Abstract
Background:
Despite recent advancements, metastatic castration-resistant prostate cancer (CRPC) is not considered curative. Novel approaches for identification of therapeutic targets of CRPC are needed.
Methods:
Next-generation sequencing revealed 945–1248 miRNAs from each lethal mCRPC sample. We constructed miRNA expression signatures of CRPC by comparing the expression of miRNAs between CRPC and normal prostate tissue or hormone-sensitive prostate cancer (HSPC). Genome-wide gene expression studies and in silico analyses were carried out to predict miRNA regulation and investigate the functional significance and clinical utility of the novel oncogenic pathways regulated by these miRNAs in prostate cancer (PCa).
Results:
Based on the novel miRNA expression signature of CRPC, miR-145-5p and miR-145-3p were downregulated in CRPC. By focusing on miR-145-3p, which is a passenger strand and has not been well studied in previous reports, we showed that miR-145-3p targeted 4 key molecules, i.e., MELK, NCAPG, BUB1, and CDK1, in CPRC. These 4 genes significantly predicted survival in patients with PCa.
Conclusions:
Small RNA sequencing for lethal CRPC and in silico analyses provided novel therapeutic targets for CRPC.
Keywords: deep sequencing, microRNA, castration-resistant prostate cancer, miR-145-3p, maternal embryonic leucine zipper kinase
Prostate cancer (PCa) is the most frequently diagnosed cancer among men in developed countries (Siegel et al, 2015). Although PCa is initially responsive to androgen deprivation therapy (ADT), most PCa develops into castration-resistant prostate cancer (CRPC). Recent approvals of novel androgen receptor (AR)-targeted agents, bone-targeted agents, and chemotherapeutic agents have resulted in survival benefits in patients with CRPC; however, CRPC is not considered curative (Crawford et al, 2015). One of the main challenges in treating CRPC is controlling aggressive, lethal metastatic PCa. Therefore, identification of the molecular mechanisms of androgen-independent, metastatic signalling pathways using current genomic approaches would help to improve therapies for this disease.
MicroRNAs (miRNAs) are endogenous small RNA molecules (19–22 bases in length) that regulate protein-coding/noncoding gene expression by translational repression or mRNA cleavage in a sequence-specific manner (Bartel, 2004). Bioinformatic approaches indicate that one miRNA can regulate thousands of mRNAs, and conversely, about 60% of the protein-coding genes in the human genome are regulated by miRNAs (Lewis et al, 2005). Aberrant expression of miRNAs can induce disruption of normal RNA regulatory networks; therefore, miRNAs act as fine-tuning regulators in almost all biological processes (Bartel, 2009; Garzon et al, 2009). In cancer cells, aberrant expression of tumour-suppressive or oncogenic miRNAs causes cancer progression (Garzon et al, 2009). Identification of aberrantly expressed miRNAs and novel regulatory network searches based on miRNA regulation has facilitated elucidation of the molecular mechanisms of cancer initiation, development, and metastasis.
In this study, we aimed to identify novel therapeutic targets of lethal metastatic CRPC (mCRPC). We obtained clinically lethal CRPC tissue from patients who died of PCa after several sequential treatments, including ADT, chemotherapy, and radiotherapy. We carried out small RNA sequencing using lethal CRPC tissue to comprehensively identify novel miRNAs and investigated the functional significance and clinical utility of the novel oncogenic pathways regulated by these miRNAs in PCa.
Materials and methods
Patients and clinical prostate specimens
Prostate specimens were obtained from patients admitted to Teikyo University Chiba Medical Centre Hospital and Chiba University Hospital from 2008 to 2014. Among patients with elevated prostate-specific antigen (PSA) levels who underwent transrectal prostate needle biopsies, we evaluated 34 PCa tissues and 19 normal prostate (non-PCa) tissues. Five paired samples of PCa and corresponding normal tissues were obtained by robot-assisted radical prostatectomy. Eight metastatic CRPC specimens were obtained from 3 patients who had been heavily treated with several agents and had eventually died of metastatic CRPC. The patients’ characteristics are summarised in Supplementary Table 1. For non-PCa and PCa specimens, we obtained two needle biopsy specimens from the same region for pathological verification. Written consent for tissue donation was obtained from each patient. The protocol was approved by the Institutional Review Board of Chiba University and Teikyo University.
Construction of the miRNA expression signature of CRPC
We performed small RNA sequencing using a HiSeq 2000 (Illumina, San Diego, CA, USA) according to the manufacturer’s protocol. The read length was 50 bp, and single-end, multiplex sequencing was performed. After cleaning of reading data using the Cutadapt program (ver. 1.2.1), high quality reads were mapped to the reference genome sequence using Bowtie software (ver. 1.0.0). To annotate reads, information from miRBase (release 20.0), Rfam (ver.11.0), and iGenomes (NCI build 37.2) was used. To compare differentially expressed miRNAs between sample groups, edgeR (ver. 3.8.6) was used. P-values were adjusted for multiple testing using the Benjamini and Hochberg method. P-values with false discovery rates (FDRs) of less than 0.05 were considered significant.
Cell culture and RNA isolation
PC3 and DU145 human prostate cancer cells were obtained from the American Type Culture Collection (Manassas, VA, USA).
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol, as described previously (Goto et al, 2015; Goto et al, 2016a; Goto et al, 2016b).
Quantitative real-time reverse transcription polymerase chain reaction (RT-qPCR)
PCR was performed as previously described (Goto et al, 2015; Goto et al, 2016a; Goto et al, 2016b). The expression levels of miR-145-3p (Assay ID: 002149) and miR-145-5p (Assay ID: 002278) were analysed by TaqMan RT-qPCR (TaqMan MicroRNA Assay; Applied Biosystems) and normalised to RNU48 (Assay ID: 001006). TaqMan probes and primers were obtained from Applied Biosystems (Assay-On-Demand Gene Expression Products): BUB1 (P/N: Hs01557695_m1), CDK1 (P/N: Hs00938777_m1), MELK (P/N: Hs01106440_m1), NCAPG (P/N: Hs00254617_m1); GUSB (P/N: Hs00939627_m1), and GAPDH (P/N: Hs02758991_g1) as an internal control.
Transfection with mature miRNA
The following mature miRNA species were used in this study: Ambion Pre-miR miRNA precursor for hsa-miR-145-3p (P/N: PM13036) and hsa-miR-145-5p (P/N: PM11480) and negative control miRNA (P/N: AM17111). RNAs were incubated with OPTI-MEM (Invitrogen) and Lipofectamine RNAiMAX reagent (Invitrogen). The transfection procedures were reported previously (Goto et al, 2015; Goto et al, 2016a).
Cell proliferation, migration, and invasion assays
Cell proliferation, migration and invasion assays were carried out as previously described (Goto et al, 2015; Goto et al, 2016a; Goto et al, 2016b).
Argonaute2 (Ago2)-bound miRNA isolation by immunoprecipitation
PC3 cells were transfected with 10 nM miRNA by reverse transfection and plated in well plates at 1 × 105 cells per well. After 48 h, cells were collected, and immunoprecipitation was performed using human anti-Ago2 antibodies (microRNA Isolation Kit, Human Ago2; Wako, Osaka, Japan) according to the manufacturer’s protocol. Expression levels of miRNAs bound to Ago2 were measured by TaqMan RT-qPCR. miRNA expression data were normalised to miR-26a (Assay ID: 000405), the expression of which was not affected by miR-145-3p/5p transfection.
Identification of genes regulated by miR-145-3p
We performed a combination of in silico and genome-wide gene expression analyses. First, genes regulated by miR-145-3p were listed using the TargetScan database (release 7.0). Next, we performed genome-wide gene expression analysis using miR-145-3p-transfected PC3 cells. A SurePrint G3 Human GE 60 K Microarray (Agilent Technologies) was used for expression profiling of miRNA transfectants in comparison with mock PC3 cells(accession number: GSE77790, log2 ratio<−1.5). Next, we identified genes that were highly upregulated in mCRPC tissue compared with those in non-PCa and HSPC and highly upregulated in PCa compared with those in non-PCa tissues (accession number: GSE35988; fold change > 2 and q-value<0.05). Finally, to validate that these genes were upregulated in mCRPC clinical specimens, we used other data sets from cBioportal (http://www.cbioportal.org/) and GEO database (GSE 70770) (Ross-Adams et al, 2015; Whitington et al, 2016). HSPC data were obtained from TCGA-PRAD (Cancer Genome Atlas Research N, 2015), and mCRPC data were obtained from SU2c/PCF (Robinson et al, 2015).
