Abstract
Nucleus accumbens-associated protein-1 (NAC1), a nuclear factor of the BTB/POZ gene family, has emerging roles in cancer. In this study, we identified the NAC1-HDAC4-HIF-1α axis as an important pathway in regulating glycolysis and hypoxic adaptation in tumor cells. We show that nuclear NAC1 binds to histone deacetylase type 4 (HDAC4), hindering phosphorylation of HDAC4 at Ser246 and preventing its nuclear export that leads to cytoplasmic degradation of the deacetylase. Accumulation of HDAC4 in the nuclei results in an attenuation of HIF-1α acetylation, enhancing the stabilization and transcriptional activity of HIF-1α and strengthening adaptive response of cells to hypoxia. We also show the role of NAC1 in promoting glycolysis in a mouse xenograft model, and demonstrate that knockdown of NAC1 expression can reinforce the antitumor efficacy of bevacizumab, an inhibitor of angiogenesis. Clinical implication of the NAC1-HDAC4-HIF-1α pathway is suggested by the results showing that expression levels of these proteins are significantly correlative in human tumor specimens and associated with the disease progression. This study not only reveals an important function of NAC1 in regulating glycolysis, but also identifies the NAC1-HDAC4-HIF-1α axis as a novel molecular pathway that promotes survival of hypoxic tumor cells.
Introduction
Hypoxic microenvironment is a common feature of solid tumors, and contributes to tumor progression, therapeutic resistance and poor prognosis.1 Under hypoxia, glycolytic switch occurs, enabling adaptive growth and survival of hypoxic cells. A number of molecular pathways and mechanisms are known to have important roles in regulation of cancer metabolism. For instance, induction of hypoxia-inducible factor-1α (HIF-1α) is critical in promoting glycolysis and hypoxic adaptation.2 Although metabolic reprogramming is now considered as one of the hallmarks of cancer,3 the molecular mechanisms behind this peculiarity remain less clear. Understanding more fully on how tumor cells metabolically adapt to hypoxic microenvironment may help develop new therapeutic intervention.
Nucleus accumbens-associated protein-1 (NAC1), encoded by the NACC1 gene, is a transcription co-repressor belonging to the bric-a-brac Tramtrack Broad complex/pox virus and Zn finger (BTB/POZ) family.4, 5 The conserved BTB protein–protein interaction domain is required for NAC1 homodimerization, which has important roles in various biological processes such as maintenance of stem cell pluripotency6 and pathogenesis of human cancer.4 The implication of NAC1 in cancer was first observed in ovarian cancer. It was found that high expression of NAC1 is closely associated with cancer cell proliferation, migration and tumor recurrence,4, 7, 8 and NAC1 has been appreciated as one of the top potential ‘driver’ genes in high-grade ovarian serous carcinomas.9 Along with others, we have shown that through its transcription-dependent or -independent functions, NAC1 can inactivate the tumor-suppressor Gadd45,10, 11 promote autophagic response,12 disable cellular senescence,13 bind to actin to regulate cancer cell cytokinesis14 and induce expression of fatty acid synthase.15
Prompted by our coincidental observation that under hypoxia, the culture medium of cells with high NAC1 expression turned to be acidic much earlier than that of cells with low NAC1 expression, we sought to explore the role of NAC1 in regulating glycolysis. Herein, we report that NAC1 is a positive regulator of glycolysis and promotes survival of hypoxic cancer cells via stabilizing HIF-1α, a transcription factor that induces the expression levels of glycolytic enzymes, GLUTs and other genes involved in hypoxia adaptation.2 We show that stabilization of HIF-1α by NAC1 is mediated through histone deacetylase type 4 (HDAC4). The physical association of NAC1 with HDAC4 interferes with phosphorylation of HDAC4 at Ser246, preventing its nuclear export. Accumulation of HDAC4 in the nuclei leads to a decrease of acetylation of HIF-1α, increasing the stabilization and transcriptional activity of HIF-1α, promoting glycolysis and survival of hypoxic tumor cells. Further, targeting of NAC1 can enhance the antitumor activity of bevacizumab, an inhibitor of angiogenesis.
Results
Expression of NAC1 promotes glycolysis in hypoxic tumor cells
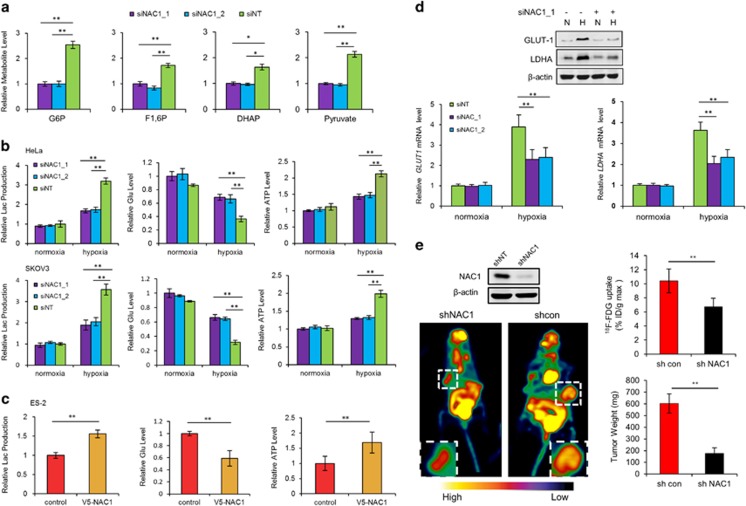
The impetus for this study came from our observation that, when tumor cells were cultured under hypoxic condition, the medium turned to be acidic (yellowish) much slower and later in the dishes containing the cancer cells subjected to silencing of NAC1 expression than in the dishes containing the control cells (Supplementary Figure S1A). To assess whether NAC1 expression affects glycolysis, we measured and compared the glycolytic intermediates in the HeLa cells with or without silencing of NAC1 expression following incubation in 1% O2 for 24 h. Figure 1a shows that as compared with the control cells transfected with a non-targeting RNA, the amounts of glucose 6-phosphate, fructose 1,6-bisphosphate, dihydroxyacetone phosphate and pyruvate, were significantly decreased in the cells transfected with the NAC1-targeted small interfering RNAs (siRNAs). Also, HeLa and SKOV3 cells with silencing of NAC1 expression showed decreased production of lactate, consumption of glucose and generation of ATP under hypoxia but not under normoxia (Figure 1b). Two NAC1-targeted siRNA sequences were tested and similar results were obtained (Supplementary Figure S2A). In addition, similar observation were made in the SV40-immortalized NAC1+/+ and NAC1−/− mouse embryo fibroblasts (Supplementary Figure S1B). By contrast, introduction of NAC1-V5, an expression vector containing the entire coding sequence of NAC1, into the ES-2 ovarian cancer cells (with low intrinsic NAC1 expression)16 (Supplementary Figure S2B) led to increases in lactate production, glucose consumption and cellular ATP under hypoxia (Figure 1c). Moreover, silencing of NAC1 expression in HeLa cells showed remarkable inhibitory effects on the hypoxia-induced expression levels of glucose transporter 1 (GLUT1) and lactate dehydrogenase-A, two important glycolytic genes (Figure 1d). Under hypoxia, the oxygen consumption rate (an indicative of OXPHOS), as measured by an extracellular metabolic flux analyzer, significantly increased in HeLa or SKOV3 cells subjected to silencing of NAC1 expression (Supplementary Figure S4), further suggesting a role for NAC1 in promoting glycolysis in hypoxic cells. To evaluate whether NAC1-mediated glycolysis is operative in vivo, we inoculated mice with the HeLa cells expressing either a control short hairpin RNA (shRNA) or NAC1-targeted shRNA, and then examined tumor growth and glucose uptake. Figure 1e shows that 18F-FDG uptake as analyzed by Micro PET, and tumor weight, were significantly decreased in the tumors with depletion of NAC1, as compared with that in the control tumors without depletion of NAC1. In addition, we observed that silencing of NAC1 expression resulted in a decrease of cell proliferation (Supplementary Figure S3A) and an increase of apoptotic cell death under hypoxia (Supplementary Figure S3B). These observations suggest an important role for NAC1 in promoting glycolysis and survival of hypoxic cells.
Figure 1.
