Abstract
Comparative gene expression analysis by qRT-PCR is commonly used to detect differentially expressed genes in studies of PCOS pathology. Impaired GC function is strongly associated with PCOS pathogenesis, and a growing body of studies has been dedicated to identifying differentially expressed genes in GCs in PCOS patients and healthy women by qRT-PCR. It is necessary to validate the expression stability of the selected reference genes across the tested samples for target gene expression normalization. We examined the variability and stability of expression of the 15 commonly used reference genes in GCs from 44 PCOS patients and 45 healthy women using the GeNorm, BestKeeper, and NormFinder statistical algorithms. We combined the rankings of the three programs to produce a final ranking based on the geometric means of their stability scores. We found that HPRT1, RPLP0, and HMBS out of 15 examined commonly used reference genes are stably expressed in GCs in both controls and PCOS patients and can be used for normalization in gene expression profiling by qRT-PCR. Future gene-expression studies should consider using these reference genes in GCs in PCOS patients for more accurate quantitation of target gene expression and data interpretation.
Introduction
Polycystic ovarian syndrome (PCOS) is characterized by ovulatory dysfunction, polycystic ovaries, and hyperandrogenism and is a common gynecological endocrinopathy and a leading cause of female reproductive failure1, 2. Its prevalence is estimated to be 6% to 18% depending on the diagnostic criteria used3–5. PCOS patients are prone to multiple metabolic disorders, including insulin resistance, type 2 diabetes mellitus, obesity, dyslipidemia, chronic low-grade inflammation, cardiovascular diseases, and psychological disturbances1–3, 6–11.
While the etiopathogenesis of PCOS remains elusive, it likely involves aberrant hormonal responses mediated by ovarian granulosa cells (GCs) during the progression of follicular development. Previous studies have reported that PCOS is associated with elevated luteinizing hormone (LH) and overexpression and overactivation of the luteinizing hormone/chorionic gonadotropin receptor (LHCGR) in GCs from PCOS patients and that this prevents follicle maturation and ovulation12–16. Impaired GC function is associated with disruption of folliculogenesis and with elevated intraovarian androgens and circulating anti-Müllerian hormone levels17–23.
Comparative gene expression profiling in normal and disease samples provides invaluable insights into the underlying mechanism of PCOS pathogenesis. Quantitative real-time PCR (qRT-PCR), microarray and RNA-seq have been widely used to measure differential gene expression of GCs from metabolically normal subjects and PCOS patients to identify the genetic regulators and their associated pathways as well as to identify biomarkers in PCOS24–35. Most studies have simply selected a reference gene, e.g. β-actin (ACTB)36, 37, 18 S ribosomal RNA (RNA18S5)38, peptidylprolyl isomerase B (PPIB)39, 40, or glyceraldehyde-3-phosphatedehydrogenase (GAPDH)41, as an endogenous internal control for target gene expression normalization without careful examination of their expression variability and stability in the tested samples. Accumulating evidence suggests that these reference genes, previously thought to be stably expressed with minimal variability across samples, are in fact variably expressed under different physiological and pathological conditions42, 43. The adoption of an inappropriate housekeeping gene could result in under- or overestimation of gene expression and subsequent misinterpretation of the data44–46. To the best of our knowledge, there are no published data that can be used to select reliable reference genes for gene expression normalization in the GCs of reproductively healthy women and PCOS patients.
Given the crucial role of GCs in PCOS pathology, we examined the expression stability and variability of the following 15 commonly used reference genes: RNA18S5, ACTB, GAPDH, ubiquitin C (UBC), β-2-microglobulin (B2M), RNA polymerase II subunit A (POLR2A), hypoxanthine phosphoribosyltransferase 1 (HPRT1), ζ-tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein (YWHAZ), hydroxymethylbilane synthase (HMBS), β-glucuronidase (GUSB), importin 8 (IPO8), phosphoglycerate kinase 1 (PGK1), ribosomal protein lateral stalk subunit P0 (RPLP0), transferrin receptor (TFRC), and ribosomal protein L13a (RPL13A) in GCs from healthy control subjects and PCOS patients. This study thus provides valuable insights into the selection of appropriate reference genes for gene expression profiling in GCs from PCOS patients.
Results
Clinical and biochemical parameters of the study subjects
The clinical and biochemical parameters of the 44 PCOS patients and 45 healthy controls are presented in Table 1. There were no significant differences in the serum concentrations of estrogen, progesterone, prolactin, or FSH for the PCOS patients and the age-matched healthy controls. As expected, the BMI of the PCOS group was significantly higher than the control group by 7.8% (PCOS: 24.20 ± 4.73 kg/m2, control: 22.45 ± 2.99 kg/m2, 95% CI: −1.771 to 9.411, t = 1.358 df = 87, p < 0.05). The PCOS group also exhibited 33.9% higher levels of testosterone compared to the control group (PCOS: 36.12 ± 17.6 ng/dl, control: 23.87 ± 7.77, 95% CI: 6.540 to 17.96, t = 4.264 df = 87, p < 0.0001). Although the FSH levels were indistinguishable between the two groups, both LH and the LH/FSH ratio were strongly increased in the PCOS group by 36.4% and 39.8%, respectively (PCOS LH: 8.10 ± 3.87 IU/L, control LH: 5.1 ± 1.84 IU/L, 95% CI: 1.688 to 4.232, t = 4.624 df = 87, p < 0.0001; PCOS LH/FSH: 1.33 ± 0.74, control LH/FSH: 0.80 ± 0.29, 95% CI: 0.2942 to 0.7658, t = 4.467, df = 87, p < 0.0001).
Table 1.
Anthropometric and clinical data of PCOS patients and healthy controls.
| Control (n = 45) | PCOS (n = 44) | p-value | |
|---|---|---|---|
| Age (years) | 29.64 ± 2.73 | 28.59 ± 3.20 | 0.09 |
| BMI (kg/m2) | 22.45 ± 2.99 | 24.20 ± 4.73 | 0.04* |
| E2 (pg/ml) | 35.27 ± 10.66 | 39.09 ± 15.49 | 0.17 |
| P (ng/ml) | 0.62 ± 0.23 | 0.67 ± 0.39 | 0.48 |
| PRL (ng/ml) | 18.66 ± 9.63 | 17.69 ± 9.51 | 0.63 |
| T (ng/dl) | 23.87 ± 7.77 | 36.12 ± 17.6 | <0.001* |
| FSH (IU/L) | 6.54 ± 1.28 | 6.26 ± 1.51 | 0.35 |
| LH (IU/L) | 5.14 ± 1.84 | 8.10 ± 3.87 | <0.001* |
| LH/FSH | 0.80 ± 0.29 | 1.33 ± 0.74 | <0.001* |
BMI: body mass index; E2: estrogen; P: progesterone; PRL: prolactin; T: testosterone; FSH: follicle stimulating hormone LH: luteinizing hormone. *p < 0.05 indicates statistical significance.
