Abstract
The Mig10/RIAM/Lpd (MRL) adapter protein Lpd regulates actin dynamics through interactions with Scar/WAVE and Ena/VASP proteins to promote the formation of cellular protrusions and to stimulate invasive migration. However, the ability of MRL proteins to interact with multiple actin regulators and to promote serum response factor (SRF) signalling has raised the question of whether MRL proteins employ alternative downstream mechanisms to drive oncogenic processes in a context-dependent manner. Here, using a Drosophila model, we show that overexpression of either human Lpd or its Drosophila orthologue Pico can promote growth and invasion of RasV12-induced cell tumours in the brain. Notably, effects were restricted to two populations of Repo-positive glial cells: an invasive population, characterized by JNK-dependent elevation of Mmp1 expression, and a hyperproliferative population lacking elevated JNK signalling. JNK activation was not triggered by reactive immune cell signalling, implicating the involvement of an intrinsic stress response. The ability to promote dissemination of RasV12-induced tumours was shared by a subset of actin regulators, including, most prominently, Chicadee/Profilin, which directly interacts with Pico, and, Mal, a cofactor for serum response factor that responds to changes in G:F actin dynamics. Suppression of Mal activity partially abrogated the ability of pico to promote invasion of RasV12 tumours. Furthermore, we found that larval glia are enriched for serum response factor expression, explaining the apparent sensitivity of glial cells to Pico/RasV12 overexpression. Taken together, our findings indicate that MRL proteins cooperate with oncogenic Ras to promote formation of glial tumours, and that, in this context, Mal/serum response factor activation is rate-limiting for tumour dissemination.
Introduction
Regulation of actin-based structures is critical for normal cell adhesion, morphology and motility.1 Correspondingly, aberrant cytoskeletal dynamics are implicated in the motility and dissemination of cancer cells.2, 3 In addition to the direct effects of actin reorganization, for example on lamellipodia-like structures at the leading edge of invasive cells,4 regulators of cytoplasmic actin also control the localization and activity of myocardin-related transcription factors (MRTF/Mal), which are transcriptional coactivators of serum response factor (SRF), by regulating the availability of monomeric (G-)actin.5 Depletion of nuclear and cytoplasmic G-actin in response to increased actin polymerization increases the rate of MRTF/Mal translocation to the nucleus, reduces the rate of nuclear export of MRTF/Mal and derepresses the expression of genes that require MRTF/Mal for transcription, leading to SRF-dependent transcription.6, 7, 8
The Mig-10/RIAM/Lamellipodin (MRL) family of adapter proteins transduce signals derived from growth factor receptors, via interactions with Ras-like GTPases and/or phospholipids, to changes in the actin cytoskeleton, increased lamellipodia protrusion, cell motility and altered cell adhesion.9, 10 Effects on the actin cytoskeleton are mediated by direct interactions with various actin regulatory proteins, including Ena/VASP, Scar/WAVE and Profilin.9, 10, 11 MRL proteins are also capable of activating SRF signalling by altering the ratio of G:F actin.12 MRL proteins are therefore good candidates for genes that drive tumour cell invasion and metastasis. Indeed, in breast cancer, Lpd is upregulated in tumours with lymph node metastases compared to lymph node-negative tumours13 and also in highly invasive MDA-MB231 breast cancer cells compared to non-invasive MCF7 breast cancer cells or normal breast tissue.14 Furthermore, increased expression and membrane localization correlate with reduced metastasis-free survival and poor prognosis in breast cancer patients.15 Mechanistically, MRL proteins promote invasive 3D breast cancer cell migration via interactions with the actin regulators Scar/Wave and Ena/VASP.15 Lpd is also part of the ‘Ras cancer signature’ as it is upregulated in human breast epithelial cells transformed with oncogenic Ras.16 The ‘Ras signature’ reflects the activation status of the Ras pathway and has been successfully used to identify patterns of pathway deregulation in human tumours and to identify clinically relevant associations with disease outcomes.16 An understanding of the functional consequences of MRL–Ras interactions in cancer development is, however, currently lacking.
Drosophila encodes only one MRL protein, called Pico, enabling the dissection of conserved cancer promoting effects of the MRL gene family in an animal model, with the potential to help guide studies in mammalian systems.17 Many biological processes related to tumorigenesis and metastasis are well conserved in flies and nearly all of the genes linked to cancer progression in humans are present in the Drosophila genome.17, 18 Here we have tested the prediction that MRL proteins might cooperate with oncogenic Ras by promoting invasiveness of RasV12-induced tumours in the larval eye disc and brain. Notably, we observed tumour overgrowth and invasion, but these cooperative effects were restricted to cells expressing the pan-glial marker Repo; loss of overexpression in glia, and not in other cell types, completely suppressed oncogenic cooperation. Notably, SRF is strongly enriched in glia providing an explanation for why glia were specifically affected. Moreover, overexpression of mal, a cofactor for SRF, or chickadee, Drosophila profilin, also cooperated strongly with oncogenic Ras to drive glial invasion. Taken together, our findings provide experimental evidence for the role of MRL proteins in the hyperproliferation and transformation of glial tumours in vivo. Furthermore, Profilin and downstream SRF signalling predominantly drive this process rather than other MRL-interactors, Ena/VASP and Scar/WAVE, as is the case in other contexts.
Results
Pico cooperates with oncogenic Ras to promote tumour dissemination
Oncogenic mutations in Ras are frequent events in early stages of cancer development, driving proliferative overgrowth and contributing to tumour formation. The Ras pathway also modulates cytoskeleton organization, cell motility and expression of metastasis signature genes,19 but cooperation between oncogenic Ras and its downstream targets are poorly understood. To test the interaction between Pico and Ras, we used a cancer model in Drosophila in which genetically defined tumours can be induced in the developing eye disc and brain.20, 21 In this model (Figure 1a), expression of the Flp gene, under the control of the eyeless promoter (eyFLP), is used to irreversibly switch on constitutive, GAL4-mediated expression of upstream activator element (UAS) target genes in the developing eye. This is achieved by FLP-mediated recombination between two Flp recombination target (FRT) sites flanking a linker cassette that otherwise silences the Actin-GAL4 (ActGAL4) driver. Once induced, GAL4 binds to and drives the expression of UAS-containing transgenes.
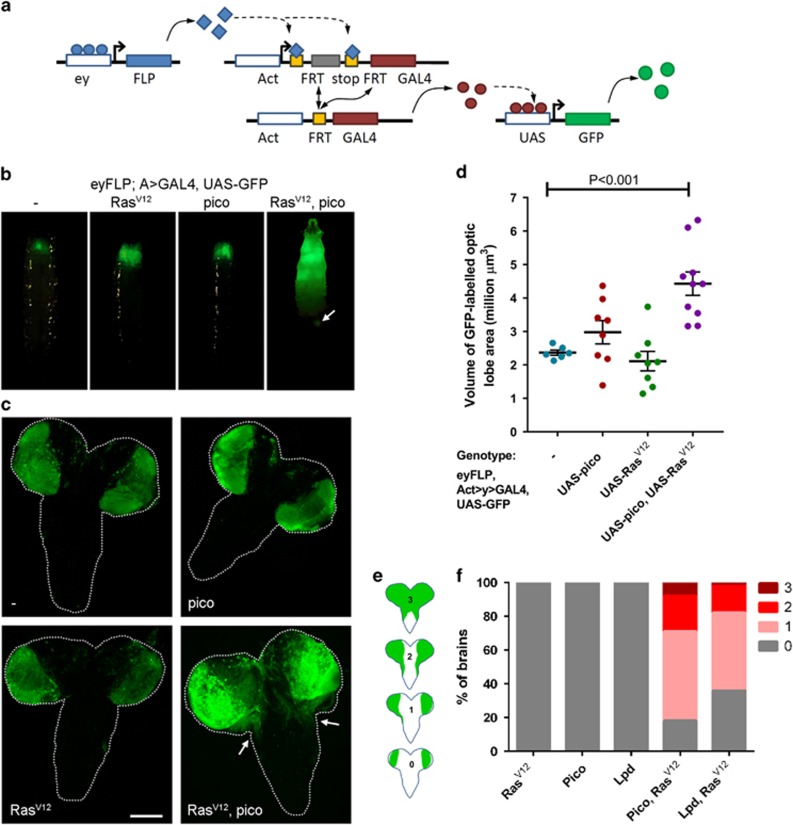
Figure 1.
