Abstract
Background:
Gene mutations in the RAS family rule out metastatic colorectal carcinomas (mCRCs) from anti-EGFR therapies.
Methods:
We report a retrospective analysis by Sequenom Massarray and fast COLD-PCR followed by Sanger sequencing on 240 mCRCs.
Results:
By Sequenom, KRAS and NRAS exons 2-3-4 were mutated in 52.9% (127/240) of tumours, while BRAF codon 600 mutations reached 5% (12/240). Fast COLD-PCR found extra mutations at KRAS exon 2 in 15/166 (9%) of samples, previously diagnosed by Sequenom as wild-type or mutated at RAS (exons 3-4) or BRAF genes. After UDG digestion results were reproduced in 2/12 analysable subclonally mutated samples leading to a frequency of true subclonal KRAS mutations of 1.2% (2.1% of the previous Sequenom wild-type subgroup). In 10 out of 12 samples, the subclonal KRAS mutations disappeared (9 out of 12) or turned to a different sequence variant (1 out of 12).
Conclusions:
mCRC can harbour coexisting multiple gene mutations. High sensitivity assays allow the detection of a small subset of patients harbouring true subclonal KRAS mutations. However, DNA changes with mutant allele frequencies <3% detected in formalin-fixed paraffin-embedded samples may be artifactual in a non-negligible fraction of cases. UDG pre-treatment of DNA is mandatory to identify true DNA changes in archival samples and avoid misinterpretation due to artifacts.
Keywords: colorectal cancer, extended RAS, BRAF, PIK3CA, mutant allele frequency, heterogeneity, artifactual DNA call, UDG treatment
The threshold of ‘precision medicine’ in oncology is continuously rising. In the past few years, many molecular players involved in oncogenesis have been exploited as potential drug targets in the ever-growing approach of patient-tailored care (Jurgensmeier et al, 2014; Stevens and Rodriguez, 2015). The results of several clinical trials (Harbeck and Wuerstlein, 2013; Landi and Cappuzzo, 2014; Khattak et al, 2015) have shown that often only specific subgroups of patients benefit from molecular-targeted treatments. Thus, the development of highly sensitive and specific companion diagnostic tests is needed to improve the characterisation of the responsive patients and to intercept molecular alterations with negative or positive predictive value (Dienstmann et al, 2015; Hovelson et al, 2015; Zhao et al, 2015). Moreover, in the current era of increasing healthcare costs colliding with shrinking budgets, the precise identification of who could benefit the most from a specific treatment is of paramount importance.
In colorectal tumours, the subset of patients amenable to targeted therapies with anti-EGFR drugs underwent few revisions (Van Cutsem et al, 2015) as new detrimental somatic point mutations have been found to hamper the response to these treatments, in addition to the more common mutations at codons 12 and 13 of KRAS. The additional mutations span codons 61, 117 and 146 of KRAS and codons 12, 13, 61, 117 and 146 of NRAS (extended RAS).
Nevertheless, both the improved understanding of tumour heterogeneity and the high sensitivity of the assays introduced in molecular diagnostics open new questions focusing on which cutoff value makes an alteration a significant marker leading to a different outcome or a different clinical response. Few and contradictory data are available on the predictive role of a low mutational load in tumour samples (defined as a low number of neoplastic cells harbouring a specific mutation) compared to wild-type lesions (Dono et al, 2013; Kimura et al, 2012; Laurent-Puig et al, 2015; Molinari et al, 2011; Normanno et al, 2015; Tougeron et al, 2013; Van Cutsem et al, 2015). To date, these questions are being explored especially in the field of lung adenocarcinomas (Wang et al, 2014) and more recently facing the issue of quantitatively monitoring the emergence of KRAS mutations in liquid biopsy samples of metastatic colorectal carcinoma (mCRC) patients treated with anti-EGFR drugs (Toledo et al, 2017).
In the present study, we investigated the real nature and true incidence of subclonal mutations detected in formalin-fixed paraffin-embedded (FFPE) samples by increasing assay sensitivities (0.1–5%).
Materials and methods
Patients
The study included 240 mCRC patients whose tumour was analysed at our institution in 2014 to select possible candidates for anti-EGFR therapy based on their mutational profile. As a local reference laboratory for the diagnostic mutational screening in solid tumours since 2010, we have been successfully participating to several EQA schemes of the European Society of Pathology and of the Italian Association of Medical Oncologists/Italian Society of Pathological Anatomy and Cytology (AIOM-SIAPEC-IAP) for KRAS and BRAF (data not shown).
The cohort was composed of 108 women and 132 men, and the median age at diagnosis was 62 (range: 34–84) and 67 years (range: 22–92), respectively. Tumour grading and staging data, according to World Health Organization and AJCC Staging criteria (VII Edition, 2010), were available for the 160 patients treated at our Institution (Table 1). The tissues analysed were mainly histological samples from primary CRCs (98%), locally resected at diagnosis (81%) or at relapse (6%). Most cases displayed moderate-to-low differentiation (92.6% G2-G3), locally advanced disease (94.5% pT3-4), lymph nodal involvement (74.4% pN1-2) and perineural and/or vascular invasion (78.4%). The series also included distant metastases (13% of the cases, including seven pulmonary, 17 liver, two bone, one peritoneal, one cerebral and one pelvic metastases).
Table 1. Clinicopathological features of tumour specimens in relation to KRAS, NRAS and BRAF gene mutation status.
| KRAS mut n (%) | NRAS mut n (%) | BRAF mut n (%) | RAS/BRAF WT n (%) | Total n (%) | |
|---|---|---|---|---|---|
|
Sex | |||||
| F | 52 (48.1) | 6 (5.6) | 10 (9.3)a | 40 (37.0) | 108 (45.0) |
| M | 61 (46.2) | 8 (6.1) | 2 (1.5) | 61 (46.2) | 132 (55.0) |
| Sub total | 113 (47.1) | 14 (5.8) | 12 (5.0) | 101 (42.1) | 240 (100) |
|
Age (years) | |||||
| Median | 64 | 68 | 65,5 | 63,5 | — |
| Range | 34–84 | 40–87 | 51–73 | 22–92 | — |
| Sub total | 101 (47.2) | 13 (6.1) | 10 (4.7) | 90 (42.0) | 214 (100) |
|
G | |||||
| 1–2 | 57 (48.7) | 5 (4.3) | 1 (0.9) | 54 (27.3) | 117 (71.8) |
| 3–4 | 21 (45.6) | 4 (8.7) | 5 (10.9)a | 16 (34.8) | 46 (28.2) |
| Sub total | 78 (47.8) | 9 (5.5) | 6 (3.7) | 70 (42.9) | 163 (100) |
|
pT | |||||
| 1–2 | 5 (55.5) | 1 (11.1) | 0 (0) | 3 (0) | 9 (5.5) |
| 3–4 | 73 (47.4) | 8 (5.2) | 6 (3.4) | 67 (43.5) | 154 (94.5) |
| Sub total | 78 (47.8) | 9 (5.5) | 6 (3.7) | 70 (42.9) | 163 (100) |
|
pN | |||||
| 0 | 19 (46.3) | 3 (7.3) | 2 (4.9) | 17 (41.5) | 41 (25.6) |
| 1 | 36 (56.3) | 3 (4.7) | 0 (0) | 25 (39.1) | 64 (40.0) |
| 2 | 21 (38.2) | 2 (3.6) | 4 (7.3) | 28 (50.9) | 55 (34.4) |
| Sub total | 76 (47.5) | 8 (5.0) | 6 (3.8) | 70 (43.7) | 160 (100) |
|
Vascular and/or perineural invasion | |||||
| Yes | 52 (44.8) | 6 (5.2) | 4 (3.4) | 54 (41.7) | 116 (78.4) |
| No | 17 (53.1) | 2 (6.3) | 1 (3.1) | 12 (37.5) | 32 (21.6) |
Abbreviations: mut=mutated; WT=wild-type; F=female; M=male.
