Abstract
Although autism spectrum disorders (ASDs) share a core set of nosological features, they exhibit substantial genetic heterogeneity. A parsimonious hypothesis posits that dysregulated epigenetic mechanisms represent common pathways in the etiology of ASDs. To investigate this hypothesis, we generated a novel mouse model resulting from brain-specific deletion of chromodomain helicase DNA-binding 5 (Chd5), a chromatin remodeling protein known to regulate neuronal differentiation and a member of a gene family strongly implicated in ASDs. RNA sequencing of Chd5−/− mouse forebrain tissue revealed a preponderance of changes in expression of genes important in cellular development and signaling, sociocommunicative behavior and ASDs. Pyramidal neurons cultured from Chd5−/− cortex displayed alterations in dendritic morphology. Paralleling ASD nosology, Chd5−/− mice exhibited abnormal sociocommunicative behavior and a strong preference for familiarity. Chd5−/− mice further showed deficits in responding to the distress of a conspecific, a mouse homolog of empathy. Thus, dysregulated chromatin remodeling produces a pattern of transcriptional, neuronal and behavioral effects consistent with the presentation of ASDs.
Introduction
Autism spectrum disorders (ASDs) are characterized by impairments in sociocommunicative interaction, repetitive patterns of behavior and insistence on sameness.1 Although ASDs are highly heritable,2 their genetic underpinnings are heterogeneous and incompletely understood. Nevertheless, recent evidence suggests a critical role for epigenetic dysregulation in de novo, inherited and idiopathic cases of ASD.3, 4
Chromatin remodeling constitutes a key epigenetic mechanism by which histone-associated DNA is dynamically exposed to transcriptional machinery. This mechanism regulates a panoply of neurodevelopmental processes,5 and its dysfunction has been implicated in ASDs.3, 6 One family of chromatin remodelers is encoded by the chromodomain helicase DNA-binding (CHD) genes, of which a majority of members have been identified through genome-wide association studies as risk genes for ASDs.3, 4 Notably, CHD8 represents one of the most widely replicated risk genes, and has been directly linked to ASD pathophysiology.7, 8, 9
In the central nervous system, chromodomain helicase DNA-binding 5 (Chd5) is a neuron-specific chromatin remodeler10, 11 that associates within a nucleosome-remodeling deacetylase-like complex.12 Expression of Chd5 is upregulated within the brain during early postnatal development in mice,13 where it influences neuronal differentiation,14 expression of numerous neurodevelopmental genes14, 15 as well as the suppression of tumorigenesis.16 Variants of the CHD5 gene have also been identified in the most recent and largest whole-exome-sequencing study of ASDs to date.3 These findings indicate a strong contribution of Chd5 to neurodevelopmental processes and a putative role in the etiology of ASDs. We therefore sought to investigate the role of chromatin remodeling in ASD-associated endophenotypes by generating and characterizing a novel mouse model resulting from genetic deletion of Chd5.
Materials and methods
Animals and genotyping
Chd5−/− mice were generated using conditional (Chd5fl/+) embryonic stem cells (C57BL/6 J) obtained from the EUCOMM consortium. Mice were genotyped using the following Chd5 primers: Floxed allele (forward (P1): 5′-TCTACAGAGCAAGTTCCAGGAC; reverse (P2): 5′-GACCCCAAGCTGAGGAAAACC-3′), deleted allele (forward (P1): 5′-TCTACAGAGCAAGTTCCAGGAC-3′; reverse (P3): 5′-CCGTGTTGGTTTCTCTGACTTTC-3′). Tissue was also genotyped for the nestin-Cre transgene using the following primers: forward: 5′-GCGGTCTGGCAGTAAAAACTATC-3′; reverse: 5′-GTGAAACAGCATTGCTGTCACTT-3′. TomR-GFP transgenic mice were obtained from Jackson Laboratories (#007676) and genotyped using the following primers: forward: 5′-CTCTGCTGCCTCCTGGCTTCT-3′ wild-type reverse: 5′-CGAGGCGGATCACAAGCAATA-3′ mutant reverse: 5′-TCAATGGGCGGGGGTCGTT-3′. Mice were bred and maintained according to the US National Institutes of Health guidelines for animal care and use and Institutional Animal Care and Use Committee of the University of Minnesota—Twin Cities. Mice were provided food and water ad libitum except during experimental testing, and housing lights were maintained on a 12:12-h light/dark cycle.
Microscopy
Fluorescent images confirming tomR-GFP reporter expression were acquired on a Zeiss LSM 710 scanning confocal microscope (Zeiss, Jena, Germany) using a × 10 numerical objective and ZEN microscopy software.
Western blot
Whole-brain tissue was collected rapidly, transferred to cold phosphate-buffered saline (PBS) and homogenized in cold lysis buffer. Homogenate was exposed to three freeze–thaw cycles, spun at 10 000 r.p.m. at 4 °C, and supernatant was extracted. Total protein was quantified using standard spectrophotometer methods. Forty micrograms of protein were loaded into a gel, and then transferred and incubated with Chd5 anti-rabbit primary antibody (1:1000), followed by Chd5 rabbit-anti-mouse secondary (1:500). Control antibodies included glyceraldehyde-3-phosphate dehydrogenase (Gapdh) primary (1:20 000) and secondary (1:500). Bands were visualized and quantified using the ImageQuant system (GE Healthcare Life Sciences, Pittsburgh, PA, USA).