Pico promotes spreading of RasV12-induced tumours. (a) Schematic outlining heritable overexpression of UAS transgenes following expression of eyFLP and removal of an FRT-flanked linker from Act>GAL4 reconstituting the Act-GAL4 driver. This driver then constitutively drives expression of UAS-GFP and other UAS constructs in daughter cells. (b) Images of whole larvae showing distribution of GFP expression induced in the eye-discs and optic lobes of larva of different genotypes, as indicated. Expression of GFP alone or together with the transgenes indicated was driven by flipping-out an FRT-flanked linker from an Act>GAL4 element using eyFLP (eyFLP, Act>GAL4). Overexpression of RasV12 with pico resulted in a dramatic increase in GFP-marked tissue sometimes leading to the formation of GFP foci at more distant sites (arrow). (c) Distribution of GFP expression in dissected brains showing overgrowth of the optic lobe and invasion of GFP-labelled cells into the VNC in RasV12, pico brains (VNC, arrows). Scale bar 100 μm. (d) Quantification of the volume of GFP-labelled cells in the optic lobes of the indicated genotypes, based on optical sections taken throughout the entire brain. Mean value of individual data points±s.e., is indicated. (e, f) Quantification of the invasion phenotype. (e) Individual cephalic complexes were assigned to one of four categories, depicted, based on the degree of VNC invasion observed: Type 0, no invasion of the VNC, Type I, tumour cell invasion occurring down one side of the VNC, Type II, tumour cells invading both sides of the VNC; and, Type III, significant tumour cell invasion of the VNC combined with overgrowth/fusion of the optic lobes. (f) Stacked bar chart showing the percentage of brains expressing either RasV12, pico, hLpd, pico/RasV12 or hLpd/RasV12, classified into each of the four categories (n=100 brains/genotype).
Using this approach, we examined the effect of overexpressing pico or RasV12 alone or together in GFP-labelled cells in the eye imaginal discs and optic lobes of wandering third instar larvae. Notably, coexpression of pico and RasV12 led to an accumulation of GFP-labelled cells and redistribution to more distant sites. This effect was not observed when either gene was overexpressed in isolation (Figure 1b). To quantify the effects on tissue overgrowth we captured images of optical sections through brains dissected from the different genotypes and measured the volume occupied by GFP-labelled cells. There was no significant difference in volume of GFP-labelled cells expressing pico or RasV12 alone compared to controls (Figures 1c and d). In contrast, pico and RasV12 co-overexpression resulted in a 1.9-fold increase in volume of GFP-labelled cells in the optic lobes compared to GFP alone controls, P<0.001 (Figure 1d).
Inspection of the distribution of GFP-labelled cells in the brain revealed that GFP-labelled pico/RasV12 tumour cells had invaded into the ventral nerve cord (VNC) in the majority (82/100) of cases, whereas cells expressing pico or RasV12 alone never extended beyond the optic lobe (Figure 1c). To quantitate the tumour cell invasion phenotypes produced for each of the genotypes, brains were assigned to one of four categories based on the degree of VNC invasion observed: Type 0, no invasion of the VNC; Type I, tumour cell invasion occurring down one side of the VNC only; Type II, tumour cells invading both sides of the VNC; Type III, significant tumour cell invasion of the VNC combined with fusion of the optic lobes (Figure 1e). Cephalic complexes dissected from animals expressing pico and RasV12 were entirely composed of Type 0 brains, whereas only 18% of RasV12/pico brains were found to exhibit no VNC invasion. Fifty-three percent of RasV12/pico brains were found to have mild Type I invasion, and 21 and 8% of brains were assigned to Type II and Type III categories, respectively (Figure 1f). To test functional conservation, we examined the effect of ectopic overexpression of human Lpd (hLpd) in this system. Brains expressing hLpd showed no evidence of invasion, but, like pico, hLpd was able to drive invasion of RasV12-induced tumours, which occurred in 64/100 of cases (Figure 1f).
We previously showed that pico promotes coordinated growth and proliferation in the wing imaginal discs12 prompting us to wonder whether other promoters of tissue growth could also drive the dissemination of otherwise benign RasV12 tumour cells into neighbouring tissues. To address this, we tested the effects of co-overexpressing Drosophila cyclin-D (cycD) and cyclin-dependent kinase-4 (cdk4) in our assay. There was no significant difference in volume of GFP-labelled cells in optic lobes expressing RasV12 with or without overexpressed cycD and cdk4, and GFP-labelled cells were never located outside of the eye-antennal discs/optic lobe region (Supplementary Figure S1). This is in agreement with previous reports that proliferative cues such as cycD and cdk4 do not account for presence of tumour cells in the VNC.21
Invasive Pico/RasV12 tumours are characterized by elevation of Mmp1 and extracellular matrix remodelling
Degradation of the extracellular matrix by matrix metalloproteases (MMPs) is required during tissue remodelling and during the progression of many types of cancer.22, 23 To investigate integrity of the extracellular matrix, we examined the distribution of Laminin, which is a major component both of the basement membrane underpinning the basal side of epithelial cells and of the gliovascular basal lamina of the blood brain barrier. In brains ectopically expressing either pico or RasV12, Laminin staining of the optic lobes was found to be smooth and uninterrupted. In contrast, discontinuous Laminin staining was observed around the optic lobes of RasV12/pico brains, consistent with degradation of the extracellular matrix (Figure 2a). When we examined MMP expression we found that Mmp1 was found to be largely absent in brains overexpressing either pico or RasV12. In contrast, a marked increase in Mmp1 levels was observed in cephalic complexes expressing both pico and RasV12(Figure 2b). Interestingly, Mmp1 expression was not detected in all RasV12, pico cells; Mmp1 staining was mainly observed in the marginal regions of the optic lobes and in the tumour cells that had invaded the VNC (Figure 2b).
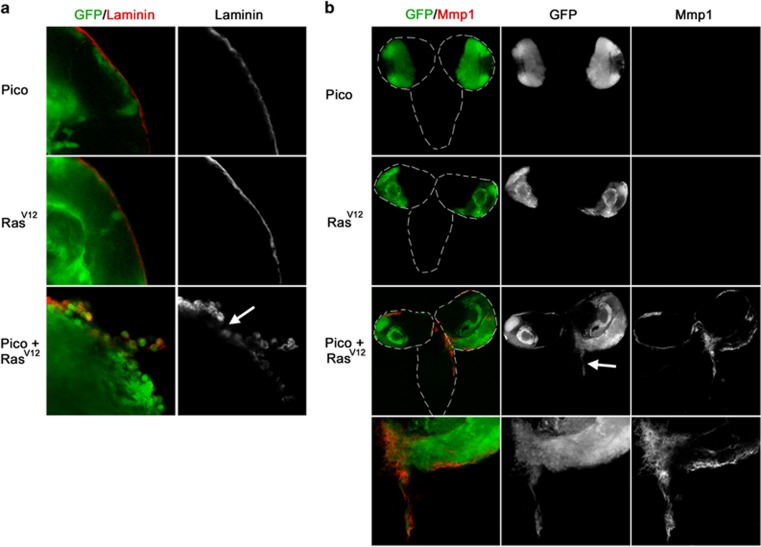
Figure 2.
Brains coexpressing RasV12 and pico display extracellular matrix degradation and ectopic expression of Mmp1. (a) Optic lobes from larvae overexpressing pico, RasV12 or RasV12, pico under the control of eyFLP, Act>GAL4, stained with anti-Laminin antibody, which labels the surface of the optic lobes. Laminin staining was found to be severely interrupted in brains coexpressing RasV12 and pico but not from brains expressing pico or RasV12 alone. (b) Distribution of the metalloproteinase Mmp1. Little or no Mmp1 staining was observed in animals expressing RasV12 or pico alone. In contrast, animals co-overexpressing RasV12 and pico had elevated Mmp1 around the edges of the optic lobes and at sites of invasion into the VNC (arrow).