Expression of NAC1 promotes glycolysis in hypoxic tumor cells. (a) HeLa cells were transfected with a non-targeting RNA (siNT) or NAC1-targeted siRNA, and exposed to hypoxia (1% O2) for 24 h. Metabolic intermediates were measured using liquid chromatography-mass spectrometry (LC-MS), and normalized to cell numbers. (b) HeLa or SKOV3 cells were transfected with a siNT or NAC1 siRNA, and then incubated under normoxia (20% O2) or hypoxia (1% O2) for 24 h. Lactate and glucose in the culture medium, and cellular ATP, were measured and normalized to cell numbers. (c) ES-2 cells were transfected with an empty vector or V5-NAC1 expression vector. Twelve hours later, cells were cultured under hypoxia for 24 h. Lactate and glucose in the culture medium, and cellular ATP, were measured and normalized to cell numbers. (d) HeLa cells were transfected with a siNT or NAC1 siRNA, followed by incubation under normoxia or hypoxia for 24 h. mRNAs of lactate dehydrogenase-A (LDHA) and GLUT1 were determined by quantitative reverse transcriptase (RT)–PCR and plotted after normalization. LDHA and GLUT1 protein were examined by western blot, with β-actin as a loading control. Bars are mean ±s.d. (n=3). (e) Glucose uptake and tumor growth in mice inoculated with Hela cells expressing a control shRNA or NAC1-targeted shRNA. 18F-FDG uptake was analyzed by Micro PET. White squares point to tumors. Data shown are mean±s.d. (n=5).
NAC1 stabilizes HIF-1α protein in hypoxic tumor cells
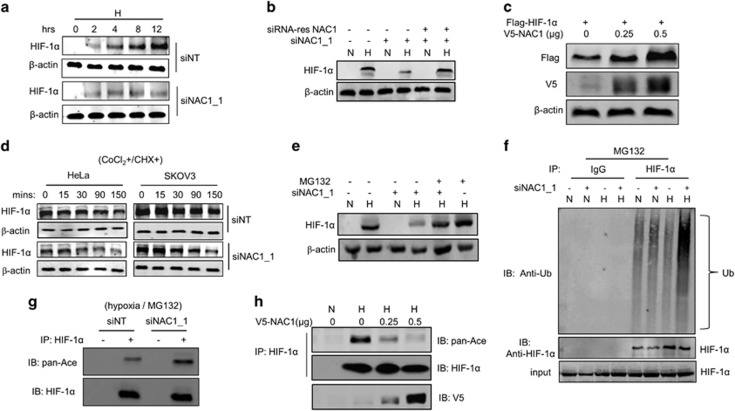
As the glycolytic genes lactate dehydrogenase-A and GLUT1 are the important transcriptional targets of HIF-1α, and their expression levels were found to be affected by NAC1 (Figure 1d) and HIF-1α re-expression could partly rescue lactate dehydrogenase-A and GLUT1 expression in the NAC1 knockdown cells (Supplementary Figure S6B), we queried whether the effect of NAC1 on glycolysis is mediated through HIF-1α, a transcription factor critical in cellular adaptation to hypoxia. As similar effects on glycolysis were observed with the two NAC1-targeted siRNA sequences, we thus used the NAC1 siRNA_1 in the subsequent experiments. In quantitative reverse transcriptase–PCR analyses, we did not detect any significant difference in the expression of HIF-1α mRNA between the control cells and the cells with NAC1 knockdown (Supplementary Figure S5A); however, silencing of NAC1 expression suppressed the induction of HIF-1α expression by hypoxia (Figure 2a). To validate the effect of the NAC1 siRNA, we generated a siRNA-resistant NAC1 expression plasmid. As shown in Figure 2b, expression of this siRNA-resistant NAC1 rescued the RNA interference-mediated reduction of HIF-1α in the hypoxic cells. To further confirm the effect of NAC1 on HIF-1α, we co-transfected HEK293T cells with a Flag-HIF-1α plasmid and various amounts of NAC1-V5 plasmid. Figure 2c shows that expression of HIF-1α protein was increased in the cells with ectopic expression of NAC1. The pulse-chase experiments demonstrated that silencing of NAC1 expression facilitated the turnover of HIF-1α protein (t1/2: HeLa, 50 vs 130 min; SKOV3, 65 vs 140 min) under hypoxic condition (Figure 2d). The downregulation of HIF-1α protein caused by NAC1 knockdown in the hypoxic cells could be rescued by treatment with MG132, a proteasome inhibitor (Figure 2e, Supplementary Figure S5B). Also, the ubiquitination of HIF-1α was increased in the hypoxic cells subjected to NAC1 silenced (Figure 2f). These results indicate that NAC1 has a role in impeding the ubiquitn–proteasomal degradation of HIF-1α protein.
Figure 2.
NAC1 promotes stabilization of HIF-1α protein. HeLa cells were transfected with a siNT or NAC1 siRNA, then: (a) exposed to hypoxia for different periods of time. HIF-1α protein was determined by western blot; (b) followed by transfection with a siRNA-resistant NAC1-expressing plasmid or empty vector. The cells were then exposed to hypoxia for 24 h. HIF-1α protein was determined by western blot. β-Actin was used as a loading control. (c) HEK293T cells were co-transfected with a V5-NAC1 plasmid (0.25 and 0.5 μg) and a Flag-HIF-1α plasmid for 48 h, and then HIF-1α (Flag) and NAC1 (V5) protein were examined by western blot. (d) HeLa and SKOV3 cells with or without silencing of NAC1 expression were treated with CoCl2 for 6 h, and then pulse-chased in the presence of cycloheximide (15 μg/ml). HIF-1α protein was determined by western blot. (e) HeLa cells with or without silencing of NAC1 expression were incubated under normoxia or hypoxia for 24 h, and then treated with vehicle or 5 μM MG132 for 6 h. HIF-1α protein was examined by western blot. (f) HeLa cells with or without silencing of NAC1 expression were incubated under normoxia or hypoxia for 24 h, and treated with vehicle or 5 μM MG132 for 6 h. Cell lysates were immunoprecipitated with a control IgG or anti-HIF-1α antibody. The immunoprecipitates and input were probed for Ub and HIF-1α by immunoblotting. (g) HeLa cells with or without silencing of NAC1 expression were incubated under hypoxia for 24 h, and treated with 5 μM MG132 for another 6 h. Cell lysates were prepared and immunoprecipitated with an anti-HIF-1α antibody, and immunoblotted for HIF-1α and its acetylation form (pan-Ace). (h) HEK293T cells were transfected with a V5-NAC1 plasmid (0.25 and 0.5 μg) or a control plasmid for 24 h, and incubated under normoxia or hypoxia for additional 12 h. Cell lysates were immunoprecipitated with an anti-HIF-1α antibody, and then immunoblotted for HIF-1α and its acetylation form (pan-Ace).
NAC1 affects the acetylation of HIF-1α via HDAC4
We next explored how NAC1 regulates the stability of HIF-1α protein. In GST pull-down experiments using Miz1 as a positive control,17 we did not detect any association of NAC1 with HIF-1α (Supplementary Figure S5C). Silencing of the expression of von Hippel-Lindau protein, a tumor suppressor known to target HIF-1α for degradation,18, 19 had no effect on the downregulation of HIF-1α seen in the hypoxic cells subjected to silencing of NAC1 expression (Supplementary Figure S5D), suggesting that the effect of NAC1 on HIF-1α protein is von Hippel-Lindau independent.
As acetylation of lysine has a critical role in regulating HIF-1α stability,20, 21, 22, 23, 24 we then examined whether NAC1 has any effects on HIF-1α acetylation. We found that under hypoxic condition, acetylation of HIF-1α was noticeably enhanced in the cells with silencing of NAC1 expression (Figure 2g, Supplementary Figure S5E), but decreased in the cells with ectopic expression of NAC1 (Figure 2h). These observations imply that NAC1 may regulate the turnover of HIF-1α through modulating its acetylation.
