Abstract
N-myc downstream-regulated gene 1 (NDRG1) has been reported to act as a key regulatory molecule in tumor progression-related signaling pathways, especially in tumor metastasis. However, the related mechanism has not been fully discovered yet. Herein we demonstrated that the novel molecule of cell migration and invasion, caveolin-1, has direct interaction with NDRG1 in human colorectal cancer (CRC) cells. Moreover, we discovered that NDRG1 reduces caveolin-1 protein expression through promoting its ubiquitylation and subsequent degradation via the proteasome in CRC cells. In addition, caveolin-1 mediates the suppressive function of NDRG1 in epithelial–mesenchymal transition, migration and invasion in vitro and metastasis in vivo. These results help to fulfill the potential mechanisms of NDRG1 in anti-metastatic treatment for human colorectal cancer.
Introduction
N-myc downstream-regulated gene 1 (NDRG1) is a cytoplasmic protein, which is highly conserved among multicellular organisms and ubiquitously occurs in various human tissues. In different reports referring to various human carcinomas, the NDRG1 is de-regulated.1, 2 Accumulating evidences has regarded NDRG1 as a metastasis suppressor.2, 3, 4 In colorectal cancer (CRC), NDRG1 is believed to be a favorable predictor for the prognosis and is demonstrated to regulate actin cytoskeleton re-organization and subsequent reduction of cancer cell migration;2 NDRG1 is also reported to inhibit the epithelial–mesenchymal transition (EMT).3 As a metastasis suppressor, NDRG1 is reported to be able to regulate different signaling pathways in tumor progression,1, 5, 6, 7, 8 resulting in interruption of major metastasis-associated functions, including EMT, cytoskeleton remodeling and subsequent migration and invasion.9 Although some molecular pathways explained the function of NDRG1 have been partially elucidated, more straightforward targets and partners of NDRG1 still need further exploration.
Caveolae is a small invagination that transports and processes diverse extracellular signals and is implicated in cellular trafficking, as well as signal transduction.10, 11, 12, 13 In response to various stimuli, lots of signaling molecules and receptors localize in caveolae making it a ‘launching platform’ for intracellular signaling cascades.10, 14, 15, 16 As essential structural constituent of caveolae, caveolin-1 (cav1) is not only able to interact with but also able to regulate different molecules recruited in caveolae, thereby representing a key checkpoint for the cell signaling regulation in cancer.12, 13
Cav1 has been regarded as having a key role in tumor progression, which influences many key capabilities in cancer progression, such as unlimited replicative potential, resistance to antigrowth signals and enhanced tissue invasion and metastasis as well as acquisition of multidrug resistance.17, 18 Although the precise effect of cav1 remains unclear as both the loss and overexpression of cav1 have been reported in various malignancies,19, 20 accumulating evidences have indicated that cav1 expression favors cancer cell migration, invasion and metastasis.21, 22, 23
Considering the special localization and function of cav1, for the first time, we identified the relationship between NDRG1 and cav1, two versatile proteins in signal regulation and having key roles in CRC progression. Our results demonstrate that NDRG1 interacts with cav1 and reduces cav1 protein expression through promoting its ubiquitylation and subsequent degradation via the proteasome in CRC cells. In addition, cav1 mediates the suppressive function of NDRG1 in EMT, migration and invasion in vitro as well as metastasis in vivo.
Results
NDRG1 suppresses migration and invasion of CRC cell lines
We first established HT29, SW480 and SW1116 CRC cell lines that constitutively and stably overexpress exogenous NDRG1. In addition, NDRG1-knockdown models were also generated in these CRC cell lines to explore the function of endogenous NDRG1 (Figure 1a). These three cell lines exhibiting different cell aggressiveness were enrolled to access the consistency of our data between different cell lines. In our former study, we discovered that NDGR1 showed the ability to inhibit the migration and invasion capability of HT29 and HCT116 cells in vitro.9 We first repeated these findings in our previously designed HT29 cell models as well as in the newly generated SW480 and SW1116 cell models. As shown in transwell assay (Supplementary Figure 1), overexpression of NDRG1 in HT29, SW480 and SW1116 cells resulted in approximately 42–97% reduction of migratory capacity (P<0.001) and approximately 94–97% reduction of invasive capacity (P<0.001), respectively. On the other hand, NDRG1 knockdown in these cells led to significant 3.9–4.1-fold (P<0.001) and 4.1–4.9-fold (P<0.001) increase of migratory and invasive capacity, respectively (Supplementary Figure 1). The results re-confirmed that NDRG1 inhibits migration and invasion in CRC cells and also demonstrated the effectiveness of the newly generated models, which can be applied to the subsequent experiments aiming at exploring the mechanisms and molecular effectors of NDRG1.
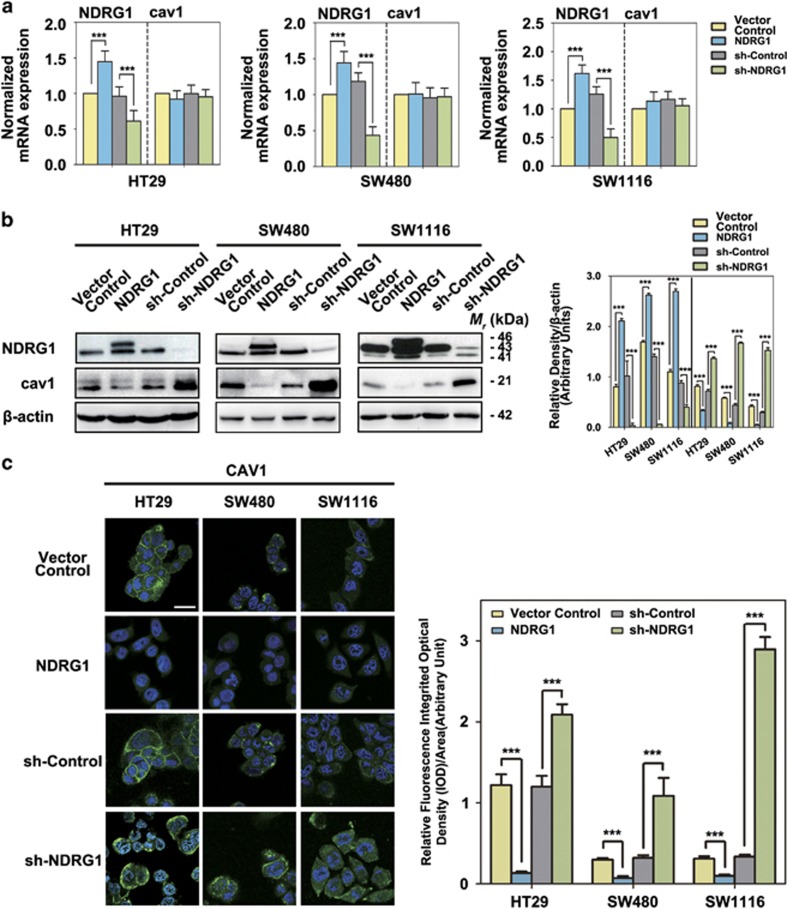
Figure 1.
Regulation of caveolin-1 expression by NDRG1 in CRC cells. (a) RT-qPCR results of the NDRG1 and caveolin-1 mRNA expressions in NDRG1 overexpression and NDRG1 knockdown HT29, SW480 and SW1116 cells compared with their relative control cells (that is, Vector Control and sh-Control, respectively). The values in histograms is represented by mean±s.d. (three experiments). ***P<0.001, relative to the respective control cells. (b) Western blots for the indicated proteins in NDRG1 overexpression and NDRG1 knockdown HT29, SW480 and SW1116 cells compared with their relative control cells (that is, Vector Control and sh-Control, respectively). Densitometry analyses for NDRG1 and cav1 are expressed relative to the loading control, β-actin. Results are typical of three to five experiments and the values in histograms is represented by mean±s.d. (three to five experiments). ***P<0.001, relative to the respective control cells. (c) Merged images were taken to show immunofluorescence staining of caveolin-1 (green) accompanied by the cell nucleus (blue) stained by DAPI in NDRG1 overexpression and NDRG1 knockdown HT29, SW480 and SW1116 cells relative to the Vector Control and sh-Control cells, respectively. Scale bars, 20 μm. Fluorescence quantification was performed by comparing the integrated optical density (IOD)/area value of cav1 to the IOD/area value of the nucleus (DAPI) in the same image. Results are typical of three to five images from different visual fields and the histogram values are mean±s.d. (three to five images). ***P<0.001, relative to the respective control cells.
NDRG1 reduces caveolin-1 protein expression in CRC cell lines
Cav1 is a multifunctional protein located at cell membrane caveolae and acts as a platform to sequester and/or inhibit the activity of various signaling molecules.10, 17 Several papers reported that migration capability is affected by alterations of cav1 expression levels in a controversial way.22, 24 We analyzed the mRNA and protein expressions of cav1 after NDRG1 overexpression and knockdown in our CRC cell models. The qRT-PCR indicated that the mRNA level of cav1 was not significantly changed when NDRG1 overexpressed/knocked down in HT29, SW480 and SW1116 cells (Figure 1a). However, immunoblots showed that the cav1 protein expression was significantly decreased (59–94%, P<0.05–0.001) when NDRG1 was overexpressed (Figure 1a). Moreover, NDRG1 knockdown significantly (P<0.001) activated the cav1 expression by 1.9–5.2-fold, respectively (Figure 1a). Furthermore, immunofluorescence staining of cav1 showed a significant (P<0.01–0.001) decrease or increase in staining intensity in the NDRG1 overexpression and knockdown cells, respectively (Figure 1b). These data suggested that NDRG1 reduced cav1 expression by regulating its protein expression.