JNK activation is required for Pico/RasV12-mediated MMP expression and tumour cell spreading
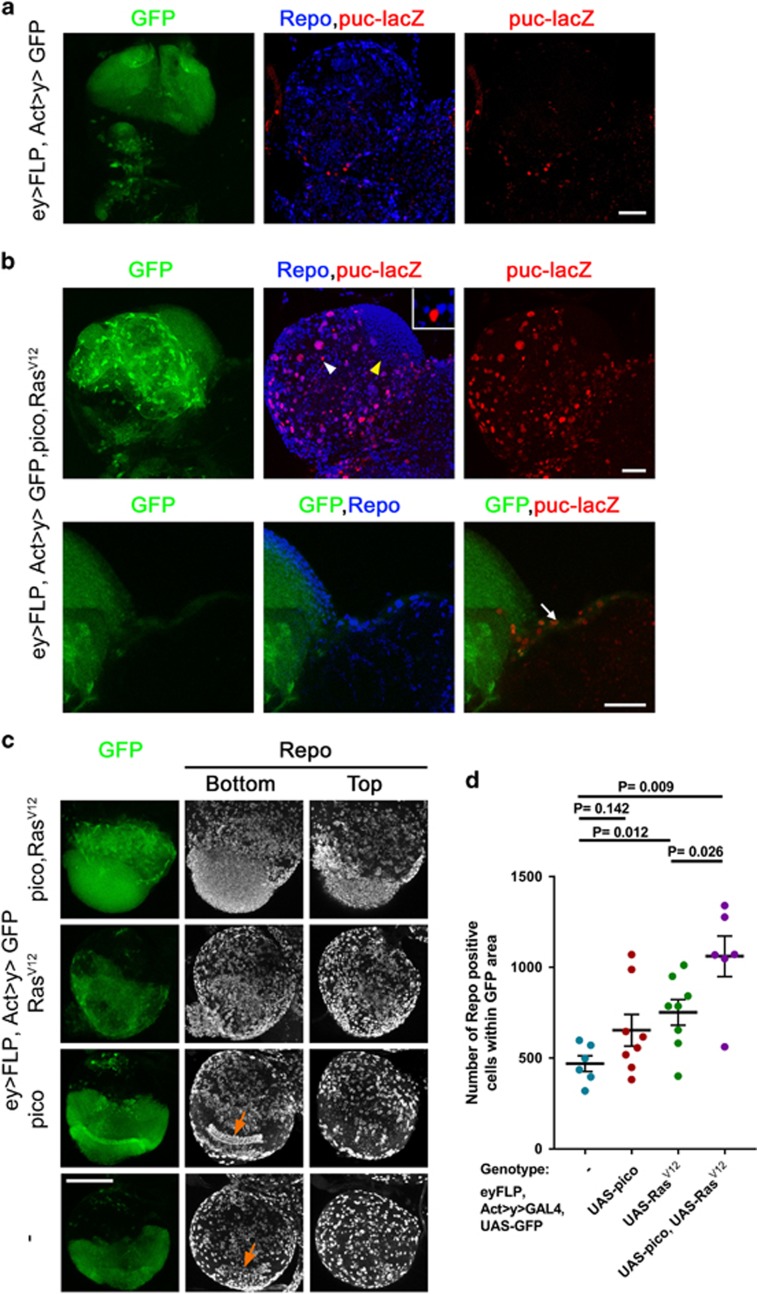
Studies of RasV12 tumours with impaired cell polarity (for example due to mutations in the tumour suppressor gene scrib) have revealed that JNK activation is critical for Mmp1 upregulation and tumour cell invasion of the VNC.24 To assess the state of JNK signalling in RasV12/pico brains, we monitored the levels of puckered, a downstream target of JNK (Martín-Blanco et al.25) using a lacZ enhancer trap (puc-lacZ). We observed limited puc-lacZ staining in brains expressing RasV12 or pico alone, but in RasV12/pico brains we observed a significant increase in the number of puc-lacZ-positive nuclei (P<0.01) indicative of elevated JNK activation in these cells (Figures 3a and b). Not every cell showed puc-lacZ-positive nuclei indicating that JNK activation was not a necessary outcome of RasV12/pico overexpression (Figure 3a).
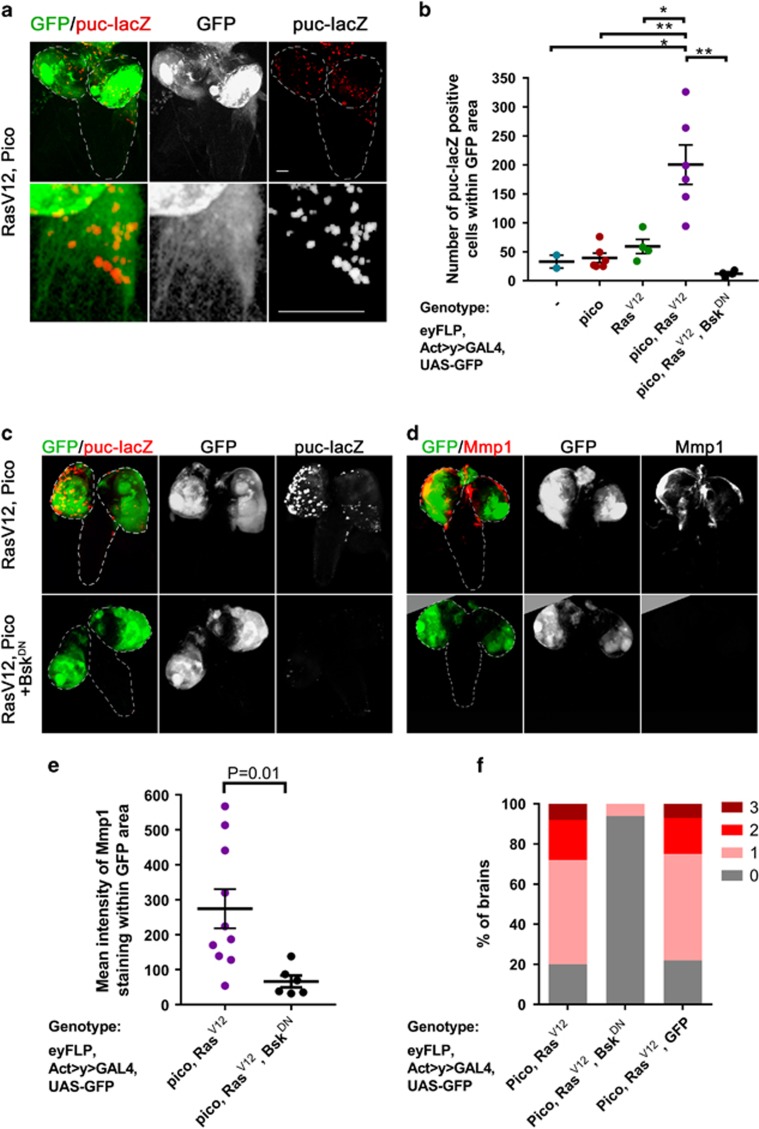
Figure 3.
Mmp1 accumulation and invasion into VNC is dependent on JNK activation. (a) Co-overexpression of RasV12 and pico in GFP-labelled tumours (green) leads to JNK activation in some tumour cells, based on β-galactosidase staining to detect puc-lacZ (red). Top panels show low magnification images of brains, lower panels show magnified images of tumours invading into VNC, which are enriched in puc-lacZ staining. Scale bars, 100 μm. (b) Quantitation of number of puc-lacZ positive foci in GFP-labelled areas of the optic lobes from the indicated genotypes. Mean value of individual data points±s.e., is indicated. (b, c) Blockade of JNK activation with dominant-negative Bsk/JNK (BskDN) suppresses activation of the JNK pathway. (c) Representative images showing puc-lacZ induction in RasV12 pico tumours and suppression of this effect by BskDN: the top row of images were taken from representative RasV12 pico larval brains; the bottom row were taken from siblings coexpressing BskDN. (d, e) Blockade of JNK activation with dominant-negative Bsk/JNK (BskDN) suppresses the induction of Mmp1. (d) Representative images of RasV12 pico tumours with or without BskDN. (e) Quantitation of mean intensity of Mmp1 within GFP-labelled RasV12 pico tumours in the presence or absence of BskDN. Mean value of individual data points±s.e., is indicated. (f) BskDN suppresses RasV12/pico-mediated invasion into the VNC, whereas an additional ‘inert’ UAS element (UAS-GFP) does not. Graph summarizing extent of invasion in the different genotypes (n=100 brains of each type) according to the scale introduced in Figure 1, with 3 being the most severe and 0 corresponding to no invasion.
To determine the requirement for JNK signalling in RasV12/pico-mediated metastasis we tested the effect of coexpressing a dominant-negative form of the Drosophila JNK, encoded by basket (bskDN). Ectopic overexpression of bskDN strongly suppressed JNK activation as monitored with puc-lacZ (Figures 3b and c). Strikingly, overexpression of bskDN also reduced Mmp1 levels 4.1-fold (P<0.01) in GFP-labelled tumour cells (Figures 3d and e) and also almost completely blocked RasV12/pico-mediated tumour cell invasion of the VNC (Figure 3f). In the absence of bskDN, evidence of spreading was observed in 80/100 cases of RasV12/pico tumours, whereas in siblings coexpressing bskDN, invasion was only evident in 6/100 cases (Fisher’s exact test, P<0.0001). Using our scale of the extent of invasion (Figure 1d) the average stage score of invasion in RasV12/pico larvae was 1.16±0.08 (mean ±s.e.m.), but this was significantly reduced by coexpression of bskDN to 0.06 ±0.02, (Student’s t-test, P<0.001). The effect of bskDN was not due to titration of GAL4 in these experiments because substitution of the UAS-bskDN element with UAS-GFP restored the invasive capability of RasV12/pico (Figure 3f). Taken together, these data indicate that pico cooperates with oncogenic Ras to promote JNK activation, and that JNK activation is essential for invasion of RasV12/pico tumours.