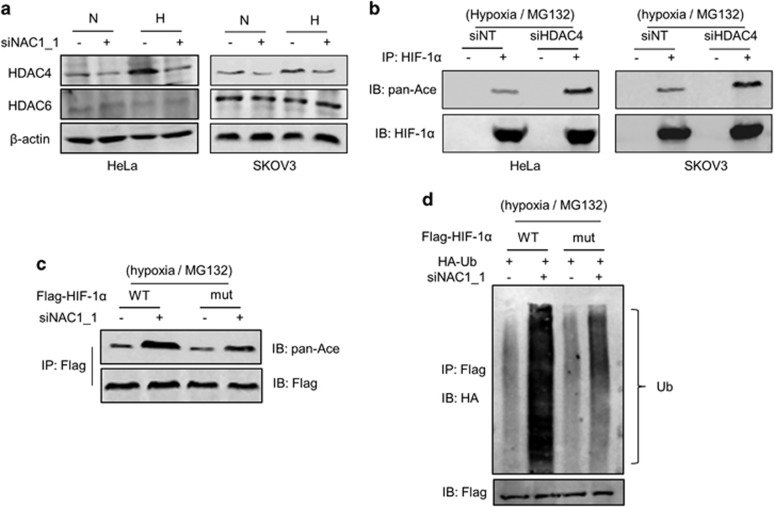
It has been reported that the class II histone deacetylases, HDAC4 and HDAC6, participate in regulating lysine acetylation of HIF-1α.23, 24 Also, it has been shown that NAC1 can interact with HDAC4.8, 25 Therefore, we explored whether the effect of NAC1 on acetylation of HIF-1α is mediated through these histone deacetylases. Figure 3a shows that silencing of NAC1 expression led to a decrease in the amount of HDAC4 in the hypoxic HeLa or SKOV3 cells, but did not affect HDAC6 level. We therefore focused on the role of HDAC4 in the NAC1-regulated acetylation and stability of HIF-1α. Figure 3b shows that silencing of HDAC4 expression resulted in an increase in the acetylation of HIF-1α. To further demonstrate the involvement of HDAC4, using the site-directed mutagenesis method, we generated a HIF-1α mutant in which the lysine sites affected by HDAC423 were altered to arginine. Figure 3c shows that in the hypoxic cells, silencing of NAC1 expression caused a greater increase in acetylation of the wild-type HIF-1α than in that of the mutated HIF-1α. Increases in the ubiquitination were also greater in the wild-type HIF-1α protein than in the mutated HIF-1α protein when the cells were subjected to silencing of NAC1 expression (Figure 3d). In addition, the cells expressing the HIF-1α mutant were more resistant than those expressing the wild-type HIF-1α to the downregulation of glycolysis caused by depletion of NAC1 (Supplementary Figures S6A and B). These results support the role for the HDAC4-mediated deacetylation in the regulation of HIF-1α stability by NAC1.
Figure 3.
NAC1 affects the acetylation of HIF-1α via HDAC4. (a) HeLa or SKOV3 cells were transfected with a siNT or NAC1 siRNA, and incubated under normoxia or hypoxia for 24 h. HDAC4 and HDAC6 proteins were examined by western blot. (b) Cells with or without silencing of HDAC4 expression were incubated under hypoxia for 24 h, and treated with 5 μM MG132 for another 6 h. Cell lysates were immunoprecipitated with an anti-HIF-1α antibody, and then immunoblotted for HIF-1α and its acetylation form (pan-Ace). (c) HeLa cells with or without silencing of NAC1 expression were transfected with a wild-type Flag-HIF-1α (WT) or mutant Flag-HIF-1α (mut) plasmid for 12 h, exposed to hypoxia for 24 h, and then treated with 5 μM MG132 for another 6 h. Cell lysates were immunoprecipitated with an anti-Flag antibody, and then immunoblotted for Flag and the acetylation form of HIF-1α (pan-Ace). (d) HeLa cells with or without silencing of NAC1 expression were transfected with a HA-Ub plasmid, Flag-HIF-1α (WT) or Flag-HIF-1α (mut) plasmid for 12 h, exposed to hypoxia for 24 h, and then treated with 5 μM MG132 for another 6 h. Cell lysates were immunoprecipitated with an anti-Flag antibody, and immunoblotted for HA.
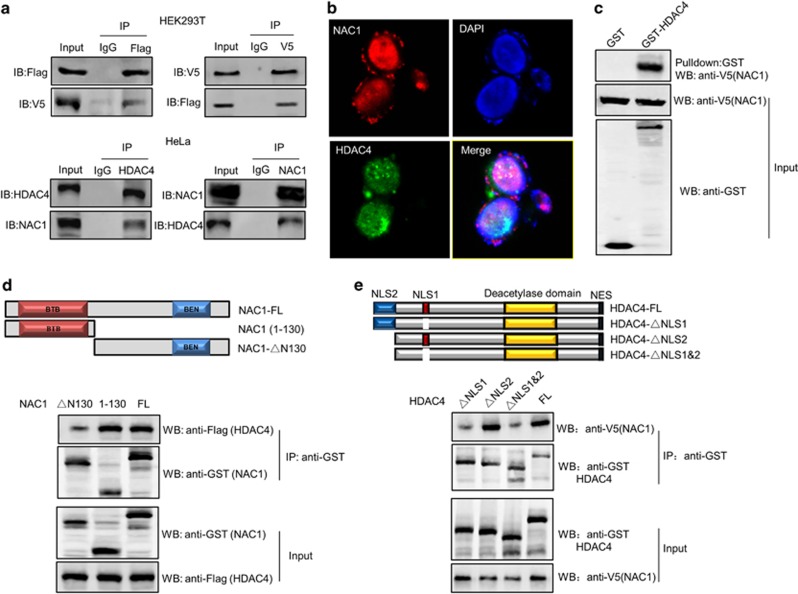
Interaction of NAC1 with HDAC4 regulates the subcellular localization and degradation of this histone deacetylase
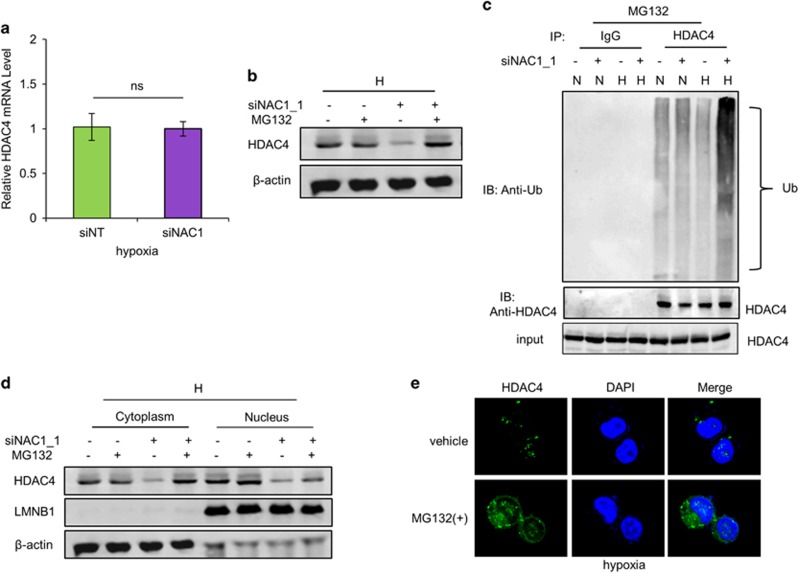
Although silencing of NAC1 expression caused a reduction of HDAC4 protein (Figure 3a), the mRNA level of HDAC4 was not affected by NAC1 depletion in the hypoxic cells, as measured by quantitative reverse transcriptase–PCR (Figure 4a). Notably, the reduction of HDAC4 protein in the hypoxic cells subjected to NAC1 depletion could be rescued by the presence of the proteasome inhibitor, MG132 (Figure 4b). Silencing of NAC1 expression also increased accumulation of the ubiquitinated HDAC4 in the hypoxic cells (Figure 4c). These results suggest a role for NAC1 in modulating the ubiquitn–proteasomal degradation of HDAC4. We further observed that NAC1 bound to HDAC4 (Figures 5a–c and Supplementary Figure S7A), and they primarily colocalized in the nuclei (Figure 5b). As the degradation of HDAC4 is controlled by its nuclear/cytoplasmic shuttling,26, 27 we hypothesized that interaction of NAC1 with HDAC4 governs its subcellular distribution, thereby modulating the degradation of the histone deacetylase. Subcellular fractionation and immunoblotting experiments showed that MG132 had a moderate effect on the level of nucleic HDAC4 in the hypoxic cells with silencing of NAC1 expression, but remarkably blocked the downregulation of the cytoplasmic HDAC4 (Figure 4d, Supplementary Figure S7B). Similar observation was made in immunostaining experiments (Figure 4e).
Figure 4.
