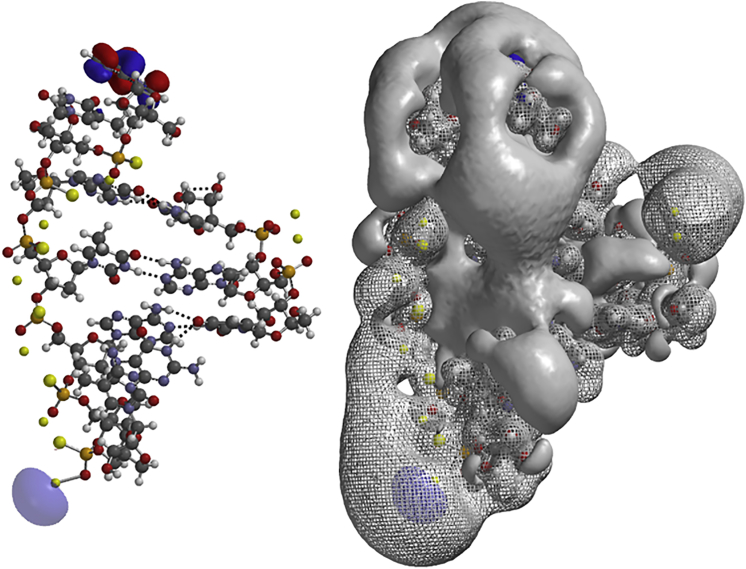
Abstract
Important oligonucleotides in anti-sense research have been investigated in silico and experimentally. This involves quantum mechanical (QM) calculations and chromatography experiments on locked nucleic acid (LNA) phosphorothioate (PS) oligonucleotides. iso-potential electrostatic surfaces are essential in this study and have been calculated from the wave functions derived from the QM calculations that provide binding information and other properties of these molecules. The QM calculations give details of the electronic structures in terms of e.g., energy and bonding, which make them distinguish or differentiate between the individual PS diastereoisomers determined by the position of sulfur atoms. Rules are derived from the electronic calculations of these molecules and include the effects of the phosphorothioate chirality and formation of electrostatic potential surfaces. Physical and electrochemical descriptors of the PS oligonucleotides are compared to the experiments in which chiral states on these molecules can be distinguished. The calculations demonstrate that electronic structure, electrostatic potential, and topology are highly sensitive to single PS configuration changes and can give a lead to understanding the activity of the molecules.
Keywords: LNA phosphorothioate, DNA/LNA oligonucleotide, diastereoisomers, Hartree-Fock calculations, iso-potential surface, anion chromatograms
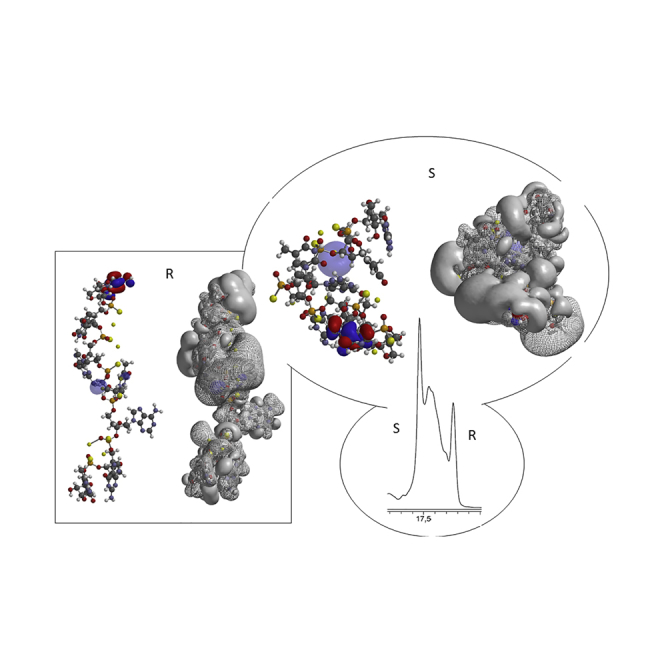
Graphical Abstract
Introduction
The pioneering work on phosphorothioate (PS) oligonucleotides was performed more than four decades ago.1, 2, 3, 4, 5 The initial work1 focused on the biological changes caused by the substitution of a native oxygen by sulfur.2, 4, 5 Replacing a non-bridging oxygen with sulfur creates a chiral center at phosphorous and produces two chiral isomers: Rp and Sp. Thus, for every PS linkage introduced in an oligonucleotide, two diastereoisomers are created. Because conventional solid-phase PS synthesis is not stereoselective, an N-mer PS oligonucleotide contains random mixtures of 2N diastereoisomers.3, 4, 6, 7, 8 Diastereoisomers exhibit different physical and chemical properties. In the pioneering stereo-regular synthesis work of Stec et al.9, it was concluded that the Sp configuration generally provided better exonuclease resistance compared to the Rp, but the Sp and not the Rp configuration was the substrate for endonucleolytic enzymes. On the other hand, Rp isomers were much better than Sp substrates for DNA-dependent RNA polymerases, RNase H, and stimulated immune responses.6, 7, 9, 10, 11
Stevenson12 and Zamecnik13 demonstrated in 1978 that synthetic oligonucleotides can be used to block translation and transcription. They used a synthetic 13-mer DNA and proved the principle that later was called antisense. At that time, it was appreciated that further nucleolytic stabilization of the native diester linkages was required for any practical therapeutic use of these molecules.11 PS oligonucleotides had been used in 1970 by CDe Clercq and co-workers14 to stabilize enzymatically produced polyribonucleotides. That work led to the selection of PSs as nucleolytic stabilizers for synthetic antisense oligonucleotides (AONs).15, 16, 17, 18, 19, 20, 21 Although non-stereodefined AONs composed of random diastereoisomeric mixtures were employed, the PS chirality issue has been continuously discussed.18, 22, 23, 24 However, firm conclusions on the necessity for producing stereo-defined AONs were not reached. For instance, it was demonstrated that PS chirality produced a small increase of the binding affinity to RNA targets,24 and also that chirality had little influence on protein binding.22 Other physical properties were also demonstrated to be dependent on stereochemistry. Karwowski et al.25 demonstrated in their paper on chiral synthesis of dimeric locked nucleic acid (LNA) that high-performance liquid chromatography (HPLC) retention time was changed with the Rp and Sp isometri. It was proposed, for practical antisense drug discovery and development, that PS chirality would play only a minor role.23, 24 With the advent of new affinity-increasing synthetic nucleotides, it was thought that the exonuclease protective properties of some chiral PS combinations, the Sp form in particular, would not be needed to prevent nucleolytic degradation.26 Others pointed to a potential advance in controlling PS chirality and speculated that the modest increase in affinity and RNase H recruitment of the Rp isomers,10, 27 in combination with the increased stability against 3′ exonucleases in human serum (Sp), alone or in combination with other chemical modifications, could be useful for productive use in the design of AONs.10, 27
Controlling PS chirality has attracted attention in antisense research, but the use of stereo-defined antisense PSs has not been widely implemented. One reason for this was that other chemical modifications that produced significant improvements in potency and nuclease stability emerged.28, 29, 30 In particular, sugar modifications, examples such as the high-affinity bicyclic LNA and LNA analogs,31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47 produced new enabling results for oligonucleotide therapeutics.
However, recent work has highlighted the importance of PS chirality. Meena et al. (N.I. Meena et al., 2015, AsiaTides, conference) showed that stereo-defined diastereoisomers of the marketed antisense drug Mipomersen exhibited improved properties. Mipomersen was approved by the U.S. Food and Drug Administration (FDA) as a random mixture comprising all the possible diastereoisomers. They concluded that controlling PS chirality can provide marked advantages for oligonucleotide therapeutics (N.I. Meena et al., 2015, AsiaTides, conference). In contrast, it has also recently been reported that controlling PS chirality in the gap region (DNA-PS segment) of AON gapmers did not produce discernible benefits for therapeutic applications and that a diastereoisomeric mix of Rp and Sp is required to balance between good activity and nuclease stability.48
At this time, the importance of controlling PS chirality is still debated. In order to approach this controversy from a novel perspective, we report here a quantum mechanical (QM) study on LNA PS oligonucleotides. A work published in 201449 demonstrated that QM modeling of oligonucleotides could be used to describe and understand fundamental properties and experimental observations, e.g., chromatograms. Here, we report an extension of that work and demonstrate for LNA oligonucleotides that PS chirality influences to a great extent the central properties of potential relevance for therapeutic oligonucleotides.
In the following section, the results from quantum mechanical calculations are presented and compared to new anion exchange (AIE) chromatogram experiments. Details of the methodology are described in Materials and Methods at the end.
Results
Justification of Computational Methods
The molecular systems investigated here are computationally large oligonucleotides that exhibit many minima in their potential energy surfaces. This created the possibility of identifying many possible stable conformers. Therefore, in order to justify the results, we started by demonstrating the relevance of the methods. First, the specificity of the methods was made plausible by studying a LNA-PS trimer (5′-TTT-3′). Second, the correctness of the methods was made plausible by the comparison of a structure optimized in Hartree-Fock self-consistent field (HF-SCF) calculations on a 9-mer DNA (5′-gcgaaagct-3′) with a published crystal structure.
In some of the calculations, environment effects have been included by either adding solvent molecules explicitly or implicitly by appropriate change of dielectric parameters. These changes did not seem to alter the structure dramatically.
