Abstract
Recombination mediated by the Red pathway of bacteriophage lambda is focused towards sites of double-chain cuts. Double-chain ends created either by type II restriction enzymes acting at unmodified recognition sites or by lambda's packaging enzyme, terminase, acting at cos are utilized in a manner similar to the double-chain break repair pathway of recombination in yeast. When lambda is allowed to recombine during replicative growth, spontaneous recombination is approximately evenly distributed along the chromosome. It has been proposed that replication-allowed recombination also is initiated by double-chain ends. In order to test this hypothesis we ask if the in vivo expression of the Mu gam protein is inhibitory to Red recombination. Mu gam has been shown in vitro to bind to linearized duplex DNA and to shield bound DNA from exonucleases. The expression of Mu gam is found to be inhibitory to Red recombination whether replication is blocked or allowed. As a control we ask if Mu gam inhibits Int-mediated recombination. It has been well documented that the Int pathway of recombination does not involve any double-chain breaks and, consistent with this, the Int pathway is not inhibited by Mu gam. We suggest that the in vivo expression of Mu gam or other similar activities may be a generally useful way to determine if those processes that respond to an artificially introduced double-chain cut normally involve double-chain ends.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akroyd J. E., Clayson E., Higgins N. P. Purification of the gam gene-product of bacteriophage Mu and determination of the nucleotide sequence of the gam gene. Nucleic Acids Res. 1986 Sep 11;14(17):6901–6914. doi: 10.1093/nar/14.17.6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akroyd J., Barton B., Lund P., Maynard Smith S., Sultana K., Symonds N. Mapping and properties of the gam and sot genes of phage mu: their possible roles in recombination. Cold Spring Harb Symp Quant Biol. 1984;49:261–266. doi: 10.1101/sqb.1984.049.01.030. [DOI] [PubMed] [Google Scholar]