NDRG1 suppresses migration and invasion through inhibiting caveolin-1
It is reported that cav1 acts not only as a tumor suppressor but also as a metastatic promoter.17 Regarding the tumor suppressor function of cav1, its expression is inhibited in several human tumors, including lung,25, 26, 27 breast28 and ovarian carcinomas.29 On the contrary, cav1 presence has also been reported to be associated with elevated metastasis, poor prognosis and poor patient survival in prostate cancer; while re-capturing of cav1 was also reported to be related to reversal of the transformed phenotype.30, 31, 32, 33 Thus, whether cav1 takes effects to preventing or promoting tumor progression appears to depend on the cellular settings. Considering that the functional fluctuation of cav1 is assumed to be associated to the regulation from its interaction partners,17 it is possible that the suppressive effect of NDRG1 on cell migration and invasion may be mediated by downregulation of cav1.
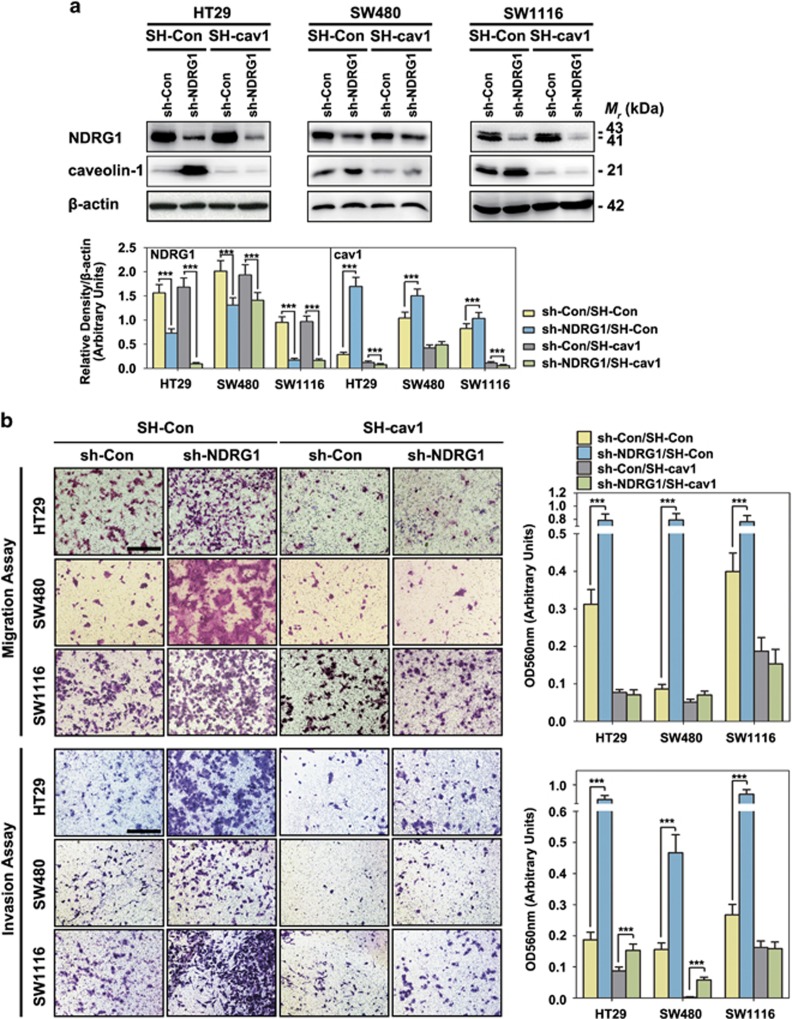
After cav1 was silenced (Figure 2a), we assessed the effect of NDRG1/cav1 downregulation on cell migration/invasion. Knocking down NDRG1 alone led to a significant increase of migratory/invasive capacity (Figure 2b, sh-NDRG1/SH-Con vs sh-Con/SH-Con), whereas knocking down cav1 alone resulted in a significant (P<0.001) decrease of migratory as well as invasive capacity (Figure 2b, sh-Con/SH-cav1 vs sh-Con/SH-Con). Notably, in sh-NDRG1/SH-cav1 groups, the increased enhanced migratory and invasive capacity caused by silencing NDRG1 was totally abolished (Figure 2b, sh-NDRG1/SH-cav1 vs sh-NDRG1/SH-Con). In fact, the migration and invasion results in sh-NDRG1/SH-cav1 group was similar to that in sh-Con/SH-cav1 group (Figure 2b).
Figure 2.
NDRG1 suppresses cancer cell migration and invasion through inhibiting caveolin-1 expression: sh-NDRG1 and SH-cav1 results. (a) The sh-NDRG1 knockdown and sh-Control HT29, SW480 and SW11116 cells were incubated with or without caveolin-1-specific shRNA (SH-cav1). Whole-cell lysates were extracted and immunoblotting was performed to assess NDRG1 and caveolin-1 expression in cells transfected with sh-Con/SH-Con, sh-NDRG1/SH-Con, sh-Con/SH-cav1 and sh-NDRG1/SH-cav1, respectively. ***P<0.001, relative to the respective control cells. (b) Transwell assays: cells were seeded in the upper chamber of insert and the migrated/invaded cells were examined after 24~48 h. Data represent the means±s.d. from three independent experiments. Representative photos of stained cells are shown with the original magnification of × 100; scale bar, 50 μm.
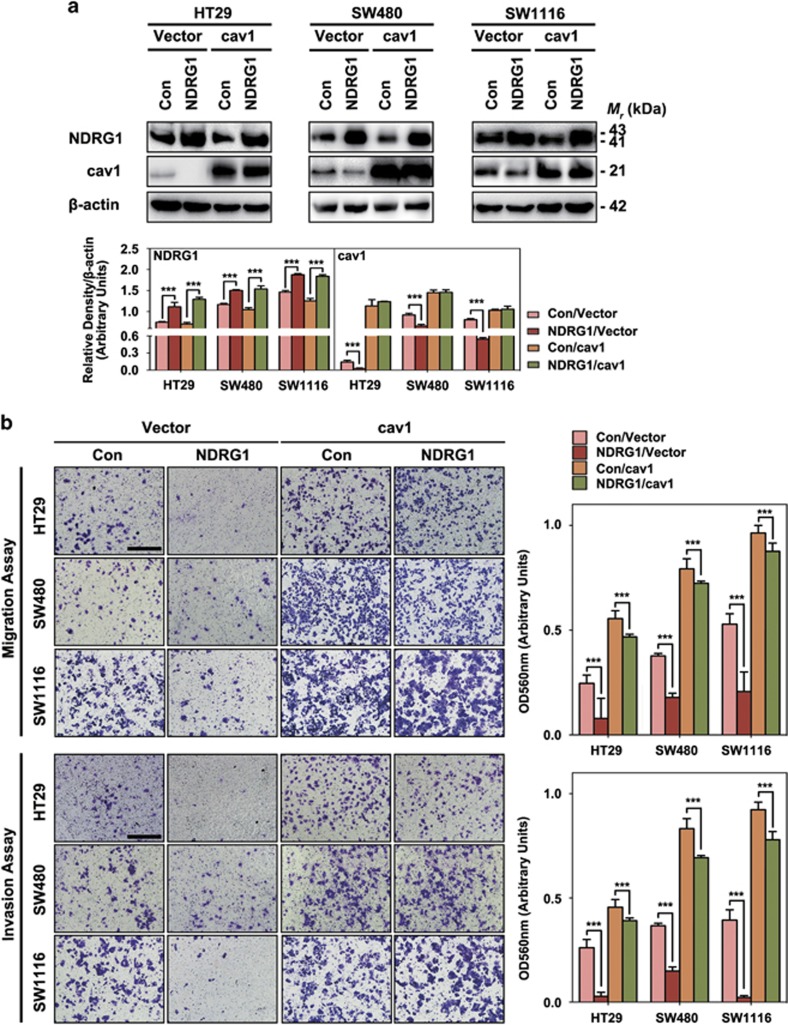
Furthermore, after cav1 was overexpressed (Figure 3a), we assessed the effect of NDRG1/cav1 overexpression on cell migration and invasion. NDRG1 overexpression alone led to a significant decrease of migratory/invasive capacity (Figure 3b, NDRG1/Vector vs Con/Vector), whereas overexpressing cav1 alone resulted in a significant (P<0.001) increase of migratory as well as invasive capacity (Figure 3b, Con/cav1 vs Con/Vector). Notably, in NDRG1/cav1 groups, the decreased enhanced migratory and invasive capacity caused by overexpression of NDRG1 was partially reversed (Figure 3b, lane NDRG1/cav1 vs NDRG1/SH-Vector).
Figure 3.
NDRG1 suppresses cancer cell migration and invasion through inhibiting caveolin-1 expression: NDRG1 and cav1 overexpression results. (a) The NDRG1 overexpression and relative control HT29, SW480 and SW11116 cells were incubated with or without caveolin-1-specific overexpression transfection (cav1). Whole-cell lysates were extracted and immunoblotting was performed to assess NDRG1 and caveolin-1 expression in cells transfected with Con/Vector, NDRG1/Vector, Con/cav1 and NDRG1/cav1, respectively. ***P<0.001, relative to the respective control cells. (b) Transwell assays: cells were seeded in upper chamber of insert and the migrated/invaded cells were examined after 24~48 h. Data represent the means±s.d. from three independent experiments. Representative photos of stained cells are shown with the original magnification of × 100; scale bar, 50 μm.