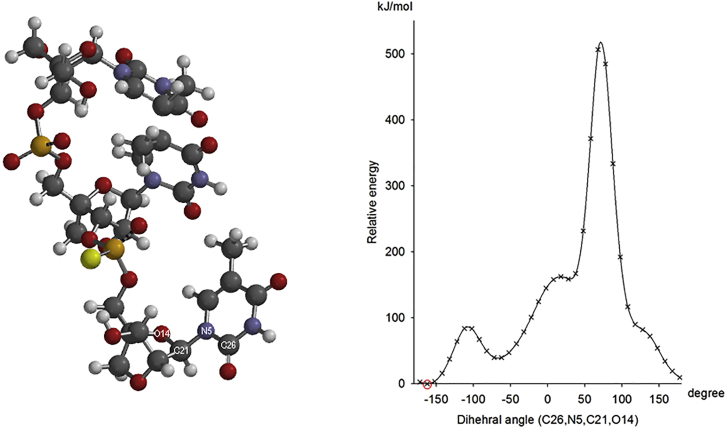
The trimer (5′-TTT-3′) was built as described in the Materials and Methods, and its structure was optimized by HF-SCF calculations (Figure 1). The two thymine nucleobases at the 5′ end interacted by stacking, whereas the thymine nucleobase at the 3′ end was displaced from the two other bases. On this background, we then chose to investigate how sensitive the energy of the molecule was to the rotation of the nucleobase at the 3′ end. The energy was calculated as a function of the rotation around the dihedral angle of the N-glycosidic bond of the 3′ nucleotide. The dihedral angle was rotated in steps of 10° over 360°, and the energy was calculated at each point. Figure 1 shows that the energy of the molecule is very dependent on the dihedral angle, reaching a maximal value above 500 kJ/mol relative to the energy of the most stable conformer. However, Figure 1 also shows that only a large change of the dihedral angle will lead to a different conformer and that different conformers are likely to reside in one of the three minima. Thus, this gives us confidence that the HF-SCF optimization method leads to relatively deep minima, and that a given initial condition can produce the same specific final structure. Further discussions are given in the Material and Methods section.
Figure 1.
Ab Initio HF-SCF Results for LNA-PS Trimer: 5′-TTSpT-3′
(Left) Optimized structure of 5′-TTSpT-3′. (Right) Relative energy as a function of rotation around the dihedral angle of the N-glycosidic bond (3′ nucleotide). The red circle shows the optimized structure (left).
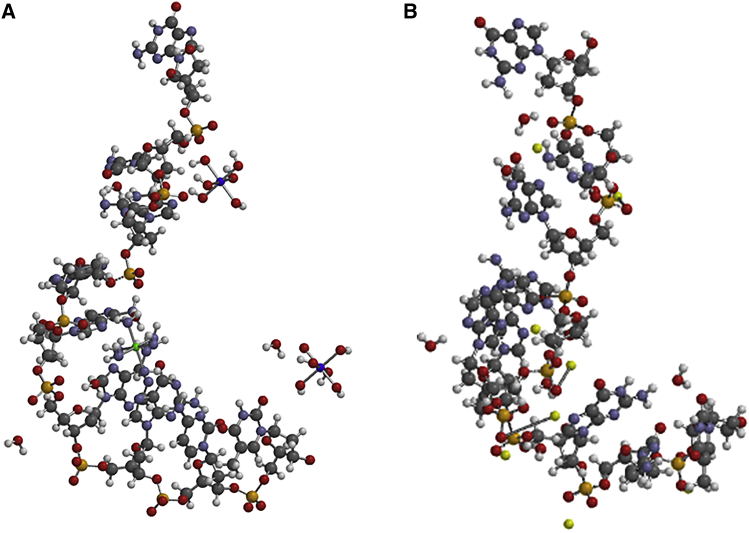
Next, justification of the structures obtained by the HF-SCF optimization method was performed by comparing a calculated structure with a crystal structure of the same compound. We chose to compare a calculated structure of the DNA 9-mer (5′-gcgaaagct-3′) with the crystal structure of the same oligonucleotide obtained from PDB, 1IXJ. The crystallographic structure was modified by removing the magnesium and cobalt ions together with their ligands. The system was made neutral by the addition of eight sodium atoms. Figure 2 shows the original crystal structure of the DNA 9-mer (A) together with the structure optimized in HF-SCF calculations (B) starting from the crystal structure. In both cases, the DNA 9-mer exhibits a distinct helical structure, with striking similarities in the helical parameters between the crystal structure and the optimized modified structure. For example, the 5′-O to 3′-O distance was 31.84 Å in the crystal structure and 28.53 Å in the optimized modified structure. In addition, it was found that the N-glycosidic dihedral angles of the last three bases, -gct-3′, varied less than 10° between the two structures, providing a basis for the similar stacking pattern. In this way, the optimized structure produced in HF-SCF calculations is very similar to the structure of the same molecule obtained from X-ray crystallographic measurements. We also produced some molecular dynamics (MD) simulations using the Metropolis algorithm,50 which gave less accurate results. For the MD simulations, the MMFFaqua force field (Merck Pharma) from Spartan, see Materials and Methods, was utilized. This has also been used to generate low-energy structures of the oligomers, leading to very similar minima, but this was besides the focus of the paper.
Figure 2.
Calculated and Crystal Structure of 9-mer DNA Diester: 5′-ggtaaacca-3′
(A) Crystal structure (1IXJ) with ligand ions magnesium and cobalt. (B) HF-SCF optimized structure of the sodium salt starting from the crystal structure without magnesium and cobalt ligand ions.
In view of the results obtained from the DNA-PS trimer and the DNA 9-mer, both the specificity and correctness of the results of HF-SCF optimizations were demonstrated. This makes us confident in using the chosen quantum mechanics methods when studying the DNA-PS and LNA-PS oligonucleotides.
HF-SCF Calculations of 8-mer DNA- and LNA-PS: 5′-cacactcc-3′ and 5′-CACACTCC-3′
The structure of the 8-mer DNA-PS with all-phosphorus stereo centers Rp (all-Rp) is shown in Figure 3A. The structure of the corresponding compound with all-phosphorous stereo centers Sp (all-Sp) is shown in Figure 3B. The all-Sp compound, Figure 3B, exhibits a helical, rather straight structure, whereas the all-Rp compound exhibits a globular, “ball-like” structure. This is reflected in the short 5′-O to 3′-O distance of only 10.5 Å for the all-Rp compound as compared to the 5′-O to 3′-O distance of 34.1 Å for the all-Sp compound (Table 1). Thus, PS chirality has a major influence on the structure of the 8-mer DNA-PS compound.
Figure 3.
Ab Initio HF-SCF Optimized Structures and Electrostatic Potentials of 8-mer DNA PS Oligonucleotides: 5′-CACACTCC-3′
(A) DNA-PS with all phosphorous in Rp configuration. (B) All phosphorous in Sp configuration.
Table 1.
DNA, in Small Letters, and LNA, in Capital Letters, Oligonucleotides with Sequence 5′-CACACTCC-3′ and Specific Chirality of the Phosphorous Atoms
| DNA/LNA Oligonucleotides | Relative Energy (eV) | Dipole (Debye) | Density, 0.002 electrons/au3 | Electrostatic Potential, 83.68 kJ/mol | Electrostatic Potential −83.68 kJ/mol | Distance (5′-O to 3′-O) (Å) | Structure | HOMO (eV) | LUMO (eV) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Design and Sequence (5′ to 3′) | Area (Å2) | Accessible Area (Å2) | Area (Å2) | Accessible Area (Å2) | Area (Å2) | Accessible Area (Å2) | ||||||
| cacactcc: 7Rp | 0.00 | 53.72 | 1,909 | 794 | 2,358 | 1,111 | –a | 411 | 10.5 | non helical | −7.67 | −0.75 |
| CACACTCC: 7Rp | 1.03b | 14.64 | 2,143 | 1,008 | 2,548 | 1,182 | 1,369 | 292 | 33.6 | helical | −8.35 | −0.05 |
| C-Sp-ACACTCC: 6Rp | 0.98b | 11.53 | 2,142 | 991 | 2,577 | 1,205 | 1,324 | 317 | 31.7 | helical | −8.36 | −0.07 |
| CACA-Sp-CTCC: 6Rp | 0.72b | 16.90 | 2,130 | 964 | 2,459 | 1,067 | 1,222 | 262 | 25.7 | helical | −8.35 | −0.15 |
| CACACTC-Sp-C: 6Rp | 1.03b | 14.66 | 2,141 | 1,017 | 2,554 | 1,203 | 1,374 | 296 | 35.1 | helical | −8.40 | −0.05 |
| cacactcc: 7Sp | 2.50c | 61.18 | 1,961 | 918 | 2,412 | 1,212 | 1,315 | 370 | 34.1 | helical | −8.34 | −0.82 |
| CACACTCC: 7Sp | 1.07b | 63.81 | 2,107 | 989 | 2,527 | 1,119 | 1,372 | 459 | 35.3 | helical | −8.03 | −0.25 |
| C-Rp-ACACTCC: 6Sp | 0.00 | 52.57 | 2,056 | 907 | 2,320 | 974 | –a | 372 | 27.4 | helical | −8.07 | 0.14 |
| CACA-Rp-CTCC: 6Sp | 0.86b | 68.39 | 2,101 | 991 | 2,560 | 1,147 | 1,369 | 476 | 36.6 | helical | −8.02 | −0.28 |
| CACACTC-Rp-C: 6Sp | 1.06b | 62.35 | 2,104 | 987 | 2,528 | 1,114 | 1,372 | 443 | 35.0 | helical | −7.91 | −0.25 |
The results are derived in HF-SCF calculations, with the basis set as 6-31G*. The table includes the structures of the oligonucleotides, their relative energies, their dipole moments, and the distances between 5′-O and 3′-O. Also included are the energies of the HOMOs and LUMOs as well as the areas and accessible areas of the charge density and electrostatic potentials.
The potential was truncated.
Energies are relative to that of C-Rp-ACACTCC 6Sp.
Energy is relative to that of cacactcc 7Rp.