BRAF mutations are significantly more prevalent in female patients (F) and G3-G4 tumours rather than in males (M) and G1-G2 tumours (P=0.013).
The study was submitted to and approved by the Ethic Institutional Review Board (IRB) responsible for ‘Biobanking and use of human tissues for experimental studies’ of the Department of Medical Sciences at the University of Turin. The IRB approved a verbal consent procedure due to the retrospective design of the study, which had no effect on their care.
Material selection
We retrieved the 240 DNA samples that were earlier banked when the diagnostic analysis was performed on one representative FFPE tumour block. For six selected cases additional FFPE samples corresponding to alternative tumour blocks of the primitive tumour and/or regional lymph-node/distant metastases were submitted to DNA extraction.
In all cases a selection of the tumour area was performed as previously described (Mariani et al, 2015). Morphological adequacy criteria were derived from the Biogate portal (https://testbiomolecolari.it/) of the AIOM-SIAPEC-IAP. Specimens were recorded as adequate when tumour enrichment was higher than the 50% and at least 100 neoplastic cell were present.
DNA extraction
DNA extraction from FFPE tissues was performed as previously described (Mariani et al, 2015) and concentrations/purity were measured by a Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA).
Mass spectrometry technology for DNA sequencing
The DNAs were amplified according to the manufacturer’s instructions on a Labcycler (SensoQuest GmbH, Germany), using the CE-IVD Myriapod Colon Status kit (Diatech Pharmacogenetics, Jesi, Italy) based on a multiplex-PCR. The amplified DNAs were submitted to a Shrimp Alkaline Phosphatase digestion (SAP reaction) to remove the excess of free nucleotides. Finally, the purified DNAs underwent a reaction of primer extension (iPLEX) with oligonucleotides flanking each gene position under study and with dideoxynucleotide terminators of known mass to discriminate wild-type from mutated genotypes, by using a matrix-assisted laser desorption ionisation time-of-flight mass spectrometry. Amplified DNAs were dispensed on a supplied solid chip prior to be scanned on a MassARRAY instrument (Sequenom Inc, San Diego, CA, USA).
The Myriapod Colon Status kit identifies 58, 54, 23 and 66 nucleotide substitutions in the KRAS, NRAS, BRAF and PIK3CA genes, respectively. Among them, the mutations related to the clinical response to anti-EGFR treatments are those affecting codons 12, 13, 59, 61, 117 and 146 in both KRAS and NRAS, while the mutations at codon 600 of BRAF have a prognostic significance. This assay also explores mutations at multiple codons of the PIK3CA gene.
Fast COLD-PCR followed by Sanger Sequencing for codons 12 and 13 of KRAS
The fast COLD-PCR for the exon 2 of KRAS followed by Sanger Sequencing was applied to all samples considered wild-type by Sequenom at exon 2 of KRAS and to a subset of those KRAS (exon 2) mutated. The assay was used: (1) to confirm the Sequenom results; (2) to identify the presence and incidence of minor tumour clones undetectable by the standard methods; (3) to determine the real nature of the subclonal mutations. In brief, 30 ng of DNA/reaction were added to fast COLD-PCR reagents and preliminary submitted to treatment at 37 °C for 30′ in the presence or absence of 0.5 U of Uracil-DNA-Glycosylase (UDG, Thermo Fisher) (Do and Dobrovic, 2012). The protocol of the fast COLD-PCR analysis was derived from Mancini et al (Mancini et al, 2010) in absence of fluorophores in the reagent mix, under conditions modified as follows: 20 cycles of standard PCR (95.0 °C 8'', 60.0 °C 30'', 72.0 °C 30'') followed by 35 cycles of COLD-PCR (82.5 °C 8'', 58.0 °C 30'', 72.0 °C 30''). The denaturation step was set at a critical dissociation temperature, favouring the amplification of the mutated sequences. The amplified products were then submitted to Sanger Sequencing.
PCR followed by Sanger Sequencing for KRAS, NRAS and BRAF mutational analysis
Targeted-PCRs were followed by Sanger Sequencing (Sanger et al, 1977) to confirm the somatic gene mutations with clinical significance demonstrated by Sequenom. Sanger Sequencing was also applied to PCR products of the fast COLD-PCR.
Reagent mixes for targeted-PCRs contained 70 ng of genomic DNA, 1 × buffer, 1.5 mM of MgCl2, 200 μM of dNTPs, 1.2 U of AmpliTaq Gold DNA Polymerase (Invitrogen, Carlsbad, CA, USA) and 0.5 μM of each couple of primers, in a final volume of 50 μl. KRAS and NRAS primers were designed using the software Primer3 (http://frodo.wi.mit.edu/primer3/) on the human genomic sequence (http://www.ncbi.nlm.nih.gov/gene) (ID:3845 and ID:7812, respectively) and are available upon request. The BRAF primers were derived from Moroni et al (Moroni et al, 2005), and generated amplicons of 228 bp. PCR conditions consisted of a unique touchdown scheme (an annealing decrease of 3.0 °C each 3 cycles starting from 64.0 °C down to 57.0 °C) that was repeated for 44 cycles. The purified products of both targeted- and fast COLD-PCR (see above) were then submitted to a cycle sequencing reaction, as previously described (Mariani et al, 2015). The Chromas software (www.technelysium.com.au) was used for sequence analysis.
Sensitivities of the assays
The declared sensitivity of the Myriapod Colon Status kit varied from 2.5 to 10%, depending on the mutation under study. Homemade reactions were designed to determine the sensitivities of Sanger Sequencing when preceded by a standard targeted-PCR (KRAS and BRAF) or by a fast COLD-PCR (KRAS). Mutated DNA extracted from specific cell lines were progressively diluted in blood-derived DNA of a healthy donor. Briefly, the A549 cell line, homozygous for the KRAS p.G12S (p.Gly12Ser) mutation, and the HT29, heterozygous for the BRAF p.V600E (p.Val600Glu) mutation, were purchased from the ATCC (LGC Standards S.r.l., Sesto San Giovanni, Milan, Italy) and tested at different dilution in the wild-type sample DNA. The described mutation at KRAS was detected by Sanger Sequencing at a sensitivity near to 10% and 0.1% starting from products of standard targeted-PCRs and fast COLD-PCRs, respectively (Supplementary Figure 1A). The sensitivity for the BRAF mutation was set at 5% (Supplementary Figure 1B).