NAC1 regulates subcellular localization and degradation of HDAC4 in hypoxic cells. (a) HeLa cells were transfected with a siNT or NAC1 siRNA, followed by exposure to hypoxia for 24 h. HDAC4 mRNA was measured by quantitative reverse transcriptase (RT)–PCR, and plotted after normalization. Bars are mean±s.d. (n=3). (b) HeLa cells with or without silencing of NAC1 were exposed to hypoxia for 24 h, and treated with vehicle or 5 μM MG132 for another 6 h. HDAC4 protein was examined by western blot. (c) HeLa cells with or without silencing of NAC1 expression were incubated under normoxia or hypoxia for 24 h, and treated with 5 μM MG132 for another 6 h. Cell lysates were immunoprecipitated with a control IgG or anti-HDAC4 antibody, followed by immunoblotting for Ub. (d, e) HeLa cells with or without silencing of NAC1 expression were incubated under hypoxic condition for 24 h, and then treated with vehicle or 5 μM MG132 for another 6 h. (d) HDAC4 protein in the nuclear and cytoplasmic fractions was detected by western blot. β-Actin, a loading control for the cytoplasmic fraction; LMNB1, loading control for the nuclear fraction. (e) Immunofluorescent detection of HDAC4 was carried out using an anti-HDAC4 antibody and Alexa Fluor 488 goat anti-rabbit secondary antibody (green). Blue, cells were counterstained with 4,6-diamidino-2-phenylindole (DAPI).
Figure 5.
NAC1 physically associates with HDAC4. (a) (Top) HEK293T cells were transfected with a Flag-HDAC4 plasmid (left) or V5-NAC1 plasmid (right). The lysates of these cells were immunoprecipitated with a control IgG or anti-Flag (HDAC4) (left) or anti-V5 (NAC1) (right) antibody. The immunoprecipitates and input were examined by immunoblotting with the respective antibodies. (Bottom) Lysates prepared from HeLa cells were immunoprecipitated with a control IgG or anti-HDAC4 or anti-NAC1 antibody. The immunoprecipitates were analyzed for NAC1 and HDAC4 by immunoblotting with the respective antibodies. (b) HeLa cells were fixed, and analyzed for NAC1 and HDAC4 by immunofluorescent detection, as described in ‘Materials and methods’ section. Blue, cells were counterstained with 4,6-diamidino-2-phenylindole (DAPI). (c) GST pull-down assay was carried out using immobilized control GST or GST–HDAC4 fusion proteins on MagneGST glutathione particles, as described in ‘Materials and Methods’ section. (d) (Top) Schematic representation of GST–NAC1 fusion protein (FL) and the deletion mutants (1-130 and Δ130). (Bottom) The bacterial-expressed GST–NAC1 or deletion mutants bound to MagneGST glutathione particles was incubated with the lysates of the Flag-HDAC4-transfected HEK293T cell. The interaction between NAC1 and HDAC4 was determined by GST pull-down assay. (e) (Top) Schematic representation of GST-HDAC4 (FL) and the deletion mutants (ΔNSL1, ΔNSL2, and ΔNSL1 and NSL2). (Bottom) The bacterial-expressed GST–HDAC4 or the NLS deletion mutants bound to MagneGST glutathione particles was incubated with the lysates of the V5-NAC1-transfected HEK293T cell. The interaction between HDAC4 and NAC1 was determined by GST pull-down assay.
Identification of the binding motifs required for NAC1–HDAC4 interaction
In order to identify the binding domains required for physical association between NAC1 and HDAC4, we generated the bacterial-expressed GST–NAC1 and deletion mutants (Figure 5d). Using these constructs in GST pull-down assay, we demonstrated that the 1–130 aa structural motif at the N terminus of NAC1 was necessary for its binding to HDAC4 (Figure 5d). HDAC4 has two nuclear localization signals (NLSs): major NLS (NLS1 residue 244–279) and minor NLS (NLS2 residue 1–117).28 To determine the binding site on HDAC4, we carried out the reciprocal experiment. The bacterial-expressed GST–HDAC4 or the NLS deletion mutants bound to MagneGST glutathione particles was incubated with the lysates of the NAC1-transfected HEK293T cells, and then analyzed by immunoblotting. Figure 5e shows that NLS1 on HDAC4 is critical for its binding to NAC1. Thus, the 1–130 aa region of NAC1 and the NLS1 on HDAC4 are the motifs required for the NAC1–HDAC4 binging. We also tested the effects of the NAC1 mutants on glycolysis by transfecting them into ES-2 cells, and then measuring lactate production and glucose consumption under hypoxic condition. We found that ectopic expression of NAC1 or the mutants containing the 1–130 aa motif led to increases in lactate production and glucose consumption, whereas the NAC1 mutant lacking the 1–130 aa region did not (Supplementary Figure S7C). Thus, the 1–130 aa region of NAC1, which interacts with HDAC4, is essential for promoting glycolysis in cancer cells.
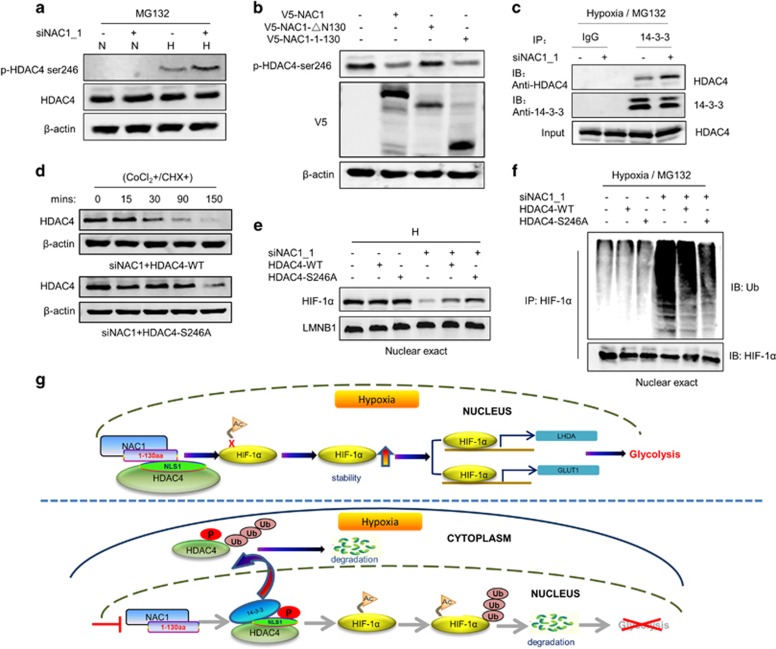
It has been shown that phosphorylation at Ser246 in the NLS1 of HDAC4 promotes its nucleic export and cytoplasmic localization;29 therefore, we further determined whether association of NAC1 with HDAC4 affects Ser246 phosphorylation in the NLS1. The results showed that in the hypoxic cells subjected to silencing of NAC1 expression, phosphorylation of HDAC4 at Ser246 was increased, as compared with that in the cells without silencing of NAC1 expression (Figure 6a, Supplementary Figure S8A). Figure 6b shows that introduction of the wild-type or NAC1 construct containing the 1–130 aa residues inhibited phosphorylation of HDAC4 at Ser246, but the mutant lacking the 1–130 aa sequences did not, suggesting that association of NAC1 with HDAC4 interferes with the phosphorylation of the deacetylase. As the cytoplasmic localization of HDAC4 is also facilitated by binding of its phosphorylated form to 14-3-3 protein,29 we examined the effect of NAC1 on the interaction between HDAC4 and 14-3-3. We showed that silencing of NAC1 expression increased the binding of HDAC4 to 14-3-3 (Figure 6c). Furthermore, the HeLa cells expressing the HDAC4 S246A, a HDAC4 mutant in which serine 246 was replaced with alanine, were insensitive to the HDAC4 degradation triggered by NAC1 knockdown (Figure 6d). Figure 6e shows that in the hypoxic cells with silencing of NAC1 expression, HDAC4 S246A mutant caused a greater increase in the amount of nuclear HIF-1α than the wild-type HDAC4. Ubiquitination level of the nuclear HIF-1α was lower in the cells expressing the HDAC4 S246A mutant than that in the wild-type HDAC4 cells (Figure 6f). Also, as compared with the wild-type HDAC4, the HDAC4 S246A mutant restored glycolysis to a greater extent in the hypoxic cells (Supplementary Figures S8B and C). These results support the concept that interaction of NAC1 with HDAC4 hinders the binding of HDAC4 to 14-3-3 and subsequent HDAC4 degradation through inhibiting phosphorylation of HDAC4 at Ser246, thereby preventing HIF-1α degradation in the nuclear compartment to promote glycolysis (Figure 6g).
Figure 6.