All these data indicated that cav1 promoted migration/invasion of colon cancer cells, whereas NDRG1 inhibits cell migration and invasion through reducing cav1 expression.
NDRG1 regulates EMT through modulating caveolin-1 expression
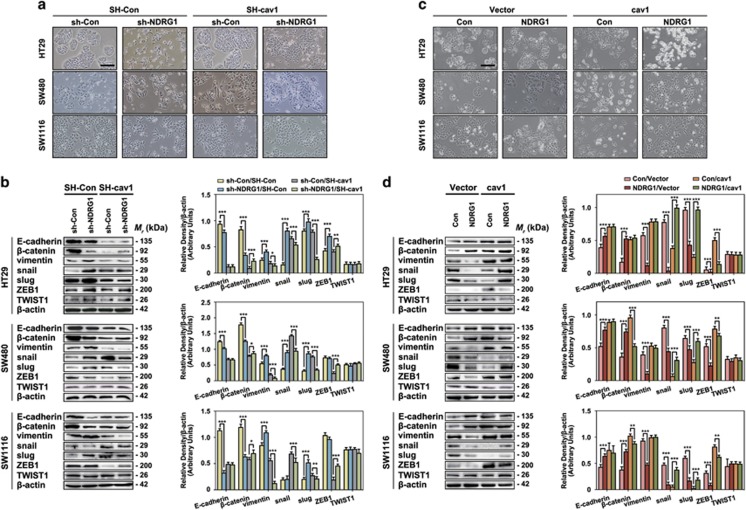
As shown in Figures 1c and 4a, sh-NDRG1 induced an obviously morphological change to a more aggressive phenotype (elongation and a reduction in cell–cell contacts), which is in agreement with our previous reports that NDRG1 expression facilitates to maintain an epithelial phenotype in cancer cells.7 Moreover, as expected, when caveolin-1 expression was suppressed, the fibroblast-like sh-NDRG1 cells experienced a transition back to an epithelial-like phenotype (cells lost polarization and clustered together), indicating that NDRG1 might inhibit EMT through reducing cav1 expression in colon cancers (Figure 4a). Furthermore, morphologically, NDRG1 overexpression significantly made the cells change to an obvious epithelial type and cav1 overexpression reversed this phenomenon (Figure 4c), which is consistent with the results shown in Figure 4a.
Figure 4.
NDRG1 regulates EMT through modulating caveolin-1 expression. (a) Bright-field images were taken to show cell morphological changes after sh-NDRG1 and/or SH-caveolin-1 treatment. Scale bar, 100 μm. (b) Immunoblot results of molecular markers of EMT after sh-NDRG1 and/or SH-caveolin-1 treatment. Densitometry analysis is expressed relative to the loading control, β-actin. Results are typical of three to five experiments and the histograms represent mean±s.d. (three to five experiments). *P<0.05; **P<0.01; ***P<0.001, relative to the respective control cells. (c) Bright-field images were taken to show cell morphological changes after NDRG1 and/or caveolin-1 overexpression treatment. Scale bar, 100 μm. (d) Immunoblot results of molecular markers of EMT after NDRG1 and/or caveolin-1 overexpression treatment. Densitometry analysis is expressed relative to the loading control, β-actin. Results are typical of three to five experiments and the histograms represent mean±s.d. (three to five experiments). **P<0.01, ***P<0.001, relative to the respective control cells.
In addition, the typical signs of EMT, the downregulation of epithelial markers (E-cadherin, β-catenin), and the upregulation of mesenchymal markers (vimentin), were reported to be regulated by NDRG1 in our previous study.3, 34 As shown in Figure 4b, sh-NDRG1 as well as SH-cav1 were able to modulate the expression of EMT markers, respectively. In details, sh-NDRG1 transfection can decrease the expression of E-cadherin (50, 20 and 86% decrease) and β-catenin (83, 72 and 69% decrease), as well as to increase the expression of vimentin (1.7-, 1.5- and 1.3-fold increase). This suggested that NDRG1 was able to inhibit EMT in CRC cells. On the other hand, in the sh-NDRG1/SH-cav1 groups, the decrease of E-cadherin and β-catenin together with the increase of vimentin induced by downregulation of NDRG1 were markedly attenuated (E-cadherin expression showed almost no alteration; β-catenin expression surprisingly showed 5-, 1.2- and 1.6-fold increase, respectively; vimentin expression showed 44%, 50% and 75% reduction, respectively) compared with the corresponding control groups (sh-Con/SH-cav1; Figure 4b). Moreover, as shown in Figure 4d, NDRG1 overexpression increased the expression of E-cadherin (1.4-, 1.4- and 1.5-fold increase) and β-catenin (3.1-, 2.1- and 1.9-fold increase), as well as to decrease the expression of mesenchymal marker, vimentin (78, 75 and 49% reduction). In the Con/cav1 groups, it also resulted the increase the expression of E-cadherin and β-catenin, as well as to decrease the expression of vimentin (Figure 4d). In the NDRG1/cav1 groups, the increasing of E-cadherin and β-catenin together with the increase of vimentin induced by overexpression of NDRG1 were partially reversed (E-cadherin and vimentin expression showed no alteration; β-catenin expression showed 46 and 15% reduction in SW480 and SW1116 cells, respectively) compared with the corresponding control groups (Con/cav1; Figure 4d).
Previously, we reported that NDRG1 had the effect of inhibiting the nuclear translocation of β-catenin.3, 34 Interestingly, the subcellular fractionation assays (Supplementary Figure 2) indicated that when NDRG1 was silenced, there was a significant accumulation of β-catenin in HT29 and SW480 cells other than in SW1116 cells in the nucleus; when NDRG1 was overexpressed, there was a marked increase of membrane β-catenin in HT29, SW480 and SW1116 cells. Furthermore, when cav1 was silenced, β-catenin expression was decreased in all different fractionations in three cell lines; while in cav1 overexpressed groups, β-catenin expression was increased as well. In addition, in sh-NDRG1/SH-cav1 HT29 and SW480 cells, there was no reverse effect of β-catenin expression in all different fractionations, but in SW1116 cells, β-catenin expression was partially reversed compared with sh-Con/SH-cav1 cells, but not totally reversed to the β-catenin expression on cell membrane in sh-Con/SH-Con groups. The same result can also be found in SW1116 NDRG1/cav1 cells compared with Con/cav1 and Con/Vector cells.
More importantly, to further discover the function of NDRG1 and cav1 in regulation of EMT, the key transcription factors, snail, slug, ZEB1 and TWIST1 were also examined. The results indicated that snail, slug and ZEB1 were involved in the NDRG1/cav1-mediated EMT modulation (Figures 4b and d). In details, sh-NDRG1 transfection was able to increase the expression of snail (5.2-, 2.5-fold increase) in HT29 and SW480 cells; it was able to increase slug (1.2-, 2.8- and 2.6-fold increase) in all three cell lines; it was able to increase ZEB1 (1.7-fold increase) in HT29 cell line (Figure 4b). NDRG1 overexpression was able to reduce the expression of snail (95, 45 and 81% reduction), slug (56, 27 and 73% reduction) and ZEB1 (69, 57 and 75% reduction) in all three cell lines (Figure 4d). However, TWIST1 did not change while NDRG1 was overexpressed or silenced. This suggested that NDRG1 was able to modulate snail-, slug- and ZEB1-induced EMT in CRC cells.
Furthermore, SH-cav1 was able to increase the expression of snail (4.2-, 2.9- and 3.6-fold increase) in all three cell lines; it was able to increase slug (2.4-, and 1.4-fold increase) in SW480 and SW1116 cell lines; it was able to decrease ZEB1 (68 and 82% reduction) in SW480 and SW1116 cell lines (Figure 4b). Cav1 overexpression was able to reduce the expression of snail (51, 92 and 90% reduction) and slug (75, 57 and 97% reduction), whereas was able to increase the expression of ZEB1 (9.8-, 1.5- and 2.6-fold increase) in all three cell lines.
Moreover, sh-NDRG1/SH-cav1 was able to partially reverse the expression of snail, slug and ZEB1 compared with sh-Con/SH-cav1 groups in all three cell lines; NDRG1/Cav1 double-overexpression was also found to have the same partial reverse effect towards the expression of snail, slug and ZEB1 compared with Con/ cav1 groups in all three cell lines.
Together, these results indicate that NDRG1 regulates EMT through modulating cav1 expression. Furthermore, this effect may be taken place via snail, slug, ZEB1 modulating signaling pathways. Moreover, there might be other mechanisms involved regarding to the NDRG1-mediated β-catenin translocation and re-distribution despite the NDRG1/cav1 co-effectiveness.