Figure 3 also includes the frontier orbitals: the highest occupied molecular orbital (HOMO) (solid) and the lowest unoccupied molecular orbital (LUMO) (transparent). It is noted that the HOMO orbitals are localized on a particular nucleobase. In the case of the all-Rp compound, this is on the fourth nucleobase, adenine, counting from the 5′ end. In the case of the all-Sp compound, the HOMO is localized on the cytosine at the 3′ end of the molecule. The LUMO for each molecule is localized on one of the sodium atoms. This is consistent with the partial positive charge associated with the sodium ions, as shown in Mulliken population analyses using Mulliken charges, natural charges, and electrostatic charges, all showing that the sodium atoms have an appreciable positive charge.51
Figure 3 shows the surfaces of the electrostatic potentials, with iso-values chosen as 83.68 kJ/mol (mesh) and −83.68 kJ/mol (solid). This energy corresponds to two to three times the energy of a hydrogen bond, and, as such, is illustrative for the sites on the oligonucleotides that interact and bind by hydrogen bonding and salt bridge formation. The structural difference between the all-Rp and all-Sp compounds is also reflected in the “accessible areas.” The accessible surface area or solvent accessible surface area is an area accessible to solvent on a given bio-molecule.52, 53 It is calculated using a rolling ball of solvent with a specially chosen radius, in this case 1.00 Å, employed to probe the surface of a molecule. In the SPARTAN program, a region is considered inaccessible if the ball or sphere of solvent is centered on a line normal to the surface, and when touching a point on the surface in the region, is causing disruption of other regions. The accessible surface area of the density and of the positive electrostatic potential is larger for the all-Sp compound. In contrast, the accessible surface area of negative electrostatic potential is larger for the all-Rp compound: 411 Å2 (Rp) as compared to 370 Å2 (all-Sp).
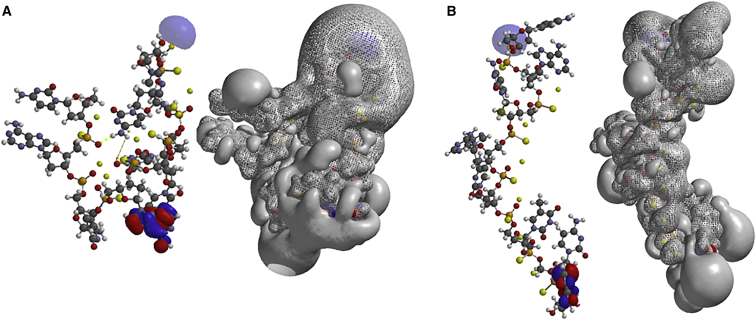
When the bridge 2′-O-CH2-4′ is added to the deoxyribose rings, the iso-sequential 8-mer LNA phosphorothioate (LNA-PS) with locked ribose rings was created. For LNA-PS nucleotides, cytosine is 5-methylated. The molecular structure and electrostatic potentials for the all-Rp LNA-PS oligonucleotide are shown in Figure 4. The corresponding results for the all-Sp LNA-PS oligonucleotide are also shown. The molecular structures and topologies of the electrostatic potentials differ considerably from those of the corresponding DNA-PS nucleotide. The most conspicuous change in the structures of the 8-mers, going from the DNA-PS oligonucleotide to the LNA-PS oligonucleotide, was that both LNAs, i.e., all Rp or all Sp, exhibit helical (helix-like) and more stretched structures. In this context, the helical structure is defined by the sugar phosphate backbone of the molecule having a spiraling turn, but does not necessarily have the bases stacked. Furthermore, going from the DNA-PS to the fully modified LNA-PS leads, for both LNAs, to increased areas of charge densities and increased areas of electrostatic potentials. This is consistent with the larger number of atoms in the LNA-PS molecules as compared to the DNA-PS molecules. It was expected that the increased rigidity in the LNA compounds would result in straighter structures with increased 5′-O to 3′-O distances. From Table 1, it is noted that the 5′-O to 3′-O distance of the all-Rp LNA-PS, 33.6 Å, is much larger than the corresponding distance for the all-Rp DNA-PS, 10.5 Å. However, the corresponding change going from the DNA to the LNA congener of the all-Sp isomers created only a modest increase from 34.1 Å to 35.3 Å.
Figure 4.
Ab Initio HF-SCF Optimized Structure and Electrostatic Potential of 8-mer LNA PS Oligonucleotide: 5′-CACACTCC-3′
All phosphorous are in either Rp (right) and Sp (left) configuration.
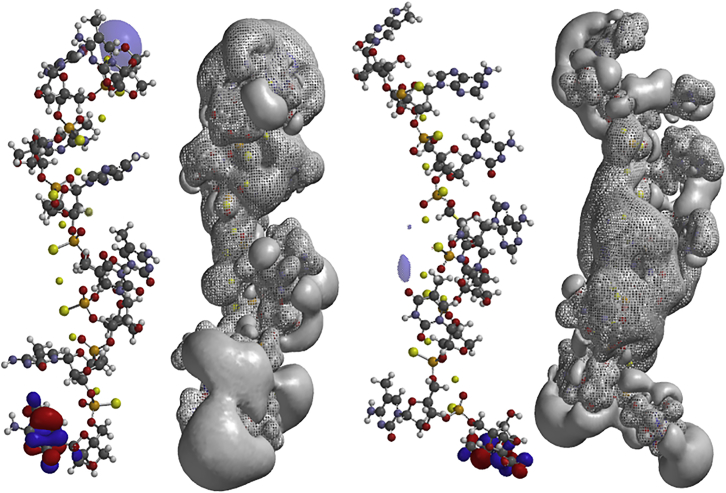
The influence of PS chirality at the phosphorous atoms was further investigated by successively exchanging a single configuration on phosphorus. One Rp center in the all-Sp and one Sp center in the all-Rp LNA-PS were exchanged for the other configuration to determine the length sensitivity of single configuration changes. In Figure 5 and Table 1, the structures of three such single PS chirality modifications are shown. The topology of the electrostatic potentials was very sensitive to a single PS configuration change, and single modifications influence the topology of the entire molecule. It was also observed that the area of the positive electrostatic potentials was approximately twice as large as that of the negative electrostatic potentials. Furthermore, the accessible surface areas of the negative electrostatic potentials were significantly larger for the all-Sp oligonucleotides or the oligonucleotides with one Sp replaced by one Rp than for the oligonucleotides with six Rps. Differences in the accessible areas are potentially important for interaction of the oligonucleotides with solvents in addition to other potential molecular interactions. Thus, the more open and stretched structure the oligonucleotides attained, the higher the accessible surface area of both polarities and, therefore, the larger the potential interaction area.
Figure 5.
Ab Initio HF-SCF Optimized Structures and Electrostatic Potentials of 8-mer LNA PS Oligonucleotides: 5′-CACACTCC-3′
(A–C) One Sp was inserted in the all-Rp LNA PS at the first (A), fourth (B), and sixth (C) PS position counted from the 5′ end.
The lengths of the oligonucleotides were also sensitive to the change in PS chirality. Insertion of a single Sp at the ends of the all-Rp LNA did not change the length significantly. However, a single Sp in the middle of the all-Rp LNA reduced the length from 33.6 Å to 25.7 Å. For the all-Sp LNA-PS, a single Rp in the middle and at the 3′ end did not change the lengths significantly. In contrast, an Rp at the 5′ end of a fully Sp LNA-PS reduced the length from 35.3 Å to 27.4 Å.
There are other parameters for monitoring the electrostatic structures of the oligonucleotides (Table 1). For example, the dipole moments also showed PS chirality sensitivity; the calculated values for the LNA-PS Rp series is much lower than those of the LNA-PS Sp series.
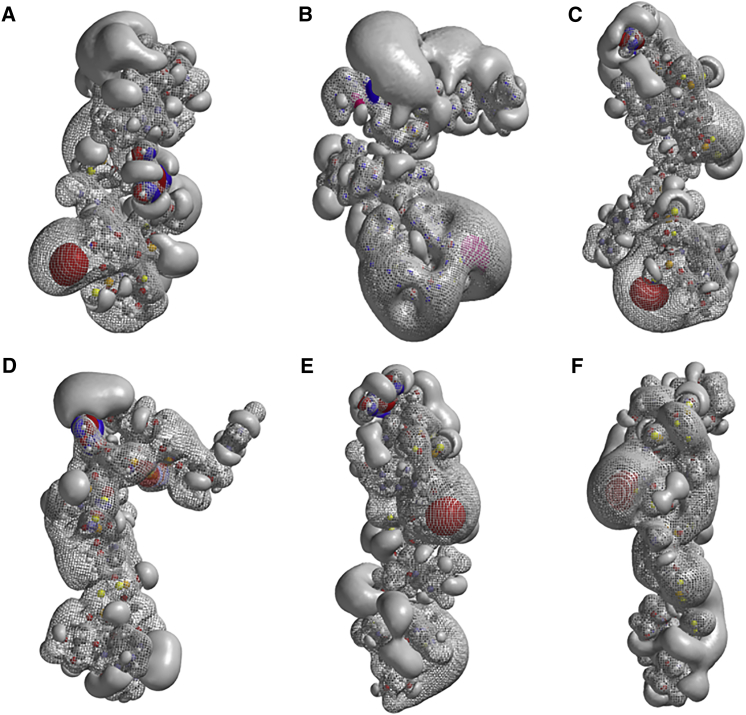
Quantum Mechanical Calculation of 7-mer-LNA-PS Gapmers: 5′-ATgtaGmC-3′
7-mer LNA-PS gapmers were constructed with either all-Rp or all-Sp configurations. The subtleties of single PS chirality changes were elucidated by studying additional 7-mer LNA-PS gapmers by replacing one Rp configuration in the all-Rp oligonucleotide with a single Sp. This was done successively, creating six 7-mer LNA-PS gapmers with all-Rp configurations but one Sp. Likewise, six 7-mer LNA-PS gapmers were created by replacing one Sp configuration with a single Rp in the all-Sp oligonucleotide. Six sodium atoms were added to the oligonucleotides, and the structures were optimized in HF-SCF calculations (Figures 6 and 7; Table 2). The all-Rp LNA-PS gapmer has a helical structure; the distance from 5′-O to 3′-O is 34.7 Å. The corresponding all-Sp LNA-PS gapmer has a “ball-like” structure (Figure 6), with a distance of 14.5 Å from 5′-O to 3′-O. This differs from the DNA 8-mer oligonucleotides, for which the all-Rp oligonucleotide is much shorter than the all-Sp oligonucleotide.
Figure 6.
Ab Initio HF-SCF Optimized Structure and Electrostatic Potential of 7-mer LNA PS Oligonucleotide 5′-AT-gta-GC-3′, with All Phosphorous in Sp Configuration
Also shown are the HOMO and LUMO.