NAC1-HDAC4 binding interferes with S246 phosphorylation of HDAC4. (a) HeLa cells were transfected with a siNT or NAC1 siRNA, and then incubated under normoxia or hypoxia for 24 h, followed by treatment with vehicle or 5 μM MG132 for another 6 h. The phosphorylation of HDAC4 was detected by western blot using an anti-phospho-HDAC4 (Ser246) antibody. (b) ES-2 cells transfected with V5-NAC1, V5-NAC1-ΔN130 or V5-NAC1-1-130 plasmids were exposed to hypoxia for 24 h. The phosphorylation of HDAC4 was detected by western blot. (c) HeLa cells were transfected with an siNT or NAC1 siRNA, and incubated under hypoxia for 24 h, followed by treatment with vehicle or 5 μM MG132 for another 6 h. Lysates of the treated cells were immunoprecipitated with a control IgG or anti-14-3-3 antibody, and the immunoprecipitates were analyzed for HDAC4 by western blot. (d) HeLa cells with silencing of NAC1 expression were transfected with a wild-type Flag-HDAC4 plasmid or a phosphorylation mutant, Flag-HDAC4 (S246A). Twenty-four hours later, the cells were treated with CoCl2 for 6 h, and then pulse-chased for HDAC4 protein in the presence of cycloheximide (15 μg/ml). HDAC4 protein was detected by western blot. (e, f) HeLa cells with or without silencing of NAC1 expression were transfected with the Flag-HDAC4 (WT) or Flag-HDAC4 (S246A) plasmid for 12 h, and exposed to hypoxia for additional 24 h. (e) Nuclear HIF-1α was detected by western blot, with LMNB1 as a loading control for the nuclear fraction. (f) Following 6-h treatment with 5 μM MG132, nuclear extracts were prepared and immunoprecipitated with an anti-HIF-1α antibody. The immunoprecipitates were then subjected to immunoblotting for Ub. (g) Proposed model for the role of NAC1-HDAC4-HIF-1α in promoting glycolysis in hypoxic cells.
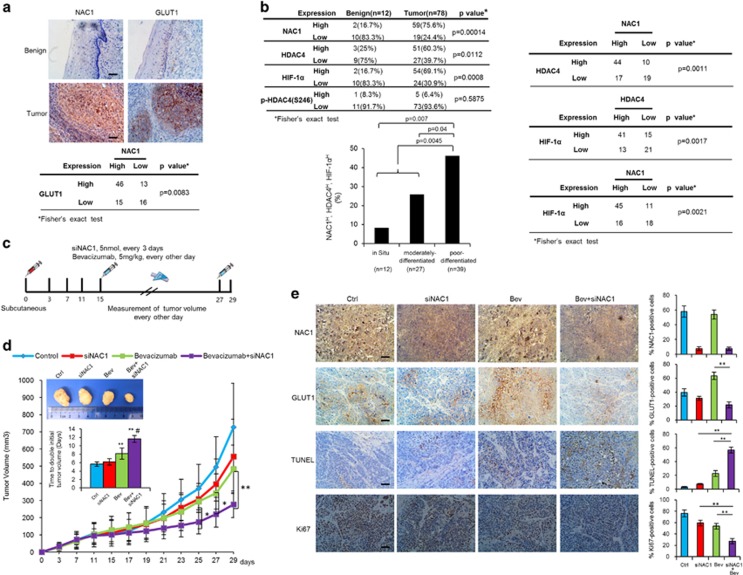
Expression of NAC1, HDAC4, HIF-1α and GLUT1 in human tumor samples
To evaluate the clinical relevance of above findings, we analyzed 78 human cervical cancer specimens and 12 adjacent normal tissues. We observed remarkably stronger staining of NAC1 and GLUT1 proteins in the tumor tissues, and statistical analyses indicated a significantly positive correlation between NAC1 and GLUT1 expression levels (Figure 7a). Furthermore, as compared with adjacent normal tissue, tumor specimens exhibited much higher staining for NAC1, HDAC4 and HIF-1α (Figure 7b). The phosphorylated HDAC4 (Ser246) level was low in both of tumor and adjacent normal tissues (Figure 7b and Supplementary Figure S8D). Statistical analyses showed that NAC1 expression was positively correlated with HDAC4 and HIF-1α expression (Figure 7b and Supplementary Figure S8D). The percentage of patients showing high expression levels of NAC1, HDAC4 and HIF-1α was increased with progression of the disease, and there was a significant difference between cervical carcinoma in situ, moderately differentiated cervical carcinoma and poor-differentiated cervical carcinoma (Figure 7b).
Figure 7.
Evidence for the role of NAC1-HDAC4-HIF-1α pathway in cancer development, progression and therapy. (a, b) Expression of NAC1, HDAC4, p-HDAC4 (S246), HIF-1α, and GLUT1 in human cervical cancer. IHC analysis of NAC1/GLUT1 (a) and NAC1/HDAC4/ p-HDAC4 (S246)/HIF-1α (b) in 12 benign tissues and 78 cervical cancer specimens. The scale bar represents 50 μm. Fisher’s exact test was used to analyze the correlation of NAC1 expression with HDAC4 and HIF-1α expression. Analyses of the association of NAC1, HDAC4 and HIF-1α expression with cancer progression were performed using Fisher’s exact test. (c–e) Silencing of NAC1 expression enhances the antitumor efficacy of bevacizumab in a mouse tumor model. (c) Experimental schedule/regimen. (d) The mice inoculated with Hela cells were divided into four treatment groups: (1) control; (2) siNAC1; (3) bevacizumab; (4) bevacizumab+siNAC1. Tumor sizes were measured every other day. The volumes of tumor and the time to double the size of the tumor volume at the initiation of treatment are shown. **P<0.01; Bev or Bev+siNAC1 vs Ctrl; #P<0.01, Bev+siNAC1 vs Bev. Bars are mean±s.d. (n=5). (e) The tumor specimens were stained for NAC1, GLUT1 and Ki67 proteins, and subjected to terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL). Bars are mean±s.d. (n=5). The scale bar represents 50 μm.
Silencing of NAC1 expression reinforces the antitumor efficacy of bevacizumab
As tumor can upregulate glycolysis to antagonize antiangiogenic therapy,30 we tested the effect of silencing of NAC1 expression on antitumor efficacy of bevacizumab, an angiogenesis inhibitor, in mice bearing the xenografts of HeLa cells. Multiple injection of NAC1 siRNA effectively suppressed expression of NAC1 (Supplementary Figure S9B). We observed that the mice receiving NAC1 siRNA or bevacizumab alone showed moderate reduction in tumor growth, as compared with the vehicle-treated mice (Figure 7d). Depletion of NAC1 had no effects on angiogenesis in tumors (Supplementary Figure S9B). Notably, combination of NAC1 siRNA with bevacizumab was significantly more effective in inhibiting tumor growth than bevacizumab alone, as evidenced by the significant differences in the tumor volume and the time to double the size of the tumor volume at the initiation of treatment (Figure 7d). The amounts of the GLUT1-positive cells in the tumor specimens were decreased in the mice receiving NAC1-targeted siRNA (Figure 7e), suggesting a link between perturbation of glycolysis and sensitizing effect of targeting NAC1. Concomitantly, combined treatment of NAC1 siRNA with bevacizumab caused a significant increase in the terminal deoxynucleotidyl transferase dUTP nick end labeling-positive cells as compared with either NAC1 siRNA or bevacizumab alone (Figure 7e). By contrast, immunohistochemistry (IHC) staining for Ki67, a marker of cell proliferation, was lower in the tumors receiving the combined treatment than in those receiving NAC1 siRNA or bevacizumab alone (Figure 7e).
Discussion
Although NAC1 has been considered as a tumor promoter and several possible oncogenic roles reported,4, 7, 9, 10, 11, 12, 13 how precisely it contributes to malignant phenotypes remains incompletely understood. The main finding of this study is that NAC1 acts as an activator of glycolysis, which is vital for survival of tumor cells in hypoxic microenvironment. As oxygen deficiency is a common feature of tumor microenvironment that promotes tumor progression including invasion, metastasis and therapeutic resistance, and altered bioenergetics is considered a new target for cancer therapy,31 the results reported here may not only help understand better the function and oncogenic potential of NAC1, but also have implications in developing new strategies to cancer prevention and treatment.