TNF-mediated immune response is not required for pico/RasV12-mediated invasion
Accumulating evidence suggests that diversion of host immunity can contribute to the acquisition of invasive behaviour. In Drosophila, inflammatory responses, mediated by Eiger/TNF-producing haemocytes, trigger JNK activation leading to invasive behaviour of RasV12-induced tumours26 (Figure 4a). The mechanism of haemocyte recruitment to tumours is not well understood. To test the involvement of the immune response in RasV12/pico brains, we examined whether there was accumulation of haemocytes at sites of tumour invasion. Although we observed haemocyte recruitment to a proportion of RasV12/pico brains, haemocyte number was not correlated with presence or severity of cellular invasion; some invasive tumours lacked associated haemocytes (Figure 4b). To test whether pico-mediated metastasis is driven by diversion of the host immune response mediated by TNF/eiger, we tested the effect of RasV12/pico overexpression in an eiger null genetic background. Loss of eiger modestly suppressed invasion in RasV12/pico brains (Figure 4c); whereas 80/100 RasV12/pico animals showed GFP-labelled cells in the VNC, invasion was observed in 67/100 RasV12/pico animals lacking eiger (eiger3/eiger3), which was at the borderline of significance (Fisher’s exact test, P=0.054). However, there was little effect on the average stage of invasion, which reduced from 1.16±0.08 to 1.14±0.10 (Student’s t-test, P=0.88) when eiger was absent. Taken together this indicates that pico does not primarily promote invasive behaviour through diversion of a TNF-mediated immune response.
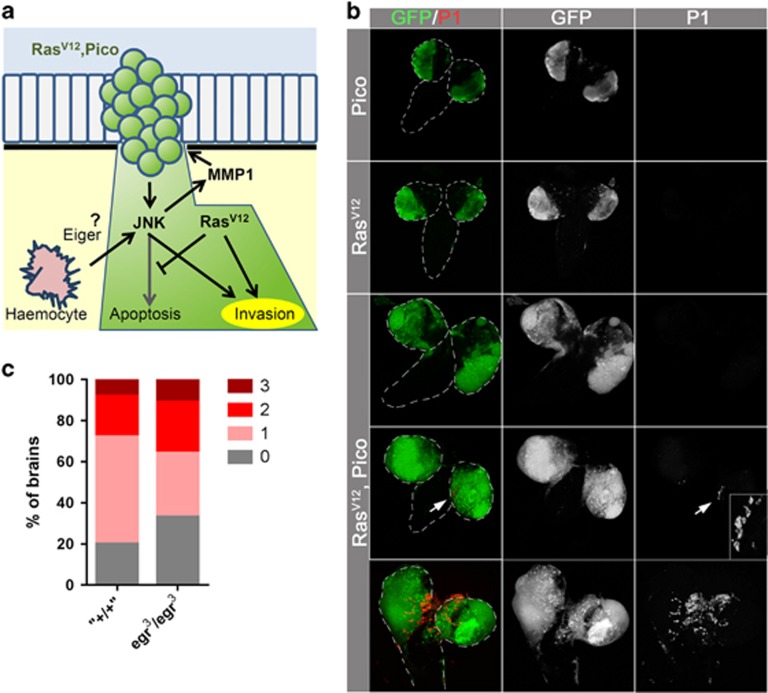
Figure 4.
Invasion is not driven by haemocyte recruitment and TNF/Eiger-dependent signalling. (a) Possible model of extrinsic signalling from haemocytes to JNK activation in tumours. In the absence of RasV12, activated JNK leads to cell death, whereas in its presence, cell death is suppressed and JNK promotes Mmp1 expression. Consequently, JNK and RasV12 cooperate to drive tumour cell invasion. TNF/Eiger-secreting haemocytes that are recruited to sites of certain primary tumours, for example scrib−/− RasV12, have been reported to be capable of providing extrinsic cues that trigger JNK activation, raising the possibility this is also the case for pico RasV12 tumours. (b) Images of dissected brains showing distribution of haemocytes, as detected with an anti-NimC1 P1 antibody. Haemocytes were not detected in brains expressing pico or RasV12 alone. In pico or RasV12 brains, haemocytes were sometimes observed at sites of invasion, but this was not a necessary outcome. (c) Graph summarizing extent of invasion in the different genotypes (n=100 brains of each type).
Pico cooperates with activated Ras to drive distinct oncogenic outcomes in glia
When we looked at distribution of JNK activation more closely in RasV12 pico brains it was apparent that many puc-lacZ positive cells decorated the surface of the optic lobes. This non-random distribution made us wonder whether JNK activation was restricted to specific cell types. Indeed, we found the combination of eyFLP with AyGAL4 was capable of driving expression in a range of cells in the larval optic lobes including glia (Figure 5a), consistent with previous reports showing expression in neuroblasts, lamina and medulla neurons, neurophils and medulla cortex glia.27 When we looked at the distribution of neuronal and glial markers in RasV12 pico tumours we found that 96±3% of puc-lacZ positive cells in GFP-labelled tumours also stained for the pan-glial marker Repo (n=4). GFP-labelled cells invading into the VNC were of this type (Figure 5b, arrow); puc-lacZ staining in these cells is consistent with our genetic data indicating a requirement for JNK to mediate Mmp1 expression and extracellular matrix breakdown (see Figure 3). Although rare, we did observe a few Repo-negative GFP-labelled cells with puc-lacZ staining, although interestingly these were typically juxtaposed directly next to Repo-positive glial cells (Figure 5b, white arrowhead and inset, see Discussion). We also observed a distinct Repo-positive population consisting of many small cells that were puc-lacZ negative, located in a region of the optic lobe that had appeared to have overproliferated (Figure 5b, yellow arrowhead; Figure 5c). When we counted the number of Repo-positive cells in GFP-labelled tumours within the optic lobes (Figures 5c and d), we found that RasV12 overexpression led to a 1.6-fold increase in glial number in GFP-labelled regions compared to GFP only controls (P<0.01). The increase in glial number was not matched by a significant increase in GFP-labelled tumour volume (Figure 1), most likely because many RasV12-overexpressing cells were small,28 suggesting RasV12-expressing glia move more quickly through the cell cycle without an accompanying increase in mass. Co-overexpression of pico significantly enhanced the effect of RasV12 (Figure 5d), leading to a 2.3-fold increase in glial number compared to GFP alone controls (P<0.05). Taken together, the data above indicate that ectopic RasV12 and pico cooperate to promote overproliferation of one glial cell population in the developing optic lobe without the activation of JNK, while promoting JNK activation and cell invasion in another glial population.
Figure 5.
Distinct oncogenic effects in glial populations. (a, b) Optic lobes from third instar larvae, orientated with the VNC to the right of each image. GFP-expressing regions are in green, glia are marked with Repo in blue and JNK activation is marked with puc-lacZ in red. (a) GFP-only expressing control showing that reconstitution of Actin-GAL4 after flipping out an FRT-flanked linker with eyFLP drives GFP expression heritably in glial lineages marked with Repo, as well as other lineages. Some cells are also labelled with puc-lacZ, although the majority of these lie outside the GFP-labelled area. (b) Coexpression of RasV12 and pico results in two distinct effects observable in Repo +ve glia (top panels): activation of JNK, marked with puc-lacZ expression, and accumulation of glial cells in a region proximal to the VNC (yellow arrowhead), largely lacking puc-lacZ expression. Inset is a magnified image of a Repo –ve cell staining positive for puc-lacZ (white arrowhead). Magnified images (bottom) show a GFP and puc-lacZ labelled population that has invaded into the VNC (arrow). Scale bars, 50 μm. (c) Anti-Repo staining showing the effect of RasV12 and pico co-overexpression on glial distribution and number in optic lobes. Repo-stained images are 2D projections of confocal z-stacks from the bottom and top of the same optic lobe. Bottom sections reveal stereotypical arrangement of glia (arrows) in control optic lobes and those overexpressing pico, which is lost upon expression of RasV12. Scale bar, 100 μm. (d) Graph showing quantification of number of Repo-positive glia in GFP-labelled areas of the optic lobes from the indicated genotypes. Mean value of individual data points±SE is indicated.