Expressions and negative correlation of NDRG1 and caveolin-1 in CRC tissues
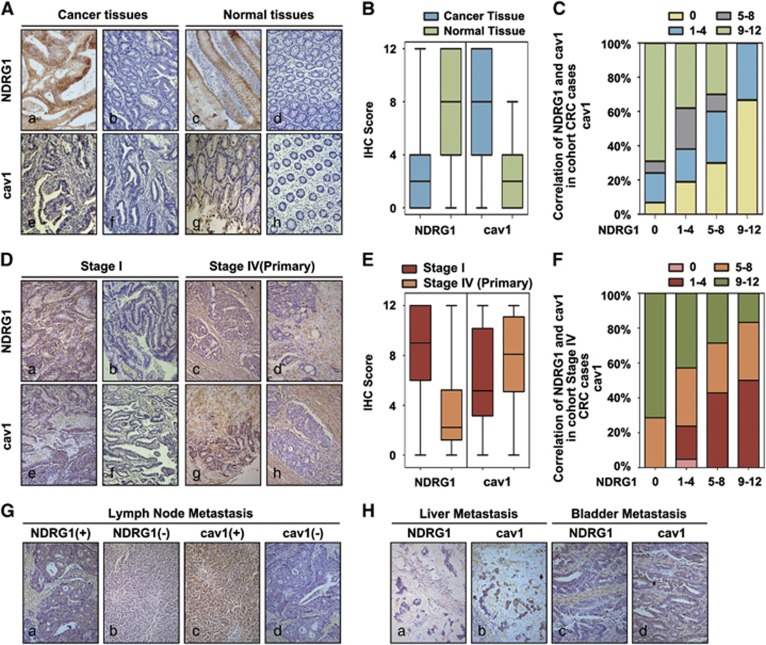
In our previous study, NDRG1 expression was markedly decreased in most CRC tissues and was negatively correlated with tumor stage, tumor differentiation and metastasis.2 Moreover, according to our literature review, there was no consensus reached on whether the cav1 expression is enhanced or inhibited in CRC. Several studies have shown that cav1 expression is reduced in human colon cancers,35, 36 which is necessary for CRC progression, whereas others have shown that cav1 is increased in colon cancer samples.37, 38 Considering our findings that NDRG1 inhibited cav1 expression in vitro, we further assessed NDRG1 and cav1 protein expressions in cancer tissues and paired normal tissues in a cohort of 64 cases of CRC patients (Figure 4). The significant lower expressions of NDRG1 were identified in cancer tissues compared with their corresponding adjacent normal tissues (Figures 5A and B). Moreover, our result showed that cav1 expression in CRC tissues was increased as compared with those in the corresponding normal tissues of the same case (Figures 5A and B). In the meantime, the increase of cav1 was also identified in the same cancer region where NDRG1 was decreased (Figure 5A). Moreover, the correlation of NDRG1 and cav1 in the cancer tissues was further investigated. As shown in Figure 5C, low NDRG1 expression is always associated with high cav1 expression, and vice versa (P<0.05).
Figure 5.
Negative correlation of NDRG1 with caveolin-1 expression in human CRC tissues. (A) Representative IHC images of CRC samples and adjacent normal tissues for the low and high expressions of NDRG1 and caveolin-1; magnification: × 100. (B) Scores for NDRG1 and caveolin-1 expression are shown as box plots, with the horizontal lines representing the median; the bottom and top of the boxes representing the 25th and 75th percentiles, respectively; and the vertical bars representing the range of data. We compared CRC tissues with matched adjacent normal tissues. (C) Box plot of NDRG1 and caveolin-1 expression in the CRC samples. The subjects were divided into two groups based on NDRG1 expression scores in the tumors, representing negative (score 0), low (scores 1–4), high (scores 5–8) and strong (score 9–12) expression of NDRG1. Data were analyzed by one-way ANOVA with Games–Howell’s correction. (D) Representative IHC images of stage I and stage IV CRC samples for the low and high expressions of NDRG1 and caveolin-1; magnification: × 100. (E) Scores for NDRG1 and caveolin-1 expression are shown as box plots in stage I and stage IV CRC samples, with the horizontal lines representing the median; the bottom and top of the boxes representing the 25th and 75th percentiles, respectively; and the vertical bars representing the range of data. We compared CRC tissues with matched adjacent normal tissues. (F) Box plot of NDRG1 and caveolin-1 expression in stage IV CRC samples. The subjects were divided into two groups based on NDRG1 expression scores in the tumors, representing negative (score 0), low (scores 1–4), high (scores 5–8) and strong (score 9–12) expression of NDRG1. Data were analyzed by one-way ANOVA with Games–Howell’s correction. (G) Representative IHC images of metastatic lymph node samples for the low and high expressions of NDRG1 and caveolin-1; magnification: × 100. (H) Representative IHC images of liver and bladder metastasis for the classical expressions of NDRG1 and caveolin-1; magnification: × 100.
We also evaluated the expression of NDRG1 and cav1 in stage I (no metastasis) and stage IV (distance metastasis) CRC cases (Figure 5D). The results indicated that the significant higher expressions of NDRG1 were identified in stage I cases compared with those in stage IV cases (Figure 5E). On the contrary, cav1 expression in stage I cases was lower than those in primary tumors in stage IV cases (Figure 5E). Moreover, the correlation of NDRG1 and cav1 in the cancer tissues was further investigated in cohort stage IV cases. As shown in Figure 5F, NDRG1 expression had a negative correlation to the expression of cav1 in primary stage IV tumors. What is worth to notice is that we have examined NDRG1 and cav1 expressions in several metastatic tissues (Figures 5G and H), and it indicated that in metastatic tissues, including lymph nodes, liver and bladder metastasis, low NDRG1 expression was also associated with high cav1 expression.
Collectively, these results suggest that NDRG1 expression has negative correlation with cav1 expression in CRC tissues.
Caveolin-1 is indispensable for the NDRG1-modulated anti-metastasis effect in vivo
Greater metastatic potential is often related to enhanced cancer migratory ability. Although the absence of NDRG1 expression was reported to be associated with activated migratory capacity of CRC cells, no direct evidence has been offered to support such a role in vivo. Therefore, we assessed whether NDRG1 could modulate the metastatic potential of CRC cells in vivo, and we wondered whether cav1 mediated the suppressive function of NDRG1 in vivo.
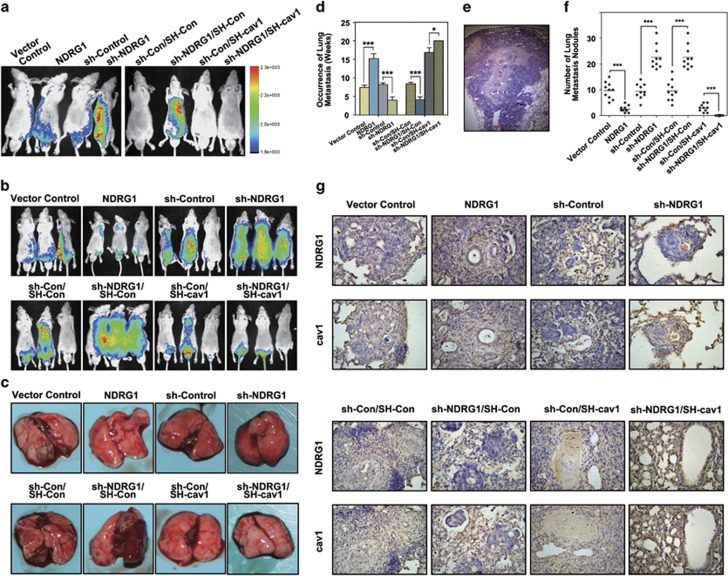
GFP-luciferase-labeled NDRG1, sh-NDRG1 HT29 cells and their relative control cells were tail-vein injected into nude mice and luciferase photon fluxes were monitored for metastasis each week (Figures 6a and b). As shown in Figure 6d, sh-NDRG1-transfected HT29 cells had evidence of earliest metastasis by ~4 weeks after the injection. However, mice injected with Vector Control and sh-Control cells did not develop any lung metastasis until 8–9 weeks after the tail-vein injection. NDRG1 overexpression cells showed metastasis in lung tissue in 5–6 weeks after the injection. After 10 weeks, almost 100% of the nude mice injected with sh-NDRG1 cells developed lung metastasis. Moreover, GFP-luciferase-labeled sh-Con/SH-Con, sh-NDRG1/SH-Con, sh-Con/SH-cav1 and sh-NDRG1/SH-cav1 HT29 cells were also tail-vein injected into nude mice and luciferase photon fluxes were monitored for metastasis each week (Figures 6a and b). As shown in Figure 6d, sh-cav1 transfected cells showed metastasis in lung tissue in 5–6 weeks after the injection. Notably, there was no detectable lung metastasis within the monitored 20 weeks after the injection of sh-NDRG1/SH-cav1 cells. All the mice were killed 20 weeks after the injection (the sh-NDRG1/SH-Con group died one after another since 5 weeks after the injection). The numbers and sizes of metastases in hematoxylin- and eosin-stained sections of lungs were counted at the time of killing (Figures 6c and e). In the sh-NDRG1 groups, a dramatically larger number of metastases in the lung tissues were found (25±7, n=10; Figure 6f). In addition, these tumor nodules tend to diffusely distribute into the whole pulmonary tissue (Figure 6e). Conversely, the NDRG1 overexpression cells developed limited numbers of metastases in the lung (Figures 6c and f). The Sh-Con/SH-cav1 cells resulted in less metastatic nodules as compared with the control group (Figures 6c and f). The Sh-NDRG1/SH-cav1 cells resulted in no metastatic nodules after 20 weeks (Figures 6c and f).
Figure 6.
Caveolin-1 is indispensable for the increased metastasis resulting from NDRG1 depletion in vivo. (a) Mice were imaged at 3 weeks after injection using Xenogen. Four representative mice were imaged and the color scale depicting the photon fluxes emitted from the tumor cells. (b) Representative images taken at 8 weeks after injection. (c) The representative images show macroscopic observations of metastases in lung at the time of killing. (d) The time of first tumor appearance of each experimental group. Results are collected and analyzed from 10 nude mice per group and the histograms represent mean±s.d. ***P<0.001, relative to the respective control cells. (e) Metastases in hematoxylin and eosin (H&E)-stained lung sections; magnification: × 40. (f) Metastases were counted on five lobes of the lung in all animal groups. The results are collected and analyzed from 10 nude mice per group and the histograms represent mean±s.d. number of metastatic nodules. ***P<0.001, relative to the respective control cells. (g) The representative IHC staining images of NDRG1 and caveolin-1 in lung metastatic tissues; magnification: × 40.