We demonstrate that NAC1 has critical roles in promoting glycolysis and survival of tumor cells under hypoxia (Figures 1 and7a, Supplementary Figures S1 and S3), and show that this previously unappreciated function of NAC1 is independent of its transcription-regulatory activity but is achieved through HDAC4-mediated stabilization of HIF-1α (Figures 2g and h, 3b,3d, 4d and e, 6e and f and Supplementary Figure S7B). Transcription-independent function of NAC1 has also been reported by others,14 and our study provides another supportive example. Based on this study, we propose the NAC1-HDAC4-HIF-1α axis as a new pathway in regulating glycolysis (Figure 6g); however, involvement of other signaling molecules that interact with NAC1 or HDAC4 should not be ruled out, as both of these two proteins are multi-functional. For instance, NAC1 was reported to interact with Parkin and contribute to Parkinson’s disease.32
The stability and activity of HIF-1α are regulated by post-translational modifications including hydroxylation, phosphorylation and acetylation,21, 33, 34 and that five lysine residues in the HIF-1α N terminus have a key role in HIF-1α protein stability in the context of HDAC4.23 Indeed, the use of the HIF-1α mutant with replacement of these lysine residues by arginine supports the role of acetylation in the NAC1-promoted stabilization of HIF-1α (Figure 3d), which may represent another regulatory mechanism for this transcription factor. Furthermore, our study found that the BTB/POZ domain of NAC1, corresponding to 1–130 aa at the N terminus, is required for its binding with the major NLS1 (residue 244–279) at the N terminus of HDAC4, and this binding hampers the phosphorylation of HDAC4 at S246 (Figures 5d and e; 6a–c, and Supplementary Figure S8A). Interaction of NAC1 with HDAC4 and the role of this interaction in NAC1-mediated transcriptional repression have been reported previously.25 Here, for the first time we have identified the critical motifs required for this interaction, and demonstrated the effect of this interaction on HDAC4 phosphorylation at S246. We did not test whether NAC1 affects phosphorylation at other sites such as Ser467 and Ser632, which may also have a role in regulating cytoplasmic localization of HDAC4.35, 36
NAC1 has been reported to have a role in regulating the stemness and differentiation of stem cells through its interaction with the factors like Nanog, Oct4, Tcf3 and Sox2.6, 37, 38 As altered energy metabolism can impact fate of stem cells,39 the NAC1-promoted glycolysis may contribute to its role in regulating differentiation of stem cells.
Targeting angiogenesis, which is closely associated with hypoxia,40, 41 is now considered a promising therapeutic strategy against solid neoplasm. Nevertheless, the decrease of blood supply caused by inhibition of angiogenesis induces hypoxic response including glycolysis,42, 43, 44 and induction of hypoxic response diminishes the efficacy of angiogenesis inhibitors. Thus, hypoxic microenvironment that causes metabolic reprogramming is a key factor contributing to therapeutic resistance. Our experiments showed that silencing of NAC1 expression could inhibit bevacizumab-mediated glycolysis, reverse therapeutic resistance and enhance the efficacy of the angiogenesis inhibitor bevacizumab (Figures 7d and e), suggesting that targeting NAC1 may represent a potential effective approach to improving antiangiogenic therapy in human cancer.
In summary, this study uncovers a new function of NAC1 in promoting glycolysis and survival of hypoxic tumor cells, and demonstrates that this previously unrecognized function of NAC1 is mediated through the HDAC4-HIF-1α axis. These findings provide a new important mechanism accounting for the oncogenic role of NAC1, and underscore the potential of targeting NAC1 as a novel strategy to cancer prevention and treatment.
Materials and methods
Cell lines and culture
The human ovarian cancer cell lines SKOV3, A2780 and ES-2, the human cervical cancer cell line, HeLa, and the human embryonic kidney HEK293T cells were purchased from ATCC (Manassas, VA, USA). The identity of these cell lines was recently verified by STR analysis. NAC1+/+ and NAC1−/− mouse embryo fibroblasts were derived from NAC1 knockout mouse embryo and the wild-type littermate.13 HeLa, A2780 and ES-2 cell lines were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal bovine serum, 100 units/ml of penicillin and 100 mg/ml of streptomycin. SKOV3 cell line was cultured in RPMI-1640 medium supplemented with 10% heat-inactivated fetal bovine serum, 100 units/ml of penicillin and 100 mg/ml of streptomycin. Cells were cultured at 37 °C in a humidified atmosphere of 20% O2/5% CO2 (normoxia) or 1% O2/5% CO2 (hypoxia). All cultures were monitored routinely and found to be free of contamination by mycoplasma or fungi, discarded after 3 months, and new lines propagated from frozen stocks.
Reagents and antibodies
The following antibodies were used in western blot, co-immunoprecipitation, immunofluorescence or IHC: anti-NAC1 (NB110-77345, Novus, Littleton, CO, USA), anti-HDAC4 (ABE262, Millpore, Billerica, MA, USA), anti-HIF-1α (#3716), anti-p-HDAC4 Ser246 (#3443), anti-Flag (#8146), anti-V5 (#13202), anti-GLUT1 (#12939), anti-lactate dehydrogenase-A (#2012), anti-14-3-3 (#8312), anti-von Hippel-Lindau (#68547), anti-GST (#2622), and anti-Myc (#2278) (Cell Signaling Technology, Danvers, MA, USA), anti-β-actin (sc-47778), anti-ubiquitin (sc-8017), anti-CD31(sc-376764) and anti-pan-Ace (Ace-K) (sc-8649) (Santa Cruz, Dallas, TX, USA), anti-HDAC6 (YT2118, Immunoway, Plano, TX, USA), mouse IgG (A7028) and rabbit IgG (A7016, Beyotime, Shanghai, China). MG132, CoCl2 and cycloheximide were purchased from Sigma-Aldrich Corporation (Sigma-Aldrich, St Louis, MI, USA). The primers and oligo DNA used in this study are listed in Supplementary Table 1.
siRNA, shRNA and plasmid transfection
Transfection of siRNA and plasmids was conducted using lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's protocol. NAC1 shRNA plasmid was synthesized by Santa Cruz, and transfection of shRNA plasmid was conducted following the manufacturer's protocol. The transfected cells were selected with puromycin (5 μg/ml) for 2 weeks. V5-NAC1 plasmid was described previously.13 Flag-HDAC4, Flag-HIF-1α and His-ubiquitin plasmids were purchased from Invitrogen. To generate the 5KR mutant of HIF-1α, we replaced the acetylation sites Lys-10, Lys-11, Lys-12, Lys-19 and Lys-21 with arginine. The HDAC4 mutant was generated by replacing the phosphorylation site Ser246 with alanine. Site-directed mutagenesis was performed using the QuikChange Kit (Stratagene, San Diego, CA, USA). Transfection of the plasmid was carried out using lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
Quantitative real-time PCR
Total RNA was prepared using TRIzol reagent (Roche, Basel, Switzerland). First-strand complementary DNA was synthesized using Omniscript reverse transcription kit (Qiagen, Hilden, Germany) with random primers. Quantitative reverse transcriptase–PCR was performed on ABI 7500 using Brilliant II SYBR Green QPCR master mix (Stratagene) and primers. After 40 cycles, data were collected and analyzed using the 7500 software (ABI, Waltham, MA, USA).
Measurement of glucose, lactate and ATP
Glucose and lactate were measured using a fluorescence-based glucose or lactate assay kit (Bio Vision, Milpitas, CA, USA). For measurement of intracellular ATP, ATP lite 1-step (PerkinElmer, Waltham, MA, USA) was used. These assays were performed following the manufacturer’s protocols.
Measurement of glycolytic intermediates
Cells (5 × 106) were washed twice with phosphate-buffered saline, and then lysed in 80% (vol/vol) methanol at −78 °C to extract intracellular polar metabolites. Cell debris was removed by centrifugation at 4 °C, and the supernatants containing metabolites were evaporated using a refrigerated Speed Vac (Thermo Scientific, Waltham, MA, USA). LC/MS/MS-based metabolomics analysis was performed as previously described.45
Oxygen consumption rate
Cellular OXPHOS was monitored using a Seahorse XF24 Extracellular Flux Analyzer (Seahorse Bioscience Inc., North Billerica, MA, USA), as previously described.46 Briefly, 1–2 × 104 cells were seeded in 24-well plates and incubated overnight. Before measurements, cells were washed, then immersed in un-buffered media and incubated in the absence of CO2 for 1 h.