Overexpression of RasV12 Pico in glia is necessary for an increase in tumour volume and cell invasion
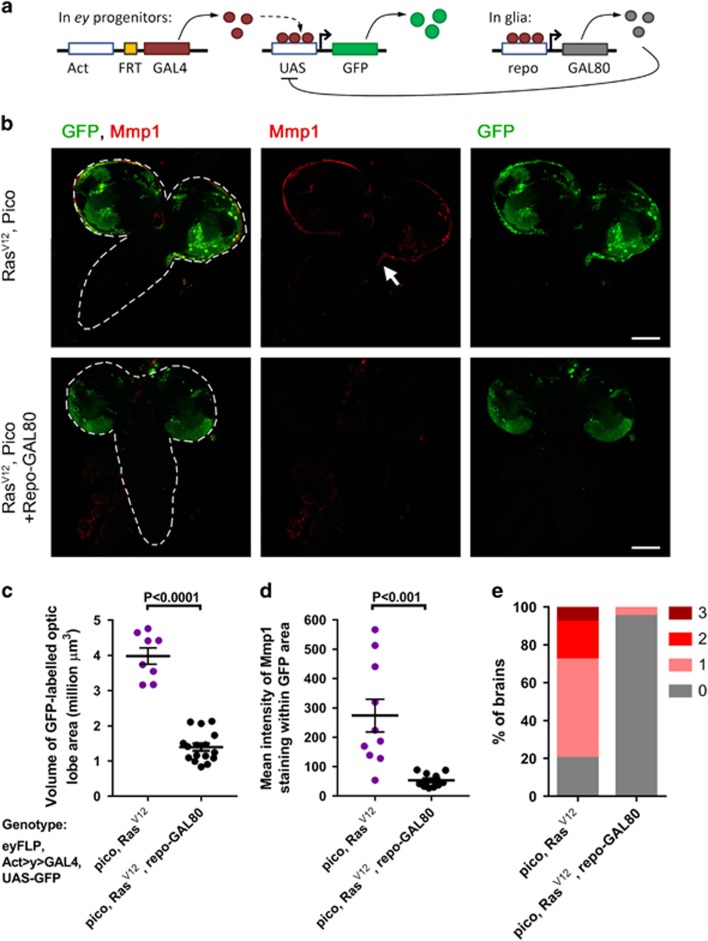
To test if the tumour overgrowth and invasion phenotypes we had observed in the optic lobe were due to ectopic expression of RasV12 and pico in eyFLP-expressing Repo+ glia, we repeated our experiments in a repo-GAL80 background to block GAL4-mediated expression specifically in repo-positive glia but not in other cell types (Figure 6a). When we measured the volume of GFP-labelled tumours in RasV12/pico optic lobes from animals with (n=8) or without repo-GAL80 (n=16), we found that repo-GAL80 reduced the mean tumour volume 2.9-fold (P<0.001; Figure 6c). The mean intensity of Mmp1 staining in GFP-labelled tumours was also reduced 5.1-fold (P<0.001; Figure 6d). Correspondingly, there was a significant reduction in the instances of invasion into the VNC to 5/100 of cases (Fisher’s exact test, P<0.0001) and a corresponding reduction in the average stage score of invasion to 0.05±0.02 (P<0.0001; Figure 6e). As an additional test, we further validated these findings by using a more restricted eyeless-driven FLPase, ey(3.5)FLP, which does not drive substantive expression in the optic lobes of the brain,29 (Supplementary Figure S2). Overexpression of RasV12/pico with ey(3.5)FLP did not replicate the growth and invasion phenotypes observed with eyFLP, consistent with our observations that overexpression in glia was required (Supplementary Figure S2). Expression of RasV12 specifically in GFP-labelled glia with repo-GAL4 was pupal lethal but led to overgrowth and extension of the larval VNC (mean VNC length 132% of control, Student’s t-test P<0.05, n=10). Coexpression of RasV12 with pico led to lethality at the wandering larval stages and extension of the VNC was significantly enhanced (to 187% of control, P<0.01, n=9), again consistent with a cooperative interaction in glia (Supplementary Figure S3). As part of the ‘Ras signature’, Lpd is implicated to act downstream of oncogenic Ras in human breast epithelial cells prompting us to test this possibility in our system. pico knockdown by RNAi did not significantly modify the extension of larval VNC exhibited in brains overexpressing RasV12 (P>0.05, Supplementary Figure S3), so we conclude that pico is not limiting for RasV12 in this context.
Figure 6.
Transcriptional blockade in glia blocks cooperation between RasV12 and pico. (a) Schematic showing suppression of GAL4-mediated overexpression in glia using repo-GAL80. (b) Brains (outlined with dashed line in first set of panels) overexpressing RasV12, pico with or without repo-GAL80, showing distribution of GFP-labelled cells (green) and Mmp1 (red). Animals co-overexpressing RasV12, pico displayed invasion of Mmp1-expressing cells into the VNC (arrow). Mmp1 staining and invasion were suppressed in siblings containing repo-GAL80; optic lobes of these animals were also reduced in size. Scale bars, 100 μm. (c) Measurements of the volume of GFP-labelled RasV12, pico optic lobe tumours with or without repo-GAL80. (d) Measurements of mean intensity of Mmp1 staining in RasV12, pico tumours with or without repo-GAL80. (c, d) Mean value of individual data points±SE is indicated. (e) Stacked bar chart summarizing extent of invasion in the different genotypes (n=100 brains of each type).
SRF is enriched in larval glia in the CNS
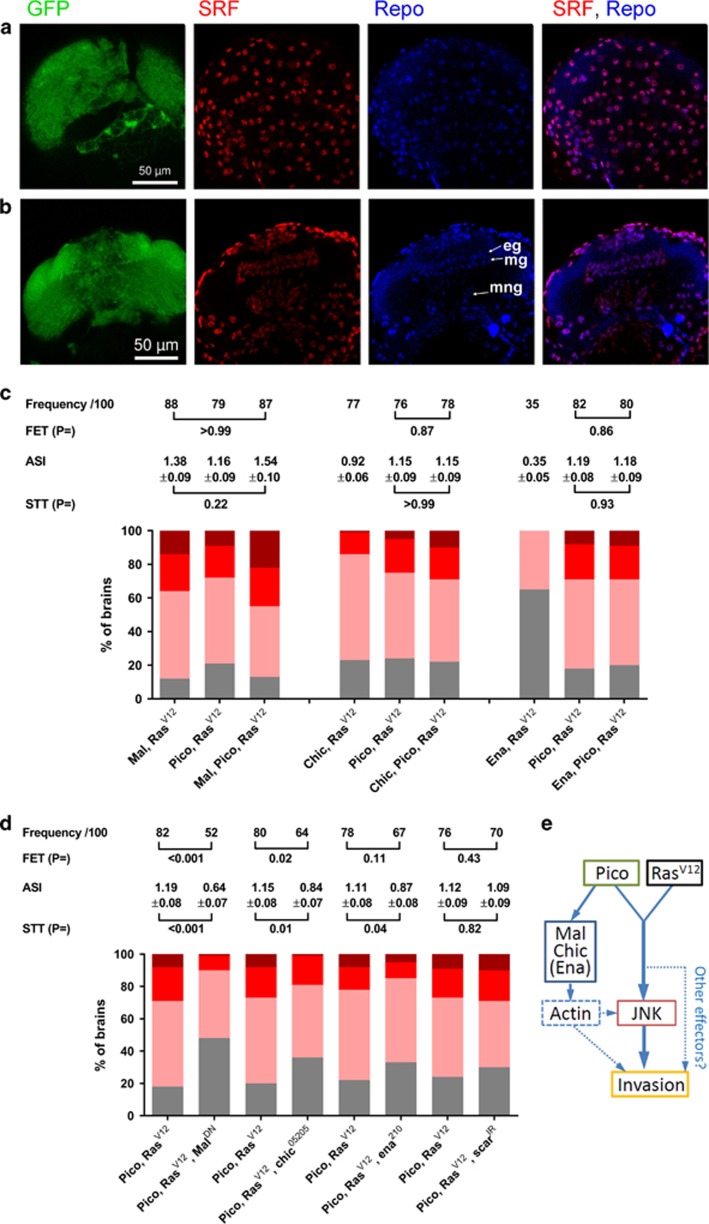
Why should glia be particularly sensitive to coexpression of ectopic pico and RasV12? We recently demonstrated that overexpression of pico reduces the ratio of G:F-actin and is capable of inducing activation of SRF signalling in vitro.12 This prompted us to question whether the cooperation between pico and RasV12 was mediated by SRF signalling. Although SRF is expressed throughout the adult brain,30 where it plays roles in sleep and visual memory,30, 31 we wondered whether SRF expression is spatially regulated in the CNS earlier in development, as it is in other tissues such as the wing imaginal disc (data not shown). Notably, we detected strong anti-SRF antibody staining in glia from third instar optic lobes (Figures 7a and b). SRF staining was evident both in Repo positive surface glia (Figure 7a) and in other glial types (Figure 7b).