Statistics
Pearson’s χ2-test corrected if necessary for the Fisher’s exact test was applied to 2 × 2 Contingency Tables to compare the mutation frequencies: (1) among patients with different characteristics; (2) among tumour DNAs analysed with different degree of enrichment, and with alternative protocols of PCR before sequencing (www.openepi.com). P-values<0.05 were considered statistically significant.
Results
KRAS/NRAS/BRAF/PIK3CA mutation frequencies by Sequenom in mCRCs
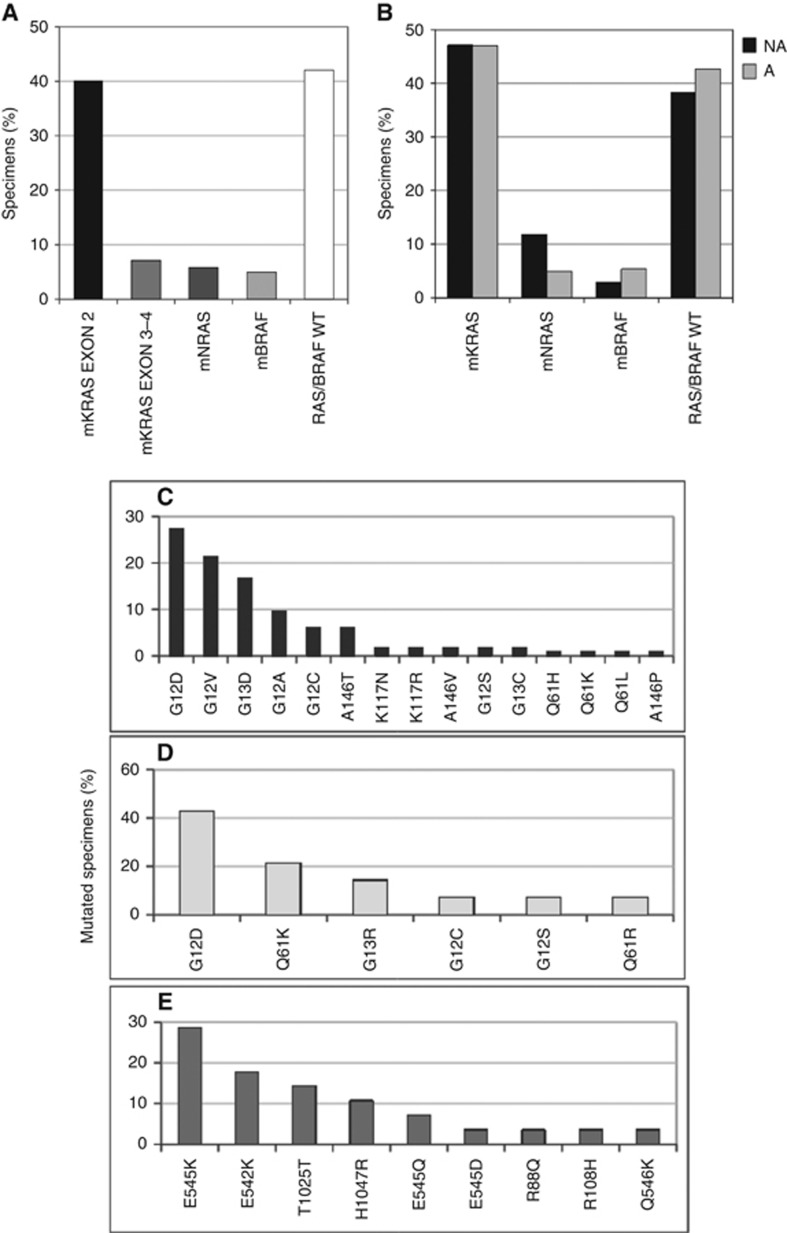
By Sequenom analysis we demonstrated a point mutation (exon 2 of KRAS) in 40.0% (96/240) of mCRC patients (Figure 1A), in line with results previously obtained by pyrosequencing (Mariani et al, 2015). The extended survey identified 43 additional patients (43/240, 17.9%) with further tumour DNA mutations, mutually exclusive and distributed as follows: 17/240 (7.1%) at exons 3-4 of KRAS, 14/240 (5.8%) at exons 2-3-4 of NRAS and 12/240 (5.0%) at codon 600 of BRAF. Overall, the ‘extended RAS’ mutated samples represented 21.5% (31/144) of those samples defined as wild-type until the ASCO guideline update (Allegra et al, 2016). Finally, by Sequenom, for the codons included in the Colon cancer kit the remaining 101/240 (42.1%) patients were considered RAS/BRAF wild type.
Figure 1.
Distribution of gene mutations and allelic variants occurring at KRAS, NRAS, BRAF and PIK3CA genes in tumour DNAs of mCRC patients by Sequenom analysis. (A) Distribution of mutated (m) and wild-type (WT) specimens at exon 2 of KRAS, at exons 3 and 4 of KRAS, at exons 2, 3 and 4 of NRAS and at codon 600 of BRAF. (B) Distribution of mCRC patients with tumour DNA sequences either WT or mutated (m) at the KRAS, NRAS and BRAF genes, according to a tumour/normal cell ratio below (NA, not adequate) and above (A, adequate) the 50% cutoff. Panels C–E represent the allelic variant distribution belonging to the mutated KRAS (C), NRAS (D) and PIK3CA (E) genes.
The same protocol allowed the investigation of additional point mutations occurring at the PIK3CA gene, albeit considered not significant at present for therapy decision-making. We found 28/240 (11.7%) patients harbouring a PIK3CA mutation, alone (9/28, 32.1%) or in conjunction with KRAS (15/28, 53.6%), NRAS (3/28, 10.7%) or BRAF (1/28, 3.6%) mutations.
In 34 samples, we were not able to macrodissect tumour areas for their enrichment at levels above 50% before DNA extraction. In this subset of samples, somatic mutations of KRAS, NRAS and BRAF demonstrated by Sequenom occurred in 16/34 (47.1%), 4/34 (11.8%), 1/34 (2.9%) specimens, respectively. The remaining samples (38.2%) were considered wild type. Of note, the distribution of somatic mutations was comparable (P>0.05) between tumours above and below the 50% enrichment cutoff (Figure 1B) and therefore it did not represent a possible bias.