Expression levels of the reference genes
The expression levels of the 15 candidate reference genes in all 44 PCOS patients and all 45 controls were evaluated based on their qRT-PCR threshold cycle value (Ct). As shown in Fig. 1, their Ct values exhibited a dynamic expression range from the lowest of 13.81 ± 0.54 for RNA18S5 to the highest of 28.25 ± 1.13 for HMBS with a median Ct value of 21.35 ± 1.12. Among the 89 samples, RNA18S5 and ACTB were the most abundant genes, followed by GAPDH, while the least abundant gene was HMBS. The assessed reference genes showed significant inter-individual variations across the samples, with SD values ranging from 0.54 for RNA18S5 to 1.26 for UBC. Thus it is clear that a vigorous test is required to evaluate the reliability of the 15 candidate reference genes for use as internal controls for comparative gene expression normalization in GCs from PCOS patients.
Figure 1.
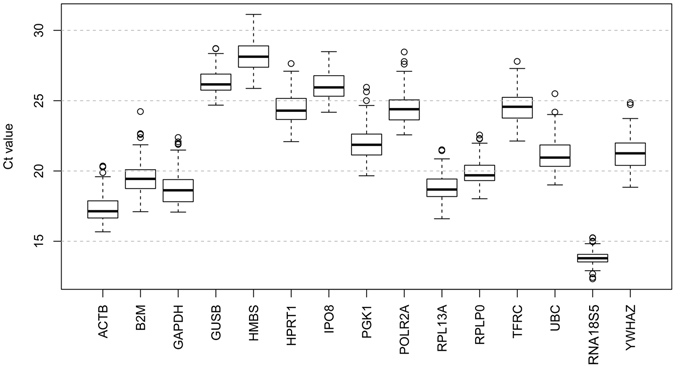
Distribution of threshold cycle (Ct) values for the 15 candidate reference genes. Box-and-whisker plots showing the range of Ct values for each reference gene. The black center line indicates the median Ct. The extended upper and lower hinges indicate 75th and 25th percentiles. The whiskers indicate the largest or smallest Ct values falling within 1.5 times the interquartile range from the upper and lower hinges. Small circles indicate the outliers.
Analysis of reference gene expression stability
We used three common algorithms – GeNorm, NormFinder, and BestKeeper – to individually examine the expression stability of the 15 candidate reference genes (Please refer to the Methods).
GeNorm analysis
GeNorm computed the M-values using a pairwise comparison approach to grade the 15 reference genes based on the similarity of these expression profiles rather than the minimal variation across the 89 samples. Lower M-values indicated greater expression stability. The computed M-values of all of the reference genes were below the defined cut-off value of 1.5, which are considered stably expressed by the algorithm (Fig. 2A). Following the step-wise exclusion of the least stably expressed gene with the highest M-value, the stability values were recalculated. In the pooled group, RPL13A and RLPP0 with the lowest M-values of 0.27 were the most stably expressed gene pair, followed by ACTB (0.41), HPRT1 (0.45), YWHAZ (0.46), GAPDH (0.49), PGK1 (0.52), HMBS (0.53), UBC (0.55), IPO8 (0.56), B2M (0.58), POLR2A (0.60), GUSB (0.62), TRFC (0.64), and RNA18S5 with the highest M-value of 0.67 making it the least stable gene across all samples.
Figure 2.
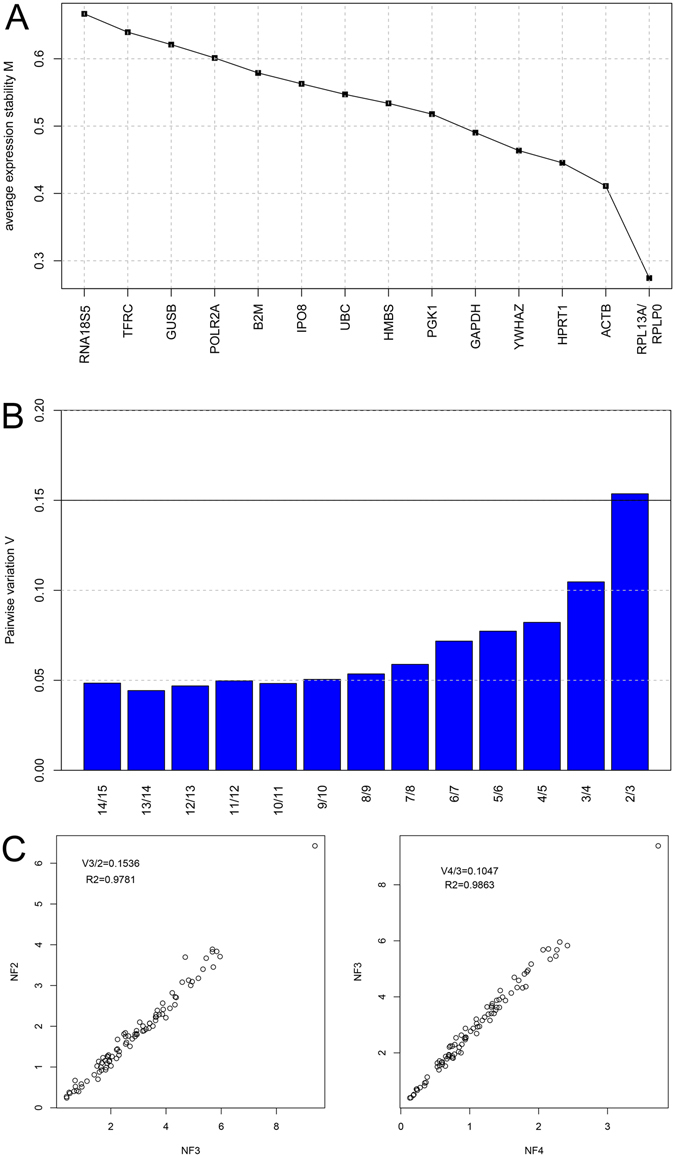
Gene expression stability analysis by GeNorm. (A) The mean expression stability value (M) of the 15 candidate reference genes. (B) Determination of the optimal number of reference genes for normalization using pairwise variation (V n/n+1) analysis by GeNorm. (C) Selected scatterplots of the normalization factor (NF) before (X-axis) and after (Y-axis) inclusion of an (n + 1)th control gene (r 2 = Spearman coefficient) at which the V-value defines the pair-wise variation between two sequential normalization factors.
We also computed the optimal number of reference genes based on the pairwise variation value (Vn/n+1). GeNorm defines a pairwise variation of 0.15 as the cutoff value, below which (Vn/n+1 < 0.15) the inclusion of an additional reference gene for normalization is not needed. As shown in Fig. 2B and C, the V2/3 value is just over the cutoff threshold of 0.15 with an expression ratio of 0.1536 (r 2 = 0.978), while the V3/4 value was below the cutoff with an expression ratio of 0.1047 (r 2 = 0.986), indicating that the top three ranked reference genes RPL13A, RLPP0, and ACTB (with the lowest M-values) were suggested for gene expression normalization in GCs from PCOS patients.