Immunohistochemistry
Tissue specimens were immunostained with an Ultra-Vision Detection System (Thermo Scientific) following the manufacturer’s protocol. Primary rabbit polyclonal antibodies against budding uninhibited by benzimidazoles 1 (BUB1; 1:50, sc-365685; Santa Cruz Biotechnology), cyclin-dependent kinase 1 (CDK1; 1:100, HPA003387; Sigma-Aldrich), maternal embryonic leucine zipper kinase (MELK; 1:200, HPA017214; Sigma-Aldrich), and non-SMC condensin I complex subunit G (NCAPG; 1:150, ab56382; Abcam) were used for immunochemistry. The procedures were described previously (Goto et al, 2015; Goto et al, 2016a; Goto et al, 2016b).
The cancer genome atlas prostate adenocarcinoma (TCGA-PRAD) and other human PCa data analysis
To explore the clinical significance of miRNAs and target genes, we used the GEO database and the RNA sequencing database in TCGA-PRAD (Cancer Genome Atlas Research N, 2015) and SU2C/PCF (Robinson et al, 2015). The gene expression and clinical data were retrieved from cBioportal (http://www.cbioportal.org/, the provisional data were downloaded on September 20, 2016). The normalised mRNA expression values in the RNA sequencing data were processed and provided as Z-scores. We also employed the gene expression microarray data (GSE 21034 and GSE 21036). Visualisation of gene expression heatmaps and clustering was carried out by MeV (Saeed et al, 2003) and Subio Platform (Subio Inc., Amami, Japan). Pathway analysis was performed using the Enrichr program (Kuleshov et al, 2016).
Statistical analysis
The relationships between 2 groups and the numerical values obtained by RT-qPCR were analysed using Mann-Whitney U-tests. Spearman’s rank test was used to evaluate the correlations between the expression of miRNAs or genes. The relationships among more than 3 variables and numerical values were analysed using Bonferroni-adjusted Mann-Whitney U-tests. A multivariate Cox proportional hazards model was used to establish independent factors for disease-free survival. Survival analysis was carried out using the Kaplan–Meier method and log-rank tests with JMP software (version 12, SAS Institute Inc., Cary, NC, USA); all other analyses were performed using Expert StatView (version 5, SAS Institute Inc.).
Results
Small RNA sequences of non-PCa, HSPC, and lethal mCRPC
First, we sequenced 14 small RNA libraries. We used 5 non-PCa specimens (nos. 40–44; Supplementary Table 1), 5 HSPC specimens (nos. 1-5) and 4 mCRPC specimens (nos. 62, 63, 65, and 66) for deep sequencing. We obtained 11,690,828–21,610,206 clean reads and 7,733,236–18,921,965 mapped reads (Supplementary Table 2). Focusing on already annotated miRNAs, we detected 945–1248 miRNAs from each sample. To elucidate the molecular mechanisms underlying lethal mCRPC, we compared the expression of miRNAs between mCRPC and non-PCa or HSPC and sorted the miRNAs by FDR from the most downregulated miRNAs in mCRPC (Table 1). miR-145-5p, a tumour-suppressive miRNA that has been extensively studied in various types of cancers, is listed as the top miRNA in these signatures. By carefully observing these downregulated miRNAs in the mCRPC expression signature, we found that the complementary strand of miR-145-5p, i.e., miR-145-3p, was also one of the top miRNAs. Because miR-145-3p is a passenger strand and its function is not yet clear, we focused on miR-145-3p for further analysis.
Table 1. Significantly downregulated miRNAs in CRPC.
| miRNA | Locus | Log FC | FDR |
|---|---|---|---|
|
CRPC vs non-PCa | |||
| miR-145-5p | 5q32 | −4.645 | 1.82E-31 |
| miR-221-3p | Xp11.3 | −4.220 | 6.96E-26 |
| miR-31-5p | 9p21.3 | −6.354 | 1.89E-24 |
| miR-145-3p | 5q32 | −3.930 | 8.42E-19 |
| miR-143-5p | 5q32 | −4.198 | 3.94E-18 |
| miR-222-3p | Xp11.3 | −4.495 | 8.07E-18 |
| miR-221-5p | Xp11.3 | −3.769 | 3.03E-12 |
| miR-143-3p | 5q32 | −3.010 | 1.02E-10 |
| miR-125b-5p | 11q24.1; 21q21.1 | −3.234 | 1.48E-10 |
| miR-150-5p | 19q13.33 | −3.454 | 1.75E-10 |
| miR-6510-3p | 17q12 | −6.536 | 1.81E-10 |
| miR-99a-5p | 21q21.1 | −2.790 | 7.83E-10 |
| miR-100-5p | 11q24.1 | −3.157 | 4.32E-09 |
| miR-204-5p | 9q21.13 | −4.540 | 2.11E-08 |
| miR-205-5p | 1q32.2 | −8.408 | 2.89E-08 |
| miR-187-3p | 18q12.1 | −4.257 | 3.35E-08 |
| miR-1247-5p | 14q32.31 | −4.560 | 3.69E-08 |
| miR-490-3p | 7q33 | −6.626 | 1.08E-07 |
| miR-125b-1-3p | 11q24.1 | −2.774 | 1.90E-07 |
| miR-27b-3p | 9q22.33 | −2.382 | 3.59E-07 |
| miR-455-5p | 9q33.1 | −2.461 | 5.97E-07 |
| miR-100-3p | 11q24.1 | −3.859 | 1.28E-05 |
| miR-1271-5p | 5q35.2 | −2.082 | 1.37E-05 |
| miR-328-3p | 16q22.1 | −2.112 | 1.55E-05 |
| miR-196b-5p | 7p15.2 | −1.740 | 2.59E-05 |
| miR-24-3p | 9q22.33; 19p13.12 | −1.699 | 5.98E-05 |
| let-7c-3p | 21q21.1 | −4.152 | 6.34E-05 |
| miR-184 | 15q25.1 | −4.483 | 8.77E-05 |
| miR-133a-5p | 18q11.1; 20q13.33 | −4.170 | 8.77E-05 |
| miR-125b-2-3p | 21q21.1 | −2.145 | 8.77E-05 |
| miR-873-5p | 9p21.1 | −3.599 | 0.000153 |
| miR-9-5p | 1q22; 5q14.3; 15q26.1 | −2.006 | 0.000153 |
| miR-504-5p | Xq26.3 | −2.390 | 0.000185 |
| miR-27b-5p | 9q22.33 | −2.251 | 0.000208 |