RNA sequencing
Total RNA was extracted from the frontal cortex brain tissue of naive, 8-week-old male mice matched with same-sex littermates (three wild types; three Chd5−/−). After collection, tissue was rapidly immersed in RNAlater (Qiagen, Valencia, CA, USA) and total RNA was isolated using the RNeasy Mini Kit (Qiagen, 74104). RNA integrity numbers were calculated using a bioanalyzer to be >7 for all samples. Extracted mRNA was purified with poly-T oligo-attached magnetic beads, heat-fragmented, and both strands synthesized and purified. The 3′ ends were then polyadenylated and RNA sequencing (RNA-Seq) libraries were prepared from samples using the Illumina (San Diego, CA, USA) TruSeq protocol, and then barcoded and sequenced using a 50-bp paired-end run on the Illumina HiSeq 2500. Roughly 20 million paired-end reads were generated for each run. Raw RNA-Seq data were uploaded to the Minnesota Supercomputer Institute Galaxy server. Quality control of raw data was completed using FastQC, and all samples had base sequence quality values (Phred) >30 with no over-represented sequences. FastQ files were then aligned to the mm10_canonical mouse genome and mapped using Tophat.17 A list of assembled transcripts for each replicate was then analyzed using Cufflinks18 to estimate abundances (represented by fragments per kilo bases per million mapped reads) and tested for differential expression. Transcriptional differences between wild-type and Chd5−/− replicates were then analyzed in cummeRbund. The lists of significantly differentially regulated transcripts (P<0.05, false-discovery rate<5%) were analyzed using the Ingenuity Pathway Analysis software (Qiagen).
Quantitative reverse transcriptase polymerase chain reaction
Total RNA was obtained from frontal cortex brain tissue of naive, 8-week-old male mice matched with same-sex littermates using the same protocol as RNA-Seq. Isolated RNA was processed for DNAase digestion and reverse-transcribed using the GoScript reverse transcriptase (Promega, Madison, WI, USA) according to the manufacturer’s manuals. The complementary DNA was diluted 1:7 before preparing the quantitative reverse transcriptase polymerase chain reaction (qRT-PCR), which was then run using the GoTaq qPCR master mix (Promega). All PCR primers were designed at qPrimerDepot (http://mouseprimerdepot.nci.nih.gov/) and purchased through the University of Minnesota’s Genomics Center. See Supplementary Table 1 for primer information. Mouse Gapdh mRNA was used as the endogenous control. For each sample, a duplex PCR reaction was set up containing a target gene primer set and a Gapdh primer set. We conducted three replicate PCR reactions for each sample. The reactions were incubated in a 96-well plate at 95 °C for 10 min, followed by 45 cycles of 95 °C for 15 s and 60±2 °C for 1 min on the CFX96 Real-Time System (Bio-Rad, Hercules, CA, USA). The relative levels of gene expression were determined according to the standard curve methods described in the Bio-Rad company manual. The expression value of the target gene in each well was first normalized by the expression value of the Gapdh gene in the same well. The median of three repeated reactions was used to represent the relative quantity of the target gene.
Neuronal culturing and morphology analysis
Frontal cortex tissue was dissected from postnatal day (PND) 0 to 1 pups (four per genotype matched from independent litters), dissociated, sparsely transfected with a green fluorescent protein (GFP)-encoded construct and plated on poly-l-lysine-coated glass coverslips (~100 000 cells per 35 mM plate) using plating media from Invitrogen (Carlsbad, CA, USA): MEM Eagle (Invitrogen, #14175095), fetal bovine serum (Invitrogen, #16140071), 20% glucose, sodium pyruvate (Invitrogen, #11360070), glutamine (Invitrogen, #25030081) and penicillin/streptomycin (Invitrogen, #15140122). Two hours after plating, media were replaced with maintenance media (Neurobasal medium (Invitrogen, #21103049), B-27 (Invitrogen, #17504044), 200 mM glutamine (Invitrogen, #25030081) and penicillin/streptomycin (Invitrogen, #15140122)), and half the media was subsequently replenished twice per week. At 12 days in vitro, primary neuronal cultures were fixed with 4% paraformaldehyde/PIPES, HEPES, EGTA, and Magnesium Sulphate Heptahydrate (PHEM)/0.12M sucrose, and then blocked with 3% radioimmunoassay grade bovine serum albumin in 1 × PBS for 1 h at room temperature. Cells were then permeabilized with 0.2% Triton-X in PBS at room temperature and blocked again with 3% radioimmunoassay grade bovine serum albumin in PBS. Cells were stained with a primary anti-MAP2 antibody (1:1000; #ab11268; Abcam, Cambridge, UK) in 1% radioimmunoassay grade bovine serum albumin/PBS overnight at 4 °C, followed by secondary (1:100) and 1 × 4,6-diamidino-2-phenylindole (DAPI) counterstain for 30 min at room temperature. Finally, cells were mounted with DABCO mountant, sealed with nail polish and stored at 4 °C. Fluorescent imaging of cells was performed on a Zeiss Axiovert 200 microscope using a × 20 objective and Micro-Manager microscopy software. Images of 15 randomly selected pyramidal neurons were acquired per culture. Cells were analyzed for dendritic arborization using the ImageJ Sholl Analysis plug-in19 with a 10 μM radius interval.
