Abstract
Background:
Telomerase (human telomerase reverse transcriptase (hTERT)) is considered a hallmark of cancer. The aim of our study was to evaluate the feasibility of the detection of hTERT transcripts in serum as a ‘pan-cancer’ diagnostic method.
Methods:
Human telomerase reverse transcriptase mRNA levels were determined in serum and serum-derived exosomes from 133 patients with different malignancies and 45 healthy controls. In four patients hTERT mRNA levels were measured in different clinical stages.
Results:
Human telomerase reverse transcriptase transcript was absent in all controls and was variably detected in 67.5% of patients with all cancer types. A correlation between hTERT transcript levels and the clinical course was found in several cases.
Conclusions:
Human telomerase reverse transcriptase mRNA levels may reflect the tumour burden and the clinical status of the patient. In patients with detectable levels, this assay may potentially serve as a diagnostic and follow-up ‘pan-cancer’ marker. Owing to the large variety of patients and small sample size in each diagnosis, the statistical power is limited and will be explored further in larger groups.
Keywords: telomerase, hTERT, exosomes
Reliable biomarkers for diagnosis and follow-up of cancer are in a dire need. An attractive currently developing approach is ‘liquid biopsy’, namely, the analysis of circulating tumour cells, cell-free nucleic acid or extracellular vesicles obtained mainly from the peripheral blood. Although attractive, this approach has various limitations and relies on identification of specific mutations in each cancer type. We surmised that measurement of human telomerase reverse transcriptase (hTERT) mRNA can be used as a possible diagnostic marker whose advantage is being specific to cancer on the one hand and common to all malignancies on the other, designating it a pan-cancer marker.
Telomerase, considered a hallmark of cancer (Phatak and Burger, 2007), is repressed in almost all human somatic cells but is activated in more than 90% of cancer cells. Its mRNA levels (hTERT) correlate with its activity and were suggested as a marker for cancer detection and progression (Hiyama and Hiyama, 2003; Shay and Wright, 2011). Several recent studies demonstrated the existence of hTERT mRNA in the serum of patients with active cancer of breast, colon and hepatocellular carcinoma (Miura et al, 2005, 2007, 2010; Terrin et al, 2008; Pucciarelli et al, 2012), follicular lymphoma (Dasi et al, 2001) and prostate cancer (March-Villalba et al, 2012). Since our preliminary results showed that measurement of serum hTERT mRNA yields very low and unstable concentrations of the transcript, we tried a more reliable, exosomal-based method.
Exosomes are nanosized endocytic vesicles secreted by most cell types into various body fluids including serum, having important roles in intercellular communications (Thery, 2011). Cell-free mRNA is stabilised in exosomes and is protected to a considerable extent from digestion by extracellular RNases (Halicka et al, 2000; Tsui et al, 2002; El-Hefnawy et al, 2004; Garcia et al, 2008).
Human telomerase reverse transcriptase mRNA was found along with other mRNA molecules in profiling of exosomal content derived from cancer cell lines (Batagov et al, 2011). We recently reported that hTERT mRNA is present in exosomes derived from all examined cancer cell lines and from the sera of human cancer patients as well (Gutkin et al, 2016).
Owing to the established importance of telomerase in cancer and previous studies reporting its existence in the serum of cancer patients, we assumed that it can be used as a reliable ‘pan-cancer’ marker for the early detection of cancer or relapse in all malignancies. We measured the concentrations of exosomal hTERT mRNA in the sera of patients with a wide variety of cancer types and evaluated its correlations to tumour characteristics. Furthermore, we followed the exosomal hTERT transcript levels during a clinical course of several patients.
Materials and methods
Population
After signing an informed consent approved by the local IRB (Helsinki committee of Rabin Medical Center), 10 ml blood in a serum separation tubes was obtained from 45 healthy volunteers and 133 consecutive untreated cancer patients in Davidoff Cancer Center. Several patients were followed during treatment to observe the kinetics of hTERT mRNA. The tubes were centrifuged at 2500 r.p.m. for 10 min; the serum was collected, divided to 1 ml aliquots and kept in −20 °C for exosomes and mRNA isolation.