| miR-3943 | 7p14.1 | −4.655 | 0.000328 |
| miR-548ba | 2p16.3 | −4.650 | 0.000431 |
| miR-30e-3p | 1p34.2 | −1.524 | 0.000431 |
| miR-214-3p | 1q24.3 | −1.859 | 0.000658 |
| miR-125a-5p | 19q13.41 | −2.004 | 0.000681 |
| miR-628-5p | 15q21.3 | −2.953 | 0.001013 |
| miR-455-3p | 9q33.1 | −2.068 | 0.001769 |
| miR-31-3p | 9p21.3 | −4.305 | 0.00182 |
| miR-199a-5p | 19p13.2; 1q24.3 | −1.433 | 0.002757 |
| miR-133a-3p | 18q11.1; 20q13.33 | −4.055 | 0.002817 |
| miR-874-3p | 5q31.2 | −1.646 | 0.003569 |
| miR-28-3p | 3q28 | −1.278 | 0.00539 |
| miR-23b-3p | 9q22.33 | −1.773 | 0.005713 |
| let-7e-3p | 19q13.41 | −1.874 | 0.006138 |
| miR-889-3p | 14q32.31 | −1.878 | 0.007487 |
| miR-142-5p | 17q22 | −1.503 | 0.007749 |
| miR-452-3p | Xq28 | −2.343 | 0.007917 |
| miR-199b-5p | 9q34.12 | −1.839 | 0.008345 |
| miR-452-5p | Xq28 | −1.670 | 0.009339 |
| miR-1260b | 11q21 | −1.873 | 0.010403 |
| miR-139-5p | 11q13.4 | −1.411 | 0.013082 |
| miR-205-3p | 1q32.2 | −4.128 | 0.01419 |
| miR-224-5p | Xq28 | −1.642 | 0.014227 |
| miR-23a-3p | 19p13.12 | −1.072 | 0.014881 |
| miR-199b-3p | 9q34.12 | −1.488 | 0.016875 |
| miR-199a-3p | 19p13.2; 1q24.3 | −1.488 | 0.016905 |
| miR-1269a | 4q13.2 | −4.294 | 0.017298 |
| miR-944 | 3q28 | −2.426 | 0.017298 |
| miR-136-5p | 14q32.31 | −1.714 | 0.018059 |
| miR-4634 | 5q35.2 | −1.960 | 0.018527 |
| miR-224-3p | Xq28 | −2.339 | 0.03156 |
| miR-6511b-3p | 16p13.3; 16p13.11 | −2.237 | 0.03156 |
| let-7c-5p | 21q21.1 | −1.701 | 0.035011 |
| miR-887-3p | 5p15.1 | −1.345 | 0.035011 |
| miR-1260a | 14q24.3 | −2.617 | 0.035037 |
| miR-1247-3p | 14q32.31 | −2.996 | 0.03528 |
| miR-1298-5p | Xq23 | −3.686 | 0.036895 |
| miR-101-5p | 1p31.3 | −1.598 | 0.039947 |
| miR-222-5p | Xp11.3 | −3.853 | 0.043768 |
| miR-656-3p | 14q32.31 | −2.760 | 0.044013 |
| miR-181c-5p | 19p13.12 | −1.622 | 0.045462 |
| miR-214-5p | 1q24.3 | −1.373 | 0.047573 |
|
CRPC vs HSPC | |||
| miR-145-5p | 5q32 | −3.140 | 1.70E-05 |
| miR-125b-5p | 11q24.1; 21q21.1 | −2.800 | 7.28E-05 |
| miR-1247-5p | 14q32.31 | −4.369 | 7.54E-05 |
| miR-143-5p | 5q32 | −3.181 | 7.54E-05 |
| miR-145-3p | 5q32 | −2.876 | 1.95E-04 |
| miR-31-5p | 9p21.3 | −6.095 | 4.08E-04 |
| miR-150-5p | 19q13.33 | −3.906 | 7.55E-04 |
| miR-221-3p | Xp11.3 | −2.534 | 7.55E-04 |
| miR-143-3p | 5q32 | −2.624 | 1.39E-03 |
| miR-490-3p | 7q33 | −6.969 | 2.41E-03 |
| miR-3943 | 7p14.1 | −4.750 | 3.08E-03 |
| miR-9-5p | 1q22; 5q14.3; 15q26.1 | −5.457 | 3.87E-03 |
| miR-100-3p | 11q24.1 | −3.618 | 4.19E-03 |
| miR-100-5p | 11q24.1 | −2.442 | 4.19E-03 |
| miR-196b-5p | 7p15.2 | −2.027 | 4.57E-03 |
| miR-125b-1-3p | 11q24.1 | −2.728 | 4.93E-03 |
| miR-99a-5p | 21q21.1 | −2.097 | 4.93E-03 |
| let-7c-3p | 21q21.1 | −3.956 | 5.91E-03 |
| miR-222-3p | Xp11.3 | −2.311 | 6.28E-03 |
| miR-221-5p | Xp11.3 | −2.562 | 7.24E-03 |
| miR-9-3p | 1q22; 5q14.3; 15q26.1 | −5.538 | 1.22E-02 |
| miR-27a-5p | 19p13.12 | −3.290 | 1.22E-02 |
| miR-125b-2-3p | 21q21.1 | −2.107 | 1.41E-02 |
| miR-27b-3p | 9q22.33 | −1.790 | 1.69E-02 |
| miR-142-5p | 17q22 | −2.176 | 2.26E-02 |
| miR-1260b | 11q21 | −2.482 | 2.30E-02 |
| miR-214-3p | 1q24.3 | −1.811 | 3.02E-02 |
| miR-671-3p | 7q36.1 | −1.868 | 3.35E-02 |
| miR-205-5p | 1q32.2 | −4.450 | 4.96E-02 |
| miR-199b-5p | 9q34.12 | −2.022 | 4.96E-02 |
Abbreviations: FC=fold change; FDR=false discovery rate.
Expression levels of miR-145-3p in PCa specimens and cell lines
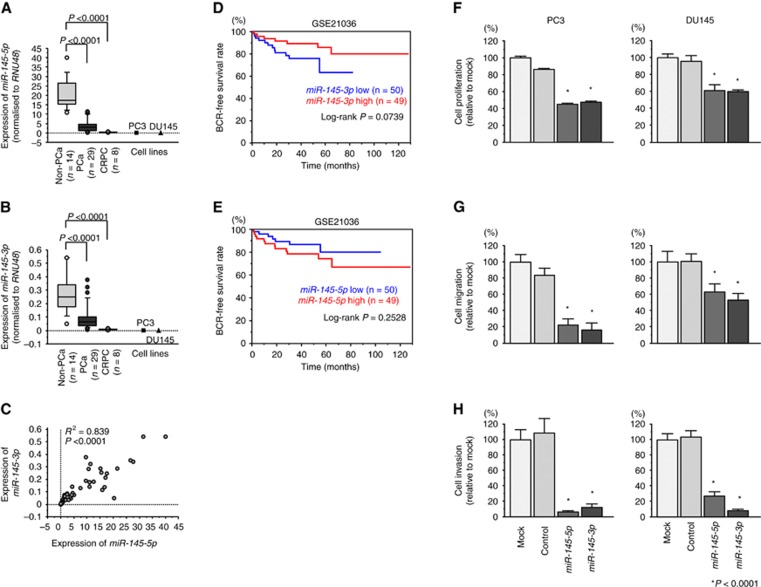
First, we validated miR-145-3p and miR-145-5p expression levels using clinical specimens (PCa: n=29, nos. 6–34; Supplementary Table 1; non-PCa: n=14, nos. 45–58; mCRPC: n=8, nos. 59–66). Patients with PCa showed relatively advanced stages, with high Gleason scores (97% of patients had a score of greater than 7) and high PSA (median: 212 ng/ml; range, 11.4–2530 ng/ml). The expression levels of miR-145-5p and miR-145-3p were significantly downregulated in PCa tissues and mCRPC tissues compared with those in non-PCa tissues (P<0.0001; Figure 1A and B). Furthermore, Spearman’s rank tests showed positive correlations between the expression of miR-145-5p and miR-145-3p (R2=0.839 and P<0.0001; Figure 1C). Validation of miR-145-3p downregulation in datasets from other research groups (GSE21036) revealed significant downregulation of miR-145-3p in mCRPC (n=8) specimens compared with that in HSPC (n=99; P<0.0001; Supplementary Figure 1A).
Figure 1.
Expression of miR-145-5p/3p in clinical prostate specimens, associations with prognosis, and functional analysis. (A) Expression levels of miR-145-5p in PCa clinical specimens and cell lines. RNU48 was used for normalisation. (B) Expression levels of miR-145-3p in PCa clinical specimens and cell lines. RNU48 was used for normalisation. (C) Correlations among the relative expression levels of miR-145-5p and miR-145-3p. (D) Kaplan-Meier analysis of miR-145-3p expression and progression-free survival (GSE 21036). (E) Kaplan-Meier analysis of miR-145-5p expression and progression-free survival (GSE 21036). (F–H) Cell proliferation, migration and invasion assays. *P<0.0001. The bars indicate SDs.