Open field and novel object recognition
The open field and novel object recognition tasks were conducted as previously described.20 Six- to eight-week-old male mice were handled for three consecutive days (5 min per day). On the first day of testing, individual mice were introduced to the open field arena (50 × 50 × 40 cm) for 20 min. Distance traveled, velocity and center time (defined as the center 50% of the arena area) were calculated using the TopScan software system (Clever Systems, Reston, VA, USA). On the following day of testing, individual mice were placed into the same arena with two identical (familiar) objects (solid-colored geometric shapes; for example, red cubes) and allowed free exploration for 10 min. After 1 h, mice were re-introduced to the same testing arena with one item replaced with a different (novel) object (for example, blue pyramid) and allowed free exploration for 5 min. The time spent investigating each item (defined as when the mouse’s head was orientated <1 cm to the object) was scored manually by a trained, genotype-naive experimenter using Button Box 5.0 (Behavioral Research Solutions, Madison, WI, USA). A preference score was calculated as: ((novel time)/(novel time+familiar time)).
Pup separation-induced ultrasonic vocalizations
The mouse pup separation-induced ultrasonic vocalization (USV) task was conducted as previously described.21 Prior to testing, the litter was separated from the dam for 15 min in order to obtain steady-state conditions. During this time, a heating pad was used to maintain pups’ basal temperature. Then, individual mice were removed from the litter and placed in a sound-attenuated chamber outfitted with a high-frequency microphone and data acquisition hardware (UltraSoundGate 416H, Avisoft Bioacoustics, Glienicke, Germany). USVs were recorded for 5 min (Recorder USGH, Avisoft Bioacoustics). Each mouse pup was tested on PNDs 6, 8, 10 and 12. Recording chambers were cleaned with 70% ethanol between pups. For analysis, recordings were processed using a custom-built R script. In brief, spectrograms were generated from raw WAV files and band pass-filtered (15–110 kHz). A time-varying parameter, spectral purity, was computed as the fraction of total power within a single-frequency bin, and filtered over 8 ms. Identification of a vocalization bout was based on three parameters: minimal length (>5 ms), spectral purity (>0.15 ms) and holding time (<10 ms), defined as the threshold time in which separate bouts were merged. Classification of each USV syllable was characterized by frequency and time parameters, and was defined as follows: NS, no frequency modulation (jumps), short duration (<25 ms); NL, no jumps, long duration (⩾25 ms); SJ, single jump (>7 kHz); MJ, multiple jumps.
Three-chamber social approach
The three-chamber approach task was conducted as described previously.22 Eight- to ten-week-old male mice were habituated to handling for three consecutive days (5 min per day). On the subsequent day, mice underwent three 10-min testing phases within an arena (20 × 25 × 45 cm) partitioned into three chambers containing fresh corncob bedding and inverted wire mesh cups in opposite chambers. In the first (habituation) phase, individual test mice were placed into the arena and allowed to explore freely. In the second (social approach) phase, an age- and sex-matched conspecific (unfamiliar mouse) was enclosed under a cup within one chamber. In the third (social novelty) phase, another age- and sex-matched conspecific (novel mouse) was added to an identical cup within the opposite chamber. The test mouse was restricted to the center chamber between phases. For each phase, the time spent within each chamber for each test mouse was scored by a genotype-naive experimenter using Button Box 5.0 (Behavioral Research Solutions). For the second (approach) and third (novelty) phases, the time the test mouse spent investigating each cup was also scored. Investigation was defined as when the mouse’s head was orientated approximately <1 cm to the cup. A ratio of time spent in either chamber or investigating either cup was calculated.
Social fear behaviors
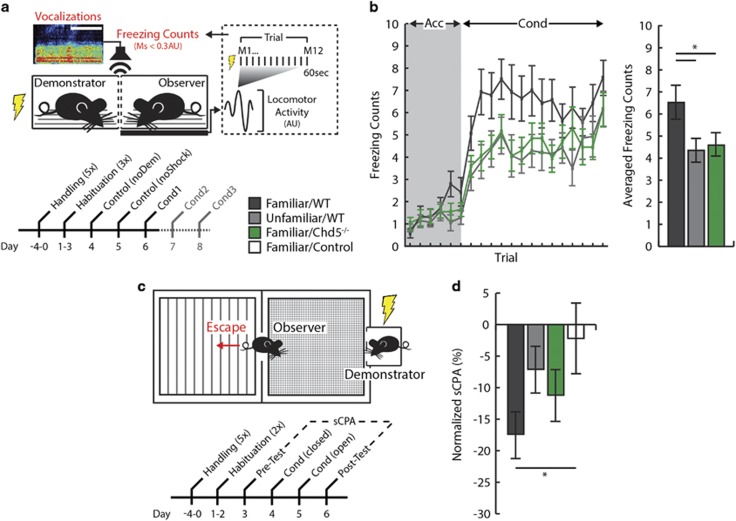
Male mice were pair-housed with littermates from the time of weaning, and then tested at 10–15 weeks of age using the socially transmitted fear paradigm. Observer mice were acclimated to handling by an experimenter for five consecutive days (5 min per day), followed by habituation to the testing context and acoustic startle over three consecutive days. All sessions commenced with a 5-min acclimation period following placement of the observer–demonstrator pair into adjacent, transparent Plexiglas cages. Activity measurements were recorded from the observer animal every 5 s using a load cell transducer, amplifier and computer equipped with Advanced Startle software (Med Associates, St. Albans, VT, USA). The unconditioned stimulus (US) was a 0.8 mA, 1.5 s scrambled foot shock delivered through the bars of the cage to the demonstrator. This shock intensity was piloted to elicit reliable auditory and visual signs of distress (that is, vocalizations and thigmotaxis behavior) from demonstrators. The conditioning protocol was composed of 15 trials, each containing 12 activity measurements preceding the US (65 s intertrial interval). The six measurements immediately preceding the US included a conditioned stimulus tone (12 kHz, 70 dB, 1 s rise time). In post hoc analyses, measurements with or without the CS did not differ; therefore, averaged activity across all 12 measurements was used. Prior to demonstrator conditioning days, observer mice underwent two consecutive days of control conditioning experiments: the first without inclusion of the demonstrator and with the US (noDem); the second with inclusion of the demonstrator and without the US (noShock). Observer–demonstrator pairs underwent three consecutive days of social conditioning. Freezing behavior was calculated as the number of measurements per trial (12 total) below a 0.3 AU activity threshold. Videos of observer mice were also recorded during conditioning sessions using infrared web cameras. Video was scored for freezing behavior by a genotype-naive experimenter using Button Box 5.0 (Behavioral Research Solutions).