Exosomes purification
Exosomes were isolated from patients’ sera by using the Total Exosome Isolation Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The purity of the exosomes was verified by the presence of specific exosomal markers (e.g., TSG101, CD9 and CD63) and the lack of non-exosomal marker (calnexin) by western blotting (Gutkin et al, 2016). Additionally, exosomes were analysed in the NanoSight tracking device (Gutkin et al, 2016).
RNA purification
RNA from exosomes was purified with Total Exosome RNA and Protein Isolation Kit (Invitrogen) according to the provided manual.
cDNA formation
mRNA was reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions.
hTERT expression by real-time PCR
The expression of hTERT was measured relatively to that of the HPRT-1 as a reference gene. Gene amplification was executed using the following sets of primers (HyLabs, Rehovot, Israel).
hTERT: forward, 5′-GTACTTTGTCAAGGTGGATGTGA-3′ and reverse, 5′-GCTGGAGGTCTGTCAAGGTAGAG-3′ HPRT-1: forward, 5′-TCAGGCAGTATAATCCAAAGATGGT-3′ and reverse, 5′-CTTCGTGGGGTCCTTTTCAC-3′.
Polymerase chain reactions were prepared with the Taqman fluorophore-labelled primers (Applied Biosystems), run and analysed on the Step One Detection System (Applied Biosystems). Reactions were performed for 50 cycles; a normal value (no expression of hTERT) was arbitrarily defined as 1 for further calculation purposes.
Tumour characteristics
For patients with solid tumours, we documented histologic features (grade of differentiation, Ki67), tumour spread (TNM staging, location of metastatic sites) and level of characteristic epithelial markers for each tumour. For patients with leukaemia, we documented white blood cell (WBC) and lactate dehydrogenase (LDH). For lymphoma patients, LDH, International Prognostic Index and the presence of bulky disease were documented and for multiple myeloma (MM) patients’ immunoglobulin levels were depicted.
Statistical analysis
The statistical analysis was generated using the SAS Software, Version 9.4, 2002–2012. Continuous variables were presented as mean±std and median (minimum–maximum), and categorical variables as (N, %). The normality of distribution for the continuous variables was assessed graphically and by Kolmogorov–Smirnov test. If deemed normal, ANOVA (or T-test for two groups) was used to compare the value of continuous variables between study groups; if not deemed normal, Wilcoxon’s test was used. χ2 and/or Fisher’s exact test were used in the analysis of categorical variables between study groups. Pearson’s correlation was used to assess the association between continuous variables. Two-sided P-values <0.05 were considered statistically significant.
Results
Demographic features of the patients
Table 1 depicts the clinical data of the patients. Human telomerase reverse transcriptase mRNAs were also measured consecutively in four patients during the course of their disease.
Table 1. The clinical data of the patients.