Figure 7.
Mal and Chic are rate limiting for tumour dissemination. (a, b) Distribution of SRF in the third instar larval optic lobes. SRF antibody staining (red) overlaps that of Repo (blue) both at the surface (a) of the optic lobes and in cross-section (b). In cross-section, staining in epithelial (eg), marginal (mg) and medulla neurophil (mng) glia is evident.(c) Cooperation between RasV12 and cytoskeletal regulators results in cancer cell invasion into the VNC. Stacked bar charts summarizing extent of invasion in the different genotypes (n=100 brains of each type) according to the scale introduced in Figure 1, with 3 being the most severe and 0 corresponding to no invasion. Shown above each group of charts is a summary of statistical tests for key pairwise combinations, as indicated, in each experiment. Frequency represents number of brains showing invasion/total number (100 in each case); FET, Fisher’s exact test; ASI, average stage score of invasion; STT, Student’s t-test. (d) malDN and chic loss of function partially suppress the effect of RasV12 pico co-overexpression. ena and scar loss of function have reduced or no effect, respectively. Results are grouped by sibling pairs (overexpressed pico RasV12±genetic modifier) and are displayed as in (c). (e) Model of cooperation between Pico and RasV12 co-overexpression. Pico and RasV12 cooperate to activate JNK, which is necessary for invasion of glia from the optic lobes into the VNC. Mal, Chic, and to a lesser extent Ena, are rate limiting for invasion. These regulators may contribute directly or indirectly to invasion via changes in the actin cytoskeleton (see Discussion).
The ability of Pico to promote tumour invasion is shared by selected actin regulatory genes
To further examine the contribution of actin dynamics and SRF to the development of invasive cell behaviours, we tested the effect of co-overexpressing oncogenic RasV12 together with Profilin/chic, which has multiple roles in the augmentation of F-actin dynamics, or with regulatory proteins that bind Pico and are known to control actin polymerization by affecting the number of free barbed ends: Enabled/ena (anti-capping factor) and SCAR (actin nucleation).32 We also tested the effect of ectopic mal, which encodes a cofactor for SRF and responds to changes in actin dynamics to induce SRF-dependent gene expression. Overexpression of any of the above factors alone in the absence of RasV12 did not induce invasive behaviour as determined by the lack of GFP-positive cells in the VNC. However, overexpression of mal, chic, ena or scar was sufficient to promote the acquisition of invasive behaviour in otherwise benign RasV12-expressing tumours (Figure 7c). Based on the percentage number of larvae showing GFP-labelled cells in the VNC, these overexpression constructs can be ranked according to their invasive potential in this system, as follows: mal (88%)> pico (79%)> chic (77%)> ena (35%)> scar (25%), where percentage of larvae with invasion into the VNC are shown in parenthesis (n=100 in each case). This is also in agreement with the average stage of invasion for RasV12-induced tumours coexpressing these regulators: mal (1.38±0.09)> pico (1.16±0.09)> chic (0.92±0.06)> ena (0.35±0.05)> scar (0.25±0.04). When we tested the effect of coexpressing RasV12 and pico together with either mal, chic or ena we found the degree of invasion observed was not significantly increased compared to the effect of pairwise combinations of these inducers or pico alone with RasV12 (Figure 7c). The lack of an additive effect suggests that these proteins may act in the same pathway to induce invasion, albeit to different extents.
MRL proteins interact directly with Profilin, Ena/VASP and the Scar/Wave complex via a number of proline-rich regions present in their C-terminal regions.9, 10, 11, 12, 33 To test whether these regions of Pico might be necessary for promoting invasion of RasV12 tumours, we expressed a truncated version of pico encoding only its central RA-PH domain (picoRA-PH). picoRA-PH failed to promote cell invasion into the VNC alone or together with coexpression of RasV12 (0/100 cases of invasion in each case), suggesting that physical interaction between Pico and its downstream effectors are important for cooperation with oncogenic Ras. To explore the requirement of chic, ena and scar for invasion driven by RasV12 pico we combined RasV12 pico with the following loss-of-function alleles or RNAi: chic05205, ena210 and scar RNAi (scarIR, VDRC-21908). Notably, chic dominantly suppressed the ability of RasV12 pico to drive invasion when one copy of the gene was mutated (Figure 7d), significantly reducing both the number of cases of tumour invasion in siblings (from 80/100 to 64/100 cases; Fisher’s exact test, P=0.02) and the average stage of invasion from 1.15±0.08 to 0.84±0.07 (Student’s t-test, P=0.01). Loss of one copy of ena had a more modest effect; there was not a significant reduction in the number of cases of invasion (P=0.11) but the stage of invasion was significantly reduced (P=0.04). scar knockdown did not significantly suppress invasion of RasV12 pico cells into the VNC (Figure 7d). Notably, the same line of scar RNAi was observed to suppress the effects of pico overexpression on developmentally regulated invasive border cell migration,11 consistent with the idea that there are context-dependent mechanisms by which MRL proteins drive invasion.
Profilin, Ena/VASP and the Scar/Wave complex affect the actin cytoskeleton directly but are also capable of promoting Mal-SRF activity via altered actin dynamics. To assess the likely contribution of direct versus indirect effects on the actin cytoskeleton, we tested the effect of a dominant-negative version of Mal (malDN), which lacks its C-terminal transcription activation domain.34 Compared to RasV12/pico control animals, there was a significant reduction in the number of cases of tumour invasion in siblings coexpressing malDN (82/100 to 52/100 cases, respectively; Fisher’s exact test, P<0.0001). There was also a significant reduction in the average stage of invasion, from 1.19±0.08 to 0.64±0.07 (Student’s t-test, P<0.0001) when malDN was present (Figure 7d). Taken together this indicates that indirect effects via Mal/SRF signalling are rate-limiting for invasion of RasV12/pico tumours (Figure 7e).
Discussion
Here, we find that pico overexpression is capable of promoting distinct oncogenic behaviours in RasV12-induced tumour cells. In particular, we observed an invasive cell population showing elevation of JNK signalling, and a hyperproliferative population lacking JNK activation. These effects were restricted to glia since the affected cell populations labelled positively with the pan-glial marker, Repo, and cooperation between RasV12/pico was lost upon transcriptional repression in glia with repo-GAL80. In glia, JNK is likely to act as a proapoptotic signal as it does in epithelia—indeed, subperineurial glial cells possess a cryptic JNK-dependent apoptotic programme.35 However, any such programme must be suppressed by survival signals from oncogenic Ras as it is in other contexts.36 We found that Mmp1 expression was JNK-dependent, supporting the idea that JNK activation is subverted by tumour cells to promote invasion. Haemocytes were not always observed at sites of invading RasV12/pico tumours, although we cannot rule out that they had not been present prior to the point at which we dissected samples for analysis. Nevertheless, invasion was not significantly affected by complete loss of eiger/TNF, which is a key haemocyte-secreted cytokine capable of eliciting immune responses, including JNK activation, in tumour cells.26 One possibility is that transformed glial cells may be resistant to haemocyte attachment and/or signalling. Examination of the cell type-specific expression pattern of TNF signalling components, such as the recently identified TNF/Eiger receptor Grindelwald,37 may provide a mechanistic explanation for why glia respond differently from epithelial tumour cells to circulating immune cells. Alternatively, transformed glia may express inhibitory cell surface or secreted molecules making them refractory to the innate immune system, as is the case for human glioma cells.38
Interestingly, a small number of Repo negative cells overexpressing RasV12/pico, adjacent to Repo positive RasV12/pico tumour cells, also displayed elevated JNK activity. In addition to roles in CNS development and function, glia are considered to be primary immune cells of the CNS that survey the CNS for neuronal damage, modulating inflammatory responses and engulfing debris or foreign material.39 The JNK pathway mediates glial engulfment activity in Drosophila,40, 41 raising the intriguing possibility that RasV12/pico stimulates glial phagocytosis of tissue damage caused by premalignant tumour cells. Diversion of the glial damage response programme by carcinoma cells has previously been reported in murine organotypic brain slice co-cultures,42 stimulating local invasion in tumours resistant to glial-induced apoptosis. It will therefore be interesting to examine whether this phenomenon is JNK-dependent.