During conditioning sessions, distress vocalizations were collected using a high-frequency microphone, data acquisition hardware (UltraSoundGate 416H; Avisoft Bioacoustics) and recording software (Recorder USGH; Avisoft Bioacoustics). For analysis, recordings were processed using similar methods as described above (pup separation-induced USVs). Total duration was computed by summing the vocalizations identified across the entire conditioning session. Vocalizations were presumed to be elicited from demonstrators based on their broadband characteristics and coincidence with shock onset/offset. An experienced experimenter confirmed the accuracy of automated call detection using the generated spectrogram.
The socially induced avoidance (SIA) task represents a novel method for studying social fear learning in mice. The SIA task was conducted over six consecutive days. All days entailed a 20-min testing session within a standard conditioned place-preference arena (20 × 25 × 45 cm) at 50 lux. On two consecutive days, observer mice were habituated to the arena without inclusion of the demonstrator. The following day (pre-test), a demonstrator mouse was introduced into a small cage on one side of the arena (counterbalanced) and the observer was allowed to freely explore. On the first day of social conditioning, the observer mouse was confined to the same compartment of arena as the demonstrator. After 5-min acclimation, the demonstrator was exposed to repeated foot shocks (1.5 s, 1 mA scrambled current per 1 min; 15 total) using the Advanced Startle software (Med Associates). On the second day of social conditioning, the observer mouse was allowed to explore the entire arena while the demonstrator was conditioned using the same protocol. On the final day of testing (post-test), the identical procedure was used as the pre-test. A normalized SIA value was calculated as the change in percentage time spent on the demonstrator-containing chamber between pre- and post-test: (post-time–pre-time)/(pre-time)). Video was collected during all sessions using infrared web cameras and scored by trained, group-naive experimenters using Button Box 5.0 (Behavioral Research Solutions).
Statistical analyses
Unless otherwise stated, comparisons between different experimental groups were made using two-tailed, Student’s t-tests assuming normal distributions and equal variances. For comparisons to chance (novel object recognition, three-chamber social approach) or controls (SIA), Dunnett’s post hoc tests were applied. Repeated measure, one-way analyses of variance were used for pup USV, startle reactivity, prepulse inhibition and fear extinction data. Two-way analyses of variance were used for olfactory habituation–dishabitution data. Sample sizes were estimated based on published literature. Experimental mice were not tested on more than one behavioral task. Unless noted, all graphs are reported as means±s.e.m.
Results
Generation of brain-specific CHD5−/− mice
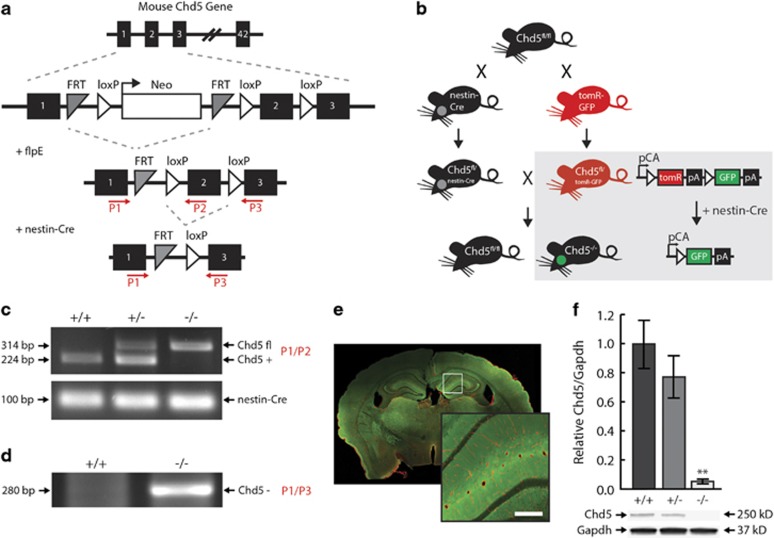
Mice harboring a conditional deletion of exon two of the Chd5 gene were first crossed with FlpE transgenic mice to remove the targeting/selection construct (Figure 1a). Conditional (Chd5fl/+) mice were then crossed with nestin-Cre transgenic mice, allowing for restricted Cre-recombinase activity and Chd5 allele deletion within neural tissue. This strategy was pursued in order to preserve Chd5 expression in the testes (a significant peripheral source of Chd5), and thereby avoid dysregulated spermatogenesis and infertility.23 A subset of Chd5fl/+ mice was also crossed with double fluorescent tomR-GFP transgenic reporter mice,24 so that Cre recombination produced GFP expression exclusively in neural tissue (Figure 1b). Chd5−/− mice were identified via PCR of genomic tail-DNA (Figure 1c), and validated via PCR of genomic brain DNA (Figure 1d) and brain fluorescence labeling (Figure 1e). Adult Chd5−/− mice show a nearly complete loss of the Chd5 protein in whole-brain extracts, whereas Chd5+/− mice show haplosufficiency and were, therefore, excluded from analyses (Figure 1f). Chd5−/− mice were embryonically viable and exhibited no gross physical abnormalities compared to littermate controls. This was expected in light of the marked increase in Chd5 protein expression during late adolescent development in the mouse.13
Figure 1.