- Akroyd J., Symonds N. Localization of the gam gene of bacteriophage mu and characterisation of the gene product. Gene. 1986;49(2):273–282. doi: 10.1016/0378-1119(86)90288-x. [DOI] [PubMed] [Google Scholar]
- BROWN A., ARBER W. TEMPERATURE-SENSITIVE MUTANTS OF COLIPHAGE LAMBDA. Virology. 1964 Oct;24:237–239. doi: 10.1016/0042-6822(64)90114-x. [DOI] [PubMed] [Google Scholar]
- CAMPBELL A. Sensitive mutants of bacteriophage lambda. Virology. 1961 May;14:22–32. doi: 10.1016/0042-6822(61)90128-3. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingery R., Echols H. Mutants of bacteriophage lambda unable to integrate into the host chromosome. Proc Natl Acad Sci U S A. 1967 Oct;58(4):1507–1514. doi: 10.1073/pnas.58.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings P. J. Measurement of restoration and conversion: its meaning for the mismatch repair hypothesis of conversion. Cold Spring Harb Symp Quant Biol. 1984;49:49–53. doi: 10.1101/sqb.1984.049.01.008. [DOI] [PubMed] [Google Scholar]
- Hradecna Z., Szybalski W. Electron micrographic maps of deletions and substitutions in the genomes of transducing coliphages lambda dg and lambda bio. Virology. 1969 Jul;38(3):473–477. doi: 10.1016/0042-6822(69)90160-3. [DOI] [PubMed] [Google Scholar]
- KAISER A. D., JACOB F. Recombination between related temperate bacteriophages and the genetic control of immunity and prophage localization. Virology. 1957 Dec;4(3):509–521. doi: 10.1016/0042-6822(57)90083-1. [DOI] [PubMed] [Google Scholar]
- Kitts P., Richet E., Nash H. A. Lambda integrative recombination: supercoiling, synapsis, and strand exchange. Cold Spring Harb Symp Quant Biol. 1984;49:735–744. doi: 10.1101/sqb.1984.049.01.083. [DOI] [PubMed] [Google Scholar]
- Kolodkin A. L., Klar A. J., Stahl F. W. Double-strand breaks can initiate meiotic recombination in S. cerevisiae. Cell. 1986 Aug 29;46(5):733–740. doi: 10.1016/0092-8674(86)90349-1. [DOI] [PubMed] [Google Scholar]