To further support the previous in vivo study, we also applied NDRG1/Vector, NDRG1/cav1 SW1116 cells and their relative control cells for tail-vein injected into nude mice (Supplementary Figure 4A). The weight of each group was monitored every 3 days, and the first time point that weight loss occurred was recorded, representing the time of first tumor appearance (Supplementary Figure 4A). NDRG1 overexpression SW1116 cells had evidence of latest occurrence of weight loss by ~35 weeks after the injection; while mice injected with Con/cav1 cells started developing weight loss from ~20 weeks after the tail-vein injection, NDRG1/cav1 double-overexpression cells showed weight loss in ~22 weeks after the injection. All the mice were killed 40 weeks after the injection. The numbers and sizes of metastases in hematoxylin- and eosin-stained sections of lungs were counted at the time of killing (Supplementary Figures 4B, C and E). In the NDRG1 overexpression group, a significant less lung metastasis was found (2.5±2, n=10; Supplementary Figure 4B). In addition, cav1 overexpressing led to earlier metastases and evolved larger amount of nodules in the lung parenchyma (Supplementary Figures 4B, C and E). Notably, cav1 overexpression cells took similar time to metastasize to the lung and developed similar amount of lung nodules to that sh-NDRG1 cells, whose cav1 expression was also high. NDRG1/cav1 double-overexpression resulted in less metastatic nodules compared with cav1 overexpression cells (Supplementary Figures 4B, C and E).
Interestingly, the cav1 expression in the lung metastases of each group was detected by immunohistochemistry (IHC). As indicated in Figure 6g, cav1 presented consistently strong positive expressions regardless of the type of injected cells, suggesting that cav1 protein experienced an accumulative process during distal metastasis. Further histological analysis shows that cav1 presented consistently strong positive expressions in the lung metastases of each group regardless of the type of injected cells. These results strongly suggest that expression of caveolin-1 can enhance the metastatic process of colon cancer cells and restore the metastatic capacity suppressed by NDRG1.
NDRG1 directly interacts with caveolin-1 in vitro
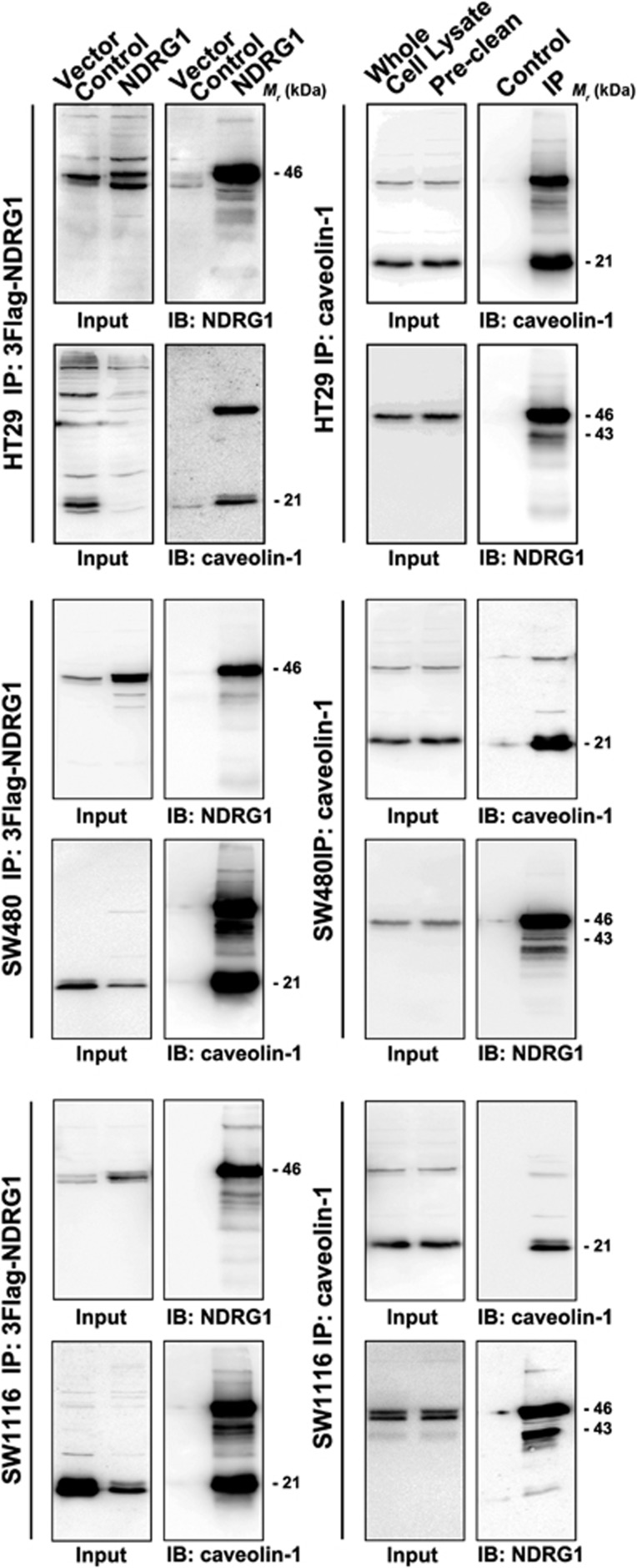
Considering that cav1 usually physically interacts with its upstream regulators and downstream effectors,11, 12, 13 in this part of experiments, we evaluated the interaction between NDRG1 and cav1 using co-immunoprecipitation assays. As shown in Figure 7, western blotting analysis on total cell lysates of HT29 cell lines (input) showed that both NDRG1 and cav1 were detected in the cells transfected with vehicle (negative control group) or 3 × Flag-tagged NDRG1 cell models. After immunoprecipitation with anti-Flag M2 affinity resin, cav1 was detected alone in the precipitates derived from the cells transfected with 3 × Flag-NDRG1, and was not detected in the negative control group. Reciprocally, after immunoprecipitation with anti-cav1 antibodies, NDRG1 was also detected in the co-immunoprecipitation complex. These results indicated that there was a direct interaction between cav1 and NDRG1 in vitro. To further confirm these results, two other colon cancer cell lines (SW480 and SW1116) were also used for co-immunoprecipitation assays. The results in these two cell lines were consistent with the results in HT29 cell line.
Figure 7.
Immunoprecipitation for NDRG1 and caveolin-1 in three CRC cell lines. Immunoprecipitation of 3 × Flag-tagged NDGR1 protein was performed using M2 beads. Caveolin-1 immunoprecipitation was performed by using protein G plus agarose beads. Caveolin-1 and NDRG1 expressions were detected by immunoblots. Caveolin-1 was detected alone in the precipitates derived from the cells transfected with 3 × Flag-NDRG1, and was not detected in the negative control group. Reciprocally, after immunoprecipitation with anti-caveolin-1 antibodies, NDRG1 was also detected in the co-immunoprecipitation (co-IP) complex. These results indicated that there was a direct interaction between caveolin-1 and NDRG1 in vitro.
NDRG1 promotes caveolin-1 ubiquitylation and degradation in vitro
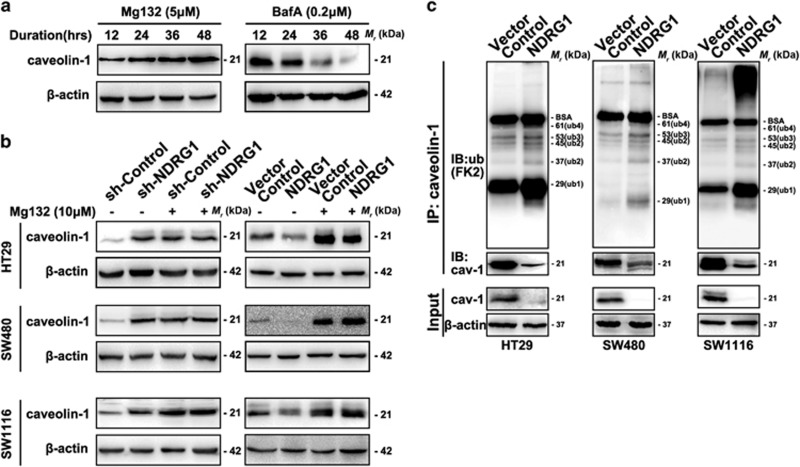
As NDRG1 physically interacts with cav1 and reduces cav1 protein expressions in CRC cell lines, we then investigated whether cav1 degradation could be modulated by NDRG1. In fact, limited amounts of reports have explored how cav1 are degraded. It has been shown that the cav1 degradation is sensitive to lysosomal inhibitors. Therefore, it is possible that it occurs in lysosomes.39 Moreover, mutants of cav1, which fail to assemble into caveolae, remain trapped in the Golgi, whereas two other members of the caveolin family (cav-2 and cav-3) are degraded through a proteasomal pathway.40, 41 Hence, the pathway of cav1 degradation was explored using both proteasome and lysosome pathway inhibitors.