Both miR-145-5p and miR-145-3p bound to Ago2
miR-145-5p is known to be a guide strand and has been shown to have a role in several types of cancers; in contrast, miR-145-3p is a passenger strand, and some previous reports have indicated that the passenger strands are degraded and do not have specific functions. Because we found that miR-145-3p was a putative key miRNA in mCRPC by RNA sequencing and confirmed that this miRNA was downregulated in mCRPC clinical specimens, we hypothesised that both miR-145-5p and miR-145-3p may be incorporated into and function as part of the RNA-induced silencing complex (RISC). To test this hypothesis, we performed immunoprecipitation with antibodies targeting Ago2, which plays a central role in the RISC. After transfection with miR-145-5p or miR-145-3p, Ago2-bound miRNAs were isolated, and RT-qPCR was carried out to determine whether miR-145-5p and miR-145-3p bound to Ago2. After transfection with miR-145-5p and immunoprecipitation by anti-Ago2 antibodies, miR-145-5p levels were significantly higher than those of mock- or miR control-transfected cells and those of miR-145-3p-transfected PC3 cells (P<0.0001; Supplementary Figure 2A). Likewise, after transfection with miR-145-3p and immunoprecipitation by anti-Ago2 antibodies, miR-145-3p levels were significantly higher than those of mock- or miR control-transfected cells and those of miR-145-5p-transfected PC3 cells (P<0.0001; Supplementary Figure 2B).
Associations between miR-145-3p expression levels and clinical parameters in PCa
Next, we investigated the clinical significance of miR-145-5p/3p in PCa. The biochemical recurrence (BCR)-free survival rate was evaluated in patients with high vs low miR-145-5p or miR-145-3p expression from data in GSE21036. Patients with lower expression of miR-145-3p tended to have shorter BCR-free intervals (P=0.0739; Figure 1D). Furthermore, patients with higher T stage (⩽ T2c: n=69 vs ⩾ T3a: n=30) had relatively lower expression of miR-145-3p (P=0.0617; Supplementary Figure 1B). However, patients with lower expression of miR-145-5p did not have shorter BCR-free interval (P=0.2528; Figure 1E). Thus, these data confirmed the clinical importance of miR-145-3p.
Effects of restoring miR-145-3p expression in PCa cells
To investigate the functional roles of miR-145-3p, we performed gain-of-function studies using miRNA transfection in PC3 and DU145 cells. Restoration of miR-145-5p or miR-145-3p significantly inhibited cell proliferation, migration, and invasion as compared with that in mock- or miR control-transfected PC3 and DU145 cells (P<0.0001; Figure 1F and H). Thus, both the guide strand and passenger strand (miR-145-3p) functioned as tumour suppressors in PCa.
To better understand the mechanisms associated with these phenotypes, we then carried out genome-wide pathway analysis for downregulated genes after transfection with miR-145-3p in PC3 cells. The most downregulated pathway was the ‘cell cycle’ pathway, followed by ‘DNA replication’. These pathways are involved in cell proliferation, which may affect cell migration and invasion (Supplementary Figure 3).
Identification of target genes regulated by miR-145-3p in mCRPC
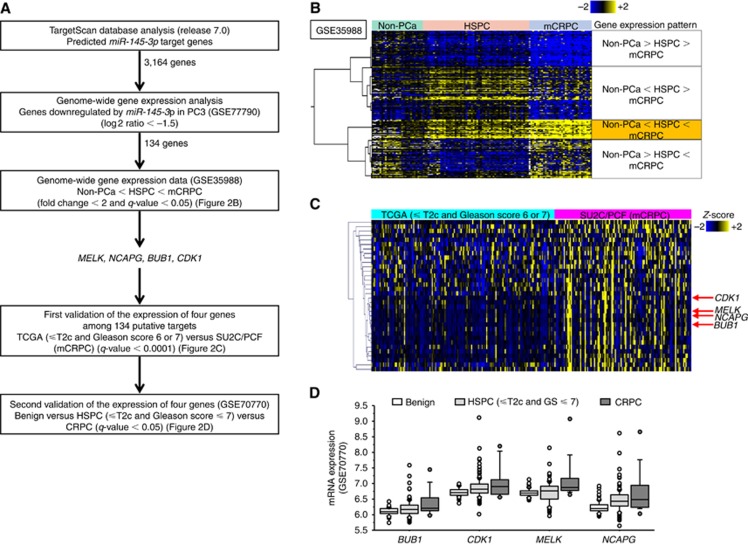
To elucidate the pathways and molecules regulated by tumour-suppressive miR-145-3p, we performed in silico and microarray analysis (Figure 2A). TargetScan analysis showed that 3,164 genes had putative target sites for miR-145-3p in their 3′ untranslated regions (UTRs). Next, we performed genome-wide gene expression analysis using PC3 cells to select genes downregulated by miR-145-3p restoration. The data were deposited in GSE 77790. A total of 134 genes were downregulated by miR-145-3p transfection (log2 ratio<-1.5). Finally, we investigated the expression statuses of these genes in non-PCa, HSPC, and mCRPC clinical specimens and examined gene expression profiles in the GEO database (GEO accession number: GSE 35988) to evaluate upregulated genes in mCRPC specimens. Among the 134 putative target genes of miR-145-3p transfectants, 4 genes were significantly upregulated in HSPC specimens compared with that in non-PCa tissues and significantly upregulated in mCRPC specimens compared with that in HSPC specimens (fold change > 2, q-value<0.05; Figure 2B). These 4 genes were MELK, NCAPG, BUB1, and CDK1. These data were validated in mCRPC tissues using another dataset from the TCGA database based on z scores. Compared with localised PCa tissue, MELK, NCAPG, BUB1, and CDK1 were highly expressed in mCRPC tissue (q-value<0.0001; Figure 2C). Furthermore, we performed second validation by another dataset (GSE70770). MELK, NCAPG, BUB1, and CDK1 genes showed progressive upregulation (non-PCa<HSPC<mCRPC; q-value<0.05; Figure 2D). Therefore, we selected MELK, NCAPG, BUB1, and CDK1 as key molecules regulated by miR-145-3p in mCRPC and subjected these genes to further analyses.
Figure 2.
Strategy for selecting putative target genes of miR-145-3p in mCRPC. (A) The TargetScan program and genome-wide gene expression analysis using miR-145-3p-transfected PC3 cells showed downregulation (log2 ratio<–1.5) in 134 genes. (B) Four genes showing progressive upregulation (non-PCa<HSPC<mCRPC; fold change > 2 and q-value<0.05) were selected as target genes. (C) The expression levels of MELK, NCAPG, BUB1, and CDK1 in mCRPC were validated in other data sets (TCGA [⩽ T2c and Gleason score 6 or 7) vs SU2C/PCF [mCRPC], q-value<0.0001). (D) Second validation by GSE70770. MELK, NCAPG, BUB1, and CDK1 genes showed progressive upregulation (non-PCa<HSPC<mCRPC; q-value<0.05).
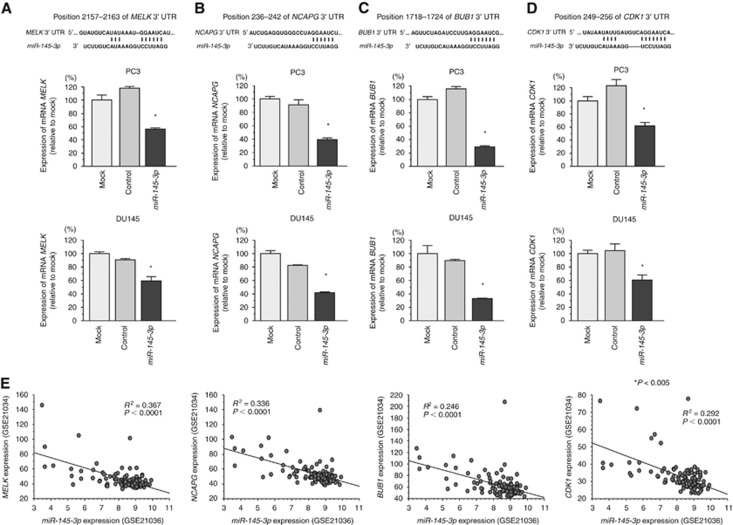
MELK, NCAPG, BUB1, and CDK1 were negatively regulated by miR-145-3p in PCa cells
To determine whether miR-145-3p restoration influenced expression of target genes, we performed RT-qPCR in PC3 and DU145 cells. MELK, NCAPG, BUB1, and CDK1 mRNAs were significantly downregulated by miR-145-3p transfection as compared with that in mock- or miR control-transfected cells (P<0.005; Figure 3A–D). Furthermore, the expression levels of miR-145-3p and MELK, NCAPG, BUB1, and CDK1 were negatively correlated (R2=0.367, P<0.0001; R2=0.336, P<0.0001; R2=0.246, P<0.0001; R2=0.292, P<0.0001, respectively; Figure 3E).