Generation of brain-specific Chd5−/− mice. (a) Chd5 gene-targeting strategy. See Materials and methods for primer (P1–3) information. (b) Breeding strategy for brain-specific Chd5−/− mice (box, schematic of tomR-GFP reporter gene expression). PCR genotyping of genomic DNA from (c) tail biopsy for (top) Chd5-targeted allele and (bottom) nestin-Cre transgene allele and (d) whole-brain extract for Chd5 null allele. (e) Representative Chd5−/− brain expressing TomR (red) in non-neural and GFP (green) in neural tissue. Inset represents magnified region. Scale bar, 100 μM. (f) Western blot of whole-brain extracts indicating the near absence of protein in Chd5−/− mice compared with Chd5+/+ mice (n=6/group). **P=0.0002. Chd5, chromodomain helicase DNA-binding 5; GFP, green fluorescent protein; PCR, polymerase chain reaction.
Transcriptional dysregulation in Chd5−/− cortex
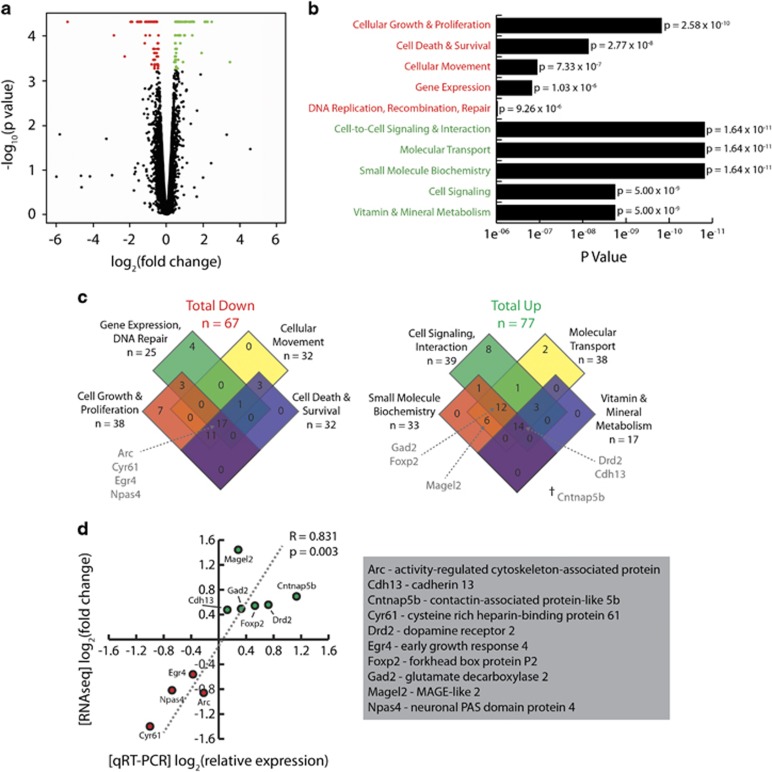
Chromatin remodeling exerts widespread effects on gene expression. To examine the transcriptional consequences of deletion of the Chd5 chromatin remodeler, we completed RNA-seq of the frontal cortex tissue collected from experiment-naive, home cage mice. A restricted subset of all (23 284) transcripts showed significantly reduced (67) or elevated (77) expression in Chd5−/− mouse cortical tissue (Figure 2a; Supplementary Table 2). The reduced transcript subset was enriched for gene ontology functions related to cellular growth and proliferation, cell death and survival, cellular movement, and gene expression (Figure 2b; Supplementary Table 3), whereas the elevated transcript subset was enriched for gene ontology functions related to cell-to-cell signaling, molecular transport, small-molecule biochemistry and vitamin/mineral metabolism (Figure 2b; Supplementary Table 4).
Figure 2.
Differential gene expression in Chd5−/− mouse cortex. (a) RNA-seq volcano plot illustration of significantly downregulated (red) and upregulated (green) gene transcripts in Chd5−/− cortical tissue. Gene transcripts with no-fold change were excluded. (b) The five most significant molecular/cellular GO terms for downregulated and upregulated gene transcripts from Ingenuity Pathway Analysis. (c) Venn diagram depictions of the most significantly (left) downregulated and (right) upregulated GO terms. Terms corresponding to each of the genes selected for qRT-PCR confirmation are indicated by arrows. †Transcript does not fall under any of the highlighted GO terms. (d) qRT-PCR confirmation of expression in a select set of transcripts (see c). Chd5, chromodomain helicase DNA-binding 5; GO, gene ontology; qRT-PCR, quantitative reverse transcriptase polymerase chain reaction; RNA-seq, RNA sequencing.
In order to confirm differential expression of transcripts identified by RNA-seq, we conducted qRT-PCR analyses on a select set of transcripts common to several molecular/cellular functions implicated in ASD pathophysiology (Figure 2c). RNA-seq and qRT-PCR values showed a statistically significant correlation, thereby validating gene expression differences in Chd5−/− mouse cortex (Figure 2d). These results confirm that deletion of the Chd5 chromatin remodeler induces targeted effects on cell signaling, development and regulation of gene expression mechanisms.