Figure 7.
Ab Initio HF-SCF Optimized Structures and Electrostatic Potentials of 7-mer LNA PS Oligonucleotides 5′-AT-gta-GC-3′
(A–F) One Rp is inserted from the 5′ end (A) to the 3′ end (F) in the all-Sp LNA PS.
Table 2.
LNA 7-mer Oligonucleotide Gapmers with Sequence 5′-ATgtaGC-3′ and Specific Chirality of the Phosphorous Atoms
| LNA Oligonucleotides |
Relative Energya (eV) |
Dipole (Debye) |
Density (0.002 electrons/au3) |
Electrostatic Potential (83.68 kJ/mol) |
Electrostatic Potential (−83.68 kJ/mol) |
Distance (5′-O to 3′-O) (Å) |
Structure |
HOMO (eV) |
LUMO (eV) |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Design and Sequence (5′ to 3′) | Area (Å2) | Accessible Area (Å2) | Area (Å2) | Accessible Area (Å2) | Area (Å2) | Accessible Area (Å2) | ||||||
| ATgtaGC: 6Rp | 0.96 | 33.68 | 1,802 | 876 | 2,159 | 1,040 | 1,135 | 214 | 34.7 | helical | −7.88 (A) | −0.39 |
| A-Sp-TgtaGC: 5Rp | 0.63 | 23.23 | 1,783 | 832 | 2,078 | 942 | 1,157 | 142 | 32.8 | helical | −7.98 (g) | −0.27 |
| AT-Sp-gta GC: 5Rp | 0.01 | 25.06 | 1,775 | 765 | 2,030 | 884 | 1,030 | 115 | 27.8 | helical | −7.97 (A) | −0.19 |
| ATg-Sp-taGC: 5Rp | 1.37 | 36.31 | 1,772 | 821 | 2,113 | 1,024 | 1,241 | 188 | 30.3 | helical | −7.82 (A) | −1.19 |
| ATgt-Sp-aGC: 5Rp | 0.73 | 29.73 | 1,793 | 853 | 2,067 | 971 | 1,102 | 185 | 22.5 | non helical | −7.91 (A) | 0.16 |
| ATgta-Sp-GC: 5Rp | 1.81 | 24.67 | 1,805 | 843 | 2,220 | 1,156 | 1,173 | 209 | 32.3 | helical | −7.69 (G) | −0.65 |
| ATgtaG-Sp-C: 5Rp | 2.39 | 34.35 | 1,812 | 856 | 2,182 | 1,067 | 1,256 | 219 | 32.9 | helical | −7.87 (A) | −0.46 |
| ATgtaGC: 6Sp | 1.26 | 33.16 | 1,659 | 668 | 2,045 | 843 | 1,100 | 240 | 14.5 | non helical | −7.88 (a) | −0.40 |
| A-Rp-TgtaGC: 5Sp | 1.24 | 27.94 | 1,739 | 747 | 2,040 | 941 | 1,166 | 183 | 24.4 | non helical | −8.14 (a) | −0.52 |
| AT-Rp-gtaGC: 5Sp | 0.00 | 61.30 | 1,759 | 820 | 2,172 | 1,140 | 1,447 | 441 | 26.3 | helical | −7.59 (g) | −0.80 |
| ATg-Rp-taGC: 5Sp | 0.68 | 5.81 | 1,734 | 756 | 1,989 | 819 | 1,150 | 116 | 20.4 | helical | −8.13 (G) | −0.27 |
| ATgt-Rp-aGC: 5Sp | 1.87 | 39.63 | 1,729 | 772 | 2,039 | 941 | 1,189 | 177 | 29.4 | helical | −7.99 (A) | −0.54 |
| ATgta-Rp-GC: 5Sp | 2.41 | 26.02 | 1,696 | 731 | 1,990 | 909 | 1,156 | 139 | 29.8 | helical | −8.18 (A) | −0.53 |
| ATgtaG-Rp-C: 5Sp | 1.62 | 30.37 | 1,763 | 762 | 1,997 | 908 | 1,115 | 239 | 30.2 | helical | −7.57 (G) | −0.60 |
The results are derived in HF-SCF calculations, with the basis set as 6-31G∗. The table includes the structures of the oligonucleotides, their relative energies, their dipole moments, and the distances between 5′-O and 3′-O. Also included are the energies of the HOMOs and LUMOs as well as the areas and accessible areas of the charge density and electrostatic potentials. The location of HOMO is indicated following the HOMO value.
All energies are relative to that of AT-Rp-gtaGC: 5Sp.
Replacement of one Rp in the all-Rp LNA-PS gapmer by a single Sp configuration leads to shorter 5′-O to 3′-O distances. The shortest distance, 22.5 Å, is observed when the Rp close to the nucleobase in the middle of the molecule, i.e., at the 3′ position of thymidine, is replaced by a Sp configuration. The shortest LNA-PS gapmer is the all-Sp compound. Replacing one Sp configuration in the all-Sp gapmer with one Rp gives rise to larger 5′-O to 3′-O distances (Table 2). In this case, the largest distance, 30.2 Å, is obtained when the Sp configuration at the 3′ end is replaced by Rp.
The fact that the maximum length variance of the fully LNA-modified 8-mers was 10.9 Å (36.6–25.7 Å) and 20.2 Å (14.5–34.7 Å) for the shorter 7-mer LNA gapmer could indicate that the more rigid LNA structure decreases the length sensitivity as a function of phosphorous chirality.
In contrast to the large differences observed in the topology of electrostatic potentials and 5′-O to 3′-O distances, the total surface areas of the charge densities and of the electrostatic potentials are not very sensitive to chirality at phosphorus atoms for the studied LNA-PS oligonucleotides. Overall, the surface areas of the positive electrostatic potentials are approximately twice as large as those of the negative electrostatic potentials. However, the accessible surface area of the negative electrostatic potential for the all-Sp 7-mer LNA-PS gapmer, 240 Å2, is larger than that of the accessible surface area of the negative electrostatic potentials for the all-Rp oligonucleotide, 214 Å2. This is in accordance with the findings for the 8-mer LNA-PS, but the difference between the values is not as significant as for the 8-mer LNA-PS.
For each of the 7-mer LNA-PS gapmers, the HOMO is localized on a single nucleobase, whereas the LUMO is localized on a sodium atom, as is the case for the 8-mer LNA-PS nucleotides. For the all-Rp gapmer, the HOMO is localized at adenine on the 5′-LNA nucleotide. This is also true for most of the gapmers, in which one Rp configuration is replaced by a Sp configuration. However, when the Rp configuration closest to the 5′ end is replaced by Sp, the HOMO is repositioned on guanine, and the HOMO in the gapmer 5′-ATgta-Sp-GC-3′ is localized on LNA-guanine. The HOMO in the all-Sp oligonucleotide is localized at adenosine, which is also the case when the Sp configuration at the 5′ end is replaced by Rp. For the remaining Sp gapmers with one Rp configuration, the HOMOs are localized on the purines DNA-g and LNA-G and A (Table 2).
As was the case for the 8-mer LNA nucleotides, the dipole moments vary considerably. The largest value, 61.30 D, was found for the compound 5′-AT-Rp-gtaGC-3′, whereas the compound 5′-ATg-Rp-taGC-3′ had a dipole moment of only 5.81 D.
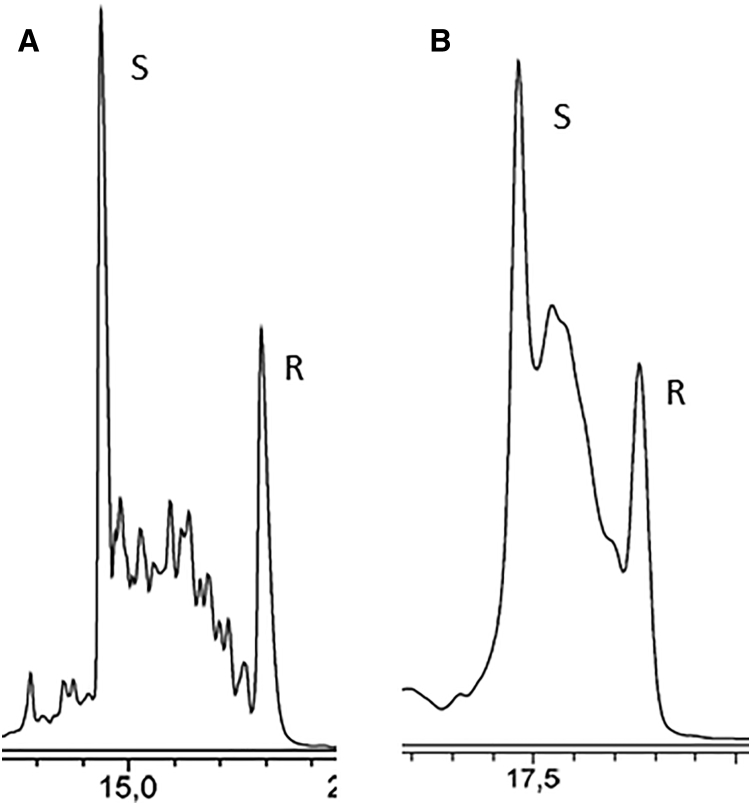
AIE Chromatograms of LNA 7-mer and 8-mer
AIE chromatography (Figure 8) was performed on the 7-gapmer and 8-mer LNAs. The stereo-defined all-Sp and all-Rp forms of both LNA PSs were co-injected with a random mixture of the same design and sequence. The chromatograms show that the stereospecific forms all-Sp (first) and all-Rp (last) constituted the two extremes relating to the retention times.
Figure 8.
AIE Chromatograms of 260 nm of Stereo-Defined and Random Mixture LNA PS Oligonucleotides
(A) LNA 8-mer (5′-CACACTCC-3′). S and R denotes all-Sp and all-Rp 8-mers, respectively. The random mixture (128 diastereoisomers) was partly resolved between the peaks of “S” and “R.” (B) LNA 7-gapmer (5′-AT-gta-GC-3′). S and R denotes all-Sp and all-Rp 7-gapmers, respectively. The random mixture (64 diastereoisomers) elutes as a broad band between the two peaks.