Figure 3.
Downregulation of MELK, NCAPG, BUB1, and CDK1 expression by miR-145-3p in PC3 and DU145 cells. (A–D) miR-145-3p binding sites in MELK mRNA. MELK mRNA expression 72 h after transfection with miR-145-3p. GUSB was used as an internal control. *P<0.0001. The bars indicate SDs. (E) The expression levels of miR-145-3p and target genes were negatively correlated in clinical PCa specimens.
Clinical significance of miR-145-3p regulation in PCa
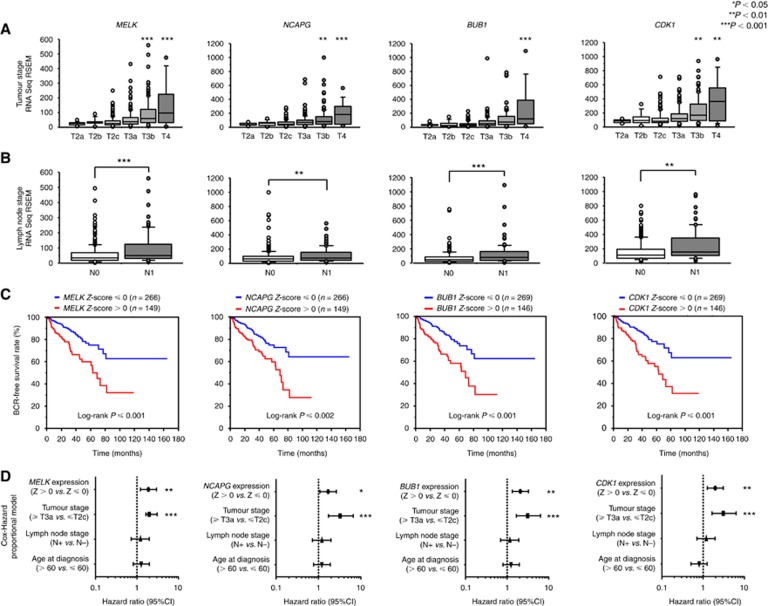
To investigate the clinical significance of MELK, NCAPG, BUB1, and CDK1 in PCa, we analysed their associations with tumour stage and lymph node stage using the TCGA-PRAD database. MELK, NCAPG, BUB1, and CDK1 were significantly upregulated as the tumour stage and lymph node stage increased (Figure 4A and B).
Figure 4.
MELK, NCAPG, BUB1, and CDK1 expression and clinical parameters. (A) The expression levels of MELK, NCAPG, BUB1, and CDK1 were significantly elevated as tumour stage increased. (B) The expression levels of MELK, NCAPG, BUB1, and CDK1 were significantly elevated in N1 stage PCa compared with that in N0 stage PCa. (C) Higher expression of MELK, NCAPG, BUB1, and CDK1 was associated with shorter BCR-free survival. (D) Multivariate Cox proportional hazards models were used to assess independent predictors of progression-free survival, including gene expression level (Z score > 0 vs Z score ⩽ 0), tumour stage (⩾ T3a vs ⩽ T2c), lymph node stage (N0 vs N1), and age at diagnosis (> 60 vs ⩽ 60). *P<0.05, **P<0.01, ***P<0.001.
Next, to examine whether the expression levels of these genes predicted BCR-free survival, patients were divided into two groups: z-score > 0 and z-score ⩽ 0 (Cerami et al, 2012; Gao et al, 2013). Higher MELK, NCAPG, BUB1, and CDK1 expression levels were associated with shorter BCR-free survival (P<0.0001, P=0.0001, P<0.0001, P<0.0001, respectively; Figure 4C).
Multivariate Cox proportional hazards models were used to assess independent predictors of BCR-free survival, including expression level of the gene (Z score > 0 vs Z score ⩽ 0), tumour stage (⩾ T3a vs ⩽ T2c), lymph node stage (N0 vs N1), and age at diagnosis (> 60 vs ⩽ 60). High MELK expression was a significant prognostic factor in patients with PCa (hazard ratio [HR]=1.889, 95% confidence interval [CI]=1.216–2.952, P=0.0047; Figure 4D). Likewise, high NCAPG (HR=1.693, 95% CI=1.084–2.650, P=0.0207), high BUB1 (HR=2.071, 95% CI=1.331–3.245, P=0.0013), and high CDK1 (HR=1.963, 95% CI=1.262–3.071, P=0.0028) were significant prognostic factors.
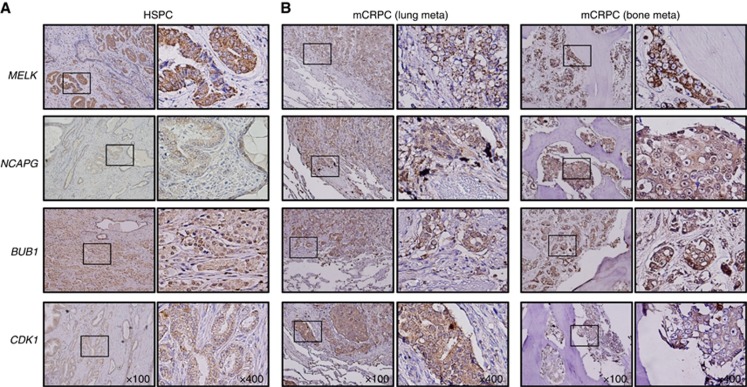
Immunohistochemical staining demonstrated high expression of MELK, NCAPG, BUB1, and CDK1 in HSPC cells (Figure 5A). Relatively higher expression of MELK, NCAPG, BUB1, and CDK1 was observed in mCRPC compared with that in HSPC (Figure 5B). Additionally, copy number amplifications or gains were observed in mCRPC compared with that in HSPC for MELK, NCAPG, BUB1, and CDK1 (P<0.0001, P=0.017, P=0.045, P<0.0001, respectively; Supplementary Figure 4).
Figure 5.
Immunohistochemical staining of MELK, NCAPG, BUB1, and CDK1 in HSPC and mCRPC specimens. (A) Immunochemical staining of MELK, NCAPG, BUB1, and CDK1 in HSPC specimens. (B) Immunochemical staining of MELK, NCAPG, BUB1, and CDK1 in mCRPC specimens.
These results suggested that MELK, NCAPG, BUB1, and CDK1 were deeply involved in the pathogenesis of CRPC. A schematic illustration shows the regulation of target genes by antitumour miR-145-3p in CRPC cells in this study (Supplementary Figure 5).
Discussion
As a fine tuner of genes, miRNAs form complicated RNA regulatory networks with mRNAs, long noncoding RNAs (lncRNAs), and other RNAs. Disruption of this network induces aberrant expression of mRNAs, promoting the development of various diseases, including cancer, metabolic diseases, and cardiovascular diseases. This suggests that we can search for novel mechanisms of disease by elucidating RNA networks based on key regulatory miRNAs. Accordingly, our group has been comprehensively analysing clinical specimens by genome-wide miRNA expression analysis and in silico analysis to reveal novel molecules that have important roles in cancer progression (Goto et al, 2015; Goto et al, 2016a; Goto et al, 2016b).
In this study, we applied next-generation RNA sequencing to lethal mCRPC specimens. Both miR-145-5p and miR-145-3p were significantly downregulated in mCRPC specimens compared with those in HSPC or non-PCa specimens. miR-145-5p is a guide strand and has been extensively studied in various types of cancers, including PCa (Ibrahim et al, 2011). Specifically, miR-145-5p functions as a post-transcriptional regulator, directly linking p53 and c-Myc expression (Sachdeva et al, 2009).