Frequency distribution and clinicopathological features of KRAS, NRAS, BRAF and PIK3CA mutated samples
KRAS was confirmed as the most frequently mutated gene in our cohort. In particular, the nucleotide substitutions affected codons 12, 13, 146 and 117 with decreasing frequency (Figure 1C). The most frequent NRAS point mutations occurred at codons 12, 61 and 13 (Figure 1D). No exon 4 KRAS mutations were detected. BRAF was mutated exclusively at codon 600 (p.V600E) (data not shown). Finally, PIK3CA mutations mainly mapped to exon 9 (18/240, 7.5%), followed by exon 20 (8/240, 3.3%) (Figure 1E).
As reported in Table 1, the BRAF gene was confirmed significantly associated with female gender (P=0.013) and with less differentiated (G3-G4) tumours (P=0.013), as previously reported (Loupakis et al, 2016). Node-negative patients resulted more frequently mutated at codon 12 than 13 (P=0.04). The opposite occurred in tumours with pN2 nodal status (P<0.05; Table 2). Among all RAS mutations, the p.G12V KRAS prevailed in G3-G4 tumours, although this correlation did not reach statistical significance (Table 3).
Table 2. Distribution of KRAS mutations according to the involved hotspot gene sequences and to clinicopathological features.
| mKRAS n (%) | mKRAS ex 2 n (%) | mKRAS ex 3-4 n (%) | mKRAS cod 12 n (%) | mKRAS cod 13 n (%) | |
|---|---|---|---|---|---|
|
Sex | |||||
| F | 52 (46.0) | 44 (44.9) | 8 (47.0) | 35 (46.7) | 9 (42.9) |
| M | 61 (54.0) | 52 (55.1) | 9 (53.0) | 40 (52.3) | 12 (57.1) |
| Subtotal | 113 (100) | 96 (100) | 17 (100) | 75 (100) | 21 (100) |
|
Age (years) | |||||
| Median | 64 | 64 | 65,5 | 64 | 63 |
| Range | 34–84 | 34–84 | 47–76 | 34–84 | 49–74 |
| Subtotal | 101 | 85 | 16 | 66 | 19 |
|
G | |||||
| 1–2 | 57 (73.1) | 48 (73.8) | 9 (69.2) | 36 (72.0) | 12 (80.0) |
| 3–4 | 21 (26.9) | 17 (26.2) | 4 (30.8) | 14 (28.0) | 3 (20.0) |
| Subtotal | 78 (100) | 65 (100) | 13 (100) | 50 (100) | 15 (100) |
|
pT | |||||
| 1–2 | 5 (6.4) | 4 (6.1) | 1 (7.7) | 3 (6.0) | 1 (6.7) |
| 3–4 | 73 (93.6) | 61 (93.9) | 12 (92.3) | 47 (94.0) | 14 (93.3) |
| Subtotal | 78 (100) | 65 (100) | 13 (100) | 50 (100) | 15 (100) |
|
pN | |||||
| 0 | 19 (25.0) | 17 (26.6) | 2 (16.7) | 16 (32.6)a | 1 (6.6)a |
| 1 | 36 (47.4) | 30 (46.8) | 6 (50.0) | 23 (46.9) | 7 (46.7) |
| 2 | 21 (27.6) | 17 (26.6) | 4 (33.3) | 10 (20.4)a | 7 (46.7)a |
| Subtotal | 76 (100) | 64 (100) | 12 (100) | 49 (100) | 15 (100) |
|
Vascular and/or perineural invasion | |||||
| Yes | 52 (75.4) | 44 (75.9) | 8 (72.7) | 32 (74.4) | 12 (80.0) |
| No | 17 (24.6) | 14 (24.1) | 3 (27.3) | 11 (25.6) | 3 (20.0) |
| Subtotal | 69 (100) | 58 (100) | 11 (100) | 43 (100) | 15 (100) |
Abbreviations: m=mutated; ex=exon; cod=codon; F=female; M=male.
The percentage of specimens KRAS mutated at the codon 12 is significantly higher in the pN0 than in the pN2 tumours (P<0.05); the opposite occurs in specimens mutated at the codon 13 of the KRAS gene.
Table 3. Distribution of the leading RAS gene mutations according to clinicopathological features.
| KRAS G12A n (%) | KRAS G12C n (%) | KRAS G12D n (%) | KRAS G12V n (%) | KRAS G13D n (%) | KRAS A146T n (%) | NRAS G12D n (%) | |
|---|---|---|---|---|---|---|---|
|
Sex | |||||||
| F | 7 (63.6) | 3 (42.9) | 14 (45.2) | 11 (45.8) | 8 (42.1) | 2 (28.6) | 4 (80.0) |
| M | 4 (36.4) | 4 (57.1) | 17 (54.8) | 13 (54.2) | 11(57.9) | 5 (71.4) | 1 (20.0) |
| Subtotal | 11 (100) | 7 (100) | 31 (100) | 24 (100) | 19 (100) | 7 (100) | 5 (100) |
|
Age (years) | |||||||
| Median | 60 | 64 | 63 | 64 | 63 | 63 | 63 |
| Range | 34–74 | 34–71 | 36–84 | 45–81 | 49–74 | 56–73 | 40–67 |
| Subtotal | 11 | 6 | 29 | 18 | 17 | 6 | 4 |
|
Tumour | |||||||
| Primary | 7 (63.6) | 6 (100) | 23 (76.7) | 18 (85.7) | 17 (89.5) | 6 (85.7) | 3 (60.0) |
| Met | 4 (36.4) | 0 (0) | 7 (13.3) | 3 (14.3) | 2 (11.5) | 1 (14.3) | 2 (40.0) |
| Subtotal | 11 (100) | 6 (100) | 30 (100) | 21 (100) | 19 (100) | 7 (100) | 5 (100) |
|
G | |||||||
| 1–2 | 8 (80.0) | 2 (66.7) | 18 (81.8) | 7(53.8) | 10 (76.9) | 4 (80.0) | 1 (50.0) |
| 3–4 | 2 (20.0) | 1 (33.3) | 4 (18.2) | 6 (46.2)a | 3 (23.1) | 1 (20.0) | 1 (50.0) |
| Subtotal | 10 (100) | 3 (100) | 22 (100) | 13 (100) | 13 (100) | 5 (100) | 2 (100) |
|
pN | |||||||
| 0 | 3 (30.0) | 1 (33.3) | 8 (38.1) | 4 (30.7) | 1 (7.7) | 1 (25.0) | 0 (0) |
| 1 | 5 (50.0) | 2 (66.7) | 8 (38.1) | 6 (46.2) | 7 (53.8) | 1 (25.0) | 1 (50.0) |
| 2 | 2 (20.0) | 0 (0) | 5 (23.8) | 3 (23.1) | 5 (38.5) | 2 (50.0) | 1 (50.0) |
| Subtotal | 10 (100) | 3 (100) | 21 (100) | 13 (100) | 13 (100) | 4 (100) | 2 (100) |
|
Vascular and/or perineural invasion | |||||||
| Yes | 8 (88.9) | 2 (66.7) | 13 (68.4) | 7 (70.0) | 10 (76.9) | 3 (75.0) | 1 (100) |
| No | 1 (11.1) | 1 (33.3) | 6 (31.6) | 3 (30.0) | 3 (23.1) | 1 (25.0) | 0 (0) |
| Subtotal | 9 (100) | 3 (100) | 19 (100) | 10 (100) | 13 (100) | 4 (100) | 1 (100) |
Abbreviations: F=female; M=male; Met=metastasis.