BestKeeper analysis
The BestKeeper applet computes the gene expression variation for individual reference genes according to the SD and CV of their Ct values across all of the samples. Figure 3 shows that GUSB was ranked the most stably expressed reference gene with a SD (±Ct) and CV (%Ct) of 0.70 and 2.67, respectively, followed by RNA18S5 (SD: 0.40, CV: 2.87), HMBS (SD: 0.90, CV: 3.19), IPO8 (SD: 0.88, CV: 3.38), TFRC (SD: 0.85, CV: 3.45), POLR2A (SD: 0.88, CV: 3.58), HPRT1 (SD: 0.88, CV: 3.59), RPLP0 (SD: 0.75, CV: 3.77), RPL13A (SD: 0.75, CV: 3.97), YWHAZ (SD: 0.88, CV: 4.12), PGK1 (SD: 0.95, CV: 4.30), ACTB (SD: 0.76, CV: 4.36), UBC (SD: 1.00, CV: 4.72), B2M (SD: 0.93, CV: 4.73), and GAPDH (SD: 0.94, CV: 5.02), which was ranked the least stable gene in GCs from PCOS.
Figure 3.
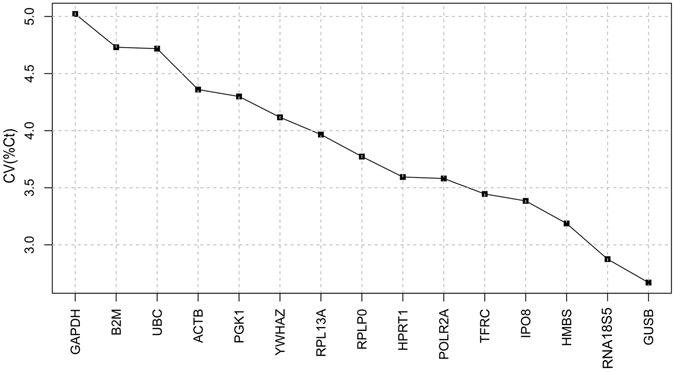
Gene expression stability analysis. Coefficient of variation (%Ct) of the 15 candidate reference genes by BestKeeper.
NormFinder analysis
We next used NormFinder, a model-based variance estimation algorithm based on the inter-group variation across samples and intra-group expression variation within the same condition to identify the optimal reference gene with minimal variability and coregulation across the samples. As shown in Fig. 4A, in contrast to the GeNorm and BestKeeper results, NormFinder ranked HPRT1 as the most stable reference gene with an M-value of 0.07, followed by YWHAZ (0.09), ACTB (0.42), HMBS (0.11), PGK1 (0.11), UBC (0.12), GAPDH (0.19), GUSB (0.2), IPO8 (0.21), POLR2A (0.22), RPLP0 (0.24), RNA18S5 (0.24), TFRC (0.25), RPL13A (0.25), and B2M, which, with the highest M-value of 0.28, was ranked as the least stable gene in GCs from PCOS patients.
Figure 4.
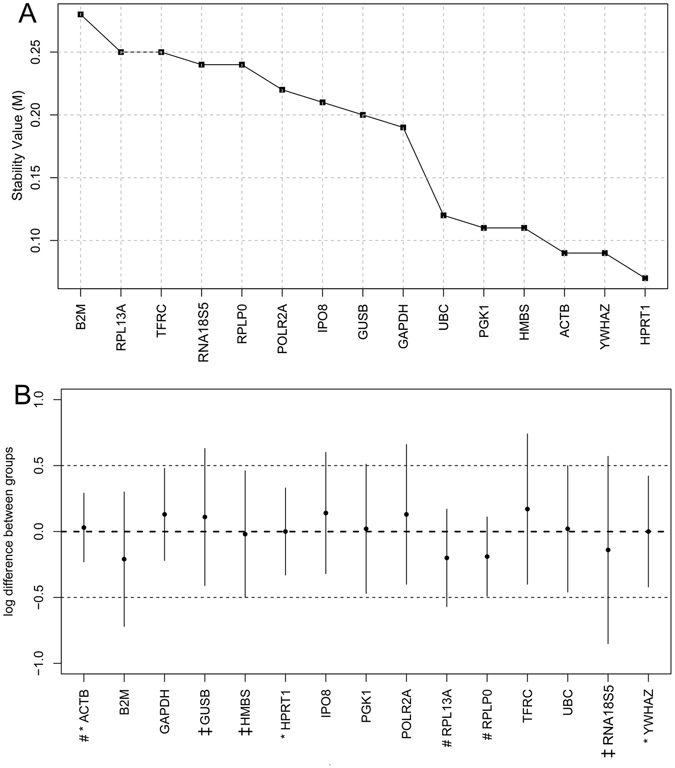
Gene expression stability analysis by NormFinder. (A) The mean expression stability value (M) of the 15 candidate reference genes by NormFinder. (B) The intergroup variation is plotted (small black circles) as the expression difference between PCOS patients and healthy controls. The vertical bars indicate intragroup variation to provide a confidence interval for the difference. The top three ranked candidate reference genes by GeNorm, BestKeeper, and NormFinder are marked by #, ‡, and *, respectively, before the gene names on the X-axis. The two thin dashed lines indicate hypothetical target genes with a log difference of 0.5 and −0.5.
Figure 4B shows the log difference between groups of the 15 reference genes ranked by the three different algorithms. The model-based NormFinder selected the most stable genes with minimal combined inter- and intragroup expression variation, whereas the pairwise comparison algorithms GeNorm and BestKeeper selected the genes with a low intragroup variation and roughly the same nonvanishing intergroup gene expression variation.
Final ranking of the 15 reference genes
Given the specific features of the three statistical algorithms, it is not surprising to see marked differences in ranking the 15 candidate reference genes. We used a previously reported method to take into consideration the three sets of results to produce a final ranking of the 15 reference genes47, 48. Briefly, the 15 reference genes were re-ranked according to their computed geometric mean from the three algorithms, and a smaller geometric mean indicated higher gene expression stability across all samples. As shown in Table 2, HPRT1 was the most stably expressed gene with the lowest geometric mean, followed by RPLP0, HMBS, YWHAZ, GUSB, ACTB, RPL13A, RNA18S5, IPO8, PKG1, GAPDH, UBS, POLR2A, TFRC, and B2M, which was the least stable. Thus HPRT1, RPLP0, and HMBS were the top three ranked reference genes for use as internal controls for qRT-PCR to normalize gene expression in GCs from PCOS patients.
Table 2.