Downregulation of miR-145-5p has been shown in several types of cancers, including PCa (Suh et al, 2011; Cui et al, 2014). The molecular mechanisms of silencing of miR-145-5p has also been investigated in various cancers (Cui et al, 2014). Past studies showed that pivotal player of tumour suppressor p53 was directly binding to p53 responsible elements in the promoter region of pre-miR-145 and p53 increased miR-145-5p expression (Sachdeva et al, 2009). Expression and antitumour roles of miR-145-5p has depended on activation of p53 in cancer cells (Spizzo et al, 2010; Ren et al, 2013). Other studies demonstrated that several transcription factors, C/EBP-1 and forkhead transcription factor family were contributed to miR-145-5p regulation (Gan et al, 2010; Sachdeva et al, 2012). Moreover, promoter region of pre-miR-145 has contained several CpG sites, suggesting hyper-methylation of the promoter region caused to the silencing of pre-miR-145 (Xia et al, 2015). In PCa cell lines (LNCaP, PC3 and DU145), expression of miR-145-5p was elevated by treatment of 5-aza-2′-deoxycytidine (Suh et al, 2011).
In PCa, reports have demonstrated the tumour-suppressive function of miR-145-5p in targeting GOLM1, ITPR2, AR, and NMT3b (Kojima et al, 2014; Larne et al, 2015; Wang et al, 2015; Xue et al, 2015). Although passenger strand miRNAs were previously thought to be nonfunctional, our results showed that passenger strands may have a function. RNA sequencing of mCRPC specimens revealed downregulation of miR-145-5p and miR-145-3p in CRPC cells, and our data indicated that miR-145-3p was incorporated into the RISC, functioning as a tumour suppressor in PCa cells. Our group has previously demonstrated the importance of passenger strand miRNAs. Both miR-145-5p and miR-145-3p regulate MTDH in lung squamous cell carcinoma and target UHRF1 in bladder cancer (Mataki et al, 2016; Matsushita et al, 2016). Furthermore, miR-139-5p and miR-139-3p co-ordinately target MMP11 in bladder cancer (Yonemori et al, 2016). Other research groups have also recently reported the importance of passenger strands (Baez-Vega et al, 2016; Simerzin et al, 2016). The involvement of passenger strand miRNAs in the regulation of cellular processes is a novel concept in RNA research.
In this study, by focusing on miR-145-3p, which had not been well studied in previous reports, we found that 4 novel genes, i.e., MELK, NCAPG, BUB1, and CDK1, may have clinical significance in PCa and could be used as therapeutic targets in CRPC.
MELK upregulation has been reported in multiple types of aggressive cancer cells, including glioblastoma and basal-like breast cancer (Visnyei et al, 2011; Speers et al, 2016). MELK functions to promote cancer cell growth by maintaining cancer stem cells and resistance to radiotherapy or chemotherapy (Ganguly et al, 2015). Additionally, recent approaches with peptide library screening have revealed that MELK directly interacts with eukaryotic translation initiation factor 4B (eIF4B) to regulate protein synthesis during mitosis (Wang et al, 2016). Other reports have suggested that MELK protein forms a complex with forkhead box M1 (FOXM1), a transcription factor that primarily regulates the cell cycle, to induce proliferation (Kato et al, 2016). MELK is reported to form a complex with another oncoprotein, c-Jun, to maintain glioblastoma stem cells. The malignant transformation of normal stem cells is accompanied by MELK overexpression (Ganguly et al, 2014). Thus, MELK is a therapeutic target for advanced refractory cancers (clinicaltrials.gov identifier: NCT01910545).
CDK1 is a key regulator of the cell cycle and forms a complex with cyclin A and cyclin B in G1/S- and G2/M-phase transitions (Asghar et al, 2015). Increased CDK1 activity is a mechanism for increasing AR expression responding to androgen deprivation in CRPC (Chen et al, 2006). Recent high-throughput RNAi screening identified checkpoint kinase 2 (CHK2) as a tumour suppressor in PCa; CHK2 negatively regulates CDK1, forming a signalling loop with AR (Ta et al, 2015). Furthermore, AR-V7 interacts with CDK1 and phosphatidylinositol-4-phosphate 5-kinase alpha to induce metastatic growth and treatment resistance (Sarwar et al, 2016). Therefore, CDK1 inhibitors may be promising novel CRPC therapeutics.
BUB1 and NCAPG have not been well studied in CRPC. BUB1 is a central component of the spindle assembly checkpoint (SAC) with budding uninhibited by benzimidazole-related 1. SAC self-monitors chromosome segregation in prometaphase mitosis to prevent aneuploidy (Elowe, 2011). Mutations in BUB1 have been reported in several types of cancers, and associations with aneuploidy have been reported (Ricke et al, 2011). Moreover, in cancer cells, BUB1 interacts with transforming growth factor (TGF)-β receptor I and II, mediating the TGF-β-dependent epithelial-mesenchymal transition (EMT) (Nyati et al, 2015). BUB1 contributes to both the cell cycle and EMT. NCAPG is a subunit of the condensin complex, a major molecular effector of chromosome condensation and division (Thadani et al, 2012). Overexpression of NCAPG has been reported in pediatric high-grade gliomas (Liang et al, 2016).
Interestingly, these 4 genes are all related to the cell cycle, particularly mitosis. The prognostic value of cell cycle proliferation genes in patients with PCa has been evaluated using the cell cycle progression (CCP) score (Cuzick et al, 2011). Researchers used 31 CCP genes to calculate the CCP score, which predicted BCR-free survival after prostatectomy. Although MELK, NCAPG, BUB1, and CDK1 were not included in these 31 genes, the clinical significance of cell cycle genes supports our study. Furthermore, HES6, a driver of androgen-independent growth of PCa, enhances E2F1 transcription factor (Ramos-Montoya et al, 2014), which consequently induces cell cycle progression. Targeting cell cycle genes in CRPC treatment is a rational option considering the clinical effects of docetaxel, which inhibits microtubule assembly, on CRPC. Recently, novel agents targeting the AR signalling axis have been shown to yield survival benefits, accompanied by the emergence of AR signalling-independent cells, including neuroendocrine PCa cells. Under these conditions, CRPC exhibits treatment-adapted phenotypes that allow cells to survive in the presence of heavy chemotherapeutics under anti-AR signalling conditions. A recent article called this condition ‘treatment-induced lineage crisis’ in metastatic CRPC, similar to triple-negative breast cancer and multidrug resistant bacteria in antibiotics (Roubaud et al, 2016). Among the 4 genes identified in this study, anti-MELK treatment is currently in phase I clinical trials. A highly potent MELK inhibitor, OTSSP167, has been shown to significantly suppress tumour growth in DU145 cell-derived xenografts in mice (Chung et al, 2012). Moreover, MELK upregulation was reported in high-grade PCa and CRPC specimens (Kuner et al, 2013; Ross-Adams et al, 2015). Additionally, MELK interacts with FOXM1, and FOXM1 and centromere protein F (CENPF) are synergistic master regulators and drivers of PCa, according to genome-wide regulatory network analyses in human and mouse PCa models (Aytes et al, 2014). OTSSP167 also inhibits BUB1 (Ji et al, 2016) and is a promising molecule for the treatment of CRPC; thus, further analysis of MELK inhibitors is needed.
Directly targeting cell cycle genes may have significant effects on CRPC treatment. This approach may lead to the development of alternative treatment paradigms. The prognostic roles and clinical significance of MELK, NCAPG, BUB1, and CDK1, as revealed in this study, warrant further analyses of these 4 genes. The past few years, a large number of preclinical studies of miRNA-based therapeutics have been reported in several cancers (Ganju et al, 2017). Among these studies, some studies have reached clinical trials (Catela Ivkovic et al, 2017; Rupaimoole and Slack, 2017). It is essential to identify the best miRNA suitable for miRNA-based therapeutics for each cancer. As for miR-145, the delivery system using disulfide linkage in polyethyleneimine-based nanoparticles to PCa has been reported (Zhang et al, 2015). These facts suggest promising future for miR-145-3p-based therapeutics in this disease.
Conclusions
Small RNA sequencing for lethal metastatic CRPC, genome-wide analysis, and in silico approaches revealed novel therapeutic targets for CRPC. This novel approach may contribute to the development of new therapeutic strategies.