| Primary site (number) | Median age, years (minimum–maximum) | Male (%) | Stage | Histology | Mean (s.d.) | Median (minimum–maximum) | % of patients with high hTERT mRNAa |
|---|---|---|---|---|---|---|---|
| Bladder (7) | 74 (55–84) | 100% | III=14.3% IV=85.7% | SCC=14.3% TCC=85.7% | 18.3 (29.8) | 3 (0.8–79.6) | 42.86 |
| Breast (16) | 60.5 (26–83) | 0% | Unk=6.2% II=6.2% III=12.6% IV=75% | Carcinoma, NOS=6.2% Adenocarcinoma=93.8% | 9.7 (19.2) | 11.5 (0.1–55.9) | 62.50 |
| CLL (9) | 75 (61–80) | 66.7% | — | — | 10.9 (13.4) | 9 (0.1–41.7) | 55.56 |
| CRC (17) | 70 (42–85) | 29.4% | III=5.9% IV=94.1% | Adenocarcinoma=100% | 14.9 (15.3) | 13.2 (0.3–53) | 70.59 |
| Endometrium and ovary (4) | 68 (65–77) | 0% | III=50% IV=50% | Adenocarcinoma=100% | 12.2 (16.4) | 5.6 (1.2–36.3) | 50.00 |
| Gastric and oesophagus (7) | 71 (62–81) | 42.6% | Unk=14.3% II=14.3% III=28.5% IV=42.9% | SCC=14.3% Adenocarcinoma=85.7% | 9.2 (9.2) | 7.1 (0.04–27.1) | 71.43 |
| Head and neck (5) | 55 (43–73) | 80% | III=40% IV=60% | Carcinoma, NOS=20% SCC=80% | 10.6 (10.2) | 11 (0.9–26.8) | 60.00 |
| Kidney (1) | 62 | 100% | IV=100% | RCC=100% | 0.24 | 0.24 | 0.00 |
| Acute leukaemia (12) | 67 (34–87) | 75% | — | ALL=16.7% AML=83.3% | 6.4 (9.9) | 1 (0.1–34.2) | 41.67 |
| Lung (19) | 64 (38–83) | 52.6% | I=5.3% III=15.8% IV=78.9% | Carcinoma, NOS=10.5% SCC=5.3% Adenocarcinoma=68.4% Small cell carcinoma=15.8% | 15.9 (20.3) | 10.3 (0.04–83.6) | 63.16 |
| Lymphoma (11) | 42 (19–84) | 63.6% | Unk=54.5% I=9.1% IV=36.4% | HL=18.2% DLBCL=27.3 Other=54.5% | 26.7 (41.2) | 4 (0.1–121.9) | 63.64 |
| Melanoma (2) | 55 (53–57) | 50% | III=50% IV=50% | Melanoma=100% | 26.5 (47.6) | 36.5 (2.9–70.2) | 50.00 |
| Multiple myeloma (3) | 74 (51–80) | 66.7% | — | — | 3.2 (2.6) | 4.5 (0.2–4.9) | 66.67 |
| Otherb (5) | 43 (32–58) | 80% | II=20% IV=80% | SCC=20% Adenocarcinoma=40% Other=40% | 10.5 (6.2) | 13.9 (0.4–14.7) | 80.00 |
| Pancreas and biliary tract (12) | 66.5 (51–91) | 33.3% | Unk=8.3% III=8.3% IV=83.4% | Carcinoma, NOS=25% Adenocarcinoma=75% | 12.2 (20.6) | 3.9 (0.2–72.9) | 58.33 |
| Prostate (3) | 81 (77–90) | 100% | IV=100% | Adenocarcinoma=100% | 0.9 (0.9) | 0.67 (0.2–1.9) | 0.00 |
Abbreviations: ALL, acute lymphocytic leukaemia; AML=acute myelocytic leukaemia; CLL=chronic lymphocytic leukaemia; CRC=colorectal cancer; DLBCL=diffuse large B-cell lymphoma; HL=Hodgkin’s lymphoma; NOS=not otherwise specified; RCC=renal cell carcinoma; RQ=relative quantification; SCC=squamous cell carcinoma; TCC=transitional cell carcinoma; Unk.=unknown.
High hTERT mRNA levels refer to RQ levels that are 3 or higher.
Other tumours included: cervix squamous cell carcinoma, hepatocellular carcinoma, testicular germ cell tumour and unknown primary.
