, at least three independent transfections were performed in duplicate in 24-well plates. Cell were grown to 75%–85% confluency in 500 μL of medium and were transfected with luciferase reporter (50 ng) and different amounts of siRNA (100 ng of stuffer DNA, plus 1 μL of siRNA stock at 1 μM, 200 nM, 40 nM, 8 nM, and 1.6 nM, and 1 μL of Lipofectamine 2000).
For qPCR analysis, the final concentration of 10 nM siRNA was transfected by RNAiMAX (Thermo Fisher Scientific) in 12-well plates. Forty-eight hours after transfection, cells were lysed with TRIzol (Thermo Fisher Scientific) for total RNA isolation.
Dual-Luciferase Reporter Assays
All reporter assays were performed using psiCheck 2.0-based, dual-luciferase reporter plasmids from Promega that express both firefly luciferase (Fluc) and Renilla luciferase (Rluc). Reporters carried complementary target sequences that were constructed by inserting annealed oligonucleotides into the XhoI/SpeI sites of the 3′ UTR of the Rluc gene in psiCheck2.2 vector.29 These reporters were used to quantify gene silencing. Forty-eight hours after transfection, cells were lysed with 100 μL of passive lysis buffer (Promega), and luciferase levels for 20 μL of lysate were determined (Dual-Luciferase Reporter Assay Kit and GloMax 96 Microplate Luminometer; Promega). Changes in expression of Rluc (target) were normalized to Fluc (internal control) and then calculated relative to the scrambled sli-siRNA control. The relative ratios of Rluc/Fluc were used to measure the efficiency of silencing. Data were averaged from at least three independent transfections, and each transfection had at least two replicates. Error bars indicate the SD.
Oligonucleotides
All oligonucleotides were synthesized by Integrated DNA Technologies. Sequences are listed in Table S1–S3.
In Vitro Transcription Template DNA Preparation
Sense and antisense oligos were first separately dissolved in water at a concentration of 100 μM. Equal volume of sense and antisense oligos were mixed and diluted to 10 μM, then annealed by dropping the tube into in a glass beaker with about 300 mL of boiled water and cooling to room temperature. The quantity of annealed product was measured by NanoDrop and diluted to 100 or 200 ng/μL.
T7 In Vitro Transcription and Purification
AmpliScribe T7-Flash Transcription Kit from Epicenter was used for all tsli-siRNA production using regular NTPs. DuraScribe T7 Transcription Kit from Epicenter was used for all 2F-tsli-siRNA production using ATP, guanosine triphosphate (GTP), 2′-F-CTP, and 2′-F-UTP. AmpliScribe T7-Flash Biotin-RNA Transcription Kit from Epicenter was used for all biotin-tsli-siRNA production using ATP, GTP, CTP, and biotin-16-UTP.
We followed manufacturer’s procedure for the above kits. The protocol was modified as follows when CIP or T4 PNK treatment is necessary: (1) for CIP treatment, in 20 μL of products from one in vitro transcription reaction before DNase treatment, we added 1 μL of DNase (supplied with T7 Transcription Kit), 1 μL of CIP, 4 μL of 10× CutSmart buffer (NEB), and water to total volume of 40 μL, and incubated at 37°C for 15 min; and (2) for T4 PNK treatment, in 20 μL of products from one in vitro transcription reaction before DNase treatment, we added 1 μL of DNase (supplied with T7 Transcription Kit), 1 μL of T4 PNK, 4 μL of 10× T4 PNK buffer (NEB), and water to total volume of 40 μL, and incubated at 37°C for 15 min.
All T7 in vitro transcription products were purified by Micro Bio-Spin P-30 Gel Columns, Tris Buffer, from Bio-Rad. Up to 40 μL of products from two in vitro transcription reactions was pooled, and 20 μL of water was added to make it total 60 μL before applying it to one spin column.
qPCR
The Bio-Rad iTaq Universal SYBR Green One-Step Kit was used for qPCR. In brief, total RNA was isolated by TRIzol followed by DNase treatment. In each reaction, 500 ng of DNase-treated total RNA was used, and all other reagents were used as specified in the protocol provided in the kit. We also followed the qPCR program suggested by the vendor. GAPDH gene was used as a normalization control to calculate ΔCt (threshold cycle) for each sample. The 2−ΔΔCt value of each sample (ΔΔCt = ΔCtsiRNA − ΔCtsiRNA ctrl) was used for RNAi knockdown measurement and converted to %. The 2−ΔΔCt value of each sample (ΔΔCt = ΔCtsiRNA-treated HEK293 cells − ΔCtuntreated HEK293 cells) was used for calculating fold changes in gene expression to measure immune response. Data were averaged from at least three replicates. Error bars indicate the SD of the mean.