As a putative mouse model of ASDs, we also compared the gene transcripts differentially expressed in the Chd5−/− mouse cortex with a set of candidate genes identified in humans with ASDs. Using the AutismKB nonsyndromic candidate gene list (2136), we discovered eight genes common to our list of 67 downregulated transcripts (Supplementary Table 5), and 15 genes common to our list of 77 upregulated transcripts (Supplementary Table 6). The overlapping list of either up- or downregulated gene transcripts was enriched highly significantly above chance (Fisher’s exact test; odds ratio=2.1; P=0.0015). This suggests that the Chd5 chromatin remodeler may directly and/or indirectly influence candidate genes that putatively give rise to ASDs.
Dendritic alterations in Chd5−/− cortex
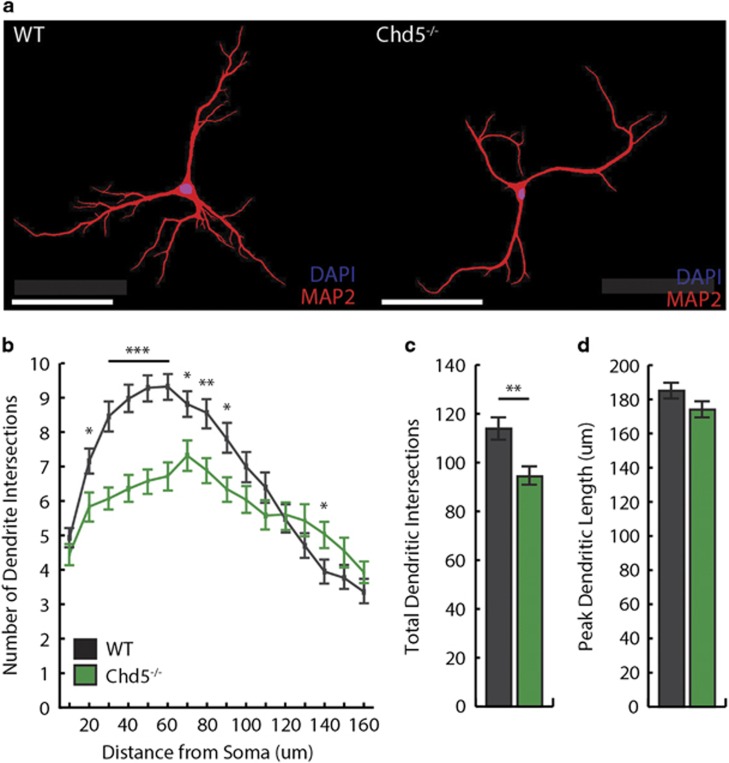
Post-mortem studies have identified cortical features of ASDs consistent with aberrant neurodevelopment.25 Whereas chromatin remodeling mediates a wide range of neurodevelopmental processes,6 Chd5 is associated specifically with early-life cortical development and neurogenesis.13, 14 Our RNA-Seq findings from cortical tissue of Chd5−/− mice implicated dysregulation of transcripts associated with cellular growth and proliferation, and cell signaling. Another set of neuron-specific chromatin remodeling proteins—nBAF53a/b—regulate dendritic arborization.26 To examine this particular feature of neurodevelopment, we dissociated and cultured cortical neurons from PND 0 to 1 mice, immunofluorescently labeled dendritic processes with MAP2,27 and quantified dendritic morphology of pyramidal neurons at 12 days in vitro via Sholl analysis (Figure 3a). In Chd5−/− pyramidal neurons, we discovered an altered profile of dendritic morphology entailing fewer intersections at sites proximal to the soma (Figures 3b and c). This reduction in arborization complexity did not appear to influence the peak dendritic length (that is, reach of dendrites from the soma; Figure 3d).
Figure 3.
Dendritic alterations in cortical pyramidal neurons of Chd5−/− mice. (a) Representative pyramidal neurons from (left) wild-type and (right) Chd5−/− mouse cortex immunofluorescently labeled with MAP2 (red) for dendritic processes and DAPI (blue) for nuclei. Scale bar, 100 μM. (b) Compared with wild type (black), Chd5−/− pyramidal neurons (green) showed significantly fewer dendritic intersections at sites more proximal to the soma (n=60 neurons, four mice per group). Compared with wild-type, Chd5−/− pyramidal neurons showed (c) significantly fewer total dendritic intersections (P=0.002) without (d) differences in peak dendritic length (P=0.066). *P<0.05, **P<0.01, ***P<0.001. Chd5, chromodomain helicase DNA-binding 5; DAPI, 4,6-diamidino-2-phenylindole.