Quantum Mechanical Calculation of an LNA-PS/RNA Duplex
The 7-mer LNA-PS gapmer, the all-Rp 5′-ATgtaGC-3′, was constructed antiparallel with a trimer RNA, 3′-cau-5′, complementary to the DNA gap of the LNA gapmer. This structure was optimized in HF-SCF calculations. The resulting structure, including the HOMO and the LUMO orbitals, is shown in Figure 9. The surfaces of the electrostatic potentials are also shown in Figure 9. Interestingly, the duplex structure was preserved from the initial calculations to the final converged structure. Both the 7-mer LNA-PS gapmer and the trimer RNA show clear stacking of the nucleobases and a W/C H-bonding motif. The lengths of the three hydrogen bonds between the guanine nucleobase on the 7-mer and cytosine on the trimer are 1.972, 1.994, and 2.107 Å. Two hydrogen bonds are identified between the thymine nucleobase on the 7-mer and adenine on the trimer, 2.006 Å and 1.992 Å. In addition, there are two hydrogen bonds between the uracil nucleobase on the trimer and adenine on the 7-mer (2.076 Å and 2.319 Å). Furthermore, the uracil nucleobase of the trimer also formed a hydrogen bond with the LNA-guanine nucleobase on the 7-mer (2.166 Å), which represents an intramolecular bonding not part of the W/C motif. The lengths of these hydrogen bonds are consistent with the published values for W/C H-bonding.54, 55 The binding energy between the 7-mer and the RNA trimer was determined by calculating the energy of the 7-mer and the RNA trimer separately, while maintaining their structures from the minimized duplex, and then subtracting the energy of the duplex. The bonding energy was calculated as 2.78 eV = 268.2 kJ/mol. This corresponds to 33.5 kJ/mol per hydrogen bond. These results indicate that the quantum optimization procedure of the duplex can produce the structures and sequence selectivity known for nucleic acids. The accuracy of the quantum calculation of the duplex structure can be tested in a similar way that Olson et al.56 and others have tested the geometry of the duplex structure, theoretically as well as experimentally. Data from our calculated duplex structure are given in Table 3 and shows great agreement to crystallographic data.56
Figure 9.
Ab Initio HF-SCF Optimized Structure of Duplex: 5′-ATgtaGC-3′ for all-Rp and RNA Trimer 5′-uac-3′
The HOMO (solid) is localized on A at the 5′ end and LUMO (transparent) was localized at a sodium atom at the 3′ end.
Table 3.
Base-Pair Geometry as Derived for the LNA-PS/RNA Duplex Obtained in HF/6-31G* Calculations
| Base Pair | d C1’....C1’ (Å) | λY (°) | λR (°) |
|---|---|---|---|
| C·G | 10.8 | 56.1 | 49.0 |
| T·A | 10.6 | 56.6 | 51.6 |
| U·A | 10.9 | 53.6 | 52.3 |
dC1’....C1’, λY, and λR are defined according to Shrake et al.53
Table 3 shows data for base-pair geometries for the 7-mer LNA-PS gapmer, the all-Rp 5′-ATgtaGC-3′, as defined by Olsson et al.53 The distances d C1’....C1’ differ at most by 0.2 Å from the values of the ideal models obtained by Olsson et al. The angles λR, N9-C1’....C1’, shown in Table 3, differ by less than 5° from those of the ideal models of Olsson et al. The largest discrepancy between λY, N1-C1’....C1’, presented in Table 3, and those of Olsson et al. is 2.1°.
Quantum Mechanical Calculation of 13-mer Oligonucleotides
HF-SCF calculations were also carried out for a large 13-mer, 5′-TCatggctgcAGC-3′, in the all-Rp form. The 13-mer was loaded with 12 sodium atoms and converged to an approximate helical structure. The distance between the 5′-O to 3′-O in the converged structure is 48.25 Å, and the dipole moment is derived as 116.3 D.
Discussion
We have employed several ways to justify the optimization methods used. First, the HF-SCF optimization was shown to be specific and the QM calculations lead to relatively deep minima. This means that fixed initial conditions will, with high probability, produce the same specific end structure. Second, it was shown that the HF-SCF optimizations can produce structures with high similarity to accepted reference models, such as X-ray crystallography. Third, biological justification was illustrated by the fact that throughout the optimization process, a DNA-RNA hybrid structure was energy minimized to a final hybrid structure obeying a Watson-Crick hybridization pattern,54, 55, 56 with a geometry (e.g., H-bond lengths) known from referenced works. Taken together, this indicates that the data have relevance in experimental chemical and biological settings.
The AIE chromatograms demonstrate expected chromatographic difference between stereo-defined diastereoisomers and random diastereoisomeric mixtures (Figure 8). In the co-injections, the stereo-defined compounds elute as sharp peaks and the random mixtures elute the many diastereoisomers (64 and 128) over a much broader range. Here, the all-Rp and all-Sp diastereoisomers exhibit extreme retention properties. Both all-Sp LNAs elute first and the all-Rp LNAs elute last. It is interesting to note that the all-Sp LNAs investigated have lower positive accessible areas of electrostatic potentials and higher negative accessible areas and vice versa for the all-Rp LNAs. This indicates that the accessible area might be a physical parameter for predicting AIE chromatography migration.
Molecular structures and electrostatic potentials (MEPs) are important factors for molecular recognition.57, 58, 59, 60 Compounds with different electrostatic potential topologies are likely to exhibit different recognition patterns in biological systems. Generally, ligand-receptor binding processes are very complex. The net binding energy is a balance between structure and MEP on the one hand and counteracting de-solvation effects and allosteric effects on the other.61, 62, 63 For instance, the binding of oligonucleotides to proteins is via hydrogen bonding, salt bridge formation, water exclusion, and hydrophobic interactions, such as nucleobase stacking with aromatic amino acids.22, 64, 65, 66, 67, 68, 69, 70, 71, 72 Protein binding can be strong and “specific” by the fact that specific sites, e.g., segments of the nucleobase sequence on the oligonucleotide, bind to specific amino acid sites on the protein. Changing a single nucleobase in a sequence of such a “specific” oligonucleotide can lead to significant Kd increase.68 Many factors play a role in understanding the properties of oligonucleotides in complex biological systems. Both binding to the fully complementary nucleic acid target and binding to mismatched nucleic acid targets must be considered. However, it is also necessary to take into account binding to other cellular biomolecules. It is well known that protein binding drives many important pharmaceutical parameters, such as toxicity, plasma half-life, tissue accumulation, cellular uptake, and activity of antisense oligonucleotides.67, 73, 74, 75, 76, 77
Small changes in chemical composition can profoundly change the structure of the oligonucleotides.77 For instance, a single change of one stereocenter in the all-Sp 7-mer oligonucleotide changes the 5′-O to 3′-O length from 14.5 Å to the range 24.4–30.2 Å, depending on which stereocenter changes configuration. It is shown here that interchanging the deoxyriboses in an 8-mer DNA-PS with LNA nucleotides caused marked differences in structure and MEP topology. For the Rp oligonucleotides (all-Rp and oligonucleotides with single Sp substitutions), the MEP topology, 5′-O to 3′-O distance, and accessible area were changed the most. For the Sp oligonucleotides (all-Sp and oligonucleotides with single Rp substitutions), these parameters were much less affected. Our experiments on AIE chromatograms support this observation, which was seen in the calculation of accessible surface area. It is too early to say if this differentiated sensitivity between Rp and Sp oligonucleotides is a general phenomenon, so at this point, we ascribe this effect to be related to these specific molecules. However, it is clearly demonstrated that modifying a single configuration in either the 8-mer fully modified LNA or the 7-mer LNA gapmer leads to profound structure and MEP changes.
The structure of the all-Sp 7-mer gapmer is “ball-like,” with a 5′-O to 3′-O of only 14.5 Å. Introduction of a single Rp stereo center straightens and doubles the length of the structure, producing a dramatically different electronic and MEP topology. Thus, the induced change by a single chirality modification in stereodefined PS oligonucleotides is “global” and propagated throughout the entire molecule (Figures 6 and 7). Because structure and MEPs are strong property determinants, a single chirality change in a stereodefined PS oligonucleotide may produce significant chemical and biological property changes. This is strongly exemplified in Table 2 and the corresponding Figure 7, where a huge difference in dipole moment appears between two 7-gapmers in Table 2 that only differs by a single chiral shift from position 2 to 3 of the R state in an otherwise all S state oligomer. The dipole moment D recorded in Table 2 decreases from 61.3 to 5.8 Debye by that shift but can be understood when looking at the potential surfaces of Figure 7B, which show a large charge separation (and D) between positive and negative surface patches compared to Figure 7C, where the charged areas are evenly distributed, making the charge separation small.
The structural, electronic, and MEP topology changes observed here are context exclusive. For instance, one could assume that the more rigid structure of the bicyclic LNA would produce more extended structures. This is supported by the fact that the all-Rp 8-mer DNA PS has a length of 10.5 Å and the iso-sequential all-Rp 8-mer LNA-PS has a length of 33.6 Å (Table 1). However, most of the Sp series (DNA and LNA) have the same lengths more or less. The measured parameters are highly compound and PS chirality specific. The QM results illustrate that some single chirality changes “globally” affect the molecule to a greater or lesser extent. It would thus be useful to determine at which position a chirality modification would create a “maximal” effect on structural, electronic, and electrostatic parameters. PS oligonucleotides are no exception to the fact that single chirality changes in stereodefined diastereoisomers have profound importance for molecular structure and properties. Although the QM structures and topologies are based on data from shorter oligonucleotides (7-mers and 8-mers), we find it plausible that longer congeners will behave in much the same way.