Final rankings of the 15 candidate reference genes.
| Ranking | GeNorm | NormFinder | BestKeeper | Final ranking |
|---|---|---|---|---|
| 1 | RPL13A/RPLP0 | HPRT1 | GUSB | HPRT1 |
| 2 | YWHAZ | RNA18S5 | RPLP0 | |
| 3 | ACTB | ACTB | HMBS | HMBS |
| 4 | HPRT1 | HMBS | IPO8 | YWHAZ |
| 5 | YWHAZ | PGK1 | TFRC | GUSB |
| 6 | GAPDH | UBC | POLR2A | ACTB |
| 7 | PGK1 | GAPDH | HPRT1 | RPL13A |
| 8 | HMBS | GUSB | RPLP0 | RNA18S5 |
| 9 | UBC | IPO8 | RPL13A | IPO8 |
| 10 | IPO8 | POLR2A | YWHAZ | PGK1 |
| 11 | B2M | RPLP0 | PGK1 | GAPDH |
| 12 | POLR2A | RNA18S5 | ACTB | UBC |
| 13 | GUSB | TFRC | UBC | POLR2A |
| 14 | TFRC | RPL13A | B2M | TFRC |
| 15 | RNA18S5 | B2M | GAPDH | B2M |
Discussion
PCOS is a heterogeneous gynecological endocrine disorder characterized by a broad spectrum of anomalies, including chronic anovulation, hyperandrogenism, and hyperinsulinism with insulin resistance, and a growing body of evidence has linked dysfunctional GCs to the pathogenicity of PCOS. To gain insight into the underlying mechanism of impaired GC functions in PCOS development, qRT-PCR has been used extensively to measure differential gene expression in normal and PCOS samples. Most researchers have chosen a reference gene for target gene expression normalization based on the assumption of its inherent expression stability across the tested samples without prior validation. The ideal reference gene requires a constant expression level across all treatments, physiological conditions, tissue/cell types, and experimental designs, but no such reference genes have been identified and they might not even exist49–56.
The expressions of the commonly used reference genes in qRT-PCR analyses might vary considerably in specific biological contexts resulting in errors in gene expression estimations. Sadek et al. reported that YWHAZ, CYCI, and ACTB were the most stably expressed reference genes out of nine examined housekeeping genes in endometrial tissues collected from PCOS patients, and the commonly used GAPDH gene was not recommended57. Milutinović et al. showed that HPRT, in comparison to ACTB, BSM, and GAPDH, was the most stable internal control for normalizations in peripheral blood mononuclear cells in normal and obese women with PCOS58. Lindholm et al. (2011) examined the expression of a panel of inflammation markers in abdominal superficial subcutaneous tissues from overweight and lean patients with PCOS. They found that RPLP0 was the best reference gene for target gene expression normalization compared to LRP10 and PP1A. The identification of reliable reference genes for expression normalization in GCs in PCOS patients has not been reported.
In this study, we evaluated 15 candidate reference genes commonly used for routine gene expression normalization in comparative gene expression profiling by qRT-PCR. Given their dynamic expression levels with significant expression variation across the samples (Fig. 1), we used three commonly used statistical programs –GeNorm, BestKeeper, and NormFinder – to analyze their Ct values and to rank them according to their M-values. GeNorm indicated that RPL13A and RPLP0 were the two most stably expressed genes with the same computed M-values, followed by ACTB (Fig. 2A). The best combination recommended by GeNorm was RPL13A, RPLP0, and ACTB (Fig. 2B,C). In contrast, BestKeeper ranked GUSB, RNA18S5, and HMBS as the top three most stable genes, while RPLP0, RPL13A, and ACTB were ranked at 8th, 9th, and 12th place, respectively. It is also notable that RNA18S5 was the least stable gene according to GeNorm (Fig. 3). NormFinder determined HPRT1 and YWHAZ to be the two most stable genes, which were ranked at 4th and 5th place, respectively, by GeNorm and at 7th and 12th, respectively, by BestKeeper (Fig. 4). It is worth noting that both GeNorm and NormFinder ranked ACTB and GAPDH, the two most commonly used reference genes, at the top position; however, they were ranked as the 12th and the least stable genes by BestKeeper (Table 2). The rankings by GeNorm and NormFinder were more consistent with each other than with the BestKeeper algorithm. Finally, we produced the final ranking by considering the results generated by the three programs. We identified HPRT1, RPLP0, and HMBS as the three most stably expressed genes for use as internal controls for comparative gene expression profiling by qRT-PCR (Table 2).
To the best of our knowledge, there is no gold standard for the optimal number of the candidate reference genes required for the expression stability study. In general, six to twelve candidate genes are commonly included for the analysis59–76. The supply of cDNA samples and the lab resources dedicated for the characterization should also be considered to make the final decision on the number of candidate genes used for the analysis.
The three identified stably expressed reference controls are specific to our experimental design. It would be of great interest to test the expression stability of these normalizers in the GCs sampled from the PCOS patients from different geographic populations or different species. Nonetheless, our statistical pipelines to identify stably expressed reference genes for comparative gene expression analysis should be broadly applied for general biomedical research.
Method
Patient recruitment
Written informed consent was obtained from all participants or their next of kins, and the study was approved by the Institutional Review Board of Reproductive Medicine of Shandong University, and all methods were performed in accordance with the relevant guidelines and regulations. Eighty-nine Chinese Han women aged 29.12 ± 3.01 years were recruited from the Reproductive Hospital Affiliated to Shandong University between January 2015 and June 2016. This study included 44 PCOS patients and 45 women with normal ovulatory function undergoing in vitro fertilization (IVF) for tubal and/or male infertility treatment. The study required no modification of our routine IVF protocol.
Clinical and biochemical measurements
PCOS was diagnosed according to the 2003 Rotterdam criteria77, 78, including any two of the following three clinical features: oligo/anovulation, clinical and/or biochemical hyperandrogenism, and polycystic ovaries on ultrasound. Women with other pathophysiological conditions associated with hyperandrogenemia, including adrenal congenital hyperplasia, Cushing’s syndrome, and androgen-secreting tumors, were excluded.
Anthropometric parameters of all subjects, including body height, and weight were measured during the first visit to the outpatient department. The body mass index (BMI) was calculated as weight (kg) divided by the square of the height (m2). Venous blood samples were collected between 8:00 a.m. and 10:00 a.m. after a 12 h overnight fast. All blood samples from PCOS patients were obtained during the early follicular phase of their menstrual cycle on days 3–5. The serum levels of follicle-stimulating hormone (FSH), luteinizing hormone (LH), total testosterone, estrogen, progesterone, and prolactin were assayed enzymatically using an automated biochemistry analyzer (Olympus 600, Clinical Chemistry Analyzer, Olympus Diagnostica GmbH, Co. Clare, Ireland).
Isolation of GCs
Cumulus GCs were isolated from all 44 PCOS patients undergoing IVF and the 45 women in the control group undergoing regular treatment for tubal and/or male factor infertility. Volunteers with a history of other gynecological or medical disorders were excluded. All women were injected with gonadotropin-releasing hormone agonist at the onset of their midluteal phase, and an ultrasound scan and serum estradiol assays were performed to monitor follicle size. When three or more follicles with a mean diameter of R1.8 cm were detected, 8,000–10,000 IU human chorionic gonadotropin (Profasi, Serono) was administered 36 hours prior to the ultrasound-guided immature oocyte retrieval procedure. After anesthetization, the oocyte retrieval was performed through a 17-gauge double-lumen aspiration needle (K-OPS-WOOD-1235, Cook Australia). The cumulus GCs around the oocytes were collected and washed twice with Dulbecco’s modified Eagle medium (DMEM) for subsequent analysis after oocyte removal in follicular aspirates using a Pasteur pipette. Red blood cells were removed with lysis buffer.