Neophobic behavior of Chd5−/− mice
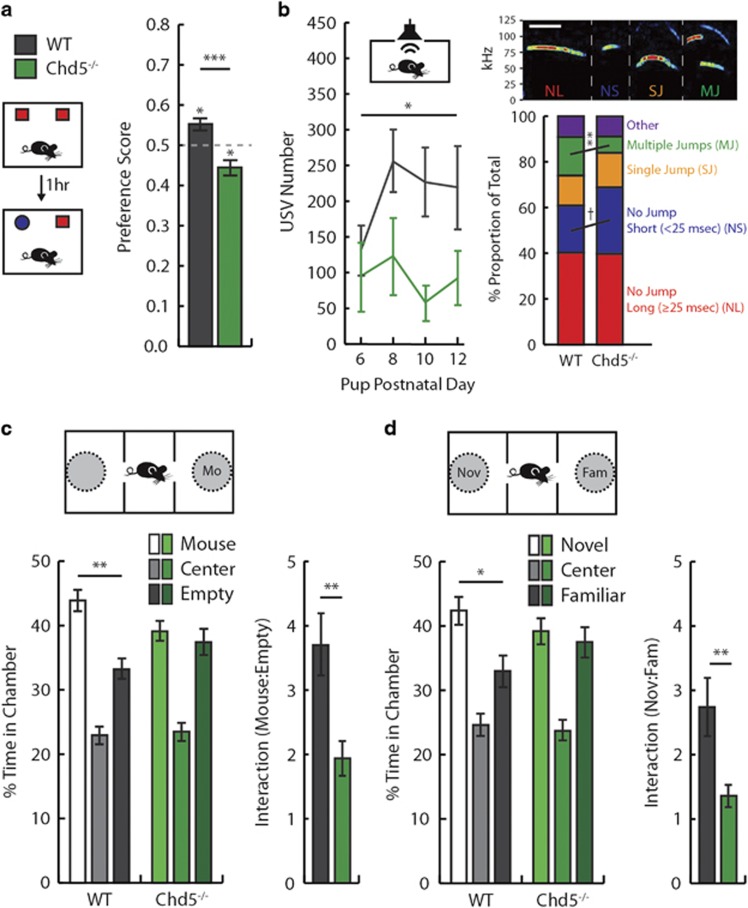
Individuals with ASDs frequently present with stereotyped motoric behavior, insistence for sameness and comorbid anxiety.1, 28 We first sought to characterize the behavior of Chd5−/− mice in standard assays for locomotor activity and anxiety. Chd5−/− mice were indistinguishable from control mice in measures of locomotor behavior, including open field velocity (Supplementary Figure 1a) and distance (Supplementary Figure 1b), and showed normal activity levels prior to startle reactivity testing (Supplementary Figure 1d). Chd5−/− mice also showed typical levels of prepulse inhibition, a measure of sensorimotor gating (Supplementary Figure 1e). However, Chd5−/− mice spent less time than their wild-type littermates within the center area of an open arena (Supplementary Figure 1c). This potential indicator of anxiety was not replicated in measures of the elevated plus-maze (Supplementary Figure 1f and g) or startle reactivity (Supplementary Figure 1d). As mice were not habituated to the open field arena prior to testing, the reduced center time of Chd5−/− mice may instead represent a neophobic behavioral response to the testing environment. To further examine neophobic behavior in Chd5−/− mice, we tested the mice in a novel object recognition task.29, 30 Both Chd5−/− and wild-type mice demonstrated equivalent time investigating two identical objects (Supplementary Figure 2a). When the same object was presented together with a novel object 1 h later, wild-type mice displayed a significant species-typical preference for the novel object (Figure 4a). In contrast, Chd5−/− mice exhibited a preference for the familiar object that was significantly different both from wild-type mice and from chance investigation. Together with the findings from the open field, these data suggest that Chd5−/− mice exhibit a proclivity for familiar environmental contexts and objects.
Figure 4.
Chd5−/− mice exhibit nosological features of autism. (a) In the NOR task, wild-type mice (black) showed a preference for the novel object (P=0.015), whereas Chd5−/− mice (green) exhibited a preference for a familiar object (P=0.02) and genotypes were significantly different from each other (P=0.0001; n=15 per group). (b; Left) Male Chd5−/− pups (n=7) produce fewer separation-induced USVs across PNDs 6–12 compared with wild types (n=14; P=0.02). (Right, top) Spectrogram examples of USV types. Scale bar, 25 ms. (Right, bottom) On PND6, Chd5−/− pups elicited a significantly smaller proportion of complex (MJ) USVs compared to wild types (P=0.01). (c; Left) Compared with an empty chamber, adult male wild-type mice spent more time in the chamber containing an enclosed, unfamiliar mouse (Mo; pairwise t-test, P=0.002), whereas Chd5−/− mice showed no preference (pairwise t-test, P=0.60; n=20 per group). (Right) Wild types investigated the conspecific-containing cage more than Chd5−/− mice (P=0.002). (d; Left) Wild types spent more time in the chamber containing an enclosed, novel conspecific (Nov) than that containing a familiar (Fam) mouse (pairwise t-test, P=0.042), whereas Chd5−/− mice showed no preference (pairwise t-test, P=0.67). (Right) Wild types investigated the novel conspecific-containing cage more than Chd5−/− mice (P=0.01). *P<0.05; **P<0.01; ***P<0.001; †P=0.06. Chd5, chromodomain helicase DNA-binding 5; PND, postnatal day; USV, ultrasonic vocalization.
Sociocommunicative abnormalities of Chd5−/− mice
Deficits in social interaction and communication are a defining feature of ASD symptomatology.1 We examined this behavioral domain in Chd5−/− mice using two standard tasks: pup separation-induced USVs and adult three-chamber social approach.31 In the pup separation-induced USV task, Chd5−/− pups produced significantly fewer USVs across early PNDs when compared with littermate wild-type mice (Figure 4b, left). Mice exhibit a complex repertoire of USVs;32 hence, we further analyzed the types of USV syllables by time/frequency characteristics. Compared with wild-type mice, Chd5−/− mice elicited a significantly smaller proportion of complex, frequency-modulated USVs (Figure 4b, right). Although our understanding of the functional significance of USV subtypes is rudimentary, these data suggest abnormal intraspecies communication in Chd5−/− mice. In the adult three-chamber social approach task, wild-type and Chd5−/− mice displayed equivalent exploration time within adjoining chambers during a habituation phase (Supplementary Figure 2b). When next exposed to an enclosed, unfamiliar conspecific mouse in one chamber, Chd5−/− mice showed no preference for the conspecific-containing chamber and spent significantly less time than wild-type mice directly investigating the mouse (Figure 4c). Following the addition of a novel conspecific in the opposite chamber, Chd5−/− mice exhibited no preference for the novel conspecific-containing chamber and significantly less time than wild-type mice directly investigating the enclosed novel conspecific (Figure 4d). Importantly, Chd5−/− mice did not show substantial deficits in recognition of olfactory cues, as indicated by robust dishabituation of investigation upon presentation of a novel odor and habituation of investigation across trials of the same odor (Supplementary Figure 2c). However, Chd5−/− showed less overall investigation of novel olfactory stimuli, a further indication of reduced engagement with novel cues in these mice.
