Abstract
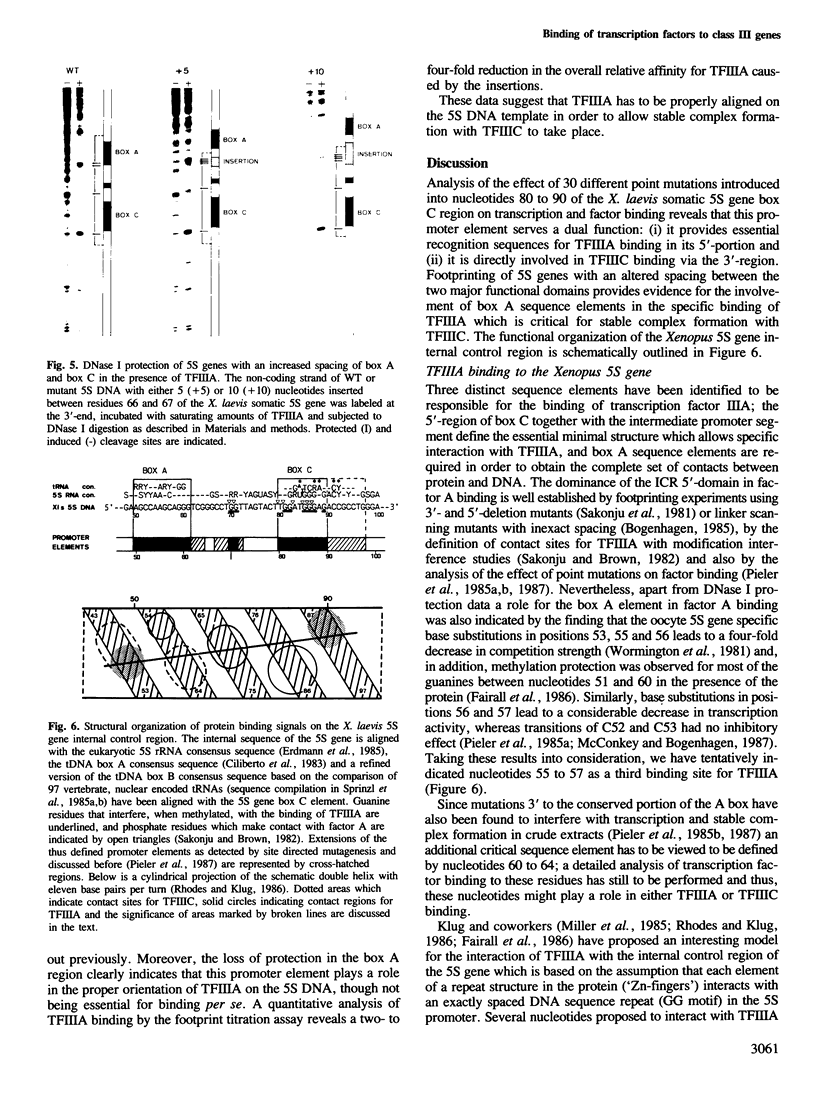
We have previously shown that of the two functional domains which constitute the Xenopus 5S gene promoter the common, conserved box A element is directly involved in the binding of the common transcription factor IIIC. Here, we describe the investigation of the role of the 5S gene specific promoter element, box C, in transcription factor binding. Analysis of 22 different single site basepair changes reveals that mutations created within the 5'-region of this segment interfere with transcription due to a reduced affinity for TFIIIA, whereas sequence alterations introduced into the 3'-region of the same element similarly inhibit transcription, but do not result in a measurable defect in TFIIIA binding. Instead, they clearly reduce the affinity for TFIIIC. DNase I protection experiments with TFIIIA on 5S mutants which have an increased spacing of box A and box C demonstrate that TFIIIA recognizes a specific box A sequence element and that the factor has to be properly aligned on the DNA template in order to allow stable complex formation with TFIIIC to proceed. The structural and functional organization of protein binding signals on the 5S gene internal control region will be discussed in regard to these results.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker R. E., Gabrielsen O., Hall B. D. Effects of tRNATyr point mutations on the binding of yeast RNA polymerase III transcription factor C. J Biol Chem. 1986 Apr 25;261(12):5275–5282. [PubMed] [Google Scholar]
- Bieker J. J., Martin P. L., Roeder R. G. Formation of a rate-limiting intermediate in 5S RNA gene transcription. Cell. 1985 Jan;40(1):119–127. doi: 10.1016/0092-8674(85)90315-0. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F. The intragenic control region of the Xenopus 5 S RNA gene contains two factor A binding domains that must be aligned properly for efficient transcription initiation. J Biol Chem. 1985 May 25;260(10):6466–6471. [PubMed] [Google Scholar]
- Bohmann D., Keller W., Dale T., Schöler H. R., Tebb G., Mattaj I. W. A transcription factor which binds to the enhancers of SV40, immunoglobulin heavy chain and U2 snRNA genes. Nature. 1987 Jan 15;325(6101):268–272. doi: 10.1038/325268a0. [DOI] [PubMed] [Google Scholar]
- Camier S., Gabrielsen O., Baker R., Sentenac A. A split binding site for transcription factor tau on the tRNA3Glu gene. EMBO J. 1985 Feb;4(2):491–500. doi: 10.1002/j.1460-2075.1985.tb03655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M. F., Gerrard S. P., Cozzarelli N. R. Analysis of RNA polymerase III transcription complexes by gel filtration. J Biol Chem. 1986 Mar 25;261(9):4309–4317. [PubMed] [Google Scholar]
- Ciliberto G., Raugei G., Costanzo F., Dente L., Cortese R. Common and interchangeable elements in the promoters of genes transcribed by RNA polymerase iii. Cell. 1983 Mar;32(3):725–733. doi: 10.1016/0092-8674(83)90058-2. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Control of eukaryotic messenger RNA synthesis by sequence-specific DNA-binding proteins. 1985 Aug 29-Sep 4Nature. 316(6031):774–778. doi: 10.1038/316774a0. [DOI] [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Erdmann V. A., Wolters J., Huysmans E., De Wachter R. Collection of published 5S, 5.8S and 4.5S ribosomal RNA sequences. Nucleic Acids Res. 1985;13 (Suppl):r105–r153. doi: 10.1093/nar/13.suppl.r105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairall L., Rhodes D., Klug A. Mapping of the sites of protection on a 5 S RNA gene by the Xenopus transcription factor IIIA. A model for the interaction. J Mol Biol. 1986 Dec 5;192(3):577–591. doi: 10.1016/0022-2836(86)90278-0. [DOI] [PubMed] [Google Scholar]