In G3-G4 tumours the percentage of the KRASp.G12V (p.Gly12Val) mutated specimens compared to the alternative KRAS-mutated specimens is higher although statistically not significant (P=0.051).
Fast COLD-PCR detection of RAS mutations with low mutant allele frequency in candidate patients for anti-EGFR-based therapies
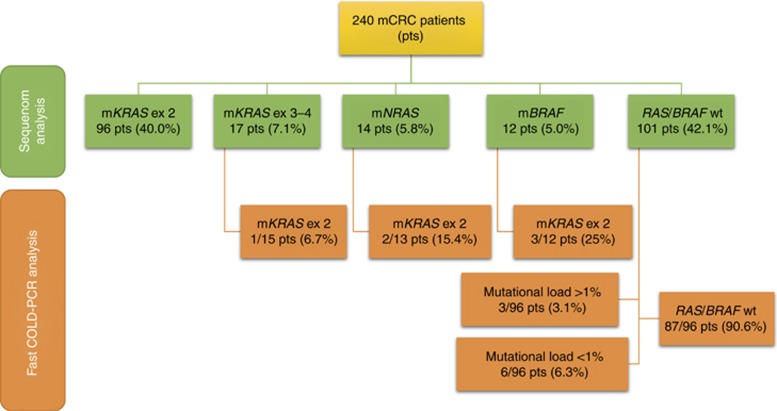
We re-analysed KRAS by fast COLD-PCR in 30/96 samples with already identified exon 2 mutations, 40/43 samples mutated either at exons 3-4 of KRAS or at NRAS or BRAF, and 96/101 wild-type. All KRAS exon 2 mutations by Sequenom were confirmed. A total of 15 additional mutations were detected (15/166, 9%), specifically 9/96 (9.4%), 1/15 (6.7%), 2/13 (15.4%) and 3/12 (25.0%) in mCRC previously categorised by Sequenom as wild-type or already mutated at KRAS (exons 3 and 4), NRAS or BRAF (Figure 2). The range of mutant allele frequency (MAF) of these subclonal mutations was comprised between 0.1% and 5%.
Figure 2.
Gene mutations according to the degree of assay sensitivities in 240 mCRC patients. Sequenom found 57.9% of specimens mutated at RAS/BRAF (mRAS/BRAF), whereas 42.1% were wild-type (WT). A more sensitive fast COLD-PCR method recruited additional mutated sequences in both wild-type samples and in mutated samples. In particular, 90.6% of the samples RAS/BRAF WT by Sequenom were confirmed by fast COLD-PCR, whereas the remaining 9.4% of DNAs were mutated at KRAS exon 2 (ex 2) with a discrete not negligible (1–5%) or low (<1%) mutational load.
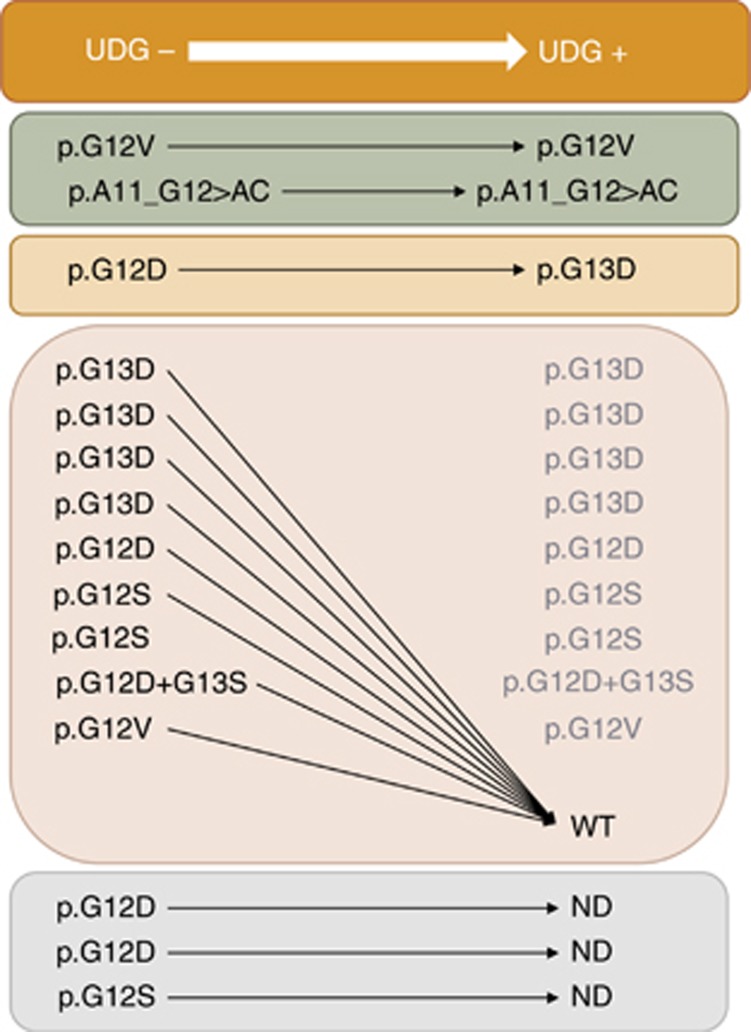
We were able to reanalyse 12/15 subclonally mutated samples after UDG digestion. Results were reproduced in two out of 12 samples, both originally wild type by Sequenom, but harbouring a mutation with a MAF >3% detected by fast-COLD-PCR. One patient had a complex mutation at position c.33_34delTGinsCT (p.A11_G12AC) and the other a mutation c.35G>T (p.G12V, p.Gly12Val), both undetectable with the Sequenom assay. Overall this leads to a frequency of true subclonal KRAS mutations of 1.2% (2.1% of the Sequenom wild-type subgroup).
In the remaining 10/12 cases (nine below and one above 3% MAF, respectively) the subclonal KRAS mutations either disappeared (9/12, including three cases harbouring a BRAF mutation) or turned to a different sequence variant at low MAF (1/12). In the latter case a possible mechanism of DNA damage other than cytosine deamination cannot be excluded (Figure 3).
Figure 3.
The fate of subclonal KRAS mutations after pre-treatment of tumour DNAs with UDG. DNAs belonging to 12/15 mCRC tumours, resulted subclonally mutated at KRAS by fast COLD-PCR in absence of UDG pre-treatment (UDG- column), had been re-analysed after UDG digestion (UDG+ column), aimed to avoid PCR artifacts due to formalin fixation injuries on DNA molecules. Eighty-three per cent of the mutated (by fast COLD-PCR, UDG-) cases became WT or changed allelic variant after UDG digestion (UDG+), thus demonstrating their artefactual nature. Only in two tumours (17% of the cases) the mutations were re-confirmed after UDG treatment (UDG+). Both mutations had a mutant allele frequency>3%. ND: not determined cases (due to lack of residual DNA).