Acknowledgments
This study was supported by JSPS KAKENHI (grant numbers (B) 16H05462, (B) 15K20071), the Futaba Electronics Memorial Foundation, the Inohana Foundation (Chiba University), the Japanese Foundation for Prostate Research, the Suzuki Urological Foundation, and a Young Research Grant from the Japanese Urological Association.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- Asghar U, Witkiewicz AK, Turner NC, Knudsen ES (2015) The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov 14(2): 130–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aytes A, Mitrofanova A, Lefebvre C, Alvarez MJ, Castillo-Martin M, Zheng T, Eastham JA, Gopalan A, Pienta KJ, Shen MM, Califano A, Abate-Shen C (2014) Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer Cell 25(5): 638–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baez-Vega PM, Echevarria Vargas IM, Valiyeva F, Encarnacion-Rosado J, Roman A, Flores J, Marcos-Martinez MJ, Vivas-Mejia PE (2016) Targeting miR-21-3p inhibits proliferation and invasion of ovarian cancer cells. Oncotarget 7(24): 36321–36337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2): 281–297. [DOI] [PubMed] [Google Scholar]
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136(2): 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N (2015) The molecular taxonomy of primary prostate cancer. Cell 163(4): 1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catela Ivkovic T, Voss G, Cornella H, Ceder Y (2017) microRNAs as cancer therapeutics: a step closer to clinical application. Cancer Lett. [DOI] [PubMed]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2(5): 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Xu Y, Yuan X, Bubley GJ, Balk SP (2006) Androgen receptor phosphorylation and stabilization in prostate cancer by cyclin-dependent kinase 1. Proc Natl Acad Sci USA 103(43): 15969–15974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Suzuki H, Miyamoto T, Takamatsu N, Tatsuguchi A, Ueda K, Kijima K, Nakamura Y, Matsuo Y (2012) Development of an orally-administrative MELK-targeting inhibitor that suppresses the growth of various types of human cancer. Oncotarget 3(12): 1629–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford ED, Higano CS, Shore ND, Hussain M, Petrylak DP (2015) Treating patients with metastatic castration resistant prostate cancer: a comprehensive review of available therapies. J Urol 194(6): 1537–1547. [DOI] [PubMed] [Google Scholar]
- Cui SY, Wang R, Chen LB (2014) MicroRNA-145: a potent tumour suppressor that regulates multiple cellular pathways. J Cell Mol Med 18(10): 1913–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzick J, Swanson GP, Fisher G, Brothman AR, Berney DM, Reid JE, Mesher D, Speights VO, Stankiewicz E, Foster CS, Moller H, Scardino P, Warren JD, Park J, Younus A, Flake DD 2nd, Wagner S, Gutin A, Lanchbury JS, Stone S (2011) Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol 12(3): 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowe S (2011) Bub1 and BubR1: at the interface between chromosome attachment and the spindle checkpoint. Mol Cell Biol 31(15): 3085–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan B, Lim C, Chu G, Hua S, Ding Z, Collins M, Hu J, Jiang S, Fletcher-Sananikone E, Zhuang L, Chang M, Zheng H, Wang YA, Kwiatkowski DJ, Kaelin WG Jr., Signoretti S, DePinho RA (2010) FoxOs enforce a progression checkpoint to constrain mTORC1-activated renal tumorigenesis. Cancer Cell 18(5): 472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly R, Hong CS, Smith LG, Kornblum HI, Nakano I (2014) Maternal embryonic leucine zipper kinase: key kinase for stem cell phenotype in glioma and other cancers. Mol Cancer Ther 13(6): 1393–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly R, Mohyeldin A, Thiel J, Kornblum HI, Beullens M, Nakano I (2015) MELK – a conserved kinase: functions, signaling, cancer, and controversy. Clin Transl Med 4: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganju A, Khan S, Hafeez BB, Behrman SW, Yallapu MM, Chauhan SC, Jaggi M (2017) miRNA nanotherapeutics for cancer. Drug Discov Today 22(2): 424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6(269): pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Calin GA, Croce CM (2009) MicroRNAs in cancer. Annu Rev Med 60: 167–179. [DOI] [PubMed] [Google Scholar]
- Goto Y, Kojima S, Kurozumi A, Kato M, Okato A, Matsushita R, Ichikawa T, Seki N (2016. a) Regulation of E3 ubiquitin ligase-1 (WWP1) by microRNA-452 inhibits cancer cell migration and invasion in prostate cancer. Br J Cancer 114(10): 1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Kojima S, Nishikawa R, Kurozumi A, Kato M, Enokida H, Matsushita R, Yamazaki K, Ishida Y, Nakagawa M, Naya Y, Ichikawa T, Seki N (2015) MicroRNA expression signature of castration-resistant prostate cancer: the microRNA-221/222 cluster functions as a tumour suppressor and disease progression marker. Br J Cancer 113(7): 1055–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Kurozumi A, Nohata N, Kojima S, Matsushita R, Yoshino H, Yamazaki K, Ishida Y, Ichikawa T, Naya Y, Seki N (2016. b) The microRNA signature of patients with sunitinib failure: regulation of UHRF1 pathways by microRNA-101 in renal cell carcinoma. Oncotarget 7(37): 59070–59086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim AF, Weirauch U, Thomas M, Grunweller A, Hartmann RK, Aigner A (2011) MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res 71(15): 5214–5224. [DOI] [PubMed] [Google Scholar]
- Ji W, Arnst C, Tipton AR, Bekier ME 2nd, Taylor WR, Yen TJ, Liu ST (2016) OTSSP167 abrogates mitotic checkpoint through inhibiting multiple mitotic kinases. PLoS ONE 11(4): e0153518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Inoue H, Imoto S, Tamada Y, Miyamoto T, Matsuo Y, Nakamura Y, Park JH (2016) Oncogenic roles of TOPK and MELK, and effective growth suppression by small molecular inhibitors in kidney cancer cells. Oncotarget 7(14): 17652–17664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Enokida H, Yoshino H, Itesako T, Chiyomaru T, Kinoshita T, Fuse M, Nishikawa R, Goto Y, Naya Y, Nakagawa M, Seki N (2014) The tumor-suppressive microRNA-143/145 cluster inhibits cell migration and invasion by targeting GOLM1 in prostate cancer. J Hum Genet 59(2): 78–87. [DOI] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma'ayan A (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44(W1): W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuner R, Falth M, Pressinotti NC, Brase JC, Puig SB, Metzger J, Gade S, Schafer G, Bartsch G, Steiner E, Klocker H, Sultmann H (2013) The maternal embryonic leucine zipper kinase (MELK) is upregulated in high-grade prostate cancer. J Mol Med (Berlin, Germany) 91(2): 237–248. [DOI] [PubMed] [Google Scholar]
- Larne O, Hagman Z, Lilja H, Bjartell A, Edsjo A, Ceder Y (2015) miR-145 suppress the androgen receptor in prostate cancer cells and correlates to prostate cancer prognosis. Carcinogenesis 36(8): 858–866. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- Liang ML, Hsieh TH, Ng KH, Tsai YN, Tsai CF, Chao ME, Liu DJ, Chu SS, Chen W, Liu YR, Liu RS, Lin SC, Ho DM, Wong TT, Yang MH, Wang HW (2016) Downregulation of miR-137 and miR-6500-3p promotes cell proliferation in pediatric high-grade gliomas. Oncotarget 7(15): 19723–19737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mataki H, Seki N, Mizuno K, Nohata N, Kamikawaji K, Kumamoto T, Koshizuka K, Goto Y, Inoue H (2016) Dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p) coordinately targeted MTDH in lung squamous cell carcinoma. Oncotarget 7(44): 72084–72098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita R, Yoshino H, Enokida H, Goto Y, Miyamoto K, Yonemori M, Inoguchi S, Nakagawa M, Seki N (2016) Regulation of UHRF1 by dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p): inhibition of bladder cancer cell aggressiveness. Oncotarget 7(19): 28460–28487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyati S, Schinske-Sebolt K, Pitchiaya S, Chekhovskiy K, Chator A, Chaudhry N, Dosch J, Van Dort ME, Varambally S, Kumar-Sinha C, Nyati MK, Ray D, Walter NG, Yu H, Ross BD, Rehemtulla A (2015) The kinase activity of the Ser/Thr kinase BUB1 promotes TGF-beta signaling. Sci Signal 8(358): ra1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Montoya A, Lamb AD, Russell R, Carroll T, Jurmeister S, Galeano-Dalmau N, Massie CE, Boren J, Bon H, Theodorou V, Vias M, Shaw GL, Sharma NL, Ross-Adams H, Scott HE, Vowler SL, Howat WJ, Warren AY, Wooster RF, Mills IG, Neal DE (2014) HES6 drives a critical AR transcriptional programme to induce castration-resistant prostate cancer through activation of an E2F1-mediated cell cycle network. EMBO Mol Med 6(5): 651–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Wang M, Guo W, Zhao X, Tu X, Huang S, Zou X, Peng X (2013) Wild-type p53 suppresses the epithelial–mesenchymal transition and stemness in PC-3 prostate cancer cells by modulating miR145. Int J Oncol 42(4): 1473–1481. [DOI] [PubMed] [Google Scholar]