- Fuhrman S. A., Engelke D. R., Geiduschek E. P. HeLa cell RNA polymerase III transcription factors. Functional characterization of a fraction identified by its activity in a second template rescue assay. J Biol Chem. 1984 Feb 10;259(3):1934–1943. [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Birnstiel M. L. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981 Dec 17;294(5842):626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- Hanas J. S., Bogenhagen D. F., Wu C. W. Binding of Xenopus transcription factor A to 5S RNA and to single stranded DNA. Nucleic Acids Res. 1984 Mar 26;12(6):2745–2758. doi: 10.1093/nar/12.6.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda B. M., Roeder R. G. Association of a 5S gene transcription factor with 5S RNA and altered levels of the factor during cell differentiation. Cell. 1980 Nov;22(1 Pt 1):119–126. doi: 10.1016/0092-8674(80)90160-9. [DOI] [PubMed] [Google Scholar]
- Kramer W., Drutsa V., Jansen H. W., Kramer B., Pflugfelder M., Fritz H. J. The gapped duplex DNA approach to oligonucleotide-directed mutation construction. Nucleic Acids Res. 1984 Dec 21;12(24):9441–9456. doi: 10.1093/nar/12.24.9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladiges W. C., Raff R. F., Brown S., Deeg H. J., Storb R. The canine major histocompatibility complex. Supertypic specificities defined by the primed lymphocyte test (PLT). Immunogenetics. 1984;19(4):359–365. doi: 10.1007/BF00345410. [DOI] [PubMed] [Google Scholar]
- Lassar A. B., Martin P. L., Roeder R. G. Transcription of class III genes: formation of preinitiation complexes. Science. 1983 Nov 18;222(4625):740–748. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- Mangin M., Ares M., Jr, Weiner A. M. Human U2 small nuclear RNA genes contain an upstream enhancer. EMBO J. 1986 May;5(5):987–995. doi: 10.1002/j.1460-2075.1986.tb04313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I. W., Lienhard S., Jiricny J., De Robertis E. M. An enhancer-like sequence within the Xenopus U2 gene promoter facilitates the formation of stable transcription complexes. Nature. 1985 Jul 11;316(6024):163–167. doi: 10.1038/316163a0. [DOI] [PubMed] [Google Scholar]
- McConkey G. A., Bogenhagen D. F. Transition mutations within the Xenopus borealis somatic 5S RNA gene can have independent effects on transcription and TFIIIA binding. Mol Cell Biol. 1987 Jan;7(1):486–494. doi: 10.1128/mcb.7.1.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H. R., Brown D. D. A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieler T., Appel B., Oei S. L., Mentzel H., Erdmann V. A. Point mutational analysis of the Xenopus laevis 5S gene promoter. EMBO J. 1985 Jul;4(7):1847–1853. doi: 10.1002/j.1460-2075.1985.tb03859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieler T., Hamm J., Roeder R. G. The 5S gene internal control region is composed of three distinct sequence elements, organized as two functional domains with variable spacing. Cell. 1987 Jan 16;48(1):91–100. doi: 10.1016/0092-8674(87)90359-x. [DOI] [PubMed] [Google Scholar]