To deepen our analysis, in six patients originally showing subclonal KRAS mutations detected in one representative sample by fast COLD-PCR only, we investigated 6–7 more blocks per patient, deriving from the primary tumour and/or regional lymph-node/distant metastases, for a total of 40 samples. Without UGD pre-treatment, 5/6 cases showed a spectrum of subclonal KRAS variants detected in 12/40 of all analysed blocks, of which only one showed the same DNA change originally detected in the representative diagnostic sample. The large majority of PCR amplified DNAs (83.3%) became KRAS wild-type and the remaining (16.7%) showed new mutation sequences, following UDG treatment.
Fast COLD-PCR detects the mutations G>T or G>A of KRAS at the codons 12 and 13. In order of frequency, the most represented artifactual mutations were c.35G>A (p.G12D, p.Gly12Asp) and c.38G>A (p.G13D, p.Gly13Asp).
Discussion
The present study (i) confirms the frequency of classical KRAS mutations (exon 2) by Sequenom and evaluates the presence of specific additional mutations, as required by the new international guidelines, in a series of 240 mCRCs; (ii) validates fast COLD-PCR method for the analysis of KRAS gene exon 2 and determines the frequency of mutations with low MAF, detectable only using highly sensitive assays; and (iii) reveals the likely artifactual nature of KRAS mutations present at very low MAF in FFPE samples, especially when below 3%.
The KRAS exon 2 mutation rate by Sequenom was 40%, in line with reported data (Rosty et al, 2013) and with results previously obtained by our group with assays of similar analytical sensitivities (Mariani et al, 2015). Of note, the ‘extended RAS’ test revealed additional mutations in 21.5% of patients who were previously considered eligible to therapies with anti-EGFR molecules. Therefore, these patients would not meet the criteria for eligibility to this targeted therapy according to the new guidelines (Allegra et al, 2016). An additional 8.3% of eligible patients were mutated at BRAF codon 600, a feature linked with a worse prognosis independently of other clinical parameters (Palomba et al, 2016; Richman et al, 2009; Souglakos et al, 2009; Therkildsen et al, 2014; Tol et al, 2010).
The Sequenom assay also allowed to explore the prevalence of PIK3CA mutations, which were identified in 11.7% of the patients, similarly to what recently reported by Palomba et al (2016). In contrast with data reported by Normanno et al (2015), we did not observe a significant association between tumours with low KRAS mutational load and co-occurrence of mutated PIK3CA sequences. This discrepancy may stem from a technical reason, as Normanno and colleagues used a targeted next-generation sequencing approach to assess the PIK3CA mutational status. A recent meta-analysis study has pointed out that PIK3CA mutations may be detrimental for the response to anti-EGFR molecules, especially those occurring at exon 20 (Huang et al, 2014). Nevertheless, this trend needs to be confirmed in clinical trials.
When we put into context our molecular data with respect to tumour features, we observed that node positive (pN2) compared to node-negative tumours were preferentially mutated at codon 13 rather than 12, supporting the hypothesis of a more aggressive behaviour of tumours mutated at codon 13. In line with this assumption, Feng et al (2015) demonstrated that KRAS mutations at codon 13 and pN2 stage are independent risk factors for distant metachronous metastases.
Interestingly, by deepening our analysis with fast COLD-PCR, we detected up to the 0.1% of mutant/wild-type DNA molecules and demonstrated that ∼10% of patients was found to harbour KRAS exon 2 mutations with low mutant allele frequency among BRAF wild-type individuals and supposed to be responsive to anti-EGFR antibodies (RAS wild-type by standard assays).
These data are in line with those reported by the CRYSTAL study analysing mCRC patients treated with fluorouracil, leucovorin and irinotecan with or without cetuximab. When tumour DNA sequences of the CRYSTAL series were analysed using a 5% mutant/wild-type cutoff, 14.7% of wild-type KRAS exon 2 patients revealed point mutations at the ‘extended RAS’ sequences. The same authors also observed that point mutations other than those occurring at exon 2 of KRAS (extended RAS) play an adverse role on progression-free and overall survival and objective responses (Van Cutsem et al, 2015). The frequency of positive samples increased up to 34.6% using an assay with a 0.1% sensitivity cutoff. Although displaying a detectable level of mutation (0.1–5%), these tumours were not associated with worse clinical responses. Regrettably, the patients harbouring a KRAS mutation with low MAF in our study were heterogeneously treated, thus precluding any clinical correlations.
Nevertheless, given that formalin fixation is known to produce several DNA injuries (Do and Dobrovic, 2015; Do et al, 2013; Lamy et al, 2011), including deamination of the cytosine to uracil which generates artifactual C:G>T:A changes, in our study we also investigated the nature of these subclonal KRAS mutations by pre-treating DNAs with UDG. This enzyme removes uracil bases belonging to cytosine deamination and creates disruptions along DNA sequence interfering with the PCR amplification of the artifactual mutated sequences only. Following UDG digestion results were reproduced in two cases only, both originally wild-type by Sequenom and found to harbour mutations with a MAF >3% by fast-COLD-PCR. This finding leads to a frequency of true subclonal KRAS mutations of 2.1% of the Sequenom wild-type tumours.
This evidence on artifactual DNA changes in FFPE samples is of utmost importance, as recently the somatic mutation load has been critically reviewed to determine the association with response or resistance to specific targeted therapies (Dono et al, 2013; Molinari et al, 2011; Van Cutsem et al, 2015). One may wonder which level of mutational load is to be considered clinically predictive of responsiveness/resistance to a specific targeted therapy. Several groups have identified KRAS mutations harbouring low MAFs in mCRC patients (Dono et al, 2013; Kimura et al, 2012; Laurent-Puig et al, 2015; Molinari et al, 2011; Normanno et al, 2015; Tougeron et al, 2013; Van Cutsem et al, 2015) (Supplementary Table 1); however, distinct cutoffs to define minor subclones have been adopted, heterogeneous cohorts (selected or unselected patients) were analysed and details about the use of UDG pre-treatment were not reported, thus leading to results that are difficult to compare. Nevertheless, based on our data and on results of other studies (Laurent-Puig et al, 2015; Normanno et al, 2015; Tougeron et al, 2013), a MAF range comprised between 1 and 3% seems to emerge as a grey area of mutational loads either with or without clinical significance. Indeed, Tougeron et al (2013) were able to show that presence of mutations with a MAF >2.3% was associated with cetuximab resistance and shorter progression-free survival. Likewise, Normanno et al (2015) reported a shorter survival for those patients with a tumour harbouring KRAS mutations with a MAF >3%. Of note, Laurent-Puig et al (2015) observed that patients with less than 1% of KRAS MAF showed similar progression-free survival and overall survival than those with wild-type KRAS tumours, thus suggesting that patients with mCRC with KRAS-mutated subclones (at least those with a KRAS-mutated subclones fraction lower or equal to 1%) had a benefit from anti-EGFR therapies.