Author Contributions
G.S. and A.D.R. conceived and designed the experiments. G.S. and A.D.R. drafted the manuscript and contributed to the final version of the manuscript.
Conflicts of Interest
The authors declare there are no competing financial interests.
Acknowledgments
This work was supported by an internal grant from City of Hope (grant no. 1000217 to A.D.R.). The authors would like to thank Louise Shively and Natalie Rosas for their help in this project.
Footnotes
Supplemental Information includes seven figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.07.008.
Contributor Information
Guihua Sun, Email: gusun@coh.org.
Arthur D. Riggs, Email: ariggs@coh.org.
Supplemental Information
References
- 1.Elbashir S.M., Lendeckel W., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zuckerman J.E., Davis M.E. Clinical experiences with systemically administered siRNA-based therapeutics in cancer. Nat. Rev. Drug Discov. 2015;14:843–856. doi: 10.1038/nrd4685. [DOI] [PubMed] [Google Scholar]
- 3.Zotti M.J., Smagghe G. RNAi technology for insect management and protection of beneficial insects from diseases: lessons, challenges and risk assessments. Neotrop. Entomol. 2015;44:197–213. doi: 10.1007/s13744-015-0291-8. [DOI] [PubMed] [Google Scholar]
- 4.Kittler R., Putz G., Pelletier L., Poser I., Heninger A.K., Drechsel D., Fischer S., Konstantinova I., Habermann B., Grabner H. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- 5.Yang D., Buchholz F., Huang Z., Goga A., Chen C.Y., Brodsky F.M., Bishop J.M. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc. Natl. Acad. Sci. USA. 2002;99:9942–9947. doi: 10.1073/pnas.152327299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hannus M., Beitzinger M., Engelmann J.C., Weickert M.T., Spang R., Hannus S., Meister G. siPools: highly complex but accurately defined siRNA pools eliminate off-target effects. Nucleic Acids Res. 2014;42:8049–8061. doi: 10.1093/nar/gku480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calegari F., Haubensak W., Yang D., Huttner W.B., Buchholz F. Tissue-specific RNA interference in postimplantation mouse embryos with endoribonuclease-prepared short interfering RNA. Proc. Natl. Acad. Sci. USA. 2002;99:14236–14240. doi: 10.1073/pnas.192559699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brummelkamp T.R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 9.Aalto A.P., Sarin L.P., van Dijk A.A., Saarma M., Poranen M.M., Arumäe U., Bamford D.H. Large-scale production of dsRNA and siRNA pools for RNA interference utilizing bacteriophage phi6 RNA-dependent RNA polymerase. RNA. 2007;13:422–429. doi: 10.1261/rna.348307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W.Y., Wang Y., Sun Y.H., Wang Y., Wang Y.P., Chen S.P., Zhu Z.Y. Efficient RNA interference in zebrafish embryos using siRNA synthesized with SP6 RNA polymerase. Dev. Growth Differ. 2005;47:323–331. doi: 10.1111/j.1440-169X.2005.00807.x. [DOI] [PubMed] [Google Scholar]
- 11.Donzé O., Picard D. RNA interference in mammalian cells using siRNAs synthesized with T7 RNA polymerase. Nucleic Acids Res. 2002;30:e46. doi: 10.1093/nar/30.10.e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Billy E., Brondani V., Zhang H., Müller U., Filipowicz W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl. Acad. Sci. USA. 2001;98:14428–14433. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sohail M., Doran G., Riedemann J., Macaulay V., Southern E.M. A simple and cost-effective method for producing small interfering RNAs with high efficacy. Nucleic Acids Res. 2003;31:e38. doi: 10.1093/nar/gng038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu X., Li T., Dang Y., Feng Y., Huang P. A novel in vitro transcription method for producing siRNAs without specific sequence requirements. Mol. Biotechnol. 2005;31:187–192. doi: 10.1385/MB:31:3:187. [DOI] [PubMed] [Google Scholar]