- Pieler T., Oei S. L., Hamm J., Engelke U., Erdmann V. A. Functional domains of the Xenopus laevis 5S gene promoter. EMBO J. 1985 Dec 30;4(13B):3751–3756. doi: 10.1002/j.1460-2075.1985.tb04144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D., Klug A. An underlying repeat in some transcriptional control sequences corresponding to half a double helical turn of DNA. Cell. 1986 Jul 4;46(1):123–132. doi: 10.1016/0092-8674(86)90866-4. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Brown D. D. Contact points between a positive transcription factor and the Xenopus 5S RNA gene. Cell. 1982 Dec;31(2 Pt 1):395–405. doi: 10.1016/0092-8674(82)90133-7. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Brown D. D., Engelke D., Ng S. Y., Shastry B. S., Roeder R. G. The binding of a transcription factor to deletion mutants of a 5S ribosomal RNA gene. Cell. 1981 Mar;23(3):665–669. doi: 10.1016/0092-8674(81)90429-3. [DOI] [PubMed] [Google Scholar]
- Segall J., Matsui T., Roeder R. G. Multiple factors are required for the accurate transcription of purified genes by RNA polymerase III. J Biol Chem. 1980 Dec 25;255(24):11986–11991. [PubMed] [Google Scholar]
- Setzer D. R., Brown D. D. Formation and stability of the 5 S RNA transcription complex. J Biol Chem. 1985 Feb 25;260(4):2483–2492. [PubMed] [Google Scholar]
- Sive H. L., Heintz N., Roeder R. G. Multiple sequence elements are required for maximal in vitro transcription of a human histone H2B gene. Mol Cell Biol. 1986 Oct;6(10):3329–3340. doi: 10.1128/mcb.6.10.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive H. L., Roeder R. G. Interaction of a common factor with conserved promoter and enhancer sequences in histone H2B, immunoglobulin, and U2 small nuclear RNA (snRNA) genes. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6382–6386. doi: 10.1073/pnas.83.17.6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Moll J., Meissner F., Hartmann T. Compilation of tRNA sequences. Nucleic Acids Res. 1985;13 (Suppl):r1–49. doi: 10.1093/nar/13.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Vorderwülbecke T., Hartmann T. Compilation of sequences of tRNA genes. Nucleic Acids Res. 1985;13 (Suppl):r51–104. doi: 10.1093/nar/13.suppl.r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman D. J., Caspers P., Geiduschek E. P. Effects of temperature and single-stranded DNA on the interaction of an RNA polymerase III transcription factor with a tRNA gene. Cell. 1985 Feb;40(2):311–317. doi: 10.1016/0092-8674(85)90145-x. [DOI] [PubMed] [Google Scholar]
- Stillman D. J., Geiduschek E. P. Differential binding of a S. cerevisiae RNA polymerase III transcription factor to two promoter segments of a tRNA gene. EMBO J. 1984 Apr;3(4):847–853. doi: 10.1002/j.1460-2075.1984.tb01895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke M. W., Roeder R. G. Multiple proteins bind to VA RNA genes of adenovirus type 2. Mol Cell Biol. 1987 Mar;7(3):1021–1031. doi: 10.1128/mcb.7.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormington W. M., Bogenhagen D. F., Jordan E., Brown D. D. A quantitative assay for Xenopus 5S RNA gene transcription in vitro. Cell. 1981 Jun;24(3):809–817. doi: 10.1016/0092-8674(81)90106-9. [DOI] [PubMed] [Google Scholar]
- Yoshinaga S. K., Boulanger P. A., Berk A. J. Resolution of human transcription factor TFIIIC into two functional components. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3585–3589. doi: 10.1073/pnas.84.11.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]