Taken together these data suggest that subclonal mutations with a very low MAF have to be taken with caution and further analysed by including an UDG digestion step in the process to truly identify mutations governing the choice of a therapeutic compound. This is also of importance for an accurate mutational tracking of patients exhibiting emergence of KRAS-mutated clones in their plasma during anti-EGFR treatments.
In conclusion, our work further highlights that mCRCs are rather heterogeneous showing coexisting multiple gene mutations, with a small subset of patients (2.1% of the Sequenom wild-type tumours) harbouring KRAS mutations missed due to intrinsic limitations of the assay (in terms of sequence coverage and technical sensitivity). In addition, we also unveil the occurrence of artifactual mutations especially when MAFs are below 3%: this evidence strongly supports the importance of UDG pre-treatment of DNA samples from FFPE to confirm the presence of true DNA changes. Especially in the context of multicenter clinical trials, a proper assessment of pathogenic mutations harboured by tumour subclones (that is, intratumour heterogeneity) is warranted to better understand their biological significance and clinical impact, in terms of response to anti-EGFR molecules and targeted therapies in general.
Acknowledgments
We are grateful to Mrs Antonietta De Felice and Dr Mariano Pace for technical assistance. This work was supported by the Ministry of University (ex 60% and 2015HAJH8E to CM and ex-60% to PC and SM), Rete Oncologica Piemonte e Valle d’Aosta (to PC) and by the Italian Association of Cancer Research (AIRC, MFAG13310 to CM).
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
The authors declare no conflict of interest.
Supplementary Material
References
- Allegra CJ, Rumble RB, Hamilton SR, Mangu PB, Roach N, Hantel A, Schilsky RL (2016) Extended RAS gene mutation testing in metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy: American Society of Clinical Oncology Provisional Clinical Opinion Update 2015. J Clin Oncol 34(2): 179–185. [DOI] [PubMed] [Google Scholar]
- Dienstmann R, Jang IS, Bot B, Friend S, Guinney J (2015) Database of genomic biomarkers for cancer drugs and clinical targetability in solid tumors. Cancer Discov 5(2): 118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do H, Dobrovic A (2012) Dramatic reduction of sequence artifacts from DNA isolated from formalin-fixed cancer biopsies by treatment with uracil- DNA glycosylase. Oncotarget 3(5): 546–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do H, Dobrovic A (2015) Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin Chem 61(1): 64–71. [DOI] [PubMed] [Google Scholar]
- Do H, Wong SQ, Li J, Dobrovic A (2013) Reducing sequence artifacts in amplicon-based massively parallel sequencing of formalin-fixed paraffin-embedded DNA by enzymatic depletion of uracil-containing templates. Clin Chem 59(9): 1376–1383. [DOI] [PubMed] [Google Scholar]
- Dono M, Massucco C, Chiara S, Sonaglio C, Mora M, Truini A, Cerruti G, Zoppoli G, Ballestrero A, Truini M, Ferrarini M, Zupo S (2013) Low percentage of KRAS mutations revealed by locked nucleic acid polymerase chain reaction: implications for treatment of metastatic colorectal cancer. Mol Med 18: 1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Liang L, Ren L, Chen J, Wei Y, Chang W, Zhu D, Lin Q, Zheng P, Xu J (2015) A specific KRAS codon 13 mutation is an independent predictor for colorectal cancer metachronous distant metastases. Am J Cancer Res 5(2): 674–688. [PMC free article] [PubMed] [Google Scholar]
- Harbeck N, Wuerstlein R (2013) Optimal sequencing of anti-HER2 therapy throughout the continuum of HER2-positive breast cancer: evidence and clinical considerations. Drugs 73(15): 1665–1680. [DOI] [PubMed] [Google Scholar]
- Hovelson DH, McDaniel AS, Cani AK, Johnson B, Rhodes K, Williams PD, Bandla S, Bien G, Choppa P, Hyland F, Gottimukkala R, Liu G, Manivannan M, Schageman J, Ballesteros-Villagrana E, Grasso CS, Quist MJ, Yadati V, Amin A, Siddiqui J, Betz BL, Knudsen KE, Cooney KA, Feng FY, Roh MH, Nelson PS, Liu CJ, Beer DG, Wyngaard P, Chinnaiyan AM, Sadis S, Rhodes DR, Tomlins SA (2015) Development and validation of a scalable next-generation sequencing system for assessing relevant somatic variants in solid tumors. Neoplasia 17(4): 385–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Liu Z, Deng D, Tan A, Liao M, Mo Z, Yang X (2014) Anti-epidermal growth factor receptor monoclonal antibody-based therapy for metastatic colorectal cancer: a meta-analysis of the effect of PIK3CA mutations in KRAS wild-type patients. Arch Med Sci 10(1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgensmeier JM, Eder JP, Herbst RS (2014) New strategies in personalized medicine for solid tumors: molecular markers and clinical trial designs. Clin Cancer Res 20(17): 4425–4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattak MA, Martin H, Davidson A, Phillips M (2015) Role of first-line anti-epidermal growth factor receptor therapy compared with anti-vascular endothelial growth factor therapy in advanced colorectal cancer: a meta-analysis of randomized clinical trials. Clin Colorectal Cancer 14(2): 81–90. [DOI] [PubMed] [Google Scholar]
- Kimura T, Okamoto K, Miyamoto H, Kimura M, Kitamura S, Takenaka H, Muguruma N, Okahisa T, Aoyagi E, Kajimoto M, Tsuji Y, Kogawa T, Tsuji A, Takayama T (2012) Clinical benefit of high-sensitivity KRAS mutation testing in metastatic colorectal cancer treated with anti-EGFR antibody therapy. Oncology 82(5): 298–304. [DOI] [PubMed] [Google Scholar]
- Lamy A, Blanchard F, Le Pessot F, Sesboue R, Di Fiore F, Bossut J, Fiant E, Frebourg T, Sabourin JC (2011) Metastatic colorectal cancer KRAS genotyping in routine practice: results and pitfalls. Mod Pathol 24(8): 1090–1100. [DOI] [PubMed] [Google Scholar]
- Landi L, Cappuzzo F (2014) Pharmacotherapy targeting the EGFR oncogene in NSCLC. Expert Opin Pharmacother 15(16): 2293–2305. [DOI] [PubMed] [Google Scholar]