RNA extraction and cDNA synthesis
Total RNA from isolated GCs was extracted using the Trizol Plus RNA Purification kit (Life Technologies) according to the manufacturer’s instructions. The RNA purity was confirmed using a NanoDrop 2000 (Thermo Scientific) and an A260:A280 ratio of 1.9–2.1. Total RNA (1 µg) was reverse transcribed to cDNA using the PrimeScript RT reagent Kit (Takara) and diluted with nuclease-free water to a final volume of 20 µl. The cDNAs were further diluted 1:20 with nuclease-free water for use as the DNA template for qRT-PCR.
qRT-PCR
We used an epMotion 5075LH (Eppendorf, Germany) automated liquid handling workstation for precise and accurate pipetting of qPCR reagents. The qPCR mix was prepared in a 10 µl final volume containing 5 µl SYBR Green Master Mix, 0.5 µl of primer pair solution (10 µM), 3 µl of nuclease-free water, and 1 µl of the diluted cDNA. The amplification reactions were performed in 384 well plates using PowerUp™ SYBR® Green Master Mix (ThermoFisher) on a QuantStudio™ 7 Flex Real-Time PCR System (Applied Biosystem, USA) according to the manufacturer’s instructions. The thermal cycling conditions were set as follows: uracil-DNA glycosylase (UDG) activation at 50 °C for 2 minutes, DNA polymerase activation at 95 °C for 2 minutes, 40 cycles of denaturation at 95 °C for 15 seconds and annealing/extension at 55 °C for 30 seconds. Specificity of amplification was confirmed by melting curve analysis. The qPCR primers sequences are shown in Table 3.
Table 3.
qPCR primer set of the 15 reference genes.
| Symbol | Gene name | Accession number | Forward primer (5′ → 3′) | Reverse primer (5′ → 3′) |
|---|---|---|---|---|
| RNA18S5 | 18S ribosomal RNA | NR_003286.2 | GGCGCCCCCTCGATGCTCTTAG | GCTCGGGCCTGCTTTGAACACTCT |
| ACTB | Actin beta | NM_001101.3 | CGACAGGATGCAGAAGGAG | ACATCTGCTGGAAGGTGGA |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | NM_001256799.2 | TGATGACATCAAGAAGGTGGTGAAG | TCCTTGGAGGCCATGTAGGCCAT |
| UBC | Ubiquitin C | NM_021009.6 | ATTTGGGTCGCGGTTCTTG | TGCCTTGACATTCTCGATGGT |
| B2M | Beta-2-microglobulin | NM_004048.2 | TGACTTTGTCACAGCCCAAGATA | CGGCATCTTCAAACCTCCA |
| POLR2A | RNA polymerase II subunit A | NM_000937.4 | GCACCACGTCCAATGACAT | GTGCGGCTGCTTCCATAA |
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 | NM_000194.2 | TGACACTGGCAAAACAATGCA | GGTCCTTTTCACCAGCAAGCT |
| YWHAZ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | NM_001135699.1 | ACTTTTGGTACATTGTGGCTTCAA | CCGCCAGGACAAACCAGTAT |
| HMBS | Hydroxymethyl-bilane synthase | NM_001135699.1 | GGCAATGCGGCTGCAA | GGGTACCCACGCGAATCAC |
| GUSB | Glucuronidase beta | NM_000181.3 | GTCTGCGGCATTTTGTCGG | CACACGATGGCATAGGAATGG |
| IPO8 | Importin 8 | NM_001190995.1 | TCCGAACTATTATCGACAGGACC | GTTCAAAGAGCCGAGCTACAA |
| PGK1 | Phosphoglycerate kinase 1 | NM_000291.3 | GAACAAGGTTAAAGCCGAGCC | GTGGCAGATTGACTCCTACCA |
| RPLP0 | Ribosomal protein lateral stalk subunit P0 | NM_001002.3 | AGCCCAGAACACTGGTCTC | ACTCAGGATTTCAATGGTGCC |
| TFRC | Transferrin receptor | NM_001128148.2 | GGCTACTTGGGCTATTGTAAAGG | CAGTTTCTCCGACAACTTTCTCT |
| RPL13A | Ribosomal protein L13a | NM_001270491.1 | GCCATCGTGGCTAAACAGGTA | GTTGGTGTTCATCCGCTTGC |
Statistical Analyses
The expression stability of the 15 reference genes was analyzed using the three R-based or Microsoft Excel-based algorithms – GeNorm56, NormFinder79, and BestKeeper80. GeNorm selects an ideal reference gene pair whose expression ratios exhibit minimal influence by external factors across different experimental conditions and samples. The first step in the algorithm is to determine the computed stability value (M-value) based on the average pair-wise variation between a gene and all other assessed reference genes as an indication of the stability of the reference gene expression. The gene with the highest M-value is then eliminated, and the selection process will be repeated to select the two most stably expressed genes with the lowest M-values. GeNorm was also used to determine the minimal number of reference genes for gene expression normalization. According to the pairwise variation calculation, for any gene pair with a cutoff (Vn/n+1) lower than 0.15, an additional (n + 1) reference gene will not significantly improve the normalization and thus is not required for normalization56.
NormFinder calculates the variance within and between groups to determine the gene expression stability. Based on the estimate of the intragroup and intergroup stability, a stability value will be assigned for each reference gene. The program ranks all assessed genes based on the M-values, and the gene with the lowest stability is considered the most stably expressed gene.
BestKeeper determines the expression stability according to the coefficient of correlation to the BestKeeper index, which consists of the geometric means of the Ct values of the tested gene set. The coefficient of variance (CV) and standard derivation (SD) are determined, and the reference gene with the lowest CV and SD is considered the most stably expressed gene. Genes with SD > 1 are discarded. This program considers the relationship between the BestKeeper index and the reference genes by calculating the Pearson correlation coefficient, the coefficient of determination (r 2), and the p-value.
Numerical values of the clinical characteristics of PCOS cases and controls were expressed as the mean ± SD. Independent-samples t-tests were performed using SPSS v.19.0 (SPSS Inc., Chicago, IL, USA). A value of p < 0.05 was regarded as statistically significant.
Acknowledgements
This work was supported in part by funds from the One-off Funding of CUHK for Joint Lab/Research Collaboration provided to CUHK-Shandong University Joint Laboratory on Reproductive Genetics (Project 3132970) and to the Hong Kong Branch of the Chinese National Center for Assisted Reproductive Technology and Reproductive Genetics (Project 3132987), from the Hospital for Reproductive Medicine, Shandong University provided to CUHK-Shandong University Joint Laboratory on Reproductive Genetics (Project 7104693), and from the Young Scholars Program of Shandong University (2016WLJH50).