The sensitivity that PS chirality imparts to structure and electrostatics can be expected to influence many biologically and pharmacologically relevant parameters. The data demonstrate that within a given oligonucleotide sequence, PS chirality offers a large structure and property space that produces many single diastereoisomers with different properties. This adds a new dimension to differentiate structure and activity among therapeutic oligonucleotides. Accordingly, the properties of a given selected “lead” molecule identified after classic non-stereo-selective procedures can be further optimized by selecting from among the various single diastereoisomers, those isomers with different and better pharmaceutical properties. In this way, it should be possible to identify isomers with optimized pharmaceutical properties among the thousands of compounds in a random mixture. An interesting aspect of this is that because all these single diastereoisomers are selected from the same pool, they will all share an identical nucleotide sequence and design so it will be known upfront that they bind to the RNA target with high affinity. This is an attribute that is unique for RNA therapeutics.
Concerning the docking of drug molecules to receptors, two models or views are in question: the lock-key and induced fit78 model. It seems that the induced fit models are more realistic due to the influences of the environment, although the effects of the environment are less obvious for the DNA/RNA oligomer binding.
Materials and Methods
Computational Methods
Most of the data presented here have been derived using ab initio quantum mechanical methods,79 i.e., all structural and energy properties of the molecules investigated are derived from the time-independent electronic Schrödinger equation, without any assumption of parameters and potentials. In particular, the ab initio HF-SCF80, 81 has been used, with the approximation of Born-Oppenheimer separating the nuclear and electronic motion. The HF-SCF approximation is an independent particle model, i.e., each electron is assumed to move in the field of the nuclei and the mean field of all other electrons. The results of the HF-SCF calculations are electronic configurations. In addition, the geometric structures of the DNA or LNA PS oligonucleotides are also determined.
The PS oligonucleotides are constructed using the nucleotide builder in SPARTAN ’14. The DNA molecules are constructed as single strand nucleotides with an α helix defined with a rise per base pair of 2.548 Å and a rotation per base pair of 32.70°. The LNAs are constructed by adding a 2′-O-CH2-4′ bridge across the appropriate furanose rings.37 Furthermore, to arrive at the LNA-PS oligonucleotides, one of the non-bridging oxygen atoms in the phosphodiester is replaced by a sulfur atom. This creates a chiral center on each phosphorus atom. Finally, the appropriate number of Na+ ions is added to make the oligomers electronically neutral. Molecules so constructed were optimized in HF-SCF calculations using either SPARTAN ‘14 or Gaussian 09. In SPARTAN, the calculations are converged when the maximum gradient is below 0.0003 H/bohr or the maximum change in bond length is less than 0.0012 Å. In Gaussian, the convergence criteria are that the maximum displacement is less than 0.0018 a.u., whereas the maximum force is less than 0.00045 a.u.
The wave functions for the LNA-PS oligonucleotides are approximated as Slater determinants, in which the elements are molecular orbitals expanded as linear combinations of atomic orbitals (LCAOs). The basis sets consist of Gaussian type functions, and in the present work, the basis set has been chosen as 6-31G*.
Test runs on small oligomers have also been carried out for DFT with B3LYP and, basically, they resulted in the same structural minima as that from HF-SCF but required more computer resources. In the case of 3-mers, the deviation, e.g., in dihedral angles, mounted to less than 1% going from HF-SCF to B3LYP.
Most of the QM studies in the past literature have been employing DFT methods for their studies of bio-molecules, although smaller than the oligonucleotides of this paper. Such DFTs studied give roughly similar electronic structures. One of the authors of this paper has been participating in an extensive QM study performed (Frimand et al.82) on peptides with aqueous solvents, in which different QM methods are compared. The study concluded that the various methods basically gave the same minimized electronic structures but consumed different computer resources.
The main task of calculating the electronic structures of molecules is to solve the time-independent Schrödinger equation of the multi-electron molecules within the Born-Oppenheimer approximation.
When the molecular orbitals are expanded in basis sets, the HF-SCF method gives rise to the matrix equation:79
where ccoef are the expansion coefficients of the molecular orbitals, E contains the orbital energies, and S is the overlap matrix. The elements of the Fock operator, Fμν, consist of the elements of the core Hamiltonian, Hμνcore, the Coulomb energy term Jμν and the exchange term Kμν. Thus, the elements of the F operator become:
The elements of the core Hamiltonian are:
where Δ stands for the Laplace operator and φμ is the molecular orbital expanded on the basis functions.
On the basis of the optimized wave function, it is possible to calculate the electrostatic potential experienced by a test charge at a particular point in three-dimensional space. The electrostatic potential can be illustrated as an iso-surface defined by points that have the same electrostatic potential energy relative to a test charge. Thus, both positive and negative potential energy surfaces can be derived that give information about where ligand molecules can be attracted to the oligonucleotides. The chosen iso-values for the electrostatic potentials here are 83.68 kJ/mol and −83.68 kJ/mol. This energy corresponds to two to three times the energy of a hydrogen bond and is thus reflective of intermolecular interactions/bonding found in biological systems. The iso-surfaces of the electrostatic potentials mostly show larger compact spheres covering larger areas of the molecules, and numbers of smaller spheres around polar/charged groups or atoms comprising electron lone pairs. Further, on the basis of the optimized wave functions, the electron densities have been derived. These are presented as iso-surfaces, with iso-values chosen as 0.002 electrons/a.u.3 Also calculated are iso-surfaces of the HOMO and LUMO. These surfaces yield information about electron donor/acceptor sites. Charges associated with the various atoms in the oligonucleotides can be calculated using the Mulliken approximation.51 Few simulations based on quantum mechanical optimization of larger bio-molecular structures starting from the atomic constituents have been reported in the literature. This is in contrast to classical MD simulations of DNA and protein structures and of the simulation of folding processes. In the latter, the multiple minima problem is well-documented and seen in the many conformational states that are observed in protein folding.
The quantum mechanical procedures result in optimized structures that often have helical structures, which in our test case of 1IXJ (Figure 2), came close to the crystal structure.80 Here, in this test, we start with the crystal structure as the initial structure and employ the HF-SCF and, in some cases, also the MD optimizations that are based on force fields.18
Synthesis of Oligonucleotides with Defined PS Chirality
The synthesis of chiral DNA and LNA 3′-O-oxazaphospholidine monomers was performed using previously described methods.47 The oligonucleotides 5′-AsTsgstsasGsmC-3′ (7-mer) and 5′- mCsAsmCsAsmCsTsmCsmC-3′ (8-mer) and the all-Rp and all-Sp isoforms were synthesized according to published procedures, with the exception that DCI (4,5-dicyano imidazole) was used as the activator.
AIE chromatography was performed on a Dionex Ultimade 3000 system. Column: DNA-pac PA100, 2 × 250 mm. Solvents buffer A (10 mM NaClO4, 1 mM EDTA, and 20 mM Tris-HCL, pH 7.8) and B (1 mM NaClO4, 1 mM EDTA, and 20 mM Tris-HCL, pH 7.8). Gradient 0 min. 0% B, 35 min. 35% B, 40 min. 0% B. Detection 260 nm. 50 μL injected.
Author Contributions
All authors, H.G.B., I.S., C.S., H.Ø., H.F.H., and T.K., have contributed to the paper, where, specifically, experiments were done by H.F.H. and T.K., computations were done by H.G.B. and I.S., and organization and presentation of the work were done by T.K., C.S., and H.Ø.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
The authors wish to thank Christoph Rosenbohm, Principal leader and Head of Discovery Operations, Roche Innovation Center Copenhagen, for fruitful discussions and for reviewing the manuscript. The Technical University of Denmark is acknowledged for providing the computational resources. Technical support has been provided by The Technical University of Denmark, DTU, and financial support has been provided by both DTU and Roche Innovation Center Copenhagen.