- Laurent-Puig P, Pekin D, Normand C, Kotsopoulos SK, Nizard P, Perez-Toralla K, Rowell R, Olson J, Srinivasan P, Le Corre D, Hor T, El Harrak Z, Li X, Link DR, Bouche O, Emile JF, Landi B, Boige V, Hutchison JB, Taly V (2015) Clinical relevance of KRAS-mutated subclones detected with picodroplet digital PCR in advanced colorectal cancer treated with anti-EGFR therapy. Clin Cancer Res 21(5): 1087–1097. [DOI] [PubMed] [Google Scholar]
- Loupakis F, Moretto R, Aprile G, Muntoni M, Cremolini C, Iacono D, Casagrande M, Ferrari L, Salvatore L, Schirripa M, Rossini D, De Maglio G, Fasola G, Calvetti L, Pilotto S, Carbognin L, Fontanini G, Tortora G, Falcone A, Sperduti I, Bria E (2016) Clinico-pathological nomogram for predicting BRAF mutational status of metastatic colorectal cancer. Br J Cancer 114(1): 30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini I, Santucci C, Sestini R, Simi L, Pratesi N, Cianchi F, Valanzano R, Pinzani P, Orlando C (2010) The use of COLD-PCR and high-resolution melting analysis improves the limit of detection of KRAS and BRAF mutations in colorectal cancer. J Mol Diagn 12(5): 705–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani S, Di Bello C, Bonello L, Tondat F, Pacchioni D, Molinaro L, Barreca A, Macri L, Chiusa L, di Celle PF, Cassoni P, Sapino A (2015) Flexible lab-tailored cut-offs for suitability of formalin-fixed tumor samples for diagnostic mutational analyses. PLoS One 10(4): e0121815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari F, Felicioni L, Buscarino M, De Dosso S, Buttitta F, Malatesta S, Movilia A, Luoni M, Boldorini R, Alabiso O, Girlando S, Soini B, Spitale A, Di Nicolantonio F, Saletti P, Crippa S, Mazzucchelli L, Marchetti A, Bardelli A, Frattini M (2011) Increased detection sensitivity for KRAS mutations enhances the prediction of anti-EGFR monoclonal antibody resistance in metastatic colorectal cancer. Clin Cancer Res 17(14): 4901–4914. [DOI] [PubMed] [Google Scholar]
- Moroni M, Veronese S, Benvenuti S, Marrapese G, Sartore-Bianchi A, Di Nicolantonio F, Gambacorta M, Siena S, Bardelli A (2005) Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol 6(5): 279–286. [DOI] [PubMed] [Google Scholar]
- Normanno N, Rachiglio AM, Lambiase M, Martinelli E, Fenizia F, Esposito C, Roma C, Troiani T, Rizzi D, Tatangelo F, Botti G, Maiello E, Colucci G, Ciardiello F Investigators C-G (2015) Heterogeneity of KRAS, NRAS, BRAF and PIK3CA mutations in metastatic colorectal cancer and potential effects on therapy in the CAPRI GOIM trial. Ann Oncol 26(8): 1710–1714. [DOI] [PubMed] [Google Scholar]
- Palomba G, Doneddu V, Cossu A, Paliogiannis P, Manca A, Casula M, Colombino M, Lanzillo A, Defraia E, Pazzola A, Sanna G, Putzu C, Ortu S, Scartozzi M, Ionta MT, Baldino G, Sarobba G, Capelli F, Sedda T, Virdis L, Barca M, Gramignano G, Budroni M, Tanda F, Palmieri G (2016) Prognostic impact of KRAS, NRAS, BRAF, and PIK3CA mutations in primary colorectal carcinomas: a population-based study. J Transl Med 14(1): 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM, Taylor G, Barrett JH, Quirke P (2009) KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol 27(35): 5931–5937. [DOI] [PubMed] [Google Scholar]
- Rosty C, Young JP, Walsh MD, Clendenning M, Walters RJ, Pearson S, Pavluk E, Nagler B, Pakenas D, Jass JR, Jenkins MA, Win AK, Southey MC, Parry S, Hopper JL, Giles GG, Williamson E, English DR, Buchanan DD (2013) Colorectal carcinomas with KRAS mutation are associated with distinctive morphological and molecular features. Mod Pathol 26(6): 825–834. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74(12): 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souglakos J, Philips J, Wang R, Marwah S, Silver M, Tzardi M, Silver J, Ogino S, Hooshmand S, Kwak E, Freed E, Meyerhardt JA, Saridaki Z, Georgoulias V, Finkelstein D, Fuchs CS, Kulke MH, Shivdasani RA (2009) Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer. Br J Cancer 101(3): 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens EA, Rodriguez CP (2015) Genomic medicine and targeted therapy for solid tumors. J Surg Oncol 111(1): 38–42. [DOI] [PubMed] [Google Scholar]
- Therkildsen C, Bergmann TK, Henrichsen-Schnack T, Ladelund S, Nilbert M (2014) The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: a systematic review and meta-analysis. Acta Oncol 53(7): 852–864. [DOI] [PubMed] [Google Scholar]
- Tol J, Dijkstra JR, Klomp M, Teerenstra S, Dommerholt M, Vink-Borger ME, van Cleef PH, van Krieken JH, Punt CJ, Nagtegaal ID (2010) Markers for EGFR pathway activation as predictor of outcome in metastatic colorectal cancer patients treated with or without cetuximab. Eur J Cancer 46(11): 1997–2009. [DOI] [PubMed] [Google Scholar]
- Toledo RA, Cubillo A, Vega E, Garralda E, Alvarez R, de la Varga LU, Pascual JR, Sanchez G, Sarno F, Prieto SH, Perea S, Lopez-Casas PP, Lopez-Rios F, Hidalgo M (2017) Clinical validation of prospective liquid biopsy monitoring in patients with wild-type RAS metastatic colorectal cancer treated with FOLFIRI-cetuximab. Oncotarget 8(21): 35289–35300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tougeron D, Lecomte T, Pages JC, Villalva C, Collin C, Ferru A, Tourani JM, Silvain C, Levillain P, Karayan-Tapon L (2013) Effect of low-frequency KRAS mutations on the response to anti-EGFR therapy in metastatic colorectal cancer. Ann Oncol 24(5): 1267–1273. [DOI] [PubMed] [Google Scholar]
- Van Cutsem E, Lenz HJ, Kohne CH, Heinemann V, Tejpar S, Melezinek I, Beier F, Stroh C, Rougier P, van Krieken JH, Ciardiello F (2015) Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. J Clin Oncol 33(7): 692–700. [DOI] [PubMed] [Google Scholar]
- Wang Z, Chen R, Wang S, Zhong J, Wu M, Zhao J, Duan J, Zhuo M, An T, Wang Y, Bai H, Wang J (2014) Quantification and dynamic monitoring of EGFR T790M in plasma cell-free DNA by digital PCR for prognosis of EGFR-TKI treatment in advanced NSCLC. PLoS One 9(11): e110780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Polley EC, Li MC, Lih CJ, Palmisano A, Sims DJ, Rubinstein LV, Conley BA, Chen AP, Williams PM, Kummar S, Doroshow JH, Simon RM (2015) GeneMed: an informatics hub for the coordination of next-generation sequencing studies that support precision oncology clinical trials. Cancer Inform 14(Suppl 2): 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.