Our results showed that the lysosomal inhibitor BafilomycinA1 (BafA) had no effect on cav1 degradation. Interestingly, on the contrary, BafA accelerated cav1 degradation, while the proteasomal inhibitor Mg132 markedly increased cav1 protein level in a time-dependent manner, indicating cav1 mainly degraded via proteasomal pathway in CRC cells (Figure 8a). Accordingly, Mg132 was chosen for further investigation. As shown in Figure 8b, Mg132 fully restored the cav1 protein levels that was reduced in NDRG1 overexpression cell models, whereas treating sh-Control and sh-NDRG1 cells with Mg132 resulted in a similar cav1 level. These results indicated that NDRG1 can regulate cav1 protein expression via promoting proteasomal degradation.
Figure 8.
Caveolin-1 ubiquitylation analysis. (a) Accumulation of caveolin-1 after treatment with 5 μm MG132, an inhibitor of the ubiquitin–proteasome system, and 0.2 μm lysosomal inhibitor BafilomycinA1 (BafA) were analyzed at indicated time points by western blot. (b) Caveolin-1 protein levels in Sh-Control, Sh-NDRG1, Vector Control and NDRG1-transfected HT29, SW480 and SW1116 cells treated with or without MG132 (10 μm) for 48 h, were determined. (c) CAV-1 in control cells and cells transfected with NDRG1 was immunoprecipitated using anti-cav-1 (N20) antibody after treatment with MG132 (5 μm) for 6 h. Western blots with anti-ubiquitin (FK2) antibody revealed that cav-1 ubiquitination level in immunoprecipitates prepared from cells overexpressing NDRG1 was much higher than control cells. The sizes of the various ubiquitylated forms of cav-1 (cav-1-Ub ~29 kDa; cav-1-Ub2 ~37 kDa; cav-1-Ub3 ~45 kDa and cav-1-Ub4 ~53 kDa; cav-1-Ub5~61 kDa) are indicated.
In proteasome-mediated protein degradation, protein often conjugates to form multiple small protein ubiquitin copies through isopeptide linkages.42, 43, 44 Therefore, while inhibiting the proteasome system, it can lead to the accumulation of ubiquitinated forms of proteins. To test whether NDRG1 has a role in the ubiquitylation of cav1 protein, NDRG1 overexpression cell models were chosen for immunoprecipitation assay. The results showed that anti-ubiquitin cav1 antibody (FK2) revealed multiple corresponding bands, demonstrating that NDRG1 enhanced ubiquitination of cav1 in HT29, SW480 and SW1116 cells (Figure 8c). All data suggested that the loss of cav1 upon NDRG1 expression is attributed to its ubiquitination and subsequent proteasomal degradation.
Discussion
The role of NDRG1 in inhibiting metastatic progression has been reported in a series of studies from several research teams.1, 2, 3, 4, 9 Our previous research partly elucidated the mechanisms and signaling pathways that NDRG1 may be involved in its anti-metastatic effect. However, how NDRG1 interacts with these pathways and the immediate downstream effectors of NDRG1 are still not known. In fact, available evidences have indicated that cav1 categorized into a certain group of proteins that function both as tumor suppressor/promoter of enhanced malignant behavior.45, 46, 47, 48 In our study, the tumor-promoting function of cav1 in CRC tumors was confirmed for silencing cav1 expression leading to reduced cell migration and invasion in vitro and weakened cell metastatic capacity in vivo. More importantly, we found that cav1 mediated the function of the well-known metastasis suppressor, NDRG1, because silencing cav1 was able to totally abolish the enhanced migration, invasion and metastasis due to NDRG1 depletion. Also, in patient samples, consistent with its tumor-promoting function, cav1 has been shown upregulated in cancer tissues compared with their corresponding adjacent tissues. Higher cav1 expression was always accompanied by lower NDRG1 expression in CRC tissues, which further demonstrates the negative association between the two proteins in tumor progression.
The EMT is one of the key theory for cancer migration, invasion and metastasis.49, 50 In this study, we showed that changes of both phenotype and EMT markers induced by NDRG1 depletion could be reversed by silencing cav1 expression, while cav1 overexpression can also reverse the effect of NDRG1 overexpression towards the changes of phenotype and EMT markers. This demonstrates that NDRG1 regulates EMT through modulating cav1 expression and this, in part, explains NDRG1-suppressive function in cell migration, invasion and metastasis mediated by cav1.
Previous studies in colon cancer presented conflicting data about the association of cav1 expression with tumor progression.8, 36, 37, 51 For example, the downregulation of cav1 reduced tumorigenesis in CRC cells.36 However, the cancer cells obtained from secondary metastasis, showed increased endogenous cav1 when compared with the parental primary tumor-derived cells,52 suggesting that activation of cav1 may take place during metastasis. It seems that this functional ‘switch’ can occur within the same cell. One theory to resolve this apparent ambiguity is to consider the possibility that the role of cav1 varies according to different tumor stage.13, 17, 23 In the early stages of cell transformation, promotion of cell proliferation and reduction of apoptosis was thought to be caused by the loss of cav1. Nevertheless, during tumor progression, several intracellular and extracellular changes take place at the molecular level that not only limit the ability of cav1 as a tumor suppressor, but then produce a ‘permissive’ cellular microenvironment that permits cav1 to operate in the opposite function.23 The mechanisms that contribute to such ‘permissive’ cellular environment constitute an area of great interest. Our results indicate that loss of NDRG1 in tumor progression is critical in this respect. We identified that NDRG1 directly promote cav1 degradation. Hence, loss of NDRG1 is responsible for cav1 accumulation during metastasis. Interestingly, in our animal experiment, though overexpressing and/or silencing NDRG1 obviously inhibited/enhanced metastasis, once secondary metastatic lesions formed in the lung, cav1 expression in these tumors was surprisingly high. There may exist some mechanisms that help cav1 to escape from NDRG1 control and go out of degradation pathways.
To understand the possible mechanisms for the increase in cav1 levels on loss of NDRG1, we focused on the cellular degradative machinery. We treated cav1-expressing colon cells with MG132 (classical proteasomal inhibitors)53, 54 and BafA (lysosomal inhibitor, which has been reported to inhibit CAV1 degradation in HeLa cells)55 for a period of up to 48 h. We discovered that only the proteasomal inhibitors have a positive effect on CAV1 expression. We demonstrated here cav1, which has been proved to be a long-lived protein expected to be degraded through lysosomal pathway, degraded through a proteasomal pathway. Furthermore, we found that NDRG1 enhanced cav1 ubiquitylation and subsequent proteasomal degradation. This is the first report about the involvement of NDRG1 in the ubiquitin–proteasome pathway related to tumor progression.
In conclusion, the present study shows that NDRG1 interacts with cav1, reducing cav1 protein expression by promoting cav1 ubiquitylation, which modulates EMT in the CRC cells. These results help to fulfill the potential mechanisms of NDRG1 in anti-metastatic treatment for human CRC.
Materials and methods
Cell culture
All cell lines were obtained from the American Type Culture Collection (ATCC) and grown under established conditions:3, 56 the HT29 human CRC cells cultured in McCoy’s 5A (Gibco, Grand Island, NY, USA) with 10% fetal bovine serum (Gibco); the human CRC cell lines, SW480 and SW1116, cultured in RPMI-1640 (Gibco) with 10% fetal bovine serum (Gibco).
Plasmid construction/transfection
The plasmid and short hairpin RNAs (shRNAs) used for transfection of NDRG1 were pBABE-3 × Flag-NDRG1 and pSIREN-shRNA (Clontech, Mountain View, CA, USA), respectively, as we have previously described.56 The GFP-labeled caveolin-1-specific shRNA and caveolin-1 overexpression plasmids were obtained from GeneChem Co. Ltd., Shanghai, China.
RNA extraction and real-time polymerase chain reaction
Real-time PCR was performed for quantification of RNA. In brief, we extracted total RNA from culture cells with Trizol reagent (Invitrogen, Carlsbad, CA, USA). Complementary DNA (cDNA) was synthesized using the High-Capacity cDNA Archive kit according to the manufacturer’s instructions (Applied Biosystems, Life Technologies, Carlsbad, CA, USA). Subsequently, real-time PCR was performed using the LightCycler 480 System (Roche Applied Science, Mannheim, Germany). The cycler protocol was 5 min at 95 °C, 40 cycles of 15 s at 95 °C, 60 s at 60 °C and 5 min at 72 °C. Gene of interest expression was normalized to the reference genes GAPDH. NDRG1 (sense primer: 5′-CTGCACCTGTTCATCAATGC-3′ anti-sense primer: 5′-AGAGAAGTGACGCTGGAACC-3′)12 and caveolin-1 (sense primer: 5′-GGGCAACATCTAGAAGCCCAACAA-3′ anti-sense primer 5′-CTGATGCACTGAATTCCAATCAGGAA-3′) were assessed. Fold expression was calculated using the 2−ΔΔCt method.
Cell migration and invasion assay
The transwell migration and invasion assay was performed using the Cytoselect 24-well cell Migration and Invasion Assay kit (Cell Biolabs, San Diego, CA, USA) according to the manufacturer’s instruction.56
Protein extraction and immunoblots
The preparation of cell lysates and immunoblots analysis were performed via established protocols.56, 57 The Subcellular Protein Fractionation Kit for Cultured Cells from Thermo Scientific (78840; Waltham, MA, USA) was utilized to prepare fractions according to the manufacturer's instructions. Primary antibodies: anti-NDRG1 (HPA006881), anti-β-actin (A1978) from Sigma-Aldrich (St. Louis, MO, USA); anti-caveolin-1 (N20) from Santa Cruz Biotechnology (Santa Cruz, CA, USA); anti-vimentin (#5741), anti-E-cadherin (#3195), anti-β-catenin (9562), anti-snail (#3879), anti-slug (#9585), anti-ZEB1 (#3396), anti-TWIST1 (#46702), anti-Na-K-ATPase (#3010), anti-GAPDH (#2118), anti-Histone H2A (#3636) were from Cell Signaling Technology (Beverly, MA, USA). The secondary antibodies such as horseradish peroxidase (HRP)-conjugated anti-rabbit (A6154), anti-goat (A5420) and anti-mouse (A4416) antibodies were from Sigma-Aldrich.