References
- 1.Eckstein F. Nucleoside phosphorothioates. J. Am. Chem. Soc. 1966;88:4292–4294. doi: 10.1021/ja00718a039. [DOI] [PubMed] [Google Scholar]
- 2.Eckstein F. A dinucleoside phosphorothioate. Tetrahedron Lett. 1967;8:1157–1160. [Google Scholar]
- 3.Eckstein F., Gindl H. [Uridin-2′,3′-O,O-cyclothiophosphate] Chem. Ber. 1968;101:1670–1673. doi: 10.1002/cber.19681010518. [DOI] [PubMed] [Google Scholar]
- 4.Eckstein F., Gindl H. Polyribonucleotides containing a thiophosphate backbone. FEBS Lett. 1969;2:262–264. doi: 10.1016/0014-5793(69)80037-2. [DOI] [PubMed] [Google Scholar]
- 5.Matzura H., Eckstein F. A polyribonucleotide containing alternation P=O and P=S linkages. Eur. J. Biochem. 1968;3:448–452. doi: 10.1111/j.1432-1033.1967.tb19551.x. [DOI] [PubMed] [Google Scholar]
- 6.Burgers P.M., Eckstein F. Absolute configuration of the diastereomers of adenosine 5′-O-(1-thiotriphosphate): consequences for the stereochemistry of polymerization by DNA-dependent RNA polymerase from Escherichia coli. Proc. Natl. Acad. Sci. USA. 1978;75:4798–4800. doi: 10.1073/pnas.75.10.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckstein F., Armstrong V.W., Sternbach H. Stereochemistry of polymerization by DNA-dependent RNA-polymerase from Escherichia coli: an investigation with a diastereomeric ATP-analogue. Proc. Natl. Acad. Sci. USA. 1976;73:2987–2990. doi: 10.1073/pnas.73.9.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saenger W., Eckstein F. Stereochemistry of a substrate for pancreatic ribonuclease. Crystal and molecular structure of the triethylammonium salt of uridine 2′, 3′-0, 0-cyclophosphorothioate. J. Am. Chem. Soc. 1970;92:4712–4718. [Google Scholar]
- 9.Stec W.J., Karwowski B., Boczkowska M., Guga P., Koziolkiewicz M., Sochacki M., Wieczorek M.W., Blaszczyk J. Deoxyribonucleoside 3′-O-(2-Thio- and 2-Oxo-“spiro”-4,4-pentamethylene-1,3,2-oxathiaphospholane)s: monomers for stereocontrolled synthesis of oligo(deoxyribonucleoside phosphorothioate)s and chimeric PS/PO oligonucleotides. J. Am. Chem. Soc. 1998;120:7156–7167. [Google Scholar]
- 10.Koziolkiewicz M., Krakowiak A., Kwinkowski M., Boczkowska M., Stec W.J. Stereodifferentiation--the effect of P chirality of oligo(nucleoside phosphorothioates) on the activity of bacterial RNase H. Nucleic Acids Res. 1995;23:5000–5005. doi: 10.1093/nar/23.24.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krieg A.M., Guga P., Stec W. P-chirality-dependent immune activation by phosphorothioate CpG oligodeoxynucleotides. Oligonucleotides. 2003;13:491–499. doi: 10.1089/154545703322860807. [DOI] [PubMed] [Google Scholar]
- 12.Stephenson M.L., Zamecnik P.C. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc. Natl. Acad. Sci. USA. 1978;75:285–288. doi: 10.1073/pnas.75.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zamecnik P.C., Stephenson M.L. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc. Natl. Acad. Sci. USA. 1978;75:280–284. doi: 10.1073/pnas.75.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.CDe Clercq E., Eckstein F., Sternbach H., Merigan T.C. The antiviral activity of thiophosphate-substituted polyribonucleotides in vitro and in vivo. Virology. 1970;42:421–428. doi: 10.1016/0042-6822(70)90285-0. [DOI] [PubMed] [Google Scholar]
- 15.Agrawal S., Goodchild J., Civeira M.P., Thornton A.H., Sarin P.S., Zamecnik P.C. Oligodeoxynucleoside phosphoramidates and phosphorothioates as inhibitors of human immunodeficiency virus. Proc. Natl. Acad. Sci. USA. 1988;85:7079–7083. doi: 10.1073/pnas.85.19.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsukura M., Shinozuka K., Zon G., Mitsuya H., Reitz M., Cohen J.S., Broder S. Phosphorothioate analogs of oligodeoxynucleotides: inhibitors of replication and cytopathic effects of human immunodeficiency virus. Proc. Natl. Acad. Sci. USA. 1987;84:7706–7710. doi: 10.1073/pnas.84.21.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsukura M., Zon G., Shinozuka K., Robert-Guroff M., Shimada T., Stein C.A., Mitsuya H., Wong-Staal F., Cohen J.S., Broder S. Regulation of viral expression of human immunodeficiency virus in vitro by an antisense phosphorothioate oligodeoxynucleotide against rev (art/trs) in chronically infected cells. Proc. Natl. Acad. Sci. USA. 1989;86:4244–4248. doi: 10.1073/pnas.86.11.4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stein C.A., Cohen J.S. Oligodeoxynucleotides as inhibitors of gene expression: a review. Cancer Res. 1988;48:2659–2668. [PubMed] [Google Scholar]
- 19.Stein C.A., Cohen J.S. Antisense compounds: potential role in cancer therapy. Important Adv. Oncol. 1989:79–97. [PubMed] [Google Scholar]
- 20.Stein C.A., Matsukura M., Subasinghe C., Broder S., Cohen J.S. Phosphorothioate oligodeoxynucleotides are potent sequence nonspecific inhibitors of de novo infection by HIV. AIDS Res. Hum. Retroviruses. 1989;5:639–646. doi: 10.1089/aid.1989.5.639. [DOI] [PubMed] [Google Scholar]
- 21.Stein C.A., Subasinghe C., Shinozuka K., Cohen J.S. Physicochemical properties of phosphorothioate oligodeoxynucleotides. Nucleic Acids Res. 1988;16:3209–3221. doi: 10.1093/nar/16.8.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benimetskaya L., Tonkinson J.L., Koziolkiewicz M., Karwowski B., Guga P., Zeltser R., Stec W., Stein C.A. Binding of phosphorothioate oligodeoxynucleotides to basic fibroblast growth factor, recombinant soluble CD4, laminin and fibronectin is P-chirality independent. Nucleic Acids Res. 1995;23:4239–4245. doi: 10.1093/nar/23.21.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cook P.D. Antisense medicinal chemistry. In: Crooke S.T., editor. Handbook of Experimental Pharmacology. Springer Verlag; 1998. pp. 51–101. [Google Scholar]
- 24.Kanehara H., Wada T., Mizuguchi M., Makino K. Influence of a thiophosphate linkage on the duplex stability - does Sp configuration always lead to higher stability than Rp? Nucleosides Nucleotides. 1996;15:1169–1178. [Google Scholar]
- 25.Karwowski B., Okruszek A., Wengel J., Stec W.J. Stereocontrolled synthesis of LNA dinucleoside phosphorothioate by the oxathiaphospholane approach. Bioorg. Med. Chem. Lett. 2001;11:1001–1003. doi: 10.1016/s0960-894x(01)00109-3. [DOI] [PubMed] [Google Scholar]
- 26.Levin A.A., Yu Z.R., Geary S.R. Second Edition. CRC Press; 2007. Basic Principles of the Pharmacokinetics of Antisense Oligonucleotide Drugs. [Google Scholar]
- 27.Uhlmann E. Recent advances in the medicinal chemistry of antisense oligonucleotides. Curr. Opin. Drug Discov. Devel. 2000;3:203–213. [PubMed] [Google Scholar]
- 28.Freier S.M., Altmann K.-H. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1997;25:4429–4443. doi: 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin P. Ein neuer Zugang zu 2′-O-Alkylribonucleosiden und Eigenschaften deren Oligonucleotiden. Helv. Chim. Acta. 1995;78:486–504. [Google Scholar]
- 30.Nielsen P.E., Egholm M., Berg R.H., Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 31.Braasch D.A., Corey D.R. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem. Biol. 2001;8:1–7. doi: 10.1016/s1074-5521(00)00058-2. [DOI] [PubMed] [Google Scholar]
- 32.Grünweller A., Hartmann R.K. Locked nucleic acid oligonucleotides: the next generation of antisense agents? BioDrugs. 2007;21:235–243. doi: 10.2165/00063030-200721040-00004. [DOI] [PubMed] [Google Scholar]
- 33.Kurreck J. Antisense technologies. Improvement through novel chemical modifications. Eur. J. Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- 34.Kurreck J., Wyszko E., Gillen C., Erdmann V.A. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002;30:1911–1918. doi: 10.1093/nar/30.9.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obika S., Nanbu D., Hari Y., Morio J.A.K., Doi T., Imanishi T. Stability and structural features of the duplexes containing nucleoside analogues with a fixed N-type conformation, 2′-O,4′-C-methyleneribonucleosides. Tetrahedron Lett. 1998;39:5401–5404. [Google Scholar]
- 36.Obika S., Nanbu D., Hari Y., Morio K., In Y., Ishida T., Imanishi T. Synthesis of 2′-O,4′-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed C3, -endo sugar puckering. Tetrahedron Lett. 1997;38:8735–8738. [Google Scholar]
- 37.Singh S.K., Kumar R., Wengel J. Synthesis of novel bicyclo[2.2.1] ribonucleosides: 2′-amino- and 2′-thio-LNA monomeric nucleosides. J. Org. Chem. 1998;63:6078–6079. doi: 10.1021/jo9806658. [DOI] [PubMed] [Google Scholar]
- 38.Singh S.K., Kumar R., Wengel J. Synthesis of 2′-amino-LNA: a novel conformationally restricted high-affinity oligonucleotide analogue with a handle. J. Org. Chem. 1998;63:10035–10039. [Google Scholar]
- 39.Singh S.K., Koshkin A.A., Wengel J., Nielsen P. LNA (locked nucleic acids): synthesis and high-affinity nucleic acid recognition. Chem. Commun. 1998:455–456. [Google Scholar]
- 40.Singh S.K., Wengel J. Universality of LNA-mediated high-affinity nucleic acid recognition. Chem. Commun. 1998:1247–1248. [Google Scholar]
- 41.Wahlestedt C., Salmi P., Good L., Kela J., Johnsson T., Hökfelt T., Broberger C., Porreca F., Lai J., Ren K. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc. Natl. Acad. Sci. USA. 2000;97:5633–5638. doi: 10.1073/pnas.97.10.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wengel J., Koshkin A., Singh S.K., Nielsen P., Meldgaard M., Rajwanshi V.K., Kumar R., Skouv J., Nielsen C.B., Jacobsen J.P. Lna (locked nucleic acid) Nucleosides Nucleotides. 1999;18:1365–1370. [Google Scholar]
- 43.Wengel J., Petersen M., Frieden M., Koch T. Chemistry of locked nucleic acids (LNA): design, synthesis, and bio-physical properties. Letters Peptide Sci. 2003;10:237–253. [Google Scholar]
- 44.Wengel J., Petersen M., Nielsen K.E., Jensen G.A., Håkansson A.E., Kumar R., Sørensen M.D., Rajwanshi V.K., Bryld T., Jacobsen J.P. LNA (locked nucleic acid) and the diastereoisomeric alpha-L-LNA: conformational tuning and high-affinity recognition of DNA/RNA targets. Nucleosides Nucleotides Nucleic Acids. 2001;20:389–396. doi: 10.1081/NCN-100002312. [DOI] [PubMed] [Google Scholar]