Recent work has shown the actin cytoskeleton acts both upstream and downstream of JNK43, 44, 45 and, conceptually, changes in cell tension resulting from altered actin cytoskeleton may trigger JNK as part of a stress response. We were interested to explore whether actin regulators that associate with Pico could similarly cooperate with RasV12. In breast cancer cells, the ability of Lpd to promote 3D invasion relies on its interactions with both Ena/VASP and Scar/WAVE.15 Although both ena and scar were capable of cooperating with RasV12 in our model, their effect was modest compared to the effect of chic (Drosophila Profilin). This might be because Ena and Scar are not limiting, or it might reflect a specific requirement for Chic, which was also limiting for the effect of pico/RasV12. Interestingly, in this regard, Profilin assists in coordination of actin turnover,46, 47 which is the driving force for membrane protrusion and spreading of some types of glia in the CNS.48 Recent work has also demonstrated that changes in actin dynamics driven by MRL proteins and their binding partners can activate SRF signalling.12, 49 Several lines of evidence suggest that Mal/SRF signalling is important for pico/RasV12 cooperation: first, SRF expression is enriched in glia; second, the effects of overexpression of mal were at least as potent as those of pico; third, malDN suppressed the pico-mediated invasion of RasV12-induced tumours. In mammalian cells the majority of SRF target genes encode cytoskeletal components50 and recent work in Drosophila suggest that actin itself is a key homeostatic target.51 Control of Mal/SRF activity therefore may provide a mechanism by which cytoskeletal gene expression is coordinated with cytoskeletal regulation.
In summary, our data indicate that overexpression of MRL proteins is capable of driving invasion and overproliferation of RasV12-induced glial cell tumours in an in vivo experimental model. Notably, our findings, in glia, implicate Drosophila Profilin and SRF signalling in MRL-mediated tumour dissemination, whereas interactions between Lpd and Ena/VASP and Scar/WAVE have been reported to be critical in the invasion of breast cancer cells. This points to important differences in the mechanism of action of MRL proteins depending on the cellular context. Given that SRF is capable of promoting human glioma cell migration52 and Lpd overexpression has been detected in glioma samples from patients,53 investigation into whether Lpd or SRF levels are associated with disease progression and patient outcome is warranted.
Materials and methods
Fly husbandry and genetics
Flies were reared at 25 °C under standard conditions. All initial Drosophila strains have been previously described. Third instar larvae were examined 6 days after egg laying. Genotypes are provided in a supplementary file.
Immunohistochemistry
Tissues dissected from third instar larvae were fixed and stained as Ciurciu et al.,54 with minor modifications. After fixation for 20 min in 4% (w/v) paraformaldehyde in PBS, dissected brains from third instar larvae were washed in PBS with 0.1% Triton-X (PBST), then blocked for 2 h in PBST with 5% FCS (blocking solution). Primary antibody staining was done overnight at 4°C in blocking solution, washed three times with PBST and incubated with secondary antibody in blocking solution for 2 h at room temperature. After three washes in PBST, brains were mounted in Vectasheild mounting media (Vector laboratories, Peterborough, UK). Primary antibodies were as follows: rabbit anti-Laminin (1:1000); guinea-pig anti-Repo (1:1000); rabbit anti-Repo (1:25,000); mouse anti-NimC1 P155(1:30), mouse anti-Mmp1 (1:1:1 mix of 3A6B4, 3B8D12, 5H7B11 from Developmental Studies Hybridoma Bank, Iowa, USA diluted 1:10); mouse anti-β−gal (Promega, Southampton, UK, 1:100); mouse anti-SRF (Active Motif, La Hulpe, Belgium, 1:100). Secondary antibodies were conjugated to Alexa-Fluor 555 or 633 (Invitrogen, Paisley, UK, 1:500). TO-PRO-3 Iodide (Invitrogen, 1:1000) or DAPI was used to visualize DNA.
Image acquisition and analysis
Dissected tissues were imaged on a Leica MZ10F stereomicroscope for scoring of invasion phenotypes, which was done blinded, or on Zeiss LSM710, 780 or 880 microscopes equipped with 405 nm, 488 nm, 568 nm and 633 nm lasers using either Fluor × 20 or Plan Apochromat × 40/1.3NA oil immersion objective. Confocal images were imported into OMERO56 and adjusted for brightness and contrast uniformly across entire fields where appropriate. Figures were constructed in Adobe Photoshop. Quantitative analysis of raw confocal data was conducted using Imaris version 8.2.0 (Oxford Instruments/Bitplane, Zurich, Switzerland). The GFP channel was segmented into 3D volumes (5 μm surface grain size) by absolute intensity using an automatically selected intensity threshold. To remove small unattached objects, only the two largest volumes were kept per experiment (corresponding in all cases to the optic lobes), and their volume measured. To count the number of Repo or puc-lacZ positive cells, the above volumes were used to mask the relevant intensity channel, which was then subject to spot segmentation using an estimated spot diameter of 5 μm and background subtraction. Spots were subjected to an automatically thresholded intensity filter. All automatic thresholding was visually inspected and adjusted if necessary. For quantitation of Mmp1 staining, stacks were projected in ‘z’ and then background subtracted in the Mmp1 channel. The GFP channel was used to segment, then the selection was measured in the Mmp1 channel. Whole animal micrographs were captured with a Leica ZF10 stereomicroscope or Zeiss Z.1 Lightsheet microscope (see Supplementary Methods for details).
Acknowledgments
We thank the Bloomington Stock Centre, Zhe Han, Julian Ng, Pernille Rorth, Eyal Schejter, Marcos Vidal, Sanjai Patel and the Manchester Fly Facility for Drosophila strains, B Altenhein, I Andó, the Developmental Studies Hybridoma Bank, S Baumgartner and Manzoor Bhat for antibodies, Marco Marcello in the Liverpool Centre for Cell Imaging for assistance with image acquisition, Chris Lofthouse for artwork, and Mirel Lucaci for technical assistance. This work was supported by grants from BBSRC (BB/L014947/1, BB/M012441/1), MRC (MR/K015931/1) and CRUK (C20691/A11834).