Exosomal hTERT mRNA levels
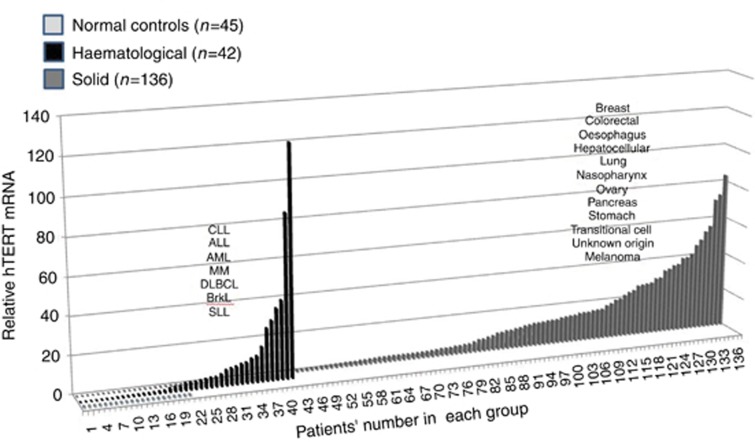
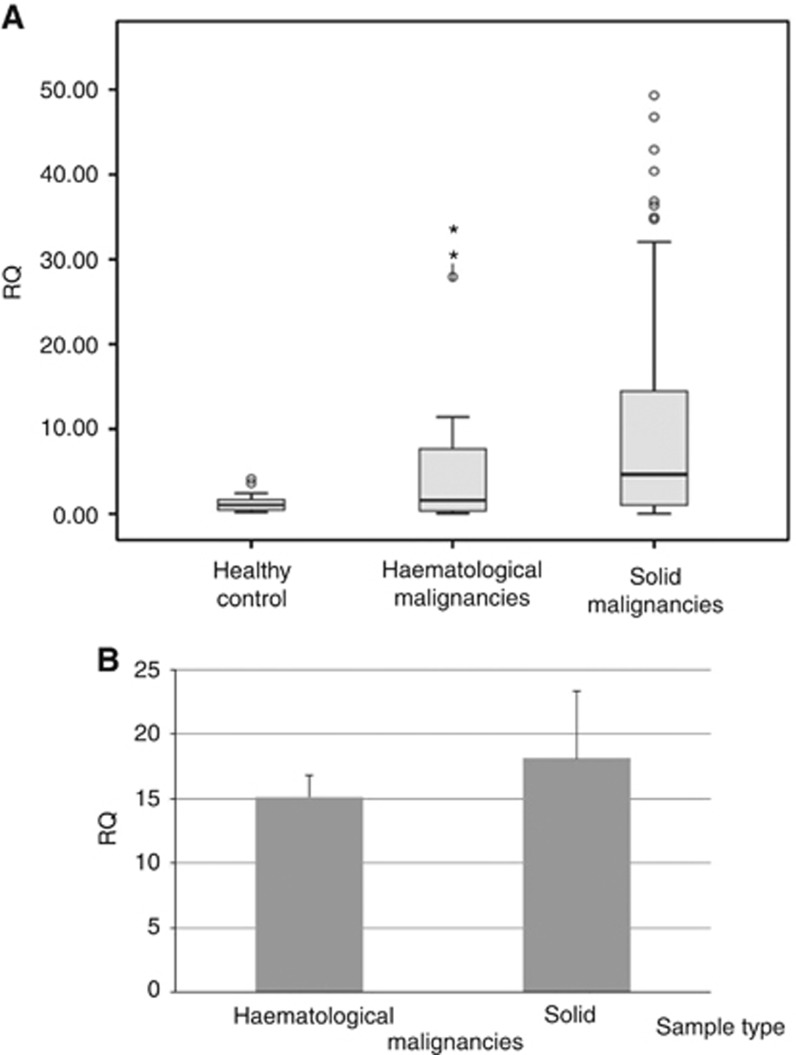
Human telomerase reverse transcripts were not detected in any one of the 45 healthy controls. Figure 1 describes the levels of the hTERT mRNA in all cancer patients. In the group of patients with solid tumours, 61 of the 98 analysed samples express hTERT mRNA in their isolated exosomes (62%). In patients with haematological malignancy, 21 of the 35 samples (60%) exhibited hTERT mRNA. Among patients with detectable hTERT, the average levels of hTERT in the solid tumours were higher than those in the haematological one (Figure 2).
Figure 1.