Immunoblotting and co-immunoprecipitation
Proteins (10–20 μg) were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and then transferred to PVDF membrane (Bio-Rad Laboratories, Hercules, CA, USA). Membranes were incubated with primary antibodies in 3% bovine serum albumin at 4 °C for overnight, followed by incubation with secondary antibodies at room temperature for 1 h. The protein signals were detected by ECL (Beyotime Biotechnology, Shanghai, China) method. For co-immunoprecipitation, appropriate antibodies were first incubated with protein A/G beads (Santa Cruz) at 4 °C for overnight, cell lysates were followed by incubation with protein A/G beads at 4 °C for 6 h. At the end of incubation, the beads were washed three times with RIPA lysis buffer, and the immunoprecipitates were eluted with a sodium dodecyl sulfate buffer and then subjected to immunoblotting.
GST pull-down and in vitro binding assay
The bacterial-expressed GST-HDAC4 FL, GST-HDAC4ΔNSL1, GST-HDAC4ΔNSL2, GST-HDAC4ΔNSL1&NSL2 or control GST bound to MagneGST glutathione particles was incubated with the lysates of the NAC1-transfected HEK293T cells for 1 h at 4 °C using Pierce (Thermo Scientific) GST Protein Interaction Pull-Down Kit. Similarly, the bacterial-expressed GST-NAC1 FL, GST-NAC1 (1-130), GST-NAC1Δ130 or control GST bound to MagneGST glutathione particles was incubated with the lysates of the HDAC4-transfected HEK293T cells. The complexes were washed, eluted with a sodium dodecyl sulfate buffer and analyzed by immunoblotting.
Imaging microPET scan
Mice were anesthetized by Matrx VMR, and 18F-FDG (185 MBq/kg) was given through the tail vein to initiate emission scan. Images were acquired on microPET (Inveon Siemens, Siemens Healthcare, Erlangen, Germany). To quantify 18F-FDG uptake on the last frame (corresponding to 40–60 min), the obtained tissue activity (counts (kBq)/ml) was divided by the injected activity in kBq per gram of body weight (185 kBq/g) to give a standardized uptake value. Animal maintenance and experimental procedures were approved by the Institutional Animal Care and Use Committee of Soochow University.
IHC analysis of tissue specimens
Paraffin-embedded cervical cancer specimens were histopathologically diagnosed by the Department of Pathology, First Affiliated Hospital of Soochow University. Use of these tissue samples was approved by the Medical Ethical Committee of Soochow University. For IHC staining, paraffin sections of 4 μm thickness on coating slides were heat-denatured with 10 mM sodium citrate buffer (pH 6.0) for 15 min for antigen retrieval, and then incubated with an anti-NAC1 antibody (1:200 dilution), anti-HDAC4 antibody (1:100 dilution), anti-p-HDAC4 (S246) (1:100 dilution), anti-HIF-1α antibody (1:100 dilution) or anti-GLUT1 (1:200 dilution). For IHC analysis on xenograft tumors, tumor specimens were either fixed in 4% paraformaldehyde or frozen in optimal cutting temperature compound (Tissue-Tek, VWR, Radnor, PA, USA). The sample slides were then incubated with anti-NAC1 antibody (1:200 dilution), anti-GLUT1 antibody (1:200 dilution), anti-CD31(1:200 dilution), anti-Ki67 antibody (1:200 dilution) or subjected to terminal deoxynucleotidyl transferase dUTP nick end labeling (Roche). The IHC staining was scored as negative/weak positive (score 0), moderate positive (score 1+) or strong positive (score 2+). The samples showing strong positive (score 2+) were defined as high expression.
Immunofluorescence staining
Cells were washed with phosphate-buffered saline, fixed with pre-cold methanol for 10 min and then permeabilized with 0.1% Triton X-100 in phosphate-buffered saline for 10 min at room temperature. Following incubation in the blocking buffer (5% bovine serum albumin) for 1 h at room temperature, the samples were incubated with an anti-NAC1 monoclonal antibody (1:100 dilution) or HDAC4 rabbit monoclonal antibody (1:100 dilution) at 4 °C for overnight. At the end of incubation, the cells were washed three times in phosphate-buffered saline and then stained with Alexa Fluor 594 goat anti-mouse IgG and Alexa Fluor 488 goat anti-rabbit IgG (1:300 dilution, Invitrogen) for 2 h at room temperature.
Animal experiments
BALB/c mice (5-week-old, female) were inoculated subcutaneously with HeLa cells (2 × 106 cells per mouse). Two weeks after inoculation, the tumor-bearing mice were divided randomly into four groups (five mice per group): (1) control; (2) NAC1 siRNA; (3) bevacizumab; and (4) NAC1 siRNA+bevacizumab. The minimum number of animals was chosen based on the convention of this type of experiment, and was consistent with valid experimental design and to obtain statistically meaningful results at the 95% confidence interval. Bevacizumab (5 mg/kg) was given intraperitoneally q2d for 2 weeks.43, 47 For delivery of cholesterol conjugated RNA, 10 nmol RNA in 0.1 ml saline was locally injected into the tumor mass q3d for 2 weeks. Tumor volumes were determined by measuring the length (L) and the width (W) of the tumors and calculating using the formula: V=L × W2/2. No blinding approach was used during this study. At the end of the experiment (on day 29), the mice were killed and tumors were surgically dissected. The tumor specimens were fixed in 4% paraformaldehyde for histopathologic examination. Animal maintenance and experimental procedures were approved by the Institutional Animal Care and Use Committee of Soochow University.
Statistical analysis
Statistical analyses were performed using Microsoft Excel software and GraphPad Prism (Graphpad, La Jolla, CA, USA). Fisher’s exact test was used to evaluate the correlation of expression levels between NAC1, HDAC4, HIF-1α and GLUT1. Tumor growth delay was analyzed by comparing the time (days) for different treatment groups to double the size of the tumor volume at the initiation of treatment. Statistical analyses of the differences in growth delay (days) between the treatment groups were performed using two-sided t-test. The results are presented as mean±s.d. *P<0.05; **P<0.01.
Acknowledgments
This work was supported by grants from National Natural Sciences Foundation of China (81473240), Special Financial Grant by China Postdoctoral Science Foundation (2015T80585), China Postdoctoral Science Foundation (2014M550308), Natural Science Foundation of Jiangsu Province of China (BK20141197), Science and Technology Foundation of Suzhou City (SYS201319) to Yi Zhang; and Natural Science Foundation of Jiangsu Province of China (BK20151209) to Cheng Ji.