- Ricke RM, Jeganathan KB, van Deursen JM (2011) Bub1 overexpression induces aneuploidy and tumor formation through Aurora B kinase hyperactivation. J Cell Biol 193(6): 1049–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G, Beltran H, Abida W, Bradley RK, Vinson J, Cao X, Vats P, Kunju LP, Hussain M, Feng FY, Tomlins SA, Cooney KA, Smith DC, Brennan C, Siddiqui J, Mehra R, Chen Y, Rathkopf DE, Morris MJ, Solomon SB, Durack JC, Reuter VE, Gopalan A, Gao J, Loda M, Lis RT, Bowden M, Balk SP, Gaviola G, Sougnez C, Gupta M, Yu EY, Mostaghel EA, Cheng HH, Mulcahy H, True LD, Plymate SR, Dvinge H, Ferraldeschi R, Flohr P, Miranda S, Zafeiriou Z, Tunariu N, Mateo J, Perez-Lopez R, Demichelis F, Robinson BD, Schiffman M, Nanus DM, Tagawa ST, Sigaras A, Eng KW, Elemento O, Sboner A, Heath EI, Scher HI, Pienta KJ, Kantoff P, de Bono JS, Rubin MA, Nelson PS, Garraway LA, Sawyers CL, Chinnaiyan AM (2015) Integrative clinical genomics of advanced prostate cancer. Cell 161(5): 1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Adams H, Lamb AD, Dunning MJ, Halim S, Lindberg J, Massie CM, Egevad LA, Russell R, Ramos-Montoya A, Vowler SL, Sharma NL, Kay J, Whitaker H, Clark J, Hurst R, Gnanapragasam VJ, Shah NC, Warren AY, Cooper CS, Lynch AG, Stark R, Mills IG, Gronberg H, Neal DE (2015) Integration of copy number and transcriptomics provides risk stratification in prostate cancer: a discovery and validation cohort study. EBioMedicine 2(9): 1133–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roubaud G, Liaw BC, Oh WK, Mulholland DJ (2016) Strategies to avoid treatment-induced lineage crisis in advanced prostate cancer. Nat Rev Clin Oncol 14(5): 269–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupaimoole R, Slack FJ (2017) MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 16(3): 203–222. [DOI] [PubMed] [Google Scholar]
- Sachdeva M, Liu Q, Cao J, Lu Z, Mo YY (2012) Negative regulation of miR-145 by C/EBP-beta through the Akt pathway in cancer cells. Nucleic Acids Res 40(14): 6683–6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY (2009) p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA 106(9): 3207–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J (2003) TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34(2): 374–378. [DOI] [PubMed] [Google Scholar]
- Sarwar M, Semenas J, Miftakhova R, Simoulis A, Robinson B, Wingren AG, Mongan NP, Heery DM, Johnsson H, Abrahamsson PA, Dizeyi N, Luo J, Persson JL (2016) Targeted suppression of AR-V7 using PIP5K1alpha inhibitor overcomes enzalutamide resistance in prostate cancer cells. Oncotarget 7(39): 63065–63081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A (2015) Cancer statistics, 2015. CA Cancer J Clin 65(1): 5–29. [DOI] [PubMed] [Google Scholar]
- Simerzin A, Zorde-Khvalevsky E, Rivkin M, Adar R, Zucman-Rossi J, Couchy G, Roskams T, Govaere O, Oren M, Giladi H, Galun E (2016) The liver-specific microRNA-122*, the complementary strand of microRNA-122, acts as a tumor suppressor by modulating the p53/mouse double minute 2 homolog circuitry. Hepatology (Baltimore, Md) 64(5): 1623–1636. [DOI] [PubMed] [Google Scholar]
- Speers C, Zhao SG, Kothari V, Santola A, Liu M, Wilder-Romans K, Evans J, Batra N, Bartelink H, Hayes DF, Lawrence TS, Brown PH, Pierce LJ, Feng FY (2016) Maternal embryonic leucine zipper kinase (MELK) as a novel mediator and biomarker of radioresistance in human breast cancer. Clin Cancer Res 22(23): 5864–5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizzo R, Nicoloso MS, Lupini L, Lu Y, Fogarty J, Rossi S, Zagatti B, Fabbri M, Veronese A, Liu X, Davuluri R, Croce CM, Mills G, Negrini M, Calin GA (2010) miR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells. Cell Death Differ 17(2): 246–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh SO, Chen Y, Zaman MS, Hirata H, Yamamura S, Shahryari V, Liu J, Tabatabai ZL, Kakar S, Deng G, Tanaka Y, Dahiya R (2011) MicroRNA-145 is regulated by DNA methylation and p53 gene mutation in prostate cancer. Carcinogenesis 32(5): 772–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta HQ, Ivey ML, Frierson HF Jr., Conaway MR, Dziegielewski J, Larner JM, Gioeli D (2015) Checkpoint kinase 2 negatively regulates androgen sensitivity and prostate cancer cell growth. Cancer Res 75(23): 5093–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thadani R, Uhlmann F, Heeger S (2012) Condensin, chromatin crossbarring and chromosome condensation. Curr Biol 22(23): R1012–R1021. [DOI] [PubMed] [Google Scholar]
- Visnyei K, Onodera H, Damoiseaux R, Saigusa K, Petrosyan S, De Vries D, Ferrari D, Saxe J, Panosyan EH, Masterman-Smith M, Mottahedeh J, Bradley KA, Huang J, Sabatti C, Nakano I, Kornblum HI (2011) A molecular screening approach to identify and characterize inhibitors of glioblastoma stem cells. Mol Cancer Ther 10(10): 1818–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BD, Ceniccola K, Yang Q, Andrawis R, Patel V, Ji Y, Rhim J, Olender J, Popratiloff A, Latham P, Lai Y, Patierno SR, Lee NH (2015) Identification and functional validation of reciprocal microRNA-mRNA pairings in African American prostate cancer disparities. Clin Cancer Res 21(21): 4970–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Begley M, Li Q, Huang HT, Lako A, Eck MJ, Gray NS, Mitchison TJ, Cantley LC, Zhao JJ (2016) Mitotic MELK-eIF4B signaling controls protein synthesis and tumor cell survival. Proc Natl Acad Sci USA 113(35): 9810–9815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitington T, Gao P, Song W, Ross-Adams H, Lamb AD, Yang Y, Svezia I, Klevebring D, Mills IG, Karlsson R, Halim S, Dunning MJ, Egevad L, Warren AY, Neal DE, Gronberg H, Lindberg J, Wei GH, Wiklund F (2016) Gene regulatory mechanisms underpinning prostate cancer susceptibility. Nat Genet 48(4): 387–397. [DOI] [PubMed] [Google Scholar]
- Xia W, Chen Q, Wang J, Mao Q, Dong G, Shi R, Zheng Y, Xu L, Jiang F (2015) DNA methylation mediated silencing of microRNA-145 is a potential prognostic marker in patients with lung adenocarcinoma. Sci Rep 5: 16901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue G, Ren Z, Chen Y, Zhu J, Du Y, Pan D, Li X, Hu B (2015) A feedback regulation between miR-145 and DNA methyltransferase 3b in prostate cancer cell and their responses to irradiation. Cancer Lett 361(1): 121–127. [DOI] [PubMed] [Google Scholar]
- Yonemori M, Seki N, Yoshino H, Matsushita R, Miyamoto K, Nakagawa M, Enokida H (2016) Dual tumor-suppressors miR-139-5p and miR-139-3p targeting matrix metalloprotease 11 in bladder cancer. Cancer Sci 107(9): 1233–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Xue X, He D, Hsieh JT (2015) A prostate cancer-targeted polyarginine-disulfide linked PEI nanocarrier for delivery of microRNA. Cancer Lett 365(2): 156–165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.