A waterfall plot describing the levels of hTERT mRNA in exosomes derived from the sera of cancer patients. Serum was separated from blood samples obtained from various patients with cancer. Exosomes were isolated and the levels of hTERT mRNA was measured by quantitative PCR (Q-PCR). Three data sets are shown: the closest line of dots represents the negative values of hTERT mRNA in healthy volunteers (light grey); the second line of bars depicts the values of hTERT mRNA in patients with haematological malignancies (black). The third group of bars represents the values of hTERT mRNA in patients with solid tumours (grey). ALL=acute lymphocytic leukaemia; AML=acute myelocytic leukaemia; Brkl= Burkit lymphoma; CLL=chronic lymphocytic leukaemia; DLBCL=diffuse large B-cell lymphoma; SLL= small lymphocytic lymphoma.
Figure 2.
Average values of the hTERT mRNA levels in cancer patients. (A) Global average of hTERT mRNA values and (B) average of patients who possessed positive values of the hTERT mRNA. RQ=relative quantification.
hTERT expression and tumour characteristics in patients with solid tumours
Although the median level of hTERT transcript for patients with stages III and IV was 8.1 (0.04–86.6) compared with 0.7 (0.4–41.8) for patients with stages I and II, this difference did not reach statistical significance (P=0.71), probably because of the low number of patients with localised disease (142 in stages III and IV compared to 14 in stages I and II). There was no correlation between T stage, nodal status and presence of distant metastases with hTERT levels. Moreover, the presence of liver, bone, lung, brain or peritoneal metastases was not related to higher hTERT level.
The median levels of hTERT transcript were 10.2 (0.3–83.6), 2.8 (0.1–33.4) and 0.4 (0.4–0.4) for poorly, moderately and well-differentiated tumours, respectively, but this difference did not reach significance (P=0.203). However, a trend for correlation between hTERT expression values and poor and moderately differentiated tumours was observed (P=0.1). No correlation was found between hTERT transcript levels and Ki67 score for breast cancer and small-cell lung cancer patients. It appears that some type of histologies, such as small-cell carcinoma, melanoma hepatocellular carcinoma and squamous cell carcinoma, had a higher level of median hTERT transcript, while in others such as acute leukaemia and transitional cell carcinoma (TCC), the median hTERT transcript levels were relatively low. However, data are limited and ANOVA test could not be used because of a large variety of histologies and low number of patients with the same type of histologies.
hTERT expression in haematological malignancies
Human telomerase reverse transcript transcript levels correlated with LDH in acute leukaemia (Pearson’s correlation coefficient=0.88, P=0.0002). In CLL and lymphoma, no correlations with LDH or WBC were found. Owing to the small number of patients with MM (n=3), no analysis was possible regarding hTERT transcript levels.
hTERT expression correlates with the clinical conditions of cancer patients
We were able to measure the expression of the hTERT mRNA over the clinical course in four patients: (1) a 73-year-old male with colorectal cancer: before treatment the levels of the hTERT mRNA were 14. After chemotherapy with good clinical response, the value of the hTERT mRNA decreased to 1. (2) A 68-year-old male with diffuse large B-cell lymphoma had hTERT mRNA levels of 26 at diagnosis. After 1 month, the levels dropped to 5.5 with good response to chemotherapy. After additional 2 months, being in complete remission, the hTERT mRNA was 1, which is considered undetectable. In two other patients, failure to respond to treatment was reflected by elevated levels of the hTERT mRNA. (3) A 64-year-old female with breast cancer had hTERT mRNA levels of 4.3 at diagnosis. After 1 month, the levels of the hTERT mRNA increased to 10. (4) The other patient was a 92-year-old male who was diagnosed with prostate cancer. At diagnosis, the exosomal hTERT mRNAs were 1.5, and after 1 month, the transcript levels increased to 10.5.
Discussion
This study is the first to show elevated concentration of hTERT mRNA in exosomes derived from the sera of cancer patients.
As expected, exosomes of people without cancer do not contain hTERT mRNA because of telomerase negativity of most human somatic cells.
The study included a large variety of patients with different cancers. Elevated expression of hTERT mRNA was found in all tumour types albeit not in all patients. The mean level of mRNA was higher in patients with solid tumours, but higher percentage of patients with haematological malignancies were positive for exosomal hTERT mRNA.