- Krasin F., Hutchinson F. Repair of DNA double-strand breaks in Escherichia coli, which requires recA function and the presence of a duplicate genome. J Mol Biol. 1977 Oct 15;116(1):81–98. doi: 10.1016/0022-2836(77)90120-6. [DOI] [PubMed] [Google Scholar]
- Kucherlapati R. S., Eves E. M., Song K. Y., Morse B. S., Smithies O. Homologous recombination between plasmids in mammalian cells can be enhanced by treatment of input DNA. Proc Natl Acad Sci U S A. 1984 May;81(10):3153–3157. doi: 10.1073/pnas.81.10.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach D. R., Stahl F. W. Viability of lambda phages carrying a perfect palindrome in the absence of recombination nucleases. 1983 Sep 29-Oct 5Nature. 305(5933):448–451. doi: 10.1038/305448a0. [DOI] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984 Jun;4(6):1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W. Fungal recombination. Microbiol Rev. 1985 Mar;49(1):33–58. doi: 10.1128/mr.49.1.33-58.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4417–4421. doi: 10.1073/pnas.80.14.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S. Deletion mutants of bacteriophage lambda. II. Genetic properties of att-defective mutants. J Mol Biol. 1971 Mar 14;56(2):385–401. doi: 10.1016/0022-2836(71)90472-4. [DOI] [PubMed] [Google Scholar]
- Resnick M. A., Martin P. The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Mol Gen Genet. 1976 Jan 16;143(2):119–129. doi: 10.1007/BF00266917. [DOI] [PubMed] [Google Scholar]
- Rossignol J. L., Nicolas A., Hamza H., Langin T. Origins of gene conversion and reciprocal exchange in Ascobolus. Cold Spring Harb Symp Quant Biol. 1984;49:13–21. doi: 10.1101/sqb.1984.049.01.004. [DOI] [PubMed] [Google Scholar]
- Russo V. E. On the physical structure of lambda recombinant DNA. Mol Gen Genet. 1973 May 28;122(4):353–366. doi: 10.1007/BF00269436. [DOI] [PubMed] [Google Scholar]