- 45.Obika S., Morio K., Nanbu D., Imanishi T. Synthesis and conformation of 3′-O,4′-C-methyleneribonucleosides, novel bicyclic nucleoside analogues for 2′,5′-linked oligonucleotide modification. Chem. Commun. 1997:1643–1644. [Google Scholar]
- 46.Seth P.P., Siwkowski A., Allerson C.R., Vasquez G., Lee S., Prakash T.P., Wancewicz E.V., Witchell D., Swayze E.E. Short antisense oligonucleotides with novel 2′-4′ conformationaly restricted nucleoside analogues show improved potency without increased toxicity in animals. J. Med. Chem. 2009;52:10–13. doi: 10.1021/jm801294h. [DOI] [PubMed] [Google Scholar]
- 47.Seth P.P., Vasquez G., Allerson C.A., Berdeja A., Gaus H., Kinberger G.A., Prakash T.P., Migawa M.T., Bhat B., Swayze E.E. Synthesis and biophysical evaluation of 2′,4′-constrained 2‘O-methoxyethyl and 2’,4′-constrained 2'O-ethyl nucleic acid analogues. J. Org. Chem. 2010;75:1569–1581. doi: 10.1021/jo902560f. [DOI] [PubMed] [Google Scholar]
- 48.Wan W.B., Migawa M.T., Vasquez G., Murray H.M., Nichols J.G., Gaus H., Berdeja A., Lee S., Hart C.E., Lima W.F. Synthesis, biophysical properties and biological activity of second generation antisense oligonucleotides containing chiral phosphorothioate linkages. Nucleic Acids Res. 2014;42:13456–13468. doi: 10.1093/nar/gku1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koch T., Shim I., Lindow M., Ørum H., Bohr H.G. Quantum mechanical studies of DNA and LNA. Nucleic Acid Ther. 2014;24:139–148. doi: 10.1089/nat.2013.0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Metropolis N., Rosenbluth A.W., Rosenbluth M.N., Teller A.H. Equation of state calculations by fast computing machines. J. Chem. Phys. 1953;21:1087–1091. [Google Scholar]
- 51.Mulliken R.S. Electronic population analysis on LCAO-MO molecular wave functions. J. Chem. Phys. 1955;23:1833–1840. [Google Scholar]
- 52.Lee B., Richards F.M. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 53.Shrake A., Rupley J.A. Environment and exposure to solvent of protein atoms. Lysozyme and insulin. J. Mol. Biol. 1973;79:351–371. doi: 10.1016/0022-2836(73)90011-9. [DOI] [PubMed] [Google Scholar]
- 54.Seeman N.C., Rosenberg J.M., Suddath F.L., Kim J.J., Rich A. RNA double-helical fragments at atomic resolution. I. The crystal and molecular structure of sodium adenylyl-3′,5′-uridine hexahydrate. J. Mol. Biol. 1976;104:109–144. doi: 10.1016/0022-2836(76)90005-x. [DOI] [PubMed] [Google Scholar]
- 55.Rosenberg J.M., Seeman N.C., Day R.O., Rich A. RNA double-helical fragments at atomic resolution. II. The crystal structure of sodium guanylyl-3′,5′-cytidine nonahydrate. J. Mol. Biol. 1976;104:145–167. doi: 10.1016/0022-2836(76)90006-1. [DOI] [PubMed] [Google Scholar]
- 56.Olson W.K., Bansal M., Burley S.K., Dickerson R.E., Gerstein M., Harvey S.C., Heinemann U., Lu X.-J., Neidle S., Shakked Z. A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol. 2001;313:229–237. doi: 10.1006/jmbi.2001.4987. [DOI] [PubMed] [Google Scholar]
- 57.Roy D., Balanarayan P., Gadre S.R. An appraisal of Poincaré-Hopf relation and application to topography of molecular electrostatic potentials. J. Chem. Phys. 2008;129:174103. doi: 10.1063/1.2999558. [DOI] [PubMed] [Google Scholar]
- 58.Roy D.K., Balanarayan P., Gadre S.R. Signatures of molecular recognition from the topography of electrostatic potential. J. Chem. Sci. 2009;121:815–821. [Google Scholar]
- 59.Muzet N., Guillot B., Jelsch C., Howard E., Lecomte C. Electrostatic complementarity in an aldose reductase complex from ultra-high-resolution crystallography and first-principles calculations. Proc. Natl. Acad. Sci. USA. 2003;100:8742–8747. doi: 10.1073/pnas.1432955100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sulea T., Purisima E.O. Profiling charge complementarity and selectivity for binding at the protein surface. Biophys. J. 2003;84:2883–2896. doi: 10.1016/S0006-3495(03)70016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burgoyne N.J., Jackson R.M. Predicting protein interaction sites: binding hot-spots in protein-protein and protein-ligand interfaces. Bioinformatics. 2006;22:1335–1342. doi: 10.1093/bioinformatics/btl079. [DOI] [PubMed] [Google Scholar]
- 62.Kenny P.W. Hydrogen bonding, electrostatic potential, and molecular design. J. Chem. Inf. Model. 2009;49:1234–1244. doi: 10.1021/ci9000234. [DOI] [PubMed] [Google Scholar]
- 63.Kraut D.A., Sigala P.A., Pybus B., Liu C.W., Ringe D., Petsko G.A., Herschlag D. Testing electrostatic complementarity in enzyme catalysis: hydrogen bonding in the ketosteroid isomerase oxyanion hole. PLoS Biol. 2006;4:e99. doi: 10.1371/journal.pbio.0040099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Max K.E., Zeeb M., Bienert R., Balbach J., Heinemann U. T-rich DNA single strands bind to a preformed site on the bacterial cold shock protein Bs-CspB. J. Mol. Biol. 2006;360:702–714. doi: 10.1016/j.jmb.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 65.Zeeb M., Max K.E., Weininger U., Löw C., Sticht H., Balbach J. Recognition of T-rich single-stranded DNA by the cold shock protein Bs-CspB in solution. Nucleic Acids Res. 2006;34:4561–4571. doi: 10.1093/nar/gkl376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harris C.R., Mackoy T., Machado A.C.D., Xu D., Rohs R., Fenley M.O. Opposites attract: shape and electrostatic complementarity in protein-DNA complexes. In: Schlick T., editor. Innovations in Biomolecular Modeling and Simulations. Royal Society of Chemistry; 2012. pp. 53–80. [Google Scholar]
- 67.Liang X.H., Sun H., Shen W., Crooke S.T. Identification and characterization of intracellular proteins that bind oligonucleotides with phosphorothioate linkages. Nucleic Acids Res. 2015;43:2927–2945. doi: 10.1093/nar/gkv143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun S., Shamoo Y. Biochemical characterization of interactions between DNA polymerase and single-stranded DNA-binding protein in bacteriophage RB69. J. Biol. Chem. 2003;278:3876–3881. doi: 10.1074/jbc.M210497200. [DOI] [PubMed] [Google Scholar]
- 69.Guvakova M.A., Yakubov L.A., Vlodavsky I., Tonkinson J.L., Stein C.A. Phosphorothioate oligodeoxynucleotides bind to basic fibroblast growth factor, inhibit its binding to cell surface receptors, and remove it from low affinity binding sites on extracellular matrix. J. Biol. Chem. 1995;270:2620–2627. doi: 10.1074/jbc.270.6.2620. [DOI] [PubMed] [Google Scholar]
- 70.Khaled Z., Benimetskaya L., Zeltser R., Khan T., Sharma H.W., Narayanan R., Stein C.A. Multiple mechanisms may contribute to the cellular anti-adhesive effects of phosphorothioate oligodeoxynucleotides. Nucleic Acids Res. 1996;24:737–745. doi: 10.1093/nar/24.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rockwell P., O’Connor W.J., King K., Goldstein N.I., Zhang L.M., Stein C.A. Cell-surface perturbations of the epidermal growth factor and vascular endothelial growth factor receptors by phosphorothioate oligodeoxynucleotides. Proc. Natl. Acad. Sci. USA. 1997;94:6523–6528. doi: 10.1073/pnas.94.12.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lai J.C., Tan W., Benimetskaya L., Miller P., Colombini M., Stein C.A. A pharmacologic target of G3139 in melanoma cells may be the mitochondrial VDAC. Proc. Natl. Acad. Sci. USA. 2006;103:7494–7499. doi: 10.1073/pnas.0602217103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lai J.C., Brown B.D., Voskresenskiy A.M., Vonhoff S., Klussman S., Tan W., Colombini M., Weeratna R., Miller P., Benimetskaya L. Comparison of d-g3139 and its enantiomer L-g3139 in melanoma cells demonstrates minimal in vitro but dramatic in vivo chiral dependency. Mol. Ther. 2007;15:270–278. doi: 10.1038/sj.mt.6300037. [DOI] [PubMed] [Google Scholar]
- 74.Stein C.A., Wu S., Voskresenskiy A.M., Zhou J.F., Shin J., Miller P., Souleimanian N., Benimetskaya L. G3139, an anti-Bcl-2 antisense oligomer that binds heparin-binding growth factors and collagen I, alters in vitro endothelial cell growth and tubular morphogenesis. Clin. Cancer Res. 2009;15:2797–2807. doi: 10.1158/1078-0432.CCR-08-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Watanabe T.A., Geary R.S., Levin A.A. Plasma protein binding of an antisense oligonucleotide targeting human ICAM-1 (ISIS 2302) Oligonucleotides. 2006;16:169–180. doi: 10.1089/oli.2006.16.169. [DOI] [PubMed] [Google Scholar]
- 76.Beltinger C., Saragovi H.U., Smith R.M., LeSauteur L., Shah N., DeDionisio L., Christensen L., Raible A., Jarett L., Gewirtz A.M. Binding, uptake, and intracellular trafficking of phosphorothioate-modified oligodeoxynucleotides. J. Clin. Invest. 1995;95:1814–1823. doi: 10.1172/JCI117860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Prakash T.P., Manoharan M., Kawasaki A.M., Fraser A.S., Lesnik E.A., Sioufi N., Leeds J.M., Teplova M., Egli M. 2′-O-[2-(methylthio)ethyl]-modified oligonucleotide: an analogue of 2′-O-[2-(methoxy)-ethyl]-modified oligonucleotide with improved protein binding properties and high binding affinity to target RNA. Biochemistry. 2002;41:11642–11648. doi: 10.1021/bi020264t. [DOI] [PubMed] [Google Scholar]
- 78.Kenakin T. Third Edition. Lippincot-Raven; 1997. Pharmacologic Analysis of Drug Receptor Interaction. [Google Scholar]
- 79.Roothaan C.C.J. New developments in molecular orbital theory. Rev. Mod. Phys. 1951;23:69–89. [Google Scholar]
- 80.Hehre W.J., Radom L., Schleyer P.v.R., Pople J. John Wiley; 1986. AB INITIO Molecular Orbital Theory. [Google Scholar]
- 81.Leach A.R. Longman; 1996. Molecular Modelling: Principles and Applications. [Google Scholar]
- 82.Frimand K., Bohr H., Jalkanen K.J., Suhai S. Structures, vibrational absorption and vibrational circular dichroism spectra of L-alanine in aqueous solution: a density functional theory and RHF study. Chem. Phys. 2000;255:165–194. [Google Scholar]