We could not find any specific characteristics of the patients who did not secrete hTERT mRNA in their exosomes. Several possible causes can be responsible for this negativity: in our previous study (Gutkin et al, 2016), we found correlations between the level of cellular telomerase activity and exosomal concentration of hTERT mRNA. In the current study, we were unable to measure the telomerase activity in tumour cells of the patients, and we found that low levels of telomerase could be a possible cause. The content of exosomes is not a passive reflection of cellular concentrations of its constituents. The general involvement of the ESCRT complex in packaging nucleic acids in exosomes has been shown recently (Colombo et al, 2013) and it may differ in various patients. Potentially, it is an interesting aspect of a future research.
More advanced disease was associated with higher hTERT transcripts levels. Unfortunately, the small number of patients prevented achievement of statistical significance for this finding.
Among patients with ovarian and endometrial adenocarcinoma, a trend was noted between higher hTERT transcript levels and higher CA125 levels. Therefore, hTERT transcript levels may have a role in early diagnosis of ovarian cancer.
In acute leukaemia high hTERT transcript levels correlated with higher LDH levels, which may represent tumour aggressiveness, but no correlation was found with WBC, which represent tumour burden. These findings support the need of further evaluation of hTERT in this population.
In four patients with detectable hTERT mRNA levels, the concentrations of hTERT mRNA reflected their clinical status, suggesting that exosomal hTERT mRNA measurement may be a useful marker for response and progression.
Our study has several limitations. First, the statistical power of this study was limited owing to a small number of patients in each cancer type. Second, data were collected from patient charts retrospectively, which may cause bias because of unknown or unrecorded confounders. Additionally, the way we have isolated the exosomes by commercial kits may have caused additional proteins to precipitate with the exosomes; therefore, better results may have been obtained by using ultracentrifugation for the purpose of exosomes’ isolation. Last, some patients received chemotherapy before the sera was collected. Although we did not find a correlation between recent chemotherapy treatment and hTERT levels in our study, the influence of chemotherapy on exosomal hTERT in not well known; therefore, the interpretation of the results in this population might be limited.
This is one of the largest study evaluating sera hTERT in cancer patients. In addition, this is the first study describing exosomal hTERT in cancer patients, using a novel method for exosome isolation and not just RNA isolation from serum, which is a much less sensitive method.
We believe that hTERT transcript levels in cancer patients’ sera may have an important role in cancer diagnosis and evaluation. The main advantage of using hTERT as tumour marker is its widespread presence in all types of cancer and thus having a potential for serving as a ‘pan-cancer’ marker.
More research is needed to establish and characterise the clinical role of exosomal hTERT mRNA measurement. It should be evaluated in specific cancer types as a diagnostic marker and biomarker for follow-up including early diagnosis of relapse and MRD. The data presented in this study attest to the feasibility of these goals.
Acknowledgments
The study was supported by research grants obtained from the Varda and Boaz Dotan Research Center in Haemato-Oncology affiliated with the CBRC at the Tel-Aviv University and from the Research Authority at the Rabin Medical Center.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
The authors declare no conflict of interest.