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Skau CT, Waterman CM. Specification of architecture and function of actin structures by actin nucleation factors. Annu Rev Biophys 2015; 44: 285–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson RP, Veltman D, Machesky LM. Actin-bundling proteins in cancer progression at a glance. J Cell Sci 2012; 125: 1073–1079. [DOI] [PubMed] [Google Scholar]
- Bravo-Cordero JJ, Hodgson L, Condeelis J. Directed cell invasion and migration during metastasis. Curr Opin Cell Biol 2012; 24: 277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Gautreau A. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat Rev Mol Cell Biol 2014; 15: 577–590. [DOI] [PubMed] [Google Scholar]
- Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol 2010; 11: 353–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse R, Copeland JW, Newsome TP, Way M, Treisman R. A role for VASP in RhoA-Diaphanous signalling to actin dynamics and SRF activity. EMBO J 2003; 22: 3050–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiropoulos A, Gineitis D, Copeland J, Treisman R. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell 1999; 98: 159–169. [DOI] [PubMed] [Google Scholar]
- Vartiainen MK, Guettler S, Larijani B, Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 2007; 316: 1749–1752. [DOI] [PubMed] [Google Scholar]
- Krause M, Leslie JD, Stewart M, Lafuente EM, Valderrama F, Jagannathan R et al. Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev Cell 2004; 7: 571–583. [DOI] [PubMed] [Google Scholar]
- Lafuente EM, van Puijenbroek AA, Krause M, Carman CV, Freeman GJ, Berezovskaya A et al. RIAM, an Ena/VASP and Profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev Cell 2004; 7: 585–595. [DOI] [PubMed] [Google Scholar]
- Law AL, Vehlow A, Kotini M, Dodgson L, Soong D, Theveneau E et al. Lamellipodin and the Scar/WAVE complex cooperate to promote cell migration in vivo. J Cell Biol 2013; 203: 673–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyulcheva E, Taylor E, Michael M, Vehlow A, Tan S, Fletcher A et al. Drosophila pico and its mammalian ortholog lamellipodin activate serum response factor and promote cell proliferation. Dev Cell 2008; 15: 680–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestier C, Cervera N, Finetti P, Esteyries S, Esterni B, Adelaide J et al. Prognosis and gene expression profiling of 20q13-amplified breast cancers. Clin Cancer Res 2006; 12: 4533–4544. [DOI] [PubMed] [Google Scholar]
- Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet 2000; 24: 227–235. [DOI] [PubMed] [Google Scholar]
- Carmona G, Perera U, Gillett C, Naba A, Law AL, Sharma VP et al. Lamellipodin promotes invasive 3D cancer cell migration via regulated interactions with Ena/VASP and SCAR/WAVE. Oncogene 2016; 35: 5155–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2006; 439: 353–357. [DOI] [PubMed] [Google Scholar]
- Bennett D, Lyulcheva E, Cobbe N. Drosophila as a potential model for ocular tumors. Ocul Oncol Pathol 2015; 1: 190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumby AM, Richardson HE. Using Drosophila melanogaster to map human cancer pathways. Nat Rev Cancer 2005; 5: 626–639. [DOI] [PubMed] [Google Scholar]
- Choi C, Helfman DM. The Ras-ERK pathway modulates cytoskeleton organization, cell motility and lung metastasis signature genes in MDA-MB-231 LM2. Oncogene 2014; 33: 3668–3676. [DOI] [PubMed] [Google Scholar]
- Brumby AM, Richardson HE. scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila. EMBO J 2003; 22: 5769–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini RA, Xu T. A genetic screen in Drosophila for metastatic behavior. Science 2003; 302: 1227–1231. [DOI] [PubMed] [Google Scholar]
- Coussens LM, Werb Z. Inflammation and cancer. Nature 2002; 420: 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A, Pastor-Pareja JC, Igaki T, Pagliarini R, Xu T. Basement membrane remodeling is essential for Drosophila disc eversion and tumor invasion. Proc Natl Acad Sci USA 2007; 104: 2721–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlirova M, Bohmann D. JNK- and Fos-regulated Mmp1 expression cooperates with Ras to induce invasive tumors in Drosophila. EMBO J 2006; 25: 5294–5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky AM et al. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev 1998; 12: 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero JB, Macagno JP, Stefanatos RK, Strathdee KE, Cagan RL, Vidal M. Oncogenic Ras diverts a host TNF tumor suppressor activity into tumor promoter. Dev Cell 2010; 18: 999–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotard C, Leung W, Salecker I. glial cells missing and gcm2 cell autonomously regulate both glial and neuronal development in the visual system of Drosophila. Neuron 2005; 48: 237–251. [DOI] [PubMed] [Google Scholar]
- Read RD, Cavenee WK, Furnari FB, Thomas JB. A drosophila model for EGFR-Ras and PI3K-dependent human glioma. PLoS Genet 2009; 5: e1000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazigou E, Apitz H, Johansson J, Loren CE, Hirst EM, Chen PL et al. Anterograde Jelly belly and Alk receptor tyrosine kinase signaling mediates retinal axon targeting in Drosophila. Cell 2007; 128: 961–975. [DOI] [PubMed] [Google Scholar]
- Donlea JM, Ramanan N, Shaw PJ. Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science 2009; 324: 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thran J, Poeck B, Strauss R. Serum response factor-mediated gene regulation in a Drosophila visual working memory. Curr Biol 2013; 23: 1756–1763. [DOI] [PubMed] [Google Scholar]
- Shekhar S, Pernier J, Carlier MF. Regulators of actin filament barbed ends at a glance. J Cell Sci 2016; 129: 1085–1091. [DOI] [PubMed] [Google Scholar]
- Hansen SD, Mullins RD. Lamellipodin promotes actin assembly by clustering Ena/VASP proteins and tethering them to actin filaments. Elife 2015; 4: e06585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Li X, Wu J, Olson EN. A myocardin-related transcription factor regulates activity of serum response factor in Drosophila. Proc Natl Acad Sci USA 2004; 101: 12567–12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohayon D, Pattyn A, Venteo S, Valmier J, Carroll P, Garces A. Zfh1 promotes survival of a peripheral glia subtype by antagonizing a Jun N-terminal kinase-dependent apoptotic pathway. EMBO J 2009; 28: 3228–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhuang Y, Han M, Xu T, Deng K. Ras promotes cell survival by antagonizing both JNK and Hid signals in the Drosophila eye. BMC Dev Biol 2009; 9: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen DS, Colombani J, Palmerini V, Chakrabandhu K, Boone E, Rothlisberger M et al. The Drosophila TNF receptor Grindelwald couples loss of cell polarity and neoplastic growth. Nature 2015; 522: 482–486. [DOI] [PubMed] [Google Scholar]
- Friese MA, Steinle A, Weller M. The innate immune response in the central nervous system and its role in glioma immune surveillance. Onkologie 2004; 27: 487–491. [DOI] [PubMed] [Google Scholar]
- Logan MA, Freeman MR. The scoop on the fly brain: glial engulfment functions in Drosophila. Neuron Glia Biol 2007; 3: 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shklover J, Mishnaevski K, Levy-Adam F, Kurant E. JNK pathway activation is able to synchronize neuronal death and glial phagocytosis in Drosophila. Cell Death Dis 2015; 6: e1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald JM, Doherty J, Hackett R, Freeman MR. The c-Jun kinase signaling cascade promotes glial engulfment activity through activation of draper and phagocytic function. Cell Death Differ 2013; 20: 1140–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang HN, van Rossum D, Sieger D, Siam L, Klemm F, Bleckmann A et al. Carcinoma cells misuse the host tissue damage response to invade the brain. Glia 2013; 61: 1331–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudrapatna VA, Bangi E, Cagan RL. A Jnk-Rho-Actin remodeling positive feedback network directs Src-driven invasion. Oncogene 2014; 33: 2801–2806. [DOI] [PubMed] [Google Scholar]
- Kulshammer E, Uhlirova M. The actin cross-linker Filamin/Cheerio mediates tumor malignancy downstream of JNK signaling. J Cell Sci 2013; 126: 927–938. [DOI] [PubMed] [Google Scholar]
- Fernandez BG, Jezowska B, Janody F. Drosophila actin-capping protein limits JNK activation by the Src proto-oncogene. Oncogene 2014; 33: 2027–2039. [DOI] [PubMed] [Google Scholar]
- Balcer HI, Goodman AL, Rodal AA, Smith E, Kugler J, Heuser JE et al. Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr Biol 2003; 13: 2159–2169. [DOI] [PubMed] [Google Scholar]
- Didry D, Carlier MF, Pantaloni D. Synergy between actin depolymerizing factor/cofilin and profilin in increasing actin filament turnover. J Biol Chem 1998; 273: 25602–25611. [DOI] [PubMed] [Google Scholar]
- Nawaz S, Sanchez P, Schmitt S, Snaidero N, Mitkovski M, Velte C et al. Actin filament turnover drives leading edge growth during myelin sheath formation in the central nervous system. Dev Cell 2015; 34: 139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro EM, Xie Z, Norovich AL, Vidaki M, Tsai LH, Gertler FB. Lpd depletion reveals that SRF specifies radial versus tangential migration of pyramidal neurons. Nat Cell Biol 2011; 13: 989–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Stewart A, Gualdrini F, East P, Horswell S, Matthews N et al. Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev 2014; 28: 943–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvany L, Muller J, Guccione E, Rorth P. The core and conserved role of MAL is homeostatic regulation of actin levels. Genes Dev 2014; 28: 1048–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv-Av A, Taller D, Attia M, Xiang C, Lee HK, Cazacu S et al. RTVP-1 expression is regulated by SRF downstream of protein kinase C and contributes to the effect of SRF on glioma cell migration. Cell Signal 2011; 23: 1936–1943. [DOI] [PubMed] [Google Scholar]
- Petryszak R, Keays M, Tang YA, Fonseca NA, Barrera E, Burdett T et al. Expression Atlas update-an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res 2016; 44: D746–D752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciurciu A, Duncalf L, Jonchere V, Lansdale N, Vasieva O, Glenday P et al. PNUTS/PP1 regulates RNAPII-mediated gene expression and is necessary for developmental growth. PLoS Genet 2013; 9: e1003885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurucz E, Markus R, Zsamboki J, Folkl-Medzihradszky K, Darula Z, Vilmos P et al. Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr Biol 2007; 17: 649–654. [DOI] [PubMed] [Google Scholar]
- Allan C, Burel JM, Moore J, Blackburn C, Linkert M, Loynton S et al. OMERO: flexible, model-driven data management for experimental biology. Nat Methods 2012; 9: 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.