Abstract
With the advent of post-genomic era, new technologies create extraordinary possibilities for diagnostics and personalized therapy, transforming todays’ medicine. Rooted in both medical genetics and clinical psychiatry, the paper is designed as an integrated source of information of the current and potential future application of emerging genomic technologies as diagnostic tools in psychiatry, moving beyond the classical concept of patient approach. Selected approaches are presented, starting from currently used technologies (next-generation sequencing (NGS) and microarrays), followed by newer options (reverse phenotyping). Next, we describe an old concept in a new light (endophenotypes), subsequently coming up with a sophisticated and complex approach (gene networks) ending by a nascent field (computational psychiatry). The challenges and barriers that exist to translate genomic research to real-world patient assessment are further discussed. We emphasize the view that only a paradigm shift can bring a fundamental change in psychiatric practice, allowing to disentangle the intricacies of mental diseases. All the diagnostic methods, as described, are directed at uncovering the integrity of the system including many types of relations within a complex structure. The integrative system approach offers new opportunity to connect genetic background with specific diseases entities, or concurrently, with symptoms regardless of a diagnosis. To advance the field, we propose concerted cross-disciplinary effort to provide a diagnostic platform operating at the general level of genetic pathogenesis of complex-trait psychiatric disorders rather than at the individual level of a specific disease.
Life is really simple, but we insist on making it complicated. Confucius
If our brains were simple enough for us to understand them, we'd be so simple that we couldn’t. Ian Stewart, The Collapse of Chaos: Discovering Simplicity in a Complex World
Introduction
Understanding of the genetic underpinnings of psychiatric disorders is expected to substantially influence the classification and management of these conditions.1 All major psychiatric diseases such as, schizophrenia (SCZ), bipolar disorder, obsessive-compulsive disorder (OCD), major depression disorder (MDD), anxiety disorders (AD), autism and attention deficit hyperactivity disorder (ADHD) are genetically complex. The polygenic architecture of psychiatric traits is determined by various combinations of interacting factors such as numerous common and rare single nucleotide polymorphisms (SNPs), small indels, copy number variations (CNVs) and large chromosomal rearrangements.2, 3, 4, 5 The Psychiatric Genomics Consortium (PGC), the largest scientific network in the history of psychiatry, using Genome-Wide Association Study (GWAS) data, has demonstrated that a considerable proportion of the heritability of mental illnesses is attributable to the aggregate effect of common genetic variants.6 In contrast to classical methods of candidate-gene studies, only genome-wide approaches could, at least partially, disentangle complex genetic architecture and offer insight into the biological background of mental diseases. National Institutes of Mental Health (NIMH) dedicated excellent research strategies to look across the human genome to foster rapid progress in the prevention and management of psychiatric conditions. Nevertheless, despite the success of genome-wide approaches, the current prospects of using these results to modify current diagnostic strategies are still limited.7 Large ongoing projects are contributing to the effort to create diagnostic genetic tests that may influence routine clinical practice in psychiatry. However, neuroscience is widely criticized for its overambitious claims in this regard, because there are unexplained disparities between psychiatry based on signs and symptoms, and neurobiological findings. The lack of scientific validity at the core of psychiatry consequently affects potential prospects for accurate scientific tests for mental disorders that could be widely translated into clinical care.
Should a given psychiatric disorder be a clinical or genetic diagnosis?
Currently, psychiatric diseases are classified by the collection of symptoms together with observed clinical phenotypes8 as outlined in Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM-5) (www.dsm5.org), being heavily criticized for its lack of biological underpinnings.9 The difficulties in assigning a symptom to a specific diagnostic category are further complicated by several factors as the ability of patients to consistently verbalize their experience and perceptive capacity of a health professional. As DSM-5 lacks scientific validity, descriptive DSM categories, originating from XIX century, cannot serve as a benchmark for molecular classification of psychiatric entities. On the other hand, the genomic studies to date have not been able to propose any alternative system, and the genetic predispositions to psychiatric disorders tend to cross these descriptive categories.10 To further explore the molecular overlap between mental disorders, NIMH launched Research Domain Criteria (RDoC) initiative. The main goal of RDoC is to develop a new framework for the major psychiatric traits, on the basis of both biological background and human behavior11 (www.nimh.nih.gov/research-priorities/rdoc/index.shtml).9 The RDoC attempts to link a phenotype to the underlying biological structure and to the genetic predispositions across current DSM-5 taxonomy. The search for a disease-specific genetic test is further complicated by extensive comorbidity across these disorders, resulting from overlapping diagnostic criteria and their descriptive character. Furthermore, in the clinical reality, an idea of well-defined psychiatric disease is rather being replaced by a large spectrum. The debate regarding new taxonomy of mental disorders build on genetic markers is ongoing, with the ultimate hope of developing a biologically based diagnostic system of these conditions.
Diagnostic genetic tests in psychiatry—at present
Currently, the only true opportunity for psychiatric diagnostic tests is the use to complement the diagnostic process in a few clinical situations:
Predicting the response or adverse effects of a drug. Pharmacogenetic markers can support therapeutic decisions in psychiatry, thus decreasing the risk of treatment failure and severe side-effects. 12 Currently, new guidelines for the use of CYP450 testing are available (www.pharmgkb.org). 13
Excluding disorders mimicking psychiatric conditions or causing symptoms specific to mental diseases (that is, certain neurometabolic or neurodegenerative diseases, gangliosidoses and other storage disorders, porphyrias). 14
Confirming a diagnosis or supporting the selection of therapy. However, only a few such tests are currently used (for example, in Fragile X syndrome, phenylketonuria, Down syndrome and 22q11 deletion syndrome). 14 The detection of a rare pathogenic variant may open new possibilities of personalized care for carriers. 15
Detecting variants associated with a higher risk for major psychiatric disorders, although this possibility is controversial. It is not known whether results of various tests alone or in combination might help determine the overall risk or affect treatment decisions. At present, search for de novo rare and disruptive mutations in genes critical for the function of the brain, particularly in extreme phenotypes and in families with several affected members, have indicated this to be a powerful approach.
Next-generation sequencing
Currently, next-generation sequencing (NGS) contributes to the global effort to identify genes affecting brain function.1 Whole-exome sequencing (WES) offers advantages over single-gene testing for the complex psychiatric traits because it simultaneously analyzes the coding regions of thousands of genes.1 Compared with the traditional genetic testing, NGS sheds new light on the genetic underpinnings of psychiatric conditions because of the amount and diversity of variants that this technology can reveal. Currently, NGS testing can be justified for use in:
neurodevelopmental disorders,
early-onset neuropsychiatric conditions,
patients without a family history of any relevant psychiatric disease (to search for de novo variants),
members of a family with an extensive history of mental illnesses (to search for transmitted variants).
Exome sequencing may uncover genetic defects associated with both rare diseases and complex traits, particularly when it is combined with sequencing of the exome-flanking non-coding regions.9, 16 The analysis of the results should refer to the available published data. Currently, a key message for the clinical application of NGS is the accuracy and caution required during data interpretation. It is imperative to use genomic databases and conduct appropriate bioinformatics analyses. Furthermore, it is important to undertake protein structural analysis and non-routine functional studies when interpreting results in undetermined cases. Moreover, the complexity of data interpretation must be made clear to patients and their families.1 Different diagnostic approaches on the basis of NGS technology are presented in Box 1.