- Shurvinton C. E., Stahl M. M., Stahl F. W. Large palindromes in the lambda phage genome are preserved in a rec+ host by inhibiting lambda DNA replication. Proc Natl Acad Sci U S A. 1987 Mar;84(6):1624–1628. doi: 10.1073/pnas.84.6.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl F. W., Kobayashi I., Stahl M. M. Distance from cohesive end site cos determines the replication requirement for recombination in phage lambda. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6318–6321. doi: 10.1073/pnas.79.20.6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl F. W., Kobayashi I., Stahl M. M. In phage lambda, cos is a recombinator in the red pathway. J Mol Biol. 1985 Jan 20;181(2):199–209. doi: 10.1016/0022-2836(85)90085-3. [DOI] [PubMed] [Google Scholar]
- Stahl F. W., Kobayashi I., Thaler D., Stahl M. M. Direction of travel of RecBC recombinase through bacteriophage lambda DNA. Genetics. 1986 Jun;113(2):215–227. doi: 10.1093/genetics/113.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl F. W., McMilin K. D., Stahl M. M., Nozu Y. An enhancing role for DNA synthesis in formation of bacteriophage lambda recombinants. Proc Natl Acad Sci U S A. 1972 Dec;69(12):3598–3601. doi: 10.1073/pnas.69.12.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
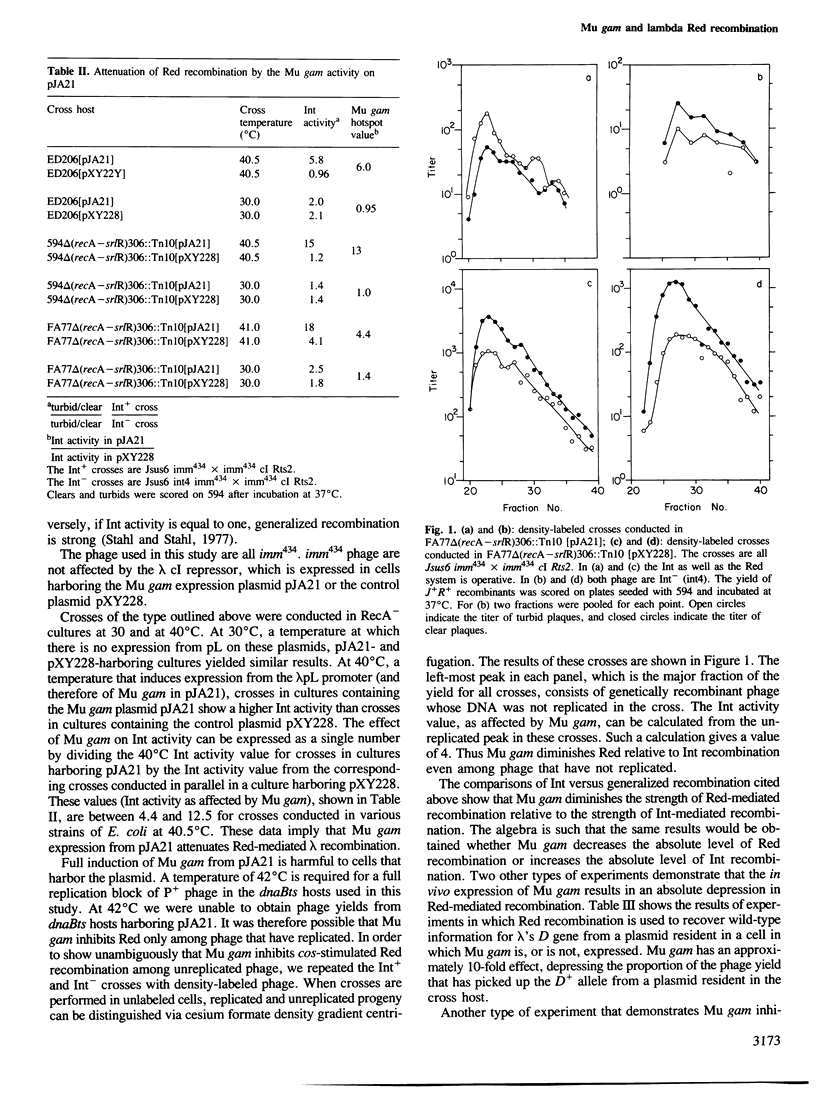
- Stahl F. W. Roles of double-strand breaks in generalized genetic recombination. Prog Nucleic Acid Res Mol Biol. 1986;33:169–194. doi: 10.1016/s0079-6603(08)60023-9. [DOI] [PubMed] [Google Scholar]
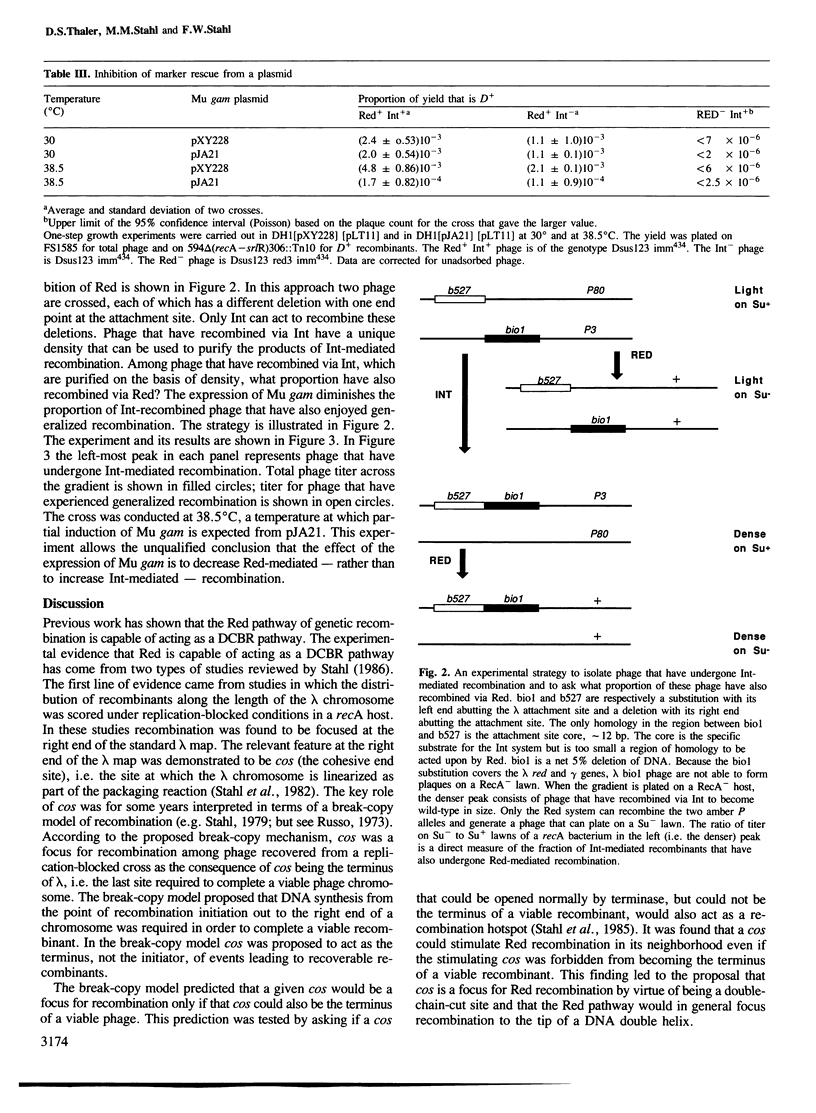
- Stahl F. W., Stahl M. M. DNA synthesis at the site of a Red-mediated exchange in phage lambda. Genetics. 1986 May;113(1):1–12. doi: 10.1093/genetics/113.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
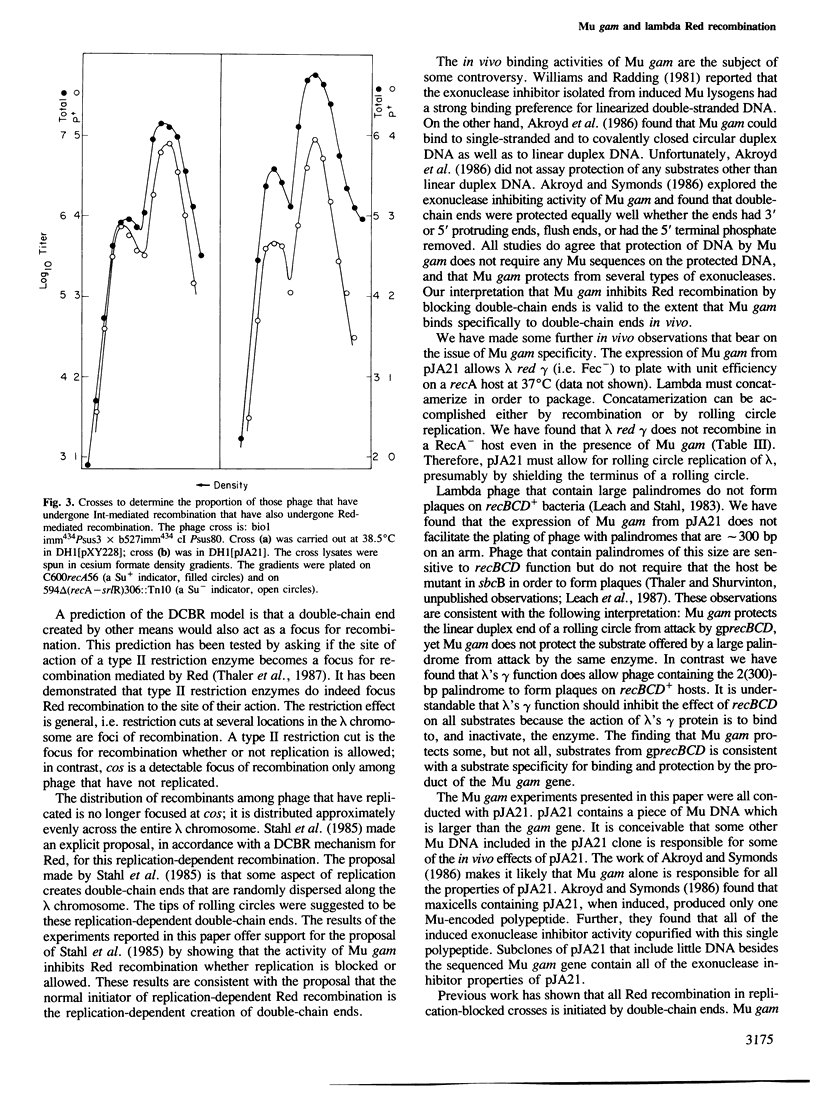