Author Contributions
L.V.Y., Z.H.A.O., S.G., L.U.G., L.E.U.N.G., C.K., X.I.O.N., Z.Q., S.U., X.W., M.A., J.L., C.H.A.N., W.Y., and L.I.U., H.B. participated in the literature search, experimental work, data mining, data analysis and interpretation, writing the manuscript, and creating the figures and tables. L.U.G. supervised the study design and critically read and edited the manuscript. All authors approved the final version for publication.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Gang Lu, Email: lugang@cuhk.edu.hk.
Hong Bin Liu, Email: humangenetics2008@hotmail.com.
References
- 1.Wild S, Pierpoint T, Jacobs H, McKeigue P. Long-term consequences of polycystic ovary syndrome: results of a 31 year follow-up study. Hum Fertil (Camb) 2000;3:101–105. doi: 10.1080/1464727002000198781. [DOI] [PubMed] [Google Scholar]
- 2.Kovacs G, Wood C. The current status of polycystic ovary syndrome. Aust N Z J Obstet Gynaecol. 2001;41:65–68. doi: 10.1111/j.1479-828X.2001.tb01296.x. [DOI] [PubMed] [Google Scholar]
- 3.Azziz R, et al. The prevalence and features of the polycystic ovary syndrome in an unselected population. J Clin Endocrinol Metab. 2004;89:2745–2749. doi: 10.1210/jc.2003-032046. [DOI] [PubMed] [Google Scholar]
- 4.Christian RC, et al. Prevalence and predictors of coronary artery calcification in women with polycystic ovary syndrome. J Clin Endocrinol Metab. 2003;88:2562–2568. doi: 10.1210/jc.2003-030334. [DOI] [PubMed] [Google Scholar]
- 5.March WA, et al. The prevalence of polycystic ovary syndrome in a community sample assessed under contrasting diagnostic criteria. Hum Reprod. 2010;25:544–551. doi: 10.1093/humrep/dep399. [DOI] [PubMed] [Google Scholar]
- 6.Ehrmann DA. Polycystic ovary syndrome. The New England journal of medicine. 2005;352:1223–1236. doi: 10.1056/NEJMra041536. [DOI] [PubMed] [Google Scholar]
- 7.Jayasena CN, Franks S. The management of patients with polycystic ovary syndrome. Nat Rev Endocrinol. 2014;10:624–636. doi: 10.1038/nrendo.2014.102. [DOI] [PubMed] [Google Scholar]
- 8.Orio F, Jr., et al. The cardiovascular risk of young women with polycystic ovary syndrome: an observational, analytical, prospective case-control study. J Clin Endocrinol Metab. 2004;89:3696–3701. doi: 10.1210/jc.2003-032049. [DOI] [PubMed] [Google Scholar]
- 9.Spritzer PM, Lecke SB, Satler F, Morsch DM. Adipose tissue dysfunction, adipokines, and low-grade chronic inflammation in polycystic ovary syndrome. Reproduction. 2015;149:R219–227. doi: 10.1530/REP-14-0435. [DOI] [PubMed] [Google Scholar]
- 10.Stepto NK, et al. Women with polycystic ovary syndrome have intrinsic insulin resistance on euglycaemic-hyperinsulaemic clamp. Hum Reprod. 2013;28:777–784. doi: 10.1093/humrep/des463. [DOI] [PubMed] [Google Scholar]
- 11.Ozcan Dag Z, Oguzturk O, Isik Y, Turkel Y, Bulcun E. Personality profile in patients with polycystic ovary syndrome. Gynecol Endocrinol. 2015;31:540–542. doi: 10.3109/09513590.2015.1018162. [DOI] [PubMed] [Google Scholar]
- 12.Ehrmann DA, Barnes RB, Rosenfield RL. Polycystic-Ovary-Syndrome as a Form of Functional Ovarian Hyperandrogenism Due to Dysregulation of Androgen Secretion. Endocrine Reviews. 1995;16:322–353. doi: 10.1210/er.16.3.322. [DOI] [PubMed] [Google Scholar]
- 13.Yong EL, Baird DT, Yates R, Reichert LE, Hillier SG. Hormonal-Regulation of the Growth and Steroidogenic Function of Human Granulosa-Cells. J Clin Endocr Metab. 1992;74:842–849. doi: 10.1210/jcem.74.4.1548349. [DOI] [PubMed] [Google Scholar]
- 14.Jakimiuk AJ, Weitsman SR, Navab A, Magoffin DA. Luteinizing hormone receptor, steroidogenesis acute regulatory protein, and steroidogenic enzyme messenger ribonucleic acids are overexpressed in thecal and granulosa cells from polycystic ovaries. J Clin Endocr Metab. 2001;86:1318–1323. doi: 10.1210/jcem.86.3.7318. [DOI] [PubMed] [Google Scholar]
- 15.Willis DS, et al. Premature response to luteinizing hormone of granulosa cells from anovulatory women with polycystic ovary syndrome: Relevance to mechanism of anovulation. J Clin Endocr Metab. 1998;83:3984–3991. doi: 10.1210/jcem.83.11.5232. [DOI] [PubMed] [Google Scholar]
- 16.Kanamarlapudi V, Gordon UD, Bernal AL. Luteinizing hormone/chorionic gonadotrophin receptor overexpressed in granulosa cells from polycystic ovary syndrome ovaries is functionally active. Reproductive Biomedicine Online. 2016;32:635–641. doi: 10.1016/j.rbmo.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Das M, et al. Granulosa cell survival and proliferation are altered in polycystic ovary syndrome. J Clin Endocr Metab. 2008;93:881–887. doi: 10.1210/jc.2007-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magoffin DA, Weitsman SR. Insulin-Like Growth-Factor-I Regulation of Luteinizing-Hormone (Lh) Receptor Messenger-Ribonucleic-Acid Expression and Lh-Stimulated Signal-Transduction in Rat Ovarian Theca-Interstitial Cells. Biol Reprod. 1994;51:766–775. doi: 10.1095/biolreprod51.4.766. [DOI] [PubMed] [Google Scholar]
- 19.Udoff L, Adashi EY. Polycystic Ovarian Disease - a New Look at an Old Subject. Curr Opin Obstet Gyn. 1995;7:340–343. [PubMed] [Google Scholar]
- 20.Pellatt L, et al. Granulosa cell production of anti-Mullerian hormone is increased in polycystic ovaries. J Clin Endocr Metab. 2007;92:240–245. doi: 10.1210/jc.2006-1582. [DOI] [PubMed] [Google Scholar]
- 21.Homburg R, et al. The relationship of serum anti-Mullerian hormone with polycystic ovarian morphology and polycystic ovary syndrome: a prospective cohort study. Hum Reprod. 2013;28:1077–1083. doi: 10.1093/humrep/det015. [DOI] [PubMed] [Google Scholar]
- 22.Homburg R, Crawford G. The role of AMH in anovulation associated with PCOS: a hypothesis. Hum Reprod. 2014;29:1117–1121. doi: 10.1093/humrep/deu076. [DOI] [PubMed] [Google Scholar]
- 23.Lan, C. W. et al. Functional microarray analysis of differentially expressed genes in granulosa cells from women with polycystic ovary syndrome related to MAPK/ERK signaling. Sci Rep-Uk5, doi:10.1038/srep14994 (2015). [DOI] [PMC free article] [PubMed]
- 24.Schmidt J, et al. Differential expression of inflammation-related genes in the ovarian stroma and granulosa cells of PCOS women. Mol Hum Reprod. 2014;20:49–58. doi: 10.1093/molehr/gat051. [DOI] [PubMed] [Google Scholar]