References
- Batagov AO, Kuznetsov VA, Kurochkin IV (2011) Identification of nucleotide patterns enriched in secreted RNAs as putative cis-acting elements targeting them to exosome nano-vesicles. BMC Genomics 12(Suppl 3): S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, Manel N, Moita LF, Théry C, Raposo G (2013) Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci 126: 5553–5565. [DOI] [PubMed] [Google Scholar]
- Dasi F, Lledó S, García-Granero E, Ripoll R, Marugán M, Tormo M, García-Conde J, Aliño SF (2001) Real-time quantification in plasma of human telomerase reverse transcriptase (hTERT) mRNA: a simple blood test to monitor disease in cancer patients. Lab Invest 81: 767–769. [DOI] [PubMed] [Google Scholar]
- El-Hefnawy T, Raja S, Kelly L, Bigbee WL, Kirkwood JM, Luketich JD, Godfrey TE (2004) Characterization of amplifiable, circulating RNA in plasma and its potential as a tool for cancer diagnostics. Clin Chem 50: 564–573. [DOI] [PubMed] [Google Scholar]
- Garcia JM, García V, Peña C, Domínguez G, Silva J, Diaz R, Espinosa P, Citores MJ, Collado M, Bonilla F (2008) Extracellular plasma RNA from colon cancer patients is confined in a vesicle-like structure and is mRNA-enriched. RNA 14: 1424–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutkin A, Uziel O, Beery E, Nordenberg J, Pinchasi M, Goldvaser H, Henick S, Goldberg M, Lahav M (2016) Tumor cells derived exosomes contain hTERT mRNA and transform nonmalignant fibroblasts into telomerase positive cells. Oncotarget 7: 59173–59188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halicka HD, Bedner E, Darzynkiewicz Z (2000) Segregation of RNA and separate packaging of DNA and RNA in apoptotic bodies during apoptosis. Exp Cell Res 260: 248–256. [DOI] [PubMed] [Google Scholar]
- Hiyama E, Hiyama K (2003) Telomerase as tumor marker. Cancer Lett (2003); 194: 221–233. [DOI] [PubMed] [Google Scholar]
- March-Villalba JA, Martínez-Jabaloyas JM, Herrero MJ, Santamaria J, Aliño SF, Dasí F (2012) Cell-free circulating plasma hTERT mRNA is a useful marker for prostate cancer diagnosis and is associated with poor prognosis tumor characteristics. PLoS ONE 7: e43470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura N, Maeda Y, Kanbe T, Yazama H, Takeda Y, Sato R, Tsukamoto T, Sato E, Marumoto A, Harada T, Sano A, Kishimoto Y, Hirooka Y, Murawaki Y, Hasegawa J, Shiota G (2005) Serum human telomerase reverse transcriptase messenger RNA as a novel tumor marker for hepatocellular carcinoma. Clin Cancer Res 11: 3205–3209. [DOI] [PubMed] [Google Scholar]
- Miura N, Maruyama S, Oyama K, Horie Y, Kohno M, Noma E, Sakaguchi S, Nagashima M, Kudo M, Kishimoto Y, Kawasaki H, Hasegawa J, Shiota G (2007) Development of a novel assay to quantify serum human telomerase reverse transcriptase messenger RNA and its significance as a tumor marker for hepatocellular carcinoma. Oncology 72(Suppl 1): 45–51. [DOI] [PubMed] [Google Scholar]
- Miura N, Osaki Y, Nagashima M, Kohno M, Yorozu K, Shomori K, Kanbe T, Oyama K, Kishimoto Y, Maruyama S, Noma E, Horie Y, Kudo M, Sakaguchi S, Hirooka Y, Ito H, Kawasaki H, Hasegawa J, Shiota G (2010) A novel biomarker TERT mRNA is applicable for early detection of hepatoma. BMC Gastroenterol 10: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatak P, Burger AM (2007) Telomerase and its potential for therapeutic intervention. Br J Pharmacol 152: 1003–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucciarelli S, Rampazzo E, Briarava M, Maretto I, Agostini M, Digito M, Keppel S, Friso ML, Lonardi S, De Paoli A, Mescoli C, Nitti D, De Rossi A (2012) Telomere-specific reverse transcriptase (hTERT) and cell-free RNA in plasma as predictors of pathologic tumor response in rectal cancer patients receiving neoadjuvant chemoradiotherapy. Ann Surg Oncol 19: 3089–3096. [DOI] [PubMed] [Google Scholar]
- Shay JW, Wright WE (2011) Role of telomeres and telomerase in cancer. Semin Cancer Biol 21: 349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrin L, Rampazzo E, De Rossi A (2008) Relationship between tumor and plasma levels of hTERT mRNA in patients with colorectal cancer: implications for monitoring of neoplastic disease. Clin Cancer Res 14: 7444–7451. [DOI] [PubMed] [Google Scholar]
- Thery C (2011) Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep (2011); 3: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui NB, Ng EK, Lo YM (2002) Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin Chem 48: 1647–1653. [PubMed] [Google Scholar]