Author contributions
Conception and design: Yi Zhang, Jin-Ming Yang; Development of methodology: Yi Zhang, Yi-Jie Ren, Xingcong Ren, Jin-Ming Yang; Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): Yi-Jie Ren, Ling-Chuan Guo, Cheng Ji, Jian Hu, Hong-Han Zhang, Qiong-Hua Xu, Wei-Dong Zhu; Analysis and interpretation of data: Yi Zhang, Zhi-Jun Ming, Jian-Xun Song, Jin-Ming Yang; Writing, review and/or revision of the manuscript: Yi Zhang, Jian-Xun Song, Xingcong Ren, Jin-Ming Yang; Administrative, technical or material support: Yi Zhang, Yun-Sheng Yuan, Jin-Ming Yang; Study supervision: Yi Zhang, Jin-Ming Yang; Other (pathology slides reviewing): Ling-Chuan Guo, Wei-Dong Zhu.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Brahimi-Horn MC, Chiche J, Pouyssegur J. Hypoxia and cancer. J Mol Med (Berl) 2007; 85: 1301–1307. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell 2012; 148: 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Nakayama N, Davidson B, Sheu JJ, Jinawath N, Santillan A et al. A BTB/POZ protein, NAC-1, is related to tumor recurrence and is essential for tumor growth and survival. Proc Natl Acad Sci USA 2006; 103: 18739–18744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Torrado R, Yamada D, Defossez PA. Born to bind: the BTB protein-protein interaction domain. Bioessays 2006; 28: 1194–1202. [DOI] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW et al. A protein interaction network for pluripotency of embryonic stem cells. Nature 2006; 444: 364–368. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Rahman MT, Rahman M, Yeasmin S, Ishikawa M, Katagiri A et al. Biological role and prognostic significance of NAC1 in ovarian cancer. Gynecol Oncol 2010; 119: 469–478. [DOI] [PubMed] [Google Scholar]
- Yeasmin S, Nakayama K, Rahman MT, Rahman M, Ishikawa M, Katagiri A et al. Biological and clinical significance of NAC1 expression in cervical carcinomas: a comparative study between squamous cell carcinomas and adenocarcinomas/adenosquamous carcinomas. Hum Pathol 2012; 43: 506–519. [DOI] [PubMed] [Google Scholar]
- Shih Ie M, Nakayama K, Wu G, Nakayama N, Zhang J, Wang TL. Amplification of the ch19p13.2 NACC1 locus in ovarian high-grade serous carcinoma. Mod Pathol 2011; 24: 638–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Nakayama N, Wang TL, Shih IeM. NAC-1 controls cell growth and survival by repressing transcription of Gadd45GIP1, a candidate tumor suppressor. Cancer Res 2007; 67: 8058–8064. [DOI] [PubMed] [Google Scholar]
- Jinawath N, Vasoontara C, Yap KL, Thiaville MM, Nakayama K, Wang TL et al. NAC-1, a potential stem cell pluripotency factor, contributes to paclitaxel resistance in ovarian cancer through inactivating Gadd45 pathway. Oncogene 2009; 28: 1941–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cheng Y, Ren X, Zhang L, Yap KL, Wu H et al. NAC1 modulates sensitivity of ovarian cancer cells to cisplatin by altering the HMGB1-mediated autophagic response. Oncogene 2012; 31: 1055–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cheng Y, Ren X, Hori T, Huber-Keener KJ, Zhang L et al. Dysfunction of nucleus accumbens-1 activates cellular senescence and inhibits tumor cell proliferation and oncogenesis. Cancer Res 2012; 72: 4262–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Fraley SI, Thiaville MM, Jinawath N, Nakayama K, Wang J et al. NAC1 is an actin-binding protein that is essential for effective cytokinesis in cancer cells. Cancer Res 2012; 72: 4085–4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda SM, Yap KL, Davidson B, Tian Y, Murthy V, Wang TL et al. Expression of fatty acid synthase depends on NAC1 and is associated with recurrent ovarian serous carcinomas. J Oncol 2010; 2010: 285191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MT, Nakayama K, Rahman M, Katagiri H, Katagiri A, Ishibashi T et al. Fatty acid synthase expression associated with NAC1 is a potential therapeutic target in ovarian clear cell carcinomas. Br J Cancer 2012; 107: 300–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead MA, Wright SC. Nac1 interacts with the POZ-domain transcription factor, Miz1. Biosci Rep 2014; 34: e00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 1999; 399: 271–275. [DOI] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 2001; 292: 464–468. [DOI] [PubMed] [Google Scholar]
- Geng H, Liu Q, Xue C, David LL, Beer TM, Thomas GV et al. HIF1alpha protein stability is increased by acetylation at lysine 709. J Biol Chem 2012; 287: 35496–35505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH, Lee YM, Chun YS, Chen J, Kim JE, Park JW. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol Cell 2010; 38: 864–878. [DOI] [PubMed] [Google Scholar]
- Jeong JW, Bae MK, Ahn MY, Kim SH, Sohn TK, Bae MH et al. Regulation and destabilization of HIF-1alpha by ARD1-mediated acetylation. Cell 2002; 111: 709–720. [DOI] [PubMed] [Google Scholar]
- Geng H, Harvey CT, Pittsenbarger J, Liu Q, Beer TM, Xue C et al. HDAC4 protein regulates HIF1alpha protein lysine acetylation and cancer cell response to hypoxia. J Biol Chem 2011; 286: 38095–38102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian DZ, Kachhap SK, Collis SJ, Verheul HM, Carducci MA, Atadja P et al. Class II histone deacetylases are associated with VHL-independent regulation of hypoxia-inducible factor 1 alpha. Cancer Res 2006; 66: 8814–8821. [DOI] [PubMed] [Google Scholar]
- Korutla L, Wang PJ, Mackler SA. The POZ/BTB protein NAC1 interacts with two different histone deacetylases in neuronal-like cultures. J Neurochem 2005; 94: 786–793. [DOI] [PubMed] [Google Scholar]
- Shimizu E, Nakatani T, He Z, Partridge NC. Parathyroid hormone regulates histone deacetylase (HDAC) 4 through protein kinase A-mediated phosphorylation and dephosphorylation in osteoblastic cells. J Biol Chem 2014; 289: 21340–21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernotta N, Clocchiatti A, Florean C, Brancolini C. Ubiquitin-dependent degradation of HDAC4, a new regulator of random cell motility. Mol Biol Cell 2011; 22: 278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AH, Yang XJ. Histone deacetylase 4 possesses intrinsic nuclear import and export signals. Mol Cell Biol 2001; 21: 5992–6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AH, Kruhlak MJ, Wu J, Bertos NR, Vezmar M, Posner BI et al. Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol Cell Biol 2000; 20: 6904–6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera LB, Bergers G. Angiogenesis: targeting vascular sprouts. Science 2014; 344: 1449–1450. [DOI] [PubMed] [Google Scholar]
- Cardenas-Navia LI, Richardson RA, Dewhirst MW. Targeting the molecular effects of a hypoxic tumor microenvironment. Front Biosci 2007; 12: 4061–4078. [DOI] [PubMed] [Google Scholar]
- Korutla L, Furlong HAt, Mackler SA. NAC1, A POZ/BTB protein interacts with Parkin and may contribute to Parkinson's disease. Neuroscience 2014; 257: 86–95. [DOI] [PubMed] [Google Scholar]
- Kaelin Jr WG, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell 2008; 30: 393–402. [DOI] [PubMed] [Google Scholar]
- Generali D, Buffa FM, Berruti A, Brizzi MP, Campo L, Bonardi S et al. Phosphorylated ERalpha, HIF-1alpha, and MAPK signaling as predictors of primary endocrine treatment response and resistance in patients with breast cancer. J Clin Oncol 2009; 27: 227–234. [DOI] [PubMed] [Google Scholar]
- Zhao X, Ito A, Kane CD, Liao TS, Bolger TA, Lemrow SM et al. The modular nature of histone deacetylase HDAC4 confers phosphorylation-dependent intracellular trafficking. J Biol Chem 2001; 276: 35042–35048. [DOI] [PubMed] [Google Scholar]
- Backs J, Song K, Bezprozvannaya S, Chang S, Olson EN. CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J Clin Invest 2006; 116: 1853–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 2008; 132: 1049–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malleshaiah M, Padi M, Rue P, Quackenbush J, Martinez-Arias A, Gunawardena J. Nac1 coordinates a sub-network of pluripotency factors to regulate embryonic stem cell differentiation. Cell Rep 2016; 14: 1181–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussaieff A, Rouleau M, Kitsberg D, Cohen M, Levy G, Barasch D et al. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab 2015; 21: 392–402. [DOI] [PubMed] [Google Scholar]
- Moeller BJ, Cao Y, Vujaskovic Z, Li CY, Haroon ZA, Dewhirst MW. The relationship between hypoxia and angiogenesis. Semin Radiat Oncol 2004; 14: 215–221. [DOI] [PubMed] [Google Scholar]
- Folkman J. Is angiogenesis an organizing principle in biology and medicine? J Pediatr Surg 2007; 42: 1–11. [DOI] [PubMed] [Google Scholar]
- Fack F, Espedal H, Keunen O, Golebiewska A, Obad N, Harter PN et al. Bevacizumab treatment induces metabolic adaptation toward anaerobic metabolism in glioblastomas. Acta Neuropathol 2015; 129: 115–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapisarda A, Hollingshead M, Uranchimeg B, Bonomi CA, Borgel SD, Carter JP et al. Increased antitumor activity of bevacizumab in combination with hypoxia inducible factor-1 inhibition. Mol Cancer Ther 2009; 8: 1867–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesange P, Poindessous V, Sabbah M, Escargueil AE, de Gramont A, Larsen AK. Intrinsic bevacizumab resistance is associated with prolonged activation of autocrine VEGF signaling and hypoxia tolerance in colorectal cancer cells and can be overcome by nintedanib, a small molecule angiokinase inhibitor. Oncotarget 2014; 5: 4709–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 2013; 339: 222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Ren X, Yuan Y, Shan Y, Li L, Chen X et al. eEF-2 kinase is a critical regulator of Warburg effect through controlling PP2A-A synthesis. Oncogene 2016; 35: 6293–6308. [DOI] [PubMed] [Google Scholar]
- Shao M, Hollar S, Chambliss D, Schmitt J, Emerson R, Chelladurai B et al. Targeting the insulin growth factor and the vascular endothelial growth factor pathways in ovarian cancer. Mol Cancer Ther 2012; 11: 1576–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.