IHC and scoring of IHC staining results
The experimental study was approved by the Ethic Committee of Shanghai Ruijin Hospital and informed consent was obtained from all the cases enrolled in this study. A cohort of 64 pairs of human CRC tissues and paired adjacent normal tissues, together with a cohort of 126 cases of different stages of CRC tissues were collected, fixed with formaldehyde and embedded with paraffin. IHC staining was performed as previously described.2 Cases with discrepant scores were re-scored by the same or additional scorers to obtain a consensus score. NDRG1 and caveolin-1 scoring was blinded to be done by different pathologists according to the German semi-quantitative scoring system.58, 59
Immunofluorescence
Immunofluorescence analysis was carried out as described previously.3, 56 The images were examined and captured using an Olympus Fluoview confocal Microscope. Rabbit mAb lgG XP isotype (Cell Signaling Technology) was used as negative control.
Co-immunoprecipitation
Immunoprecipitation was performed by an established method.57 Immunoprecipitation of 3 × Flag-tagged NDGR1 protein was performed using M2 Beads (Sigma-Aldrich). Caveolin-1 immunoprecipitation was performed by using protein G plus agarose (Santa Cruz Biotechnology). NDRG1 and cav1 proteins were detected by immunoblot analysis.
Caveolin-1 ubiquitylation assay
The cells were treated with Mg132 (5 μg/ml) for 6 h to inhibit proteasome activity and then lysed using SDS-free RIPA buffer and immunoprecipitated with anti-caveolin-1 antibody followed by protein G plus agarose. The samples were then subjected to SDS–PAGE to detect caveolin-1 and ubiquitin (FK2, Enzo Life Sciences, New York, NY, USA).
Animal experiments
Four to 6 weeks old (weighing ~20 g) BALB/c nude mice (male/female ratio 1:1) were obtained from the Shanghai Institute of Materia Medica, Chinese Academy of Science, and maintained under specific pathogen-free conditions. No statistical method was applied for the sample size estimation for the animal study, although to ensure the precision of the results, each experimental group had enrolled 8–10 nude mice in an un-randomized manner. The experimental protocol was approved by the Shanghai Medical Experimental Animal Care Commission. The cells (5 × 106 in 300 ml PBS) were tail-vein injected into the nude mice to establish the ‘experimental metastatic’ model. The photon fluxes (photons per second) emitted from tumor cells were monitored by Bioluminescence Imaging to detect the occurrence of metastasis. The bioluminescence images were acquired with the IVIS Imaging System (PerkinElmer, Akron, OH, USA). Analysis was performed with Living Image software (PerkinElmer) by measuring photon flux of chest and upper abdominal region. On the indicated post-injection day, the mice were killed and photographed. All the lung tissues were embedded in paraffin, followed by hematoxylin and eosin and IHC staining. Metastases were counted on five lobes of the lung after hematoxylin and eosin staining. No blinding was done for the animal experiments.
Statistic analysis
The data are expressed as mean±s.d. of ⩾3 independent experiments and were statistically analyzed using Student’s t-test. The results were considered significant when P<0.05.
Acknowledgments
The study is financially supported by National High-tech Research and Development Projects (863) (2012AA021103), National Natural Science Foundation of China (NSFC) (81402423, 81572818), Science and Technology Commission of Shanghai Municipality (13JC1404100, 14YF1402800).
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Liu W, Iiizumi-Gairani M, Okuda H, Kobayashi A, Watabe M, Pai SK et al. KAI1 gene is engaged in NDRG1 gene-mediated metastasis suppression through the ATF3-NFkappaB complex in human prostate cancer. J Biol Chem 2011; 286: 18949–18959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Sun J, Feng B, Ma J, Zang L, Dong F et al. The metastasis suppressor, N-myc downregulated gene 1 (NDRG1), is a prognostic biomarker for human colorectal cancer. PLoS One 2013; 8: e68206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zhang D, Yue F, Zheng M, Kovacevic Z, Richardson DR. The iron chelators Dp44mT and DFO inhibit TGF-beta-induced epithelial-mesenchymal transition via up-regulation of N-Myc downstream-regulated gene 1 (NDRG1). J Biol Chem 2012; 287: 17016–17028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Xing F, Iiizumi-Gairani M, Okuda H, Watabe M, Pai SK et al. N-myc downstream regulated gene 1 modulates Wnt-beta-catenin signalling and pleiotropically suppresses metastasis. EMBO Mol Med 2012; 4: 93–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, de la Monte S, Wands JR. The p85beta regulatory subunit of PI3K serves as a substrate for PTEN protein phosphatase activity during insulin mediated signaling. Biochem Biophys Res Commun 2010; 397: 513–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoi F, Izumi H, Kawahara A, Murakami Y, Kinoshita H, Kage M et al. N-myc downstream regulated gene 1/Cap43 suppresses tumor growth and angiogenesis of pancreatic cancer through attenuation of inhibitor of kappaB kinase beta expression. Cancer Res 2009; 69: 4983–4991. [DOI] [PubMed] [Google Scholar]
- Kovacevic Z, Chikhani S, Lui GY, Sivagurunathan S, Richardson DR. The iron-regulated metastasis suppressor NDRG1 targets NEDD4L, PTEN, and SMAD4 and inhibits the PI3K and Ras signaling pathways. Antioxid Redox Signal 2013; 18: 874–887. [DOI] [PubMed] [Google Scholar]
- Pearce LR, Sommer EM, Sakamoto K, Wullschleger S, Alessi DR. Protor-1 is required for efficient mTORC2-mediated activation of SGK1 in the kidney. Biochem J 2011; 436: 169–179. [DOI] [PubMed] [Google Scholar]
- Sun J, Zhang D, Bae DH, Sahni S, Jansson P, Zheng Y et al. Metastasis suppressor, NDRG1, mediates its activity through signaling pathways and molecular motors. Carcinogenesis 2013; 34: 1943–1954. [DOI] [PubMed] [Google Scholar]
- Okamoto T, Schlegel A, Scherer PE, Lisanti MP. Caveolins, a family of scaffolding proteins for organizing ‘preassembled signaling complexes’ at the plasma membrane. J Biol Chem 1998; 273: 5419–5422. [DOI] [PubMed] [Google Scholar]
- Parton RG, Simons K. The multiple faces of caveolae. Nat Rev Mol Cell Biol 2007; 8: 185–194. [DOI] [PubMed] [Google Scholar]
- Williams TM, Lisanti MP. Caveolin-1 in oncogenic transformation, cancer, and metastasis. Am J Physiol Cell Physiol 2005; 288: C494–C506. [DOI] [PubMed] [Google Scholar]
- Cohen AW, Hnasko R, Schubert W, Lisanti MP. Role of caveolae and caveolins in health and disease. Physiol Rev 2004; 84: 1341–1379. [DOI] [PubMed] [Google Scholar]
- Gonzalez E, Nagiel A, Lin AJ, Golan DE, Michel T. Small interfering RNA-mediated down-regulation of caveolin-1 differentially modulates signaling pathways in endothelial cells. J Biol Chem 2004; 279: 40659–40669. [DOI] [PubMed] [Google Scholar]
- Patel HH, Murray F, Insel PA. Caveolae as organizers of pharmacologically relevant signal transduction molecules. Annu Rev Pharmacol Toxicol 2008; 48: 359–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamabhai M, Anderson RG. Second cysteine-rich region of epidermal growth factor receptor contains targeting information for caveolae/rafts. J Biol Chem 2002; 277: 24843–24846. [DOI] [PubMed] [Google Scholar]
- Burgermeister E, Liscovitch M, Rocken C, Schmid RM, Ebert MP. Caveats of caveolin-1 in cancer progression. Cancer Lett 2008; 268: 187–201. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000; 100: 57–70. [DOI] [PubMed] [Google Scholar]
- Liscovitch M, Burgermeister E, Jain N, Ravid D, Shatz M, Tencer L. Caveolin and cancer: A complex relationship. In: Mattson MP (ed). Membrane Microdomain Signaling: Lipid Rafts in Biology and Medicine, chapter 9. Humana Press: Totowa, NJ, USA, 2005, pp 161–190.
- Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet 2000; 24: 227–235. [DOI] [PubMed] [Google Scholar]
- van Golen KL. Is caveolin-1 a viable therapeutic target to reduce cancer metastasis? Expert Opin Ther Targets 2006; 10: 709–721. [DOI] [PubMed] [Google Scholar]
- Navarro A, Anand-Apte B, Parat MO. A role for caveolae in cell migration. FASEB J 2004; 18: 1801–1811. [DOI] [PubMed] [Google Scholar]
- Goetz JG, Lajoie P, Wiseman SM, Nabi IR. Caveolin-1 in tumor progression: the good, the bad and the ugly. Cancer Metastasis Rev 2008; 27: 715–735. [DOI] [PubMed] [Google Scholar]
- Zhang W, Razani B, Altschuler Y, Bouzahzah B, Mostov KE, Pestell RG et al. Caveolin-1 inhibits epidermal growth factor-stimulated lamellipod extension and cell migration in metastatic mammary adenocarcinoma cells (MTLn3). Transformation suppressor effects of adenovirus-mediated gene delivery of caveolin-1. J Biol Chem 2000; 275: 20717–20725. [DOI] [PubMed] [Google Scholar]
- Ho CC, Huang PH, Huang HY, Chen YH, Yang PC, Hsu SM. Up-regulated caveolin-1 accentuates the metastasis capability of lung adenocarcinoma by inducing filopodia formation. Am J Pathol 2002; 161: 1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racine C, Belanger M, Hirabayashi H, Boucher M, Chakir J, Couet J. Reduction of caveolin 1 gene expression in lung carcinoma cell lines. Biochem Biophys Res Commun 1999; 255: 580–586. [DOI] [PubMed] [Google Scholar]
- Wikman H, Kettunen E, Seppanen JK, Karjalainen A, Hollmen J, Anttila S et al. Identification of differentially expressed genes in pulmonary adenocarcinoma by using cDNA array. Oncogene 2002; 21: 5804–5813. [DOI] [PubMed] [Google Scholar]
- Lee SW, Reimer CL, Oh P, Campbell DB, Schnitzer JE. Tumor cell growth inhibition by caveolin re-expression in human breast cancer cells. Oncogene 1998; 16: 1391–1397. [DOI] [PubMed] [Google Scholar]
- Wiechen K, Diatchenko L, Agoulnik A, Scharff KM, Schober H, Arlt K et al. Caveolin-1 is down-regulated in human ovarian carcinoma and acts as a candidate tumor suppressor gene. Am J Pathol 2001; 159: 1635–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Truong LD, Timme TL, Ren C, Wheeler TM, Park SH et al. Elevated expression of caveolin is associated with prostate and breast cancer. Clin Cancer Res 1998; 4: 1873–1880. [PubMed] [Google Scholar]
- Karam JA, Lotan Y, Roehrborn CG, Ashfaq R, Karakiewicz PI, Shariat SF. Caveolin-1 overexpression is associated with aggressive prostate cancer recurrence. Prostate 2007; 67: 614–622. [DOI] [PubMed] [Google Scholar]
- Bartz R, Zhou J, Hsieh JT, Ying Y, Li W, Liu P. Caveolin-1 secreting LNCaP cells induce tumor growth of caveolin-1 negative LNCaP cells in vivo. Int J Cancer 2008; 122: 520–525. [DOI] [PubMed] [Google Scholar]
- Yang G, Addai J, Wheeler TM, Frolov A, Miles BJ, Kadmon D et al. Correlative evidence that prostate cancer cell-derived caveolin-1 mediates angiogenesis. Hum Pathol 2007; 38: 1688–1695. [DOI] [PubMed] [Google Scholar]
- Jin R, Liu W, Menezes S, Yue F, Zheng M, Kovacevic Z et al. The metastasis suppressor NDRG1 modulates the phosphorylation and nuclear translocation of beta-catenin through mechanisms involving FRAT1 and PAK4. J Cell Sci 2014; 127: 3116–3130. [DOI] [PubMed] [Google Scholar]
- Bender F, Montoya M, Monardes V, Leyton L, Quest AF. Caveolae and caveolae-like membrane domains in cellular signaling and disease: identification of downstream targets for the tumor suppressor protein caveolin-1. Biol Res 2002; 35: 151–167. [DOI] [PubMed] [Google Scholar]
- Bender FC, Reymond MA, Bron C, Quest AF. Caveolin-1 levels are down-regulated in human colon tumors, and ectopic expression of caveolin-1 in colon carcinoma cell lines reduces cell tumorigenicity. Cancer Res 2000; 60: 5870–5878. [PubMed] [Google Scholar]
- Fine SW, Lisanti MP, Galbiati F, Li M. Elevated expression of caveolin-1 in adenocarcinoma of the colon. Am J Clin Pathol 2001; 115: 719–724. [DOI] [PubMed] [Google Scholar]
- Patlolla JM, Swamy MV, Raju J, Rao CV. Overexpression of caveolin-1 in experimental colon adenocarcinomas and human colon cancer cell lines. Oncol Rep 2004; 11: 957–963. [PubMed] [Google Scholar]
- Hill MM, Bastiani M, Luetterforst R, Kirkham M, Kirkham A, Nixon SJ et al. PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell 2008; 132: 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbiati F, Volonte D, Minetti C, Bregman DB, Lisanti MP. Limb-girdle muscular dystrophy (LGMD-1C) mutants of caveolin-3 undergo ubiquitination and proteasomal degradation. Treatment with proteasomal inhibitors blocks the dominant negative effect of LGMD-1C mutanta and rescues wild-type caveolin-3. J Biol Chem 2000; 275: 37702–37711. [DOI] [PubMed] [Google Scholar]
- Razani B, Engelman JA, Wang XB, Schubert W, Zhang XL, Marks CB et al. Caveolin-1 null mice are viable but show evidence of hyperproliferative and vascular abnormalities. J Biol Chem 2001; 276: 38121–38138. [DOI] [PubMed] [Google Scholar]
- Ratner JN, Balasubramanian B, Corden J, Warren SL, Bregman DB. Ultraviolet radiation-induced ubiquitination and proteasomal degradation of the large subunit of RNA polymerase II. Implications for transcription-coupled DNA repair. J Biol Chem 1998; 273: 5184–5189. [DOI] [PubMed] [Google Scholar]
- Treier M, Staszewski LM, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 1994; 78: 787–798. [DOI] [PubMed] [Google Scholar]
- Bregman DB, Halaban R, van Gool AJ, Henning KA, Friedberg EC, Warren SL. UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proc Natl Acad Sci USA 1996; 93: 11586–11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan TY, Zhu TN, Cui YJ, Zhang G, Song XJ, Gao DM et al. Expression of caveolin-1 is correlated with lung adenocarcinoma proliferation, migration, and invasion. Med Oncol 2015; 32: 207. [DOI] [PubMed] [Google Scholar]
- Quest AF, Lobos-Gonzalez L, Nunez S, Sanhueza C, Fernandez JG, Aguirre A et al. The caveolin-1 connection to cell death and survival. Curr Mol Med 2013; 13: 266–281. [DOI] [PubMed] [Google Scholar]
- Thompson TC, Timme TL, Li L, Goltsov A. Caveolin-1, a metastasis-related gene that promotes cell survival in prostate cancer. Apoptosis 1999; 4: 233–237. [DOI] [PubMed] [Google Scholar]
- Zhu M, LEM L, Starzec A, Stierle V, Marbeuf-Gueye C. Caveolin-1 and doxorubicin-induced P-glycoprotein modulate plasma cholesterol membrane accessibility in erythrolymphoblastic cell line. Anticancer Res 2010; 30: 3451–3458. [PubMed] [Google Scholar]
- Christiansen JJ, Rajasekaran AK. Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res 2006; 66: 8319–8326. [DOI] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest 2009; 119: 1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi B, Strugnell SS, Goetz JG, Kojic LD, Cox ME, Griffith OL et al. Phosphorylated caveolin-1 regulates Rho/ROCK-dependent focal adhesion dynamics and tumor cell migration and invasion. Cancer Res 2008; 68: 8210–8220. [DOI] [PubMed] [Google Scholar]
- Torres VA, Tapia JC, Rodriguez DA, Lladser A, Arredondo C, Leyton L et al. E-cadherin is required for caveolin-1-mediated down-regulation of the inhibitor of apoptosis protein survivin via reduced beta-catenin-Tcf/Lef-dependent transcription. Mol Cell Biol 2007; 27: 7703–7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science 1995; 268: 533–539. [DOI] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 1994; 78: 761–771. [DOI] [PubMed] [Google Scholar]
- Hayer ASM, Ritz D, Engel S, Meyer HH, Helenius A. Caveolin-1 is ubiquitinated and targeted to intralumenal vesicles in endolysosomes for degradation. J Cell Biol 2010; 191: 615–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Zhang D, Zheng Y, Zhao Q, Zheng M, Kovacevic Z et al. Targeting the metastasis suppressor, NDRG1, using novel iron chelators: regulation of stress fiber-mediated tumor cell migration via modulation of the ROCK1/pMLC2 signaling pathway. Mol Pharmacol 2013; 83: 454–469. [DOI] [PubMed] [Google Scholar]
- Kovacevic Z, Fu D, Richardson DR. The iron-regulated metastasis suppressor, Ndrg-1: identification of novel molecular targets. Biochim Biophys Acta 2008; 1783: 1981–1992. [DOI] [PubMed] [Google Scholar]
- Kok LF, Lee MY, Tyan YS, Wu TS, Cheng YW, Kung MF et al. Comparing the scoring mechanisms of p16INK4a immunohistochemistry based on independent nucleic stains and independent cytoplasmic stains in distinguishing between endocervical and endometrial adenocarcinomas in a tissue microarray study. Arch Gynecol Obstet 2010; 281: 293–300. [DOI] [PubMed] [Google Scholar]
- Koo CL, Kok LF, Lee MY, Wu TS, Cheng YW, Hsu JD et al. Scoring mechanisms of p16INK4a immunohistochemistry based on either independent nucleic stain or mixed cytoplasmic with nucleic expression can significantly signal to distinguish between endocervical and endometrial adenocarcinomas in a tissue microarray study. J Transl Med 2009; 7: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.