- Stahl F. W., Stahl M. M. Recombination pathway specificity of Chi. Genetics. 1977 Aug;86(4):715–725. doi: 10.1093/genetics/86.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L. S., Morrison P., Kolodner R. Intramolecular recombination of linear DNA catalyzed by the Escherichia coli RecE recombination system. J Mol Biol. 1985 Dec 5;186(3):515–525. doi: 10.1016/0022-2836(85)90126-3. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Thaler D. S., Stahl M. M., Stahl F. W. Double-chain-cut sites are recombination hotspots in the Red pathway of phage lambda. J Mol Biol. 1987 May 5;195(1):75–87. doi: 10.1016/0022-2836(87)90328-7. [DOI] [PubMed] [Google Scholar]
- Thomas K. R., Folger K. R., Capecchi M. R. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986 Feb 14;44(3):419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- Van Vliet F., Couturier M., De Lafonteyne J., Jedlicki E. Mu-1 directed inhibition of DNA breakdown in Escherichia coli, recA cells. Mol Gen Genet. 1978 Aug 4;164(1):109–112. doi: 10.1007/BF00267606. [DOI] [PubMed] [Google Scholar]
- Wake C. T., Vernaleone F., Wilson J. H. Topological requirements for homologous recombination among DNA molecules transfected into mammalian cells. Mol Cell Biol. 1985 Aug;5(8):2080–2089. doi: 10.1128/mcb.5.8.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigle J. Assembly of phage lambda in vitro. Proc Natl Acad Sci U S A. 1966 Jun;55(6):1462–1466. doi: 10.1073/pnas.55.6.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil J., Signer E. R. Recombination in bacteriophage lambda. II. Site-specific recombination promoted by the integration system. J Mol Biol. 1968 Jul 14;34(2):273–279. doi: 10.1016/0022-2836(68)90252-0. [DOI] [PubMed] [Google Scholar]
- White J. H., Lusnak K., Fogel S. Mismatch-specific post-meiotic segregation frequency in yeast suggests a heteroduplex recombination intermediate. Nature. 1985 May 23;315(6017):350–352. doi: 10.1038/315350a0. [DOI] [PubMed] [Google Scholar]
- Williams J. G., Radding C. M. Partial purification and properties of an exonuclease inhibitor induced by bacteriophage Mu-1. J Virol. 1981 Aug;39(2):548–558. doi: 10.1128/jvi.39.2.548-558.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]