- 25.Chronowska E. High-throughput analysis of ovarian granulosa cell transcriptome. Biomed Res Int. 2014;2014:213570. doi: 10.1155/2014/213570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nivet AL, Vigneault C, Blondin P, Sirard MA. Changes in granulosa cells’ gene expression associated with increased oocyte competence in bovine. Reproduction. 2013;145:555–565. doi: 10.1530/REP-13-0032. [DOI] [PubMed] [Google Scholar]
- 27.Martoriati A, Gerard N. Interleukin-1 (IL-1) system gene expression in granulosa cells: kinetics during terminal preovulatory follicle maturation in the mare. Reprod Biol Endocrinol. 2003;1:42. doi: 10.1186/1477-7827-1-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang F, et al. Alternative splicing of the androgen receptor in polycystic ovary syndrome. Proc Natl Acad Sci U S A. 2015;112:4743–4748. doi: 10.1073/pnas.1418216112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chazenbalk G, et al. Abnormal expression of genes involved in inflammation, lipid metabolism, and Wnt signaling in the adipose tissue of polycystic ovary syndrome. J Clin Endocrinol Metab. 2012;97:E765–770. doi: 10.1210/jc.2011-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Resende LO, et al. Single-cell expression analysis of BMP15 and GDF9 in mature oocytes and BMPR2 in cumulus cells of women with polycystic ovary syndrome undergoing controlled ovarian hyperstimulation. J Assist Reprod Genet. 2012;29:1057–1065. doi: 10.1007/s10815-012-9825-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemieux S, Lewis GF, Ben-Chetrit A, Steiner G, Greenblatt EM. Correction of hyperandrogenemia by laparoscopic ovarian cautery in women with polycystic ovarian syndrome is not accompanied by improved insulin sensitivity or lipid-lipoprotein levels. J Clin Endocrinol Metab. 1999;84:4278–4282. doi: 10.1210/jcem.84.11.6140. [DOI] [PubMed] [Google Scholar]
- 32.Long W, et al. Characterization of serum microRNAs profile of PCOS and identification of novel non-invasive biomarkers. Cell Physiol Biochem. 2014;33:1304–1315. doi: 10.1159/000358698. [DOI] [PubMed] [Google Scholar]
- 33.Salilew-Wondim D, et al. Polycystic ovarian syndrome is accompanied by repression of gene signatures associated with biosynthesis and metabolism of steroids, cholesterol and lipids. J Ovarian Res. 2015;8:24. doi: 10.1186/s13048-015-0151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wissing ML, et al. The transcriptome of corona radiata cells from individual Mcapital I, Ukrainiancapital I, Ukrainian oocytes that after ICSI developed to embryos selected for transfer: PCOS women compared to healthy women. J Ovarian Res. 2014;7:110. doi: 10.1186/s13048-014-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams J, et al. Enhanced Inflammatory Transcriptome in the Granulosa Cells of Women With Polycystic Ovarian Syndrome. J Clin Endocrinol Metab. 2016;101:3459–3468. doi: 10.1210/jc.2015-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Resende LOT, et al. Single-cell expression analysis of BMP15 and GDF9 in mature oocytes and BMPR2 in cumulus cells of women with polycystic ovary syndrome undergoing controlled ovarian hyperstimulation. J Assist Reprod Gen. 2012;29:1057–1065. doi: 10.1007/s10815-012-9825-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salilew-Wondim, D. et al. Polycystic ovarian syndrome is accompanied by repression of gene signatures associated with biosynthesis and metabolism of steroids, cholesterol and lipids. J Ovarian Res8, doi:10.1186/s13048-015-0151-5 (2015). [DOI] [PMC free article] [PubMed]
- 38.Artimani T, et al. Downregulation of adiponectin system in granulosa cells and low levels of HMW adiponectin in PCOS. J Assist Reprod Gen. 2016;33:101–110. doi: 10.1007/s10815-015-0620-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wissing, M. L. et al. The transcriptome of corona radiata cells from individual MII oocytes that after ICSI developed to embryos selected for transfer: PCOS women compared to healthy women. J Ovarian Res7, doi:10.1186/s13048-014-0110-6 (2014). [DOI] [PMC free article] [PubMed]
- 40.Teixeira FL, et al. Aberrant expression of growth differentiation factor-9 in oocytes of women with polycystic ovary syndrome. J Clin Endocr Metab. 2002;87:1337–1344. doi: 10.1210/jcem.87.3.8316. [DOI] [PubMed] [Google Scholar]
- 41.Catteau-Jonard S, et al. Anti-Mullerian Hormone, Its Receptor, FSH Receptor, and Androgen Receptor Genes Are Overexpressed by Granulosa Cells from Stimulated Follicles in Women with Polycystic Ovary Syndrome. J Clin Endocr Metab. 2008;93:4456–4461. doi: 10.1210/jc.2008-1231. [DOI] [PubMed] [Google Scholar]
- 42.Patel P, Boyd CA, Johnston DG, Williamson C. Analysis of GAPDH as a standard for gene expression quantification in human placenta. Placenta. 2002;23:697–698. doi: 10.1053/plac.2002.0859. [DOI] [PubMed] [Google Scholar]
- 43.Meller M, Vadachkoria S, Luthy DA, Williams MA. Evaluation of housekeeping genes in placental comparative expression studies. Placenta. 2005;26:601–607. doi: 10.1016/j.placenta.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 44.Nestorov J, Matic G, Elakovic I, Tanic N. Gene Expression Studies: How to Obtain Accurate and Reliable Data by Quantitative Real-Time Rt Pcr. J Med Biochem. 2013;32:325–338. doi: 10.2478/jomb-2014-0001. [DOI] [Google Scholar]
- 45.Dheda K, et al. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal Biochem. 2005;344:141–143. doi: 10.1016/j.ab.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 46.Murphy J, Bustin SA. Reliability of real-time reverse-transcription PCR in clinical diagnostics: gold standard or substandard? Expert Rev Mol Diagn. 2009;9:187–197. doi: 10.1586/14737159.9.2.187. [DOI] [PubMed] [Google Scholar]
- 47.Chen D, Pan X, Xiao P, Farwell MA, Zhang B. Evaluation and identification of reliable reference genes for pharmacogenomics, toxicogenomics, and small RNA expression analysis. J Cell Physiol. 2011;226:2469–2477. doi: 10.1002/jcp.22725. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y, Chen D, Smith MA, Zhang B, Pan X. Selection of reliable reference genes in Caenorhabditis elegans for analysis of nanotoxicity. PLoS One. 2012;7:e31849. doi: 10.1371/journal.pone.0031849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 50.Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 51.Schmittgen TD, Zakrajsek BA. Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods. 2000;46:69–81. doi: 10.1016/S0165-022X(00)00129-9. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki T, Higgins PJ, Crawford DR. Control selection for RNA quantitation. Biotechniques. 2000;29:332–337. doi: 10.2144/00292rv02. [DOI] [PubMed] [Google Scholar]