- 15.Kim D.H., Longo M., Han Y., Lundberg P., Cantin E., Rossi J.J. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat. Biotechnol. 2004;22:321–325. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- 16.Pichlmair A., Schulz O., Tan C.P., Näslund T.I., Liljeström P., Weber F., Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 17.Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 18.Hornung V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K.K., Schlee M. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 19.Schlee M., Hartmann G. The chase for the RIG-I ligand—recent advances. Mol. Ther. 2010;18:1254–1262. doi: 10.1038/mt.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlee M., Hornung V., Hartmann G. siRNA and isRNA: two edges of one sword. Mol. Ther. 2006;14:463–470. doi: 10.1016/j.ymthe.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helm M., Brulé H., Giegé R., Florentz C. More mistakes by T7 RNA polymerase at the 5′ ends of in vitro-transcribed RNAs. RNA. 1999;5:618–621. doi: 10.1017/s1355838299982328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pleiss J.A., Derrick M.L., Uhlenbeck O.C. T7 RNA polymerase produces 5′ end heterogeneity during in vitro transcription from certain templates. RNA. 1998;4:1313–1317. doi: 10.1017/s135583829800106x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nam S.C., Kang C.W. Transcription initiation site selection and abortive initiation cycling of phage SP6 RNA polymerase. J. Biol. Chem. 1988;263:18123–18127. [PubMed] [Google Scholar]
- 25.Goldgraben M.A., Russell R., Rueda O.M., Caldas C., Git A. Double-stranded microRNA mimics can induce length- and passenger strand-dependent effects in a cell type-specific manner. RNA. 2016;22:193–203. doi: 10.1261/rna.054072.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gondai T., Yamaguchi K., Miyano-Kurosaki N., Habu Y., Takaku H. Short-hairpin RNAs synthesized by T7 phage polymerase do not induce interferon. Nucleic Acids Res. 2008;36:e18. doi: 10.1093/nar/gkm1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang W., Bartel D.P. The menu of features that define primary microRNAs and enable de novo design of microRNA genes. Mol. Cell. 2015;60:131–145. doi: 10.1016/j.molcel.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu S., Jin L., Zhang Y., Huang Y., Zhang F., Valdmanis P.N., Kay M.A. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell. 2012;151:900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun G., Yeh S.Y., Yuan C.W., Chiu M.J., Yung B.S., Yen Y. Molecular properties, functional mechanisms, and applications of sliced siRNA. Mol. Ther. Nucleic Acids. 2015;4:e221. doi: 10.1038/mtna.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheloufi S., Dos Santos C.O., Chong M.M., Hannon G.J. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cifuentes D., Xue H., Taylor D.W., Patnode H., Mishima Y., Cheloufi S., Ma E., Mane S., Hannon G.J., Lawson N.D. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J.S., Maurin T., Robine N., Rasmussen K.D., Jeffrey K.L., Chandwani R., Papapetrou E.P., Sadelain M., O’Carroll D., Lai E.C. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl. Acad. Sci. USA. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y.P., Schopman N.C., Berkhout B. Dicer-independent processing of short hairpin RNAs. Nucleic Acids Res. 2013;41:3723–3733. doi: 10.1093/nar/gkt036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma H., Zhang J., Wu H. Designing Ago2-specific siRNA/shRNA to avoid competition with endogenous miRNAs. Mol. Ther. Nucleic Acids. 2014;3:e176. doi: 10.1038/mtna.2014.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imburgio D., Rong M., Ma K., McAllister W.T. Studies of promoter recognition and start site selection by T7 RNA polymerase using a comprehensive collection of promoter variants. Biochemistry. 2000;39:10419–10430. doi: 10.1021/bi000365w. [DOI] [PubMed] [Google Scholar]
- 36.Huang F. Efficient incorporation of CoA, NAD and FAD into RNA by in vitro transcription. Nucleic Acids Res. 2003;31:e8. doi: 10.1093/nar/gng008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang F., Wang G., Coleman T., Li N. Synthesis of adenosine derivatives as transcription initiators and preparation of 5′ fluorescein- and biotin-labeled RNA through one-step in vitro transcription. RNA. 2003;9:1562–1570. doi: 10.1261/rna.5106403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Padilla R., Sousa R. Efficient synthesis of nucleic acids heavily modified with non-canonical ribose 2′-groups using a mutantT7 RNA polymerase (RNAP) Nucleic Acids Res. 1999;27:1561–1563. doi: 10.1093/nar/27.6.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sousa R., Padilla R. A mutant T7 RNA polymerase as a DNA polymerase. EMBO J. 1995;14:4609–4621. doi: 10.1002/j.1460-2075.1995.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iioka H., Loiselle D., Haystead T.A., Macara I.G. Efficient detection of RNA-protein interactions using tethered RNAs. Nucleic Acids Res. 2011;39:e53. doi: 10.1093/nar/gkq1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Furset G., Sioud M. Design of bifunctional siRNAs: combining immunostimulation and gene-silencing in one single siRNA molecule. Biochem. Biophys. Res. Commun. 2007;352:642–649. doi: 10.1016/j.bbrc.2006.11.059. [DOI] [PubMed] [Google Scholar]
- 42.Joshi G., Dash P.K., Agarwal A., Sharma S., Parida M. Bifunctional siRNA containing immunostimulatory motif enhances protection against pandemic H1N1 virus infection. Curr. Gene Ther. 2015;15:492–502. doi: 10.2174/1566523215666150812120547. [DOI] [PubMed] [Google Scholar]
- 43.Matheis F., Besch R. Bifunctional siRNAs for tumor therapy. Methods Mol. Biol. 2014;1169:181–192. doi: 10.1007/978-1-4939-0882-0_17. [DOI] [PubMed] [Google Scholar]
- 44.Han Q., Zhang C., Zhang J., Tian Z. The role of innate immunity in HBV infection. Semin. Immunopathol. 2013;35:23–38. doi: 10.1007/s00281-012-0331-y. [DOI] [PubMed] [Google Scholar]
- 45.Han Q., Zhang C., Zhang J., Tian Z. Involvement of activation of PKR in HBx-siRNA-mediated innate immune effects on HBV inhibition. PLoS ONE. 2011;6:e27931. doi: 10.1371/journal.pone.0027931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ebert G., Poeck H., Lucifora J., Baschuk N., Esser K., Esposito I., Hartmann G., Protzer U. 5′ Triphosphorylated small interfering RNAs control replication of hepatitis B virus and induce an interferon response in human liver cells and mice. Gastroenterology. 2011;141:696–706. doi: 10.1053/j.gastro.2011.05.001. 706.e1–3. [DOI] [PubMed] [Google Scholar]
- 47.Khairuddin N., Gantier M.P., Blake S.J., Wu S.Y., Behlke M.A., Williams B.R., McMillan N.A. siRNA-induced immunostimulation through TLR7 promotes antitumoral activity against HPV-driven tumors in vivo. Immunol. Cell Biol. 2012;90:187–196. doi: 10.1038/icb.2011.19. [DOI] [PubMed] [Google Scholar]
- 48.Coleman T.M., Wang G., Huang F. Superior 5′ homogeneity of RNA from ATP-initiated transcription under the T7 phi 2.5 promoter. Nucleic Acids Res. 2004;32:e14. doi: 10.1093/nar/gnh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frank F., Fabian M.R., Stepinski J., Jemielity J., Darzynkiewicz E., Sonenberg N., Nagar B. Structural analysis of 5′-mRNA-cap interactions with the human AGO2 MID domain. EMBO Rep. 2011;12:415–420. doi: 10.1038/embor.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schirle N.T., Sheu-Gruttadauria J., MacRae I.J. Structural basis for microRNA targeting. Science. 2014;346:608–613. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frank F., Sonenberg N., Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 52.Huang L., Lieberman J. Production of highly potent recombinant siRNAs in Escherichia coli. Nat. Protoc. 2013;8:2325–2336. doi: 10.1038/nprot.2013.149. [DOI] [PubMed] [Google Scholar]
- 53.Qian Z.K., Xuan B.Q., Min T.S., Xu J.F., Li L., Huang W.D. Cost-effective method of siRNA preparation and its application to inhibit hepatitis B virus replication in HepG2 cells. World J. Gastroenterol. 2005;11:1297–1302. doi: 10.3748/wjg.v11.i9.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heninger A.K., Buchholz F. Production of endoribonuclease-prepared short interfering RNAs (esiRNAs) for specific and effective gene silencing in mammalian cells. CSH Protoc. 2007;2007 doi: 10.1101/pdb.prot4824. pdb.prot4824. [DOI] [PubMed] [Google Scholar]
- 55.Halic M., Moazed D. 22G-RNAs in transposon silencing and centromere function. Mol. Cell. 2009;36:170–171. doi: 10.1016/j.molcel.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gu W., Shirayama M., Conte D., Jr., Vasale J., Batista P.J., Claycomb J.M., Moresco J.J., Youngman E.M., Keys J., Stoltz M.J. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen X., Qian Y., Yan F., Tu J., Yang X., Xing Y., Chen Z. 5′-Triphosphate-siRNA activates RIG-I-dependent type I interferon production and enhances inhibition of hepatitis B virus replication in HepG2.2.15 cells. Eur. J. Pharmacol. 2013;721:86–95. doi: 10.1016/j.ejphar.2013.09.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.