Next-generation sequencing—diagnostic approaches.
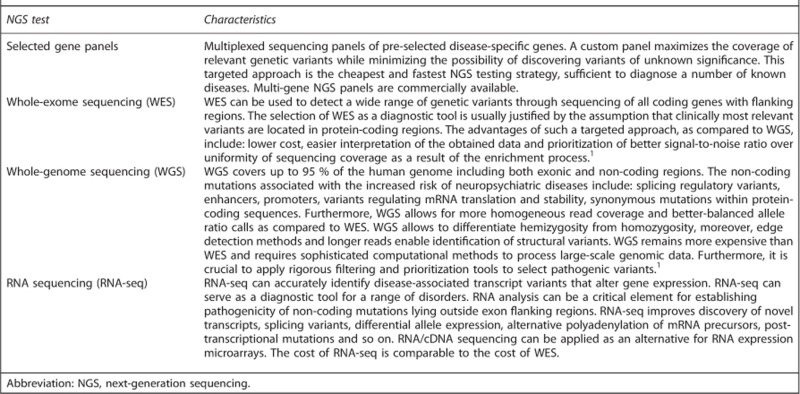
Microarrays testing in psychiatry
At present, NGS is an unprecedented source of information on single nucleotide variants, whereas microarray-based comparative genomic hybridization techniques along with cytogenetic techniques remain the most important tool for the analysis aimed at the detection of structural variants (that is, variants from >1000 bp to those involving megabases of DNA). Even though CNVs are not disease-specific, structural variants have been confirmed as risk factors for a variety of mental disorders.17 Large CNVs segregating at rare frequencies (>1 Mb and present in <1% of individuals) increase the risk for SCZ, ASD, substance use disorder (SUD), intellectual disability, ADHD and developmental delay.17, 18, 19, 20, 21, 22, 23 Mental disorders are frequently associated with CNVs of chromosome 22q11. The microdeletions in the 22q11 are found with significantly higher frequency among SCZ patients. De novo CNVs are found with higher frequency among sporadic cases, whereas inherited CNVs are enriched among familial cases.24 Liu et al. consider that the risk of developing SCZ associated with CNV of 22q11 is equal to ‘the risk for a child of two schizophrenic parents or monozygotic co-twin of an affected patient’. Accordingly, these authors assume that the increased risk for SCZ, associated with this region, is a result of the aggregate effect of several genes at this locus.25 International Schizophrenia Consortium, in a genome-wide study, detected 13 large deletions of the 22q11.2 chromosome region in the group of 3391 SCZ cases and none in 3181 non-schizophrenic individuals. Furthermore, these authors declare that around 30% of patients with 22q11.2 deletion syndrome develop a mental disease.26 Consistently with this study, Itsara et al.,27 following an analysis of 2500 individuals from 9 studies, identified 31 CNVs in the 22q11 region in patients with autism, intellectual disability and SCZ. Of note is also the finding of Walsh et al.28 who detected numerous microdeletions and microduplications in 150 individuals with SCZ and 268 ancestry-matched controls. The disrupted genes encoded proteins involved in neurodevelopment.
Finally, Xu et al. carried out a genome-wide study using a large SNP array, confirming significant association of rare de novo (but not inherited) CNVs with SCZ. They found that de novo microscopic chromosomal abnormalities were around eight times more common in non-familial cases of SCZ than in unaffected controls.29
Reverse phenotyping of psychiatric phenotypes
A promising approach in psychiatry is reverse phenotyping. This approach enables stratification of patients according to genetic markers but not according to phenotypes. This concept arose from the observation that similar phenotypes appear when specific genes are disrupted.1, 16 Moreover, it bypasses debate about a revision of the psychiatric taxonomy, does not rely on a gold standard and does not require complete understanding of the disease etiology. Such a system can coexist alongside conventional diagnostic systems. For example, this approach has identified MBD5 as a potential new candidate-gene associated with mental disorders, such as bipolar disorder.30 Accordingly, the field of biological psychiatry may need to change the way in which studies are conducted and reported. The reverse genotyping concept involves initial genotyping followed by exhaustive phenotyping on the basis of the genotype information.31 Refining the phenotype definition by reverse phenotyping may consistently improve understanding of the genetic architecture of psychiatric traits.32 Sahoo et al. have analyzed the microarray results of 38 779 individuals referred for testing because of various neuropsychiatric conditions. Of the analyzed group, 1113 individuals carried CNVs encompassing putative SCZ susceptibility loci. Of these, 1035 had a CNV of one of six recurrent loci. The authors have concluded that the results from their genotype-first analysis of SCZ susceptibility loci, the largest analysis to date, indicate that phenotypic effects of CNVs associated with SCZ are pleiotropic and further indicate the existence of common biological pathways among various neuropsychiatric disorders.33 In another study, Goede et al., after a detailed clinical evaluation of two families referred for global developmental delay and learning disability, have excluded that the presumptive causative missense variant RABL6 is the underlying cause of the disorder and have instead demonstrated that the condition is because of a homozygous INPP5E mutation.34 Reverse phenotyping, together with careful protein structural analysis and functional tests, had a crucial role in achieving the correct diagnosis. Further in-depth study of the mutational and phenotypic spectra associated with INPP5E has demonstrated that mutations in this gene lead to a range of ciliopathy-phenotypes with considerable intra-familial phenotypic variability. This study has demonstrated that reverse phenotyping enables the identification and compilation of a wide phenotypic spectrum of INPP5E-related disorders.34
Ethical issues related to genetic testing in psychiatry
Emerging genomic technologies, such as NGS of DNA fragments, created an unlimited access to DNA profile of a patient without providing adequate means to assess the clinical relevance of countless possible combinations of genetic variants.1 In contrast to the enthusiasm accompanying Human Genome Project in the 90s, the contemporary approach to the achievements of genetics seems to be ambiguous. The consequences of predictive genetic testing for psychiatric diseases provoke additional ethical questions and raise a number of sensitive human rights issues such as dignity, discrimination, traumatization and social stigma. The inability to accurately predict a phenotypic outcome is a restraint of pre-symptomatic testing for most genes responsible for mental health. Even though NGS has a potential to identify thousands of sequence variants or even unravel the whole genome, currently it is not possible to link these findings with complex neuropsychiatric traits.35 The genetic determinism and potential discrimination based on the paradigm that mental health is pre-programmed and entirely depending on the composition of individual DNA has to be redefined. In practice, because of the complex interactions between genes along with environmental conditions, phenotypic consequences of the individual’s genotype remain unpredictable and far-going interpretation of predictive genetic test results may be highly unethical.35, 36, 37 Unlike in most other medical and health care domains, ethical issues that may arise as the consequences of genetic testing in psychiatry are considered unusually problematic emphasizing the dual nature of psychiatry, that is, dependence on both, social norms and biological functions. In addition, the knowledge about genetic risk may constitute further determinant contributing to the development of a mental illness. For example, information of high biological risk of SCZ may result in higher level of expressed emotions in the patients’ environment, which is well known negative risk factor in SCZ.38
Distinct ethical dilemmas emerge with respect to prognosis, long-term medical history, prenatal testing, incidental findings and so on. The interpretation and disclosure of unsolicited findings of limited or unknown significance or variants irrelevant to the diagnostic indication remains a fundamental question. There is an agreement that patients should be informed about findings of high importance for their health and at least partly amenable. American College of Medical Genetics (ACMG) Recommendations for Reporting of Incidental Findings In Clinical Exome and Genome Sequencing is listing the genetic disorders of high prevalence, where early intervention is possible (https://www.acmg.net/docs/ACMG_Releases_Highly-Anticipated_Recommendations_on_Incidental_Findings_in_Clinical_Exome_and_Genome_Sequencing.pdf). On the other hand the patients’ right not to know should be respected accordingly to the European Convention of Human Rights and Biomedicine (Article 10.2) (https://www.coe.int/en/web/conventions/full-list/-/conventions/rms/090000168007cf98) and UNESCO Declaration of Human Genome (Article 5c) (http://portal.unesco.org/en/ev.php-URL_ID=13177&URL_DO=DO_TOPIC&URL_SECTION=201.html). Even more complex challenge is that of interpreting, communicating and disclosing of genetic testing results in children, especially concerning late-onset disorders such as neurodegenerative conditions or cancer. At present, there is no accepted standard on how to deal with such findings; ethical boards do not propose any definitive answer to these questions and vary widely on the extent of disclosure of the results to the asymptomatic patient and his/her family. Currently, it is commonly assumed that the decision concerning the disclosure of genomic findings should be made both on the basis of the recommendations of the board of experts and patient’s own choice.1 The inability to accurately predict final phenotype is also a restraint of prenatal genetic testing for psychiatric diseases and behavioral traits. Considering these difficulties, further debates about the ethical dimensions related to this issue are needed. Ultimately, every patient should be informed about strengths, limitations and challenges in the interpretation of genomic testing results. Thus, it is imperative that genetic counselors contribute to multi-disciplinary care teams effectively and efficiently implementing genetic testing to patients care. The genetic counseling before and after WES or whole-genome sequencing (WGS) is necessary, due to the potential risks of psychological harm. Genetic counselors provide the professional interpretation of testing results to translate the meaning to patients and to ascertain the preference of the patient in terms of disclosure of unsolicited findings.
Endophenotypes—will they or won’t they simplify gene identification and genetic testing of mental disorders?
The term ‘endophenotype’ is defined as a trait intermediate between genes and a disorder.39, 40 The concept of endophenotypes reflects the need to make the complex architecture of mental disorders simpler and promotes the view that psychiatric diagnoses can be distilled into essential elements, thus making genetic analysis and diagnosis easier. An endophenotype can be any type of trait, such as neurophysiological, cognitive, biochemical, neuroimaging or behavioral, that is more elementary than the disorder itself and is associated with only a few genes.39, 40 According to Gottesman and Gould,39 for a quantitative trait to be considered an endophenotype, ‘it must co-segregate with the illness within a family, and it must be independent of the clinical state’. For example, electrophysiological deficits in pre-pulse inhibition (linked to FABP7, CHRNA7) and P50 sensory gating or a decline in working memory (linked to RELN) have been considered as intermediate traits in SCZ.41 In addition, a deficit in face emotion labeling in association with CACNA1C is a commonly identified endophenotype in BD.40 In an initiative to understand the molecular background of quantitative endophenotypes in SCZ, the Consortium on the Genetics of Endophenotypes in Schizophrenia (COGS) was established.42, 43 COGS has initiated two further studies. The first study was undertaken using a family-based ascertainment strategy (COGS-1),44 which have been followed by a larger case-controlled ascertainment study (COGS-2).45 Both COGS strategies allowed to detect similar endophenotype deficits. Of 15 measures retrieved from a group of SCZ probands, their siblings, and control subjects, five distinct heritable neurocognitive factors emerged with neurobiological, genetic and treatment consequences. Significant heritability estimates for the factors ranged from 22% (episodic memory) to 39% (visual abstraction). These five endophenotypes may also be helpful as principal RDoC cognitive measures that underlie several related mental disorders.8 A further investigation with a large COGS-2 cohort is planned, in which GWAS aimed at detecting variants associated with these deficits will be performed.46 New analytical strategies that use endophenotype ranking, cumulative endophenotype loading and gene burden analyses will be adopted with the goal of identifying the genetic mechanisms of heritable deficits in SCZ. Another group, the Minnesota Center for Twin and Family Research (MCTFR), has performed a GWAS in 4900 individuals to uncover the genetic factors involved in seventeen electrophysiological endophenotypes. A wide range of genetic approaches, including biometric and genetic heritability analyses, GWAS, candidate-gene studies, rare-variant analyses of nonsynonymous SNPs in the exome and WGS, were used. Unexpectedly, for 8 of the 17 traits, associated genes were not detected. Furthermore, genes linked with the other 9 traits were not associated with any known brain function.47 These findings do not preclude the possibility of a genetic origin of these traits, but they suggest that the effect of any selected variant will be small. More promising results relating electrophysiological endophenotypes to clinical phenotypes have been obtained by the Collaborative Study of the Genetics of Alcoholism (COGA).48 This study has identified GABRA2 and CHRM2 as variants increasing the risk of alcoholism.48, 49 In this study, the use of quantitative phenotypes greatly increased the chances of detecting of genetic determinants and allowed broad genetic signals to be distilled into more elementary components reflecting variation in single genes. Recently, Mullin et al.50 have successfully deconstructed an endophenotype into lower complexity synaptic mechanisms. Using a Drosophila olfactory habituation model dependent on GABAergic interneurons, the authors have discovered an analogy between the fly model and the observed deficits related to GABAergic interneuron dysfunction and impaired sensory habituation, which constitute an endophenotype in SCZ.51 The Drosophila model was used to understand the effects of fly loss-of-function mutations affecting BLOC-1 orthologous subunits on synaptic networks.52 The authors have studied phenotypes associated with mutations in the SCZ susceptibility gene DYSB in isolation or in combination with null alleles in the DYSB network component BLOS. They have demonstrated that homozygotes or compound heterozygotes of loss-of-function alleles of DYSB or BLOS1 impair synapse structure and function morphology at the larval neuromuscular junction and impair olfactory habituation in Drosophila. Furthermore, this study has shown the differential sensitivity of examined phenotypes to the alteration in the gene dosage due to the presence of null BLOC-1 mutation. These findings suggest that an endophenotype is not just a result of the additive effects of two or more genes in a regulatory network, and further studies aimed at the deconstructing an endophenotype should consider different levels of the organization of the entire complex.50
Astonishingly, despite decades of research, much confusion remains regarding the relevance of endophenotypes as being more close to gene elements in the architecture of mental disorders. Endophenotypes are, in theory, simpler than DSM categories, and some of them really are. For example, the COGA study has successfully linked electrophysiological endophenotypes to susceptibility genes,48 whereas other complex traits might reflect the action of many variants with small effects, as demonstrated in the MCTFR project,47 or may represent a continuous spectrum. ‘Simple’ endophenotypes manifest a straightforward linear pathway from gene to phenotype, whereas ‘complex’ endophenotypes constitute much more complicated biological modules.53 Iacono et al.54 have suggested that ‘the whole concept of endophenotypes is problematic’ and should be reconsidered, particularly the concept that ‘some brain phenomena are simpler than others’. Can an EEG-measured brain wave be considered to be more fundamental than personality, which is almost impossible to quantify? Indeed, both are functions of the entire brain.54 In line with the overall trend in psychiatry to subtype and stratify, it has recently been proposed that endophenotypes may be further broken down into their components, thereby providing ‘endophenotypes for endophenotypes’ with a simpler genetic architecture, and expectedly, a higher signal-to-noise ratio in the analysis.22 Such an approach has successfully been applied in the abovementioned study of Mullin et al. In a further attempt to facilitate gene discovery, substantially larger samples, together with multiple genetic approaches linking structural to functional genomics (for example, gene expression or expression quantitative trait loci studies) and epigenetics have been proposed to improve the understanding of the genetic architecture of endophenotypes.50 Finally, it has been suggested to expand genetic studies beyond individuals of European ancestry, thus potentially improving the likelihood of finding rare variants of at least moderate effect size.22 Efforts to use endophenotypes as diagnostic tools present an important challenge for future research. It is possible that electrophysiology may not be optimal for measuring endophenotypes. There are a great number of other candidate endophenotypes, such as neuroimaging, that are probably more similar to the underlying genetics and thus hold promise for identifying phenotypes and serving as intermediate traits between a genetic marker and a disease.40 Alternatively, different multi-modality phenotypic patterns might be combined to enhance the analysis performance similarly to the successful application to Alzheimer's disease?55 The identification of endophenotypes has not led to important discoveries of new risk variants for mental diseases to date, although the more complex the structure, the more it is necessary to simplify and summarize core components to understand them. This is why we should not give up on the overall strategy just yet.
Gene networks as biomarkers of psychiatric disorders
Currently, multiple studies have emphasized the advantage of analyzing gene networks to decipher the molecular basis underlying psychiatric disorders. This concept has emerged from the idea that phenotype-specific gene networks forming multi-dimensional structures account for every complex trait.56 The lower level (single-gene signals) may provide some information, but it misses other explanations and possible causations. Higher-level processes (networks) are more informative because a phenotype is not a simple sum of additive effects of all variants underlying a condition. Gene networks, similarly to endophenotypes, can be considered to be interfaces between the genotype and phenotype. In line with such a reductionist approach, genetic networks apply to sets of genes, transcriptional regulatory networks, protein–protein interactions, metabolic networks, gene regulatory networks and interactions between these networks organized at multiple and hierarchical levels of complexity.57 The analysis of gene networks in specific pathways, with a greater emphasis on the existence of interactions among genes that are overlooked when gene variants are examined separately, appears to be a challenging concept to apply to major psychiatric disorders.58, 59 If a coherent theory explaining the association between basic DNA structure and a psychiatric phenotype does not exist, there should be an attempt to translate the predicates to another theory and to connect different levels of organization of the system, which would not require a full understanding of the inherent molecular complexity. From a theoretical point of view, this approach might represent a step toward understanding of how a change in the genotype modulates a psychiatric phenotype itself. To establish a ‘network psychiatry’, various gene networks-based approaches, focusing on a phenotype or a disease of interest should be integrated. Gene regulatory networks may serve as ‘blueprints’ of molecular interactions.60 The PGC schizophrenia pathway analysis is a good example of a success in this regard.61 Various gene network approaches are currently being used to merge genomics, epigenomics, transcriptomics and proteomics, including coding and non-coding genomic elements, eQTLs, gene expression and co-expression profiles, protein–protein interactions and metabolomics, and to serve as a structure that locates every gene within the framework of its environment, thus identifying all genetic contributors to a disorder at all levels, from a single cell to the entire brain. Transcriptional data analysis within the context of the network approach appears to be particularly important, because alternative splicing seems to occur more frequently in the brain than in other organs.62 For example, Chen et al. have tested groups of co-expressed genes for association with SCZ. Using a weighted gene co-expression network analysis, they have found that two modules of genes were differentially expressed in patients compared with controls. The gene group upregulated in the cerebral cortex was enriched in neuron differentiation and neuron development genes, and another group, altered in the cerebral cortex and cerebellum, was enriched in genes engaged in neuronal protection. The results were consequently preserved in five concomitantly analyzed expression data sets.63 Furthermore, Zhang et al.,64 using eQTL analysis together with module-level genetic signal enrichment, have found remodeling of multiple transcriptional modules in Alzheimer’s disease. The researchers have identified TYROBP as a regulatory hub. They have concluded that linking genotype with expression profiles and phenotype is very promising approach for establishing the causality. Currently, several recent influential papers have addressed network approaches in neuropsychiatric diseases. For example, Guan et al. have published an analysis of tissue-specific networks, in an approach integrating large-scale genomics data sets with tissue-specific expression profiles. Cross-network comparison predicted novel gene candidates related to ataxia in mice and identified variants predicted solely by the cerebellum network. The prediction performance was significantly improved using tissue-specific networks compared with the global functional network.65 Many powerful studies have elucidated neuropsychiatric disease mechanisms using gene networks by analyzing multiple data types, such as mRNA expression, GWAS, epigenomic data and protein–protein interactions. Cristino et al. have combined available data in gene networks associated with ASD, X-linked intellectual disability, ADHD and SCZ. They have identified ~4000 genes possibly contributing to all four disorders, which are involved in various intracellular processes, including cell-to-cell communication and neurodevelopment. They have found similarities and differences between transcriptional and post-transcriptional processes in the molecular networks that underlie the above mentioned disorders. SNP analysis confirmed that 789 of 4839 nonredundant SNPs are located near or within 435 genes, the majority within non-coding regions, in the protein–protein network. Furthermore, 113 SNP loci were found associated with at least one transcription factor binding site. The authors have proposed a hypothetical framework to explore the molecular pathomechanism of neuropsychiatric conditions and enable gene finding for further analysis. Such an approach has the potential to uncover the phenotype arising from a distinct combination of genetic alterations and to serve as a new diagnostic tool for determining the risk of the individual genotype, thereby guiding an individual therapeutic strategy.66 The abovementioned study of endophenotype deconstruction in Drosophila by Mullin et al. has tested how interactions within dynamic networks of multiple loci remodel the phenotype and has demonstrated that mechanistic decomposition of an endophenotype into more elementary elements may be better understood in light of the gene modifications encoding components of the network. This study has illustrated that specific phenotypes result from variations in the genetic control of neurodevelopmental regulatory networks.50 It is also worth to mention that a genome-wide transcriptome profile can be regarded as a quantitative endophenotype, which can be correlated to other potential endophenotypes, including MRI or behavioral phenotypes. Richiardi et al. have analyzed resting-state fMRI and transcriptomics data from 161 genes linked to ion channel activity and synaptic transmission to create computational models of such networks. They have found that resting-state functional connectivity in healthy adolescents was significantly altered by polymorphic variants of 136 genes. Similarly, the transcriptional activity of these genes correlated with axonal connectivity in mice. These results highlight that resting-state functional networks are associated with the coordinated activity of an array of genes.67 A cross-scale collaborative effort has shed new light on transcriptomics, epigenomics and proteomics of the human brain. This new approach includes a systematic view of the immensely complex molecular foundation of brain development, structure and function at different levels and was undertaken by the Allen Brain Institute (http://alleninstitute.org/media/filer_public/72/23/7223a5a9-0df1-4538-94fb-6d7d0c593f54/2016_0328_pressrelease_nature_reid.pdf) and BrainSpan (www.brainspan.org).68 These integrated gene networks could be used as reliable diagnostic, prognostic or predictive biomarkers.69 Such an approach may have special value in cases of complex disorders, depending on interactive pathways rather than specific genes.70 Because whole network-related biomarkers are considerably more complex than single/multiple gene signals, new statistical tools have emerged to compare these networks, such as Comparative Network Analysis.71 Furthermore, databases are needed to use network biomarkers efficiently; thus, we anticipate the initiation of a large-scale international collaboration to establish such a catalog. Aside from databases, network analysis tools, such as the tranSMART platform based on the open source i2b2 (informatics for integrating biology and the bedside) are necessary to allow for integration with all kind of molecular and clinical data.72 TranSMART was founded by the NIH Roadmap NCBC to give clinicians access to applications for biological and clinical data integration.72 In addition, the understanding of gene networks is necessary to foster more efficient drug design, paving the way toward personalized medicine.73 Several initiatives have been established to undertake network analysis in the human central nervous system. The NIH, together with the NIMH, has launched the new program ‘Psychiatric Gene Networks’ to support in silico approaches and experimental functional analyses of gene networks and complex pathways that confer susceptibility to severe mental illnesses (http://grants.nih.gov/grants/guide/rfa-files/RFA-MH-16-310.html#sthash.TIYGP32X.dpuf). The integration of numerous methods for an assessment of biological networks has also been proposed within the Dialogue on Reverse Engineering Assessment and Methods (DREAM) project. DREAM fosters a coordinated effort to form reverse engineered cellular networks from genomics data.74, 75, 76, 77 Similarly, the goal of NETwork-Based Analysis of Genomic variations (NETBAG) is to build a structure of numerous genetic signals in which highly interrelated genes are expected to cooperate in a phenotype. To identify affected molecular networks, an algorithm that searches for groups of disease-associated genetic variations has been developed. Several NETBAG studies have evaluated CNVs implicated in psychiatric conditions and have found interconnected modules related to synaptic function.78, 79, 80 Another interesting approach is to merge co-expression data, protein–protein interactions and genetic variants, as presented by Merging Affected Genes into Integrated networks (MAGI) (http://eichlerlab.gs.washington.edu/MAGI). MAGI has identified networks containing functionally linked genes enriched for pathogenic mutations in ASD that are co-shared with epilepsy, SCZ and intellectual disability. A major benefit of network analyses results from the knowledge on how genes combine and interact across different levels of analysis and enables integration with other data. Such a framework acting at a network level enables understanding of the mechanism by which more elementary genetic signals contribute to psychiatric phenotypes and to classify observed phenotypes quantitatively. A more systemic view of biological networks emerging in psychiatry today opens new possibilities for diagnostic molecular testing.81
Mathematical models of brain function—could computational neuroscience aid in diagnosis of mental diseases?
Mathematical modeling is emerging as an important research and diagnostic tool, substantially complementing traditional methods for studying mental diseases. Computational psychiatry approaches are currently attracting much attention82 and have been applied to many disorders, such as SCZ, anxiety, personality disorders, ASD, ADHD and addiction.82, 83, 84, 85 A variety of computational tools have been proposed to quantitatively model the complexity of genetic and environmental effects and to link these findings with the altered brain function underlying a clinical phenotype. Computational psychiatry can accommodate both categorical and dimensional (continuous) approaches, thus providing some mechanistic explanations for symptoms and cross-linking categorical diagnoses with the dimensional system.82, 86, 87 Mental illnesses affect different organizational levels in the brain, from genes and proteins to neurons and neural networks. Mathematical models consider genes, neuromediators or neuronal networks as a source of psychiatric diseases within the context that they operate, and can be used to interconnect data from different levels within a theoretical frame.82, 88 The first step of this process is to describe complex brain functions with the vocabulary of mathematics, providing probabilistic descriptions of different level networks and their co-dependence. Furthermore, recreated in silico neural networks can be used to examine behavior-related processes. This computational approach allows for both bottom-up (genotype-phenotype) and top-down (phenotype-genotype) strategies of information processing, thereby enabling the generation of simulated data and further changing various elements of the model to determine how an entire system changes in silico. The simulated circuit of neurons helps predict how these structures should behave in reality.82 All computational model predictions should be replicated using independent data sets and confirmed using experimental validation as a gold standard. According to Brodersen et al., new computational categories and dimensions may improve the classification of psychiatric diseases, support diagnostics and identify new targets for the therapy of psychiatric conditions. This group has demonstrated that such a system can be implemented in clinical practice, thus suggesting the applicability of mathematical models as a more accurate diagnostic tool for mental disorders.89 Mäki-Marttunen et al. have developed a computer model for evaluating additive effects of multiple SCZ-linked genetic variants on ion channels and calcium transporters, which affect neuronal excitability and synaptic transmission. The computational simulations enabled the prediction of how modifications in these functions change the activity of neuronal cells. Such an approach may shed light on basic pathological mechanisms underlying SCZ, which could potentially be targeted with appropriate drugs, and also may reveal new biomarkers to monitor therapeutic effects. These neuronal changes may be crucial for the development of ADHD, ASD, SCZ, BD and SUD, as risk variants related to neuronal dysfunctions in these disorders are revealed.90 According to Huys et al., the computational approach addresses the complex challenges of present-day psychiatry, promising future breakthroughs in the understanding of mental diseases, with an emphasis on clinical applications and psychiatric taxonomy. The authors present two complementary approaches toward computational psychiatry: a data-driven approach applying machine learning and a theory-driven resolution that mathematically determines relationships among variables. The review highlights the utility of combining both approaches as very promising in solving clinical problems.91 According to recent evidence, computational psychiatry is likely to discover new disease biomarkers by using mathematical modeling and computational simulation techniques to study biological systems.92 Nonetheless, it is imperative to establish the utility of computational approaches in clinical trials.
The polygenic risk scores
The polygenic architecture of psychiatric traits remains a challenge for GWAS. It became apparent that even increasingly powerful approaches, that is, nearly population-level testing together with improved statistical procedures, may not always eliminate a constant fraction of errors corrupting the data structure. The power of GWAS is limited by several factors including: genetic heterogeneity, highly quantitative phenotypes, small-effect genetic variants, alleles of low frequency, unexpected linkage disequilibrium, non-causative SNPs better associated with a phenotype than a causal variant, genetic interactions between loci and/or the environment, non-additive interaction of two or more loci, structural variants and large chromosomal rearrangements.93
Because psychiatric traits are governed by large groups of genes working together, condition-specific gene regulatory networks should explain the phenotype much better than single genetic markers. Accordingly, the research has moved toward the analysis of gene networks to develop more clinically useful information.94 Given the need for a systemic approach, a Polygenic Risk Score (PRS) analysis is regarded as a new tool helping to resolve some issues related to the polygenicity. PRS may identify the cumulative effect of common variants weighted by effect size.95 A major advantage of PRS is that it captures small genetic effects, which independently are undetectable at the original GWAS significance threshold, but collectively are associated with a given trait. Moreover, PRS can uncover shared genetic etiology among psychiatric traits. Polygenic genome-wide analysis techniques are especially powerful for a subset of patients (an endophenotype).96 However, even though the PRS is a more powerful approach than original GWAS, the accuracy achieved to date is still insufficient for clinical decision-making at an individual level. PRS, as originally rooted in GWAS, has some of its inherent limitations (sample-size dependence and the power limited by the power of initial GWAS). The most powerful method of PRS constructions is considering the effects of all SNPs together. This method appropriately models linkage disequilibrium between variants. A major benefit of such an approach results from the integration of functionally linked SNPs explaining a larger part of the genetic influences on the trait, than by taking into consideration only elementary genetic signals pre-selected on the basis of the stringent significance threshold. These systemic prediction methods can shed light on the biological background being conceptually similar to computational models in psychiatry, as they combine numerous genomic signals and draw inferences between them (gene-pathways/gene networks).97 Such polygenic score is a step toward a systemic approach that can be further extended to combine multiple data including both genetic factors and environmental influences, such as gene expression and epigenetic modifications. Such a model could explain another portion of missing heritability.
Conclusions
In this review, we have outlined multiple attempts to substantially advance genetic testing in psychiatry. Unfortunately, despite continued efforts, there is a large gap between genomic findings and their potential to be converted into valid diagnostic tests. We emphasize the view that only a paradigm shift can bring a fundamental change in psychiatric practice, allowing to disentangle the intricacies of mental diseases. The integrative system approach and concerted cross-disciplinary effort offer new opportunity to connect genetic background with specific diseases entities, or concurrently, with symptoms regardless of a diagnosis. Both gene networks and computational psychiatry are systemic algorithmic processes of connecting a disease or a symptom with a relevant cause on the basis of co-operative network-level effects. Such a holistic approach remains in accordance with the current projects of RDoC.98 However, even relevant genetic network markers can be difficult to interpret, and various combinations of genetic and environmental determinants may ultimately contribute to a particular clinical phenotype.99, 100 Because the phenotypic plasticity of psychiatric traits is very high, the genotype alone cannot be used to reliably predict the clinical outcome. Nonetheless, the phenotypic plasticity of psychiatric traits can vary, and the relationship between genotype and phenotype may be more straightforward in SCZ or BD than in MDD or ADHD. This may be the reason of GWAS decreased sensitivity, pointing to the insensitivity of predictive genetic testing in psychiatric traits with the highest phenotypic plasticity. Therefore, the respective genotype does not converge on a specific phenotype but instead results in a range of related illnesses.101 In light of previously described limitations, the potential diagnostic approach to complex traits may be based on mathematical descriptions of network complexity built from a combination of a large number of interconnected variables. Such a heuristic computational tool should include not only genomic and epigenomic information but also high-dimensional data describing the phenotype to reflect an individual brain signature. Whereas each mental state has a correlate in the brain, it should be possible to identify its biological counterpart. In parallel with quantum physics, whether we are able to create a probabilistic model of the entire universe, we should also be able to build an unambiguous model of the brain in its entirety, on the basis of a coherent ‘theory of everything’. Such models may provide insight into the manner in which fundamental biological processes, when disrupted, can alter brain function. At the extremes of the model, near the risk distribution-containing subjects of very high risk, attention on unique families and rare variants is warranted. Finally, any translational applications of new knowledge require prospective, controlled trials to demonstrate clinical applicability of all discoveries.
Acknowledgments
We acknowledge Jeremiah M. Scharf MD, PhD for his invaluable comments, which greatly improved the manuscript.
Footnotes
The authors declare no conflict of interest.
References
- Demkow U, Ploski R. Clinical Applications for Next-Generation Sequencing. Academic Press: Boston, MA, USA, 2016. [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511: 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuang MT. Genotypes, phenotypes, and the brain. A search for connections in schizophrenia. Br J Psychiatry 1993; 163: 299–307. [DOI] [PubMed] [Google Scholar]
- Tsuang MT. Genetics, epidemiology, and the search for causes of schizophrenia. Am J Psychiatry 1994; 151: 3–6. [DOI] [PubMed] [Google Scholar]
- Schreiber M, Dorschner M, Tsuang D. Next-generation sequencing in schizophrenia and other neuropsychiatric disorders. Am J Med Genet B Neuropsychiatr Genet 2013; 162B: 671–678. [DOI] [PubMed] [Google Scholar]
- Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 2013; 45: 984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencz T, Malhotra AK. Targeting the schizophrenia genome: a fast track strategy from GWAS to clinic. Mol Psychiatry 2015; 20: 820–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbert BN, Insel TR. Toward the future of psychiatric diagnosis: the seven pillars of RDoC. BMC Med 2013; 11: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel TR. The NIMH Research Domain Criteria (RDoC) Project: precision medicine for psychiatry. Am J Psychiatry 2014; 171: 395–397. [DOI] [PubMed] [Google Scholar]
- Lotan A, Fenckova M, Bralten J, Alttoa A, Dixson L, Williams RW et al. Neuroinformatic analyses of common and distinct genetic components associated with major neuropsychiatric disorders. Front Neurosci 2014; 8: 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 2013; 381: 1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butwicka A, Szymańska K, Retka W, Wolańczyk T. Neuroleptic malignant syndrome in an adolescent with CYP2D6 deficiency. Eur J Pediatr 2014; 173: 1639–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks JK, Swen JJ, Thorn CF, Sangkuhl K, Kharasch ED, Ellingrod VL et al. Clinical pharmacogenetics implementation consortium guideline for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants. Clin Pharmacol Ther 2013; 93: 40–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanska K, Szczałuba K, Lugowska A, Obersztyn E, Radkowski M, Nowakowska BA et al. The analysis of genetic aberrations in children with inherited neurometabolic and neurodevelopmental disorders. Biomed Res Int 2014; 2014: 424796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehne B, Lewis CM, Schlit T. Exome localization of complex disease association signals. BMC Genomics 2011; 12: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffan E, Semple RK. Next generation sequencing–implications for clinical practice. Br Med Bull 2011; 99: 53–71. [DOI] [PubMed] [Google Scholar]
- Green EK, Rees E, Walters JTR, Smith K-G, Forty L, Grozeva D et al. Copy number variation in bipolar disorder. Mol Psychiatry 2016; 21: 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell 2012; 148: 1223–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, Goldstein A et al. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N Engl J Med 2012; 367: 1321–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Rees E, Walters JT, Escott-Price V, Georgieva L, Richards AL et al. The penetrance of copy number variations for schizophrenia and developmental delay. Biol Psychiatry 2014; 75: 378–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams NM, Zaharieva I, Martin A, Langley K, Mantripragada K, Fossdal R et al. Rare chromosomal deletions and duplications in attention-deficit hyperactivity disorder: a genome-wide analysis. Lancet 2010; 376: 1401–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens EM, Bachman P, Glahn DC, Bearden CE. Electrophysiological endophenotypes for schizophrenia. Harv Rev Psychiatry 2016; 24: 129–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees E, Walters JT, Georgieva L, Isles AR, Chambert KD, Richards AL et al. Analysis of copy number variations at 15 schizophrenia-associated loci. Br J Psychiatry 2014; 204: 108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Heath SC, Sobin C, Roos JL, Galke BL, Blundell ML et al. Genetic variation at the 22q11 PRODH2/DGCR6 locus presents an unusual pattern and increases susceptibility to schizophrenia. Proc Nat Acad Sci USA 2002; 99: 3717–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Abecasis GR, Heath SC, Knowles A, Demars S, Chen Y-J et al. Genetic variation in the 22q11 locus and susceptibility to schizophrenia. Proc Natl Acad Sci USA 2002; 99: 16859–16864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 2008; 455: 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsara A, Cooper GM, Baker C, Girirajan, SLiJ, Absher D et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet 2009; 84: 148–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science 2008; 320: 539–543. [DOI] [PubMed] [Google Scholar]
- Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet 2008; 40: 880–885. [DOI] [PubMed] [Google Scholar]
- Hodge JC, Mitchell E, Pillalamarri V, Toler TL, Bartel F, Kearney HM et al. Disruption of MBD5 contributes to a spectrum of psychopathology and neurodevelopmental abnormalities. Mol Psychiatry 2014; 19: 368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stessman HA, Bernier R, Eichler EE. A genotype-first approach to defining the subtypes of a complex disease. Cell 2014; 156: 872–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze TG, McMahon FJ. Defining the phenotype in human genetic studies: forward genetics and reverse phenotyping. Hum Hered 2004; 58: 131–138. [DOI] [PubMed] [Google Scholar]
- Sahoo T, Theisen A, Rosenfeld JA, Lamb AN, Ravnan JB, Schultz RA et al. Copy number variants of schizophrenia susceptibility loci are associated with a spectrum of speech and developmental delays and behavior problems. Genet Med 2011; 13: 868–880. [DOI] [PubMed] [Google Scholar]
- de Goede C, Yue WW, Yan G, Ariyaratnam S, Chandler KE, Downes L et al. Role of reversephenotyping in interpretation of next generation sequencing data and a review of INPP5E related disorders. Eur J Paediatr Neurol 2016; 20: 286–295. [DOI] [PubMed] [Google Scholar]
- Frebourg T. The challenge for the next generation of medical geneticists. Hum Mutat 2014; 35: 909–911. [DOI] [PubMed] [Google Scholar]
- Hoop JG. Ethical considerations in psychiatric genetics. Harv Rev Psychiatry 2008; 16: 322–338. [DOI] [PubMed] [Google Scholar]
- Besterman AD. The ethics of genetic testing in psychiatry. Virtual Mentor 2012; 14: 460–463. [DOI] [PubMed] [Google Scholar]
- Kavanagh DJ. Recent developments in expressed emotion and schizophrenia. Br J Psychiatry 1992; 160: 601–620. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry 2003; 160: 636–645. [DOI] [PubMed] [Google Scholar]
- Hao X, Yan J, Yao X, Risacher SL, Saykin AJ, Zhang D et al. Diagnosis-guided method for identifying multi-modality neuroimaging biomarkers associated with genetic risk factors in Alzheimer's disease. Pac Symp Biocomput 2016; 21: 108–119. [PMC free article] [PubMed] [Google Scholar]
- Owen MJ, O'Donovan MC, Thapar A, Craddock N. Neurodevelopmental hypothesis of schizophrenia. Br J Psychiatry 2011; 198: 173–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkins ME, Dobie DJ, Cadenhead KS, Olincy A, Freedman R, Green MF et al. The consortium on the genetics of endophenotypes in schizophrenia: model recruitment, assessment, and endophenotyping methods for a multisite collaboration. Schizophr Bull 2007; 33: 33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow NR, Gur RE, Braff DL. Consortium on the genetics of schizophrenia (COGS) assessment of endophenotypes for schizophrenia: an introduction to this special issue of schizophrenia research. Schizophr Res 2015; 163: 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman LJ, Hellemann G, Nuechterlein KH, Greenwood TA, Braff DL, Cadenhead KS et al. Factor structure and heritability of endophenotypes in schizophrenia: findings from the consortium on the genetics of schizophrenia (COGS-1). Schizophr Res 2015; 163: 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow NR, Light GA, Sprock J, Calkins ME, Green MF, Greenwood TA. Deficient prepulse inhibition in schizophrenia detected by the multi-site consortium on the genetics in Schizophrenia. Schizophr Res 2014; 152: 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braff DL. The importance of endophenotypes in schizophrenia research. Schizophr Res 2015; 163: 1–8. [DOI] [PubMed] [Google Scholar]
- Miller MB, Basu S, Cunningham J, Eskin E, Malone SM, Oetting WS et al. The Minnesota center for twin and family research genome-wide association study twin. Res Hum Genet 2012; 15: 767–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Jones K, Saccone N, Hinrichs A, Wang JC, Goate A et al. Endophenotypes successfully lead to gene identification: results from the collaborative study on the genetics of alcoholism. Behav Genet 2006; 36: 112–126. [DOI] [PubMed] [Google Scholar]
- Hinrichs AL, Wang JC, Bufe B, Kwon JM, Budde J, Allen R et al. Functional variant in a bitter-taste receptor (hTAS2R16) influences risk of alcohol dependence. Am J Hum Genet 2006; 78: 103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullin AP, Sadanandappa MK, Ma W, Dickman DK, VijayRaghavan K, Ramaswami M et al. Gene dosage in the dysbindin schizophrenia susceptibility network differentially affect synaptic function and plasticity. J Neurosci 2015; 35: 325–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, MacDonald ML, Elswick DE, Sweet RA. The glutamate hypothesis of schizophrenia: evidence from human brain tissue studies. Ann N Y Acad Sci 2015; 1338: 38–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheli VT, Daniels RW, Godoy R, Hoyle DJ, Kandachar V, Starcevic M. Genetic modifiers of abnormal organelle biogenesis in a Drosophila model of BLOC-1 deficiency. Hum Mol Genet 2010; 19: 861–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters JTR, Owen MJ. Endophenotypes in psychiatric genetics. Mol Psychiatry 2007; 12: 886–890. [DOI] [PubMed] [Google Scholar]
- Iacono WG, Vaidyanathan U, Vrieze SI, Malone SM. Knowns and unknowns for psychophysiological endophenotypes: Integration and response to commentaries. Psychophysiology 2014; 51: 1339–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao X, Yan J, Yao X, Shannon LR, Andrew J, Saykin AJ et al. Diagnosis-guided method for identifying multi-modality neuroimaging biomarkers associated with genetic risk factors in Alzheimer’s disease. Pac Symp Biocomput 2016; 21: 108–119. [PMC free article] [PubMed] [Google Scholar]
- Emmert-Streib F, de Matos Simoes R, Mullan P, Haibe-Kains B, Dehmer M. The gene regulatory network for breast cancer: integrated regulatory landscape of cancer hallmarks. Front Genet 2014; 5: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pache RA, Aloy P. Novel framework for the comparative analysis of biological networks. PLoS ONE 2012; 7: e31220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Buchanan S, Brennand KJ, Merchant KM. Evolving toward a human-cell based and multiscale approach to drug discovery for CNS disorders. Front Pharmacol 2014; 5: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Califano A, Butte AJ, Friend S, Ideker T, Schadt E. Leveraging models of cell regulation and GWAS data in integrative network-based association studies. Nat Genet 2012; 44: 841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabáasi A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet 2011; 12: 56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium. Psychiatricgenome-wide association studyanalysesimplicateneuronal, immune and histonepathways. Nat Neurosci 2015; 18: 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 2008; 40: 1413–1415. [DOI] [PubMed] [Google Scholar]
- Chen C, Cheng L, Grennan K, Pibiri F, Zhang C, Badner JA et al. Two gene co-expression modules differentiate psychotics and controls. Mol Psychiatry 2013; 18: 1308–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell 2013; 153: 707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Gorenshteyn D, Burmeister M, Wong AK, Schimenti JC, Handel MA et al. Tissue-specific functional networks for prioritizing phenotype and disease genes. PLoS Comput Biol 2012; 8: e1002694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristino AS, Williams SM, Hawi Z, An J-Y, Bellgrove MA, Schwartz CE et al. Neurodevelopmental and neuropsychiatric disorders represent an interconnected molecular system. Mol Psychiatry 2014; 19: 294–301. [DOI] [PubMed] [Google Scholar]
- Richiardi J, Altmann A, Milazzo AC, Chang C, Chakravarty MM, Banaschewski T et al. Correlated gene expression supports synchronous activity in brain networks. Science 2015; 348: 1241–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Gandal MJ, Geschwind DH. Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat Rev Genet 2015; 16: 441–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehmer M, Mueller L, Emmert-Streib F. Quantitative network measures as biomarkers for classifying prostate cancer disease states: a systems approach to diagnostic biomarkers. PLoS ONE 2013b; 8: e77602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Islam M, Hoque M, Banik R, Roy S, Sumi S, Hassan FM et al. Comparative analysis of differential network modularity in tissue specific normal and cancer protein interaction networks. J Clin Bioinform 2013; 3: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athey BD, Braxenthaler M, Haas M, Guo Y. TranSMART: an open source and community-driven informatics and data sharing platform for clinical and translational research. AMIA Summ Trans Sci Proc 2013; 2013: 6–8. [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Basu A. Network medicine in drug design: implications for neuroinflammation. Drug Discov Today 2012; 17: 600–607. [DOI] [PubMed] [Google Scholar]
- Stolovitzky G, Monroe D, Califano A. Dialogue on reverse-engineering assessment and methods—the DREAM of high-throughput pathway inference. Ann N Y Acad Sci 2007; 1115: 1–22. [DOI] [PubMed] [Google Scholar]
- Stolovitzky G, Prill RJ, Califano A. Lessons from the DREAM2 Challenges. Ann N Y Acad Sci 2009; 1158: 159–195. [DOI] [PubMed] [Google Scholar]
- Marbach D, Schaffter T, Mattiussi C, Floreano D. Generating realistic in silico gene networks for performance assessment of reverse engineering methods. J Comput Biol 2009; 16: 229–239. [DOI] [PubMed] [Google Scholar]
- Prill RJ, Marbach D, Saez-Rodriguez J, Sorger PK, AlexopoulosLG, Xue X et al. Towards a rigorous assessment of systems biology models: the DREAM3 challenges. PLoS ONE 2010; 5: e9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Gilman SR, Chiang AH, Sanders SJ, Vitkup D. Genotype to phenotype relationships in autism spectrum disorders. Nat Neurosci 2015; 18: 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron 2011; 70: 898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman I, Rzhetsky A, Vitkup D. Network properties of genes harboring inherited disease mutations. Proc Natl Acad Sci 2008; 105: 4323–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmert-Streib F, DehmerM, Haibe-Kains B. Gene regulatory networks and their applications: understanding biological and medical problems in terms of networks. Front Cell Dev Biol 2014; 2: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams RA, Huys QJM, Roiser JP. Computational psychiatry: towards a mathematically informed understanding of mental illness. J Neurol Neurosurg Psychiatry 2016; 8: 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning M, Behrens TE, Jocham G, O’Reilly JX, Bishop SJ. Anxious individuals have difficulty learning the causal statistics of aversive environments. Nat Neurosci 2015; 18: 590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser TU, Iannaccone R, Ball J, Mathys C, Brandeis D, Walitza S et al. Role of the medial prefrontal cortex in impaired decision making in juvenile attention-deficit/hyperactivity disorder. JAMA Psychiatry 2014; 71: 1165–1173. [DOI] [PubMed] [Google Scholar]
- Montague PR, Dolan RJ, Friston KJ, Dayan P. Computational psychiatry. Trends Cogn Sci 2012; 16: 72–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Os J, Gilvarry C, Bale R, Van Horn E, Tattan T, White I et al. A comparison of the utility of dimensional and categorical representations of psychosis. UK700 Group. Psychol Med 1999; 29: 595–606. [DOI] [PubMed] [Google Scholar]
- Wang X-J, Krystal JH. Computational psychiatry. Neuron 2014; 84: 638–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read J, Bentall RP, Fosse R. Time to abandon the bio-bio-bio model of psychosis: exploring the epigenetic and psychological mechanisms by which adverse life events lead to psychotic symptoms. Epidemiol Psichiatr Soc 2009; 18: 299–310. [PubMed] [Google Scholar]
- Brodersen KH, Deserno L, Schlagenhauf F, Lin Z, Penny WD, Buhmann JM et al. Dissecting psychiatric spectrum disorders by generative embedding. Neuro Image Clin 2013; 4: 98–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäki-Marttunen T, Halnes G, Devor A, Witoelar A, Bettella F, Djurovic S et al. Functional effects of schizophrenia-linked genetic variants on intrinsic single-neuron excitability: a modeling study. Biol Psychiatry Cogn Neurosci Neuroimag 2016; 1: 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huys QJ, Maia TV, Frank MJ. Computational psychiatry as a bridge from neuroscience to clinical applications. Nat Neurosci 2016; 19: 404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodyer IM. Mathematical models as an aid for improving the validity of descriptive psychiatry. Br J Psychiatry 2012; 201: 335–336. [DOI] [PubMed] [Google Scholar]
- Korte A, Farlow A. The advantages and limitations of trait analysis with GWAS: a review. Plant Methods 2013; 9: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubovski SL. The limitations of genetic testing in psychiatry. Psychother Psychosom 2016; 85: 129–135. [DOI] [PubMed] [Google Scholar]
- Gandal MJ, Leppa V, Won H, Parikshak NN, Geschwind DH. The road to precision psychiatry: translating genetics into disease mechanisms. Nat Neurosci 2016; 19: 1397–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalley HC, Papmeyer M, Sprooten E, Romaniuk L, Blackwood DH, Glahn DC et al. The influence of polygenic risk for bipolar disorder on neural activation assessed using fMRI. Transl Psychiatry 2012; 2: e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet 2010; 42: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak MJ, Cuthbert BN. The NIMH research domain criteria initiative: background, issues, and pragmatics. Psychophysiology 2016; 53: 286–297. [DOI] [PubMed] [Google Scholar]
- Fanous AH, Kendler KS. Genetic heterogeneity, modifier genes, and quantitative phenotypes in psychiatric illness: searching for a framework. Mol Psychiatry 2005; 10: 6–13. [DOI] [PubMed] [Google Scholar]
- Patel SD, Le-Niculescu H, Koller DL, Green SD, Lahiri DK, McMahon FJ et al. Coming to grips with complex disorders: genetic risk prediction in bipolar disorder using panels of genes identified through convergent functional genomics. Am J Med Genet 2010; 153B: 850–877. [DOI] [PubMed] [Google Scholar]
- Parikshak NN, Gandal MJ, Geschwind DH. Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat Rev Genet 2015; 16: 441–458. [DOI] [PMC free article] [PubMed] [Google Scholar]