- 53.Thellin O, et al. Housekeeping genes as internal standards: use and limits. J Biotechnol. 1999;75:291–295. doi: 10.1016/S0168-1656(99)00163-7. [DOI] [PubMed] [Google Scholar]
- 54.Tricarico C, et al. Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal Biochem. 2002;309:293–300. doi: 10.1016/S0003-2697(02)00311-1. [DOI] [PubMed] [Google Scholar]
- 55.Warrington JA, Nair A, Mahadevappa M, Tsyganskaya M. Comparison of human adult and fetal expression and identification of 535 housekeeping/maintenance genes. Physiol Genomics. 2000;2:143–147. doi: 10.1152/physiolgenomics.2000.2.3.143. [DOI] [PubMed] [Google Scholar]
- 56.Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology3, RESEARCH0034 (2002). [DOI] [PMC free article] [PubMed]
- 57.Sadek KH, et al. Variation in stability of housekeeping genes in endometrium of healthy and polycystic ovarian syndrome women. Hum Reprod. 2012;27:251–256. doi: 10.1093/humrep/der363. [DOI] [PubMed] [Google Scholar]
- 58.Nestorov DVM. D. M. I. B. A. J. B. M. M. N. G. M. J. Hypoxanthine Guanine Phosphoribosyl Transferase Is the Most Stable Reference Gene for Gene Expression Analysis by Quantitative PCR in Peripheral Blood Mononuclear Cells from Women with the Polycystic Ovary Syndrome. J Med Biochem. 2014;33:356–363. [Google Scholar]
- 59.Zhang C, Li T, Hou CL, Shen XY. Selection of reference genes from Shiraia bambusicola for RT-qPCR analysis under different culturing conditions. AMB Express. 2017;7:14. doi: 10.1186/s13568-016-0314-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen Y, et al. Selection of reference genes for quantitative real-time PCR normalization in creeping bentgrass involved in four abiotic stresses. Plant Cell Rep. 2015;34:1825–1834. doi: 10.1007/s00299-015-1830-9. [DOI] [PubMed] [Google Scholar]
- 61.Xu L, et al. Validation of Reference Genes for Quantitative Real-Time PCR during Bicolor Tepal Development in Asiatic Hybrid Lilies (Lilium spp.) Front Plant Sci. 2017;8:669. doi: 10.3389/fpls.2017.00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, W. X. et al. Selection of Suitable Reference Genes for Quantitative Real-Time PCR Normalization in Three Types of Rat Adipose Tissue. Int J Mol Sci17, doi:10.3390/ijms17060968 (2016). [DOI] [PMC free article] [PubMed]
- 63.Wang H, Zhang X, Liu Q, Liu X, Ding S. Selection and evaluation of new reference genes for RT-qPCR analysis in Epinephelus akaara based on transcriptome data. Plos One. 2017;12:e0171646. doi: 10.1371/journal.pone.0171646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robledo D, et al. Analysis of qPCR reference gene stability determination methods and a practical approach for efficiency calculation on a turbot (Scophthalmus maximus) gonad dataset. BMC Genomics. 2014;15:648. doi: 10.1186/1471-2164-15-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van de Moosdijk AA, van Amerongen R. Identification of reliable reference genes for qRT-PCR studies of the developing mouse mammary gland. Sci Rep. 2016;6:35595. doi: 10.1038/srep35595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rydbirk R, et al. Assessment of brain reference genes for RT-qPCR studies in neurodegenerative diseases. Sci Rep. 2016;6:37116. doi: 10.1038/srep37116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gong H, et al. Evaluation of candidate reference genes for RT-qPCR studies in three metabolism related tissues of mice after caloric restriction. Sci Rep. 2016;6:38513. doi: 10.1038/srep38513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaluzna M, Kuras A, Pulawska J. Validation of reference genes for the normalization of the RT-qPCR gene expression of virulence genes of Erwinia amylovora in apple shoots. Sci Rep. 2017;7:2034. doi: 10.1038/s41598-017-02078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dzaki N, Ramli KN, Azlan A, Ishak IH, Azzam G. Evaluation of reference genes at different developmental stages for quantitative real-time PCR in Aedes aegypti. Sci Rep. 2017;7:43618. doi: 10.1038/srep43618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martins MQ, et al. Selection and Validation of Reference Genes for Accurate RT-qPCR Data Normalization in Coffea spp. under a Climate Changes Context of Interacting Elevated [CO2] and Temperature. Front Plant Sci. 2017;8:307. doi: 10.3389/fpls.2017.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wan PJ, et al. Reference genes for quantitative real-time PCR analysis in symbiont Entomomyces delphacidicola of Nilaparvata lugens (Stal) Sci Rep. 2017;7:42206. doi: 10.1038/srep42206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hou F, et al. Identification and validation of reference genes for quantitative real-time PCR studies in long yellow daylily, Hemerocallis citrina Borani. Plos One. 2017;12:e0174933. doi: 10.1371/journal.pone.0174933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.De Spiegelaere W, et al. Reference gene validation for RT-qPCR, a note on different available software packages. Plos One. 2015;10:e0122515. doi: 10.1371/journal.pone.0122515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ji H, et al. Selection of Reliable Reference Genes for Real-time qRT-PCR Analysis of Zi Geese (Anser anser domestica) Gene Expression. Asian-Australas J Anim Sci. 2013;26:423–432. doi: 10.5713/ajas.2012.12417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Palombella S, et al. Identification of reference genes for qPCR analysis during hASC long culture maintenance. Plos One. 2017;12:e0170918. doi: 10.1371/journal.pone.0170918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mo F, Zhao J, Liu N, Cao LH, Jiang SX. Validation of reference genes for RT-qPCR analysis of CYP4T expression in crucian carp. Genet Mol Biol. 2014;37:500–507. doi: 10.1590/S1415-47572014000400005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chang J, et al. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertility and Sterility. 2004;81:19–25. doi: 10.1016/j.fertnstert.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 78.Fauser BCJM, et al. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome (PCOS) Human Reproduction. 2004;19:41–47. doi: 10.1093/humrep/deh098. [DOI] [PubMed] [Google Scholar]
- 79.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 80.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]