Empathy deficits of Chd5−/− mice
Impairments in social cognition, including facial processing, emotional recognition and empathy, are prominent endophenotypes of ASDs.33, 34, 35 We modeled empathy in mice using two social fear paradigms, in which foot shock applied to a demonstrator mouse elicits fear behavior in an observer mouse. We first studied freezing in observer mice using a load cell transducer system, which measures force-generated locomotor activity concurrent with demonstrator conditioning (Figure 5a, top). This automated technique provides a reliable measure of freezing (MTP, IIG, JCG—under review at Nature Communications). Freezing of observers was first measured during control experiments conducted on 2 days prior to conditioning (Figure 5a, bottom). On one day, electrical current was passed through an empty demonstrator cage (noDem), and on the other the demonstrator was present but not conditioned (noShock). Average nonspecific freezing behavior was not significantly different between wild-type and Chd5−/− observers (Supplementary Figure 3). As a means of validating our paradigm as a measure of empathy, we first evaluated the effect of familiarity on freezing behavior. We found that wild-type observer mice froze during foot shock conditioning of the demonstrator mice, and—consistent with the literature36—this behavior was significantly greater for observers that were both siblings and cage mates of demonstrators (henceforth, ‘familiar/wild-type’) compared with observers that were non-sibling and non-cage mates (henceforth, ‘unfamiliar/wild-type’ Figure 5b). We next examined socially transmitted fear in Chd5−/− mice. Although basal activity levels were similar to those seen in familiar/wild-type mice, familiar/Chd5−/− observer mice exhibited significantly less freezing during conditioning of demonstrator mice (Figure 5b). This effect was not due to differences in the duration or characteristics of vocalizations emitted from demonstrator mice (Supplementary Figure 4a) or to a deficit in the capacity to acquire or express Pavlovian fear-conditioning behavior (Supplementary Figure 5). Locomotor activity measurements obtained immediately after demonstrator foot shock (measurement (M)1) indicated similar orientation responses for both genotypes and unfamiliar observer mice (Supplementary Figure 4b). This finding implies that sensory processing of cues emanating from demonstrator mice was intact in all three groups. In contrast, only familiar/wild-type mice exhibited sustained freezing behavior throughout each trial (that is, M2–12).
Figure 5.
Chd5−/− mice show deficits in behavioral measures of empathy. (a) Schematic of the socially transmitted fear paradigm (Acc, acclimation; AU, arbitrary unit; Cond, demonstrator conditioning; M, measurement; Control, non-conditioned wild-type). (b; Left) Familiar/wildtype observer mice (black, n=14) froze more than familiar/Chd5−/− (green, n=20) or unfamiliar/wild-type (gray, n=14) observers during demonstrator conditioning (right), a significant effect across all conditioning trials (unfamiliar/wild-type: P=0.03; familiar/Chd5−/− : P=0.05). (c) Schematic of the SIA paradigm. (d) Familiar/wild-type observers exhibited significant SIA compared with controls (P=0.043), whereas familiar/Chd5−/− (P=0.32) and unfamiliar/wild-type (P=0.79) observer mice did not. *P<0.05. Chd5, chromodomain helicase DNA-binding 5; SIA, socially induced avoidance.
We then investigated whether socially mediated fear could be learned in observer mice by testing them 24 h after conditioning. We did not observe a conditioned freezing response in any group. However, over two additional conditioning days, familiar/wild-type observer mice exhibited increases in activity, an effect not seen in familiar/Chd5−/− mice (Supplementary Figure 4c). This increase in activity may have indicated aversion, or escape behavior, the most common ethological response to audible frequency distress vocalizations of ground squirrels, another species of rodent.37 To evaluate this possibility further, we tested Chd5−/− mice in a SIA paradigm in which an observer mouse is exposed to a demonstrator mouse undergoing fear conditioning in one chamber of a two-sided place-preference arena (Figure 5c; MTP, IIG, JCG—under review at Nature Communications). On a second day of conditioning, when observer mice were free to move between the chambers, familiar/Chd5−/− mice showed escape behavior during acclimation; yet, this behavior was not maintained during conditioning compared with non-conditioned controls (Supplementary Figure 4d). Conversely, familiar/wild-type mice exhibited escape behavior during demonstrator conditioning, and unfamiliar/wild-type mice did not show escape behavior during either acclimation or conditioning. Side preferences were equivalent for all groups at pre-test, yet only familiar/wild-type mice exhibited a significant preference for the non-demonstrator side in the post-test, in which conditioning of the demonstrator mouse was not conducted (Supplementary Figure 4e). Lastly, a normalized score of SIA indicated that only familiar/wild-type mice exhibited significant avoidance of the demonstrator-containing side (Figure 5d). Thus, in not responding to the distress of a familiar conspecific either by freezing or by escape, Chd5−/− mice behaved similarly to wild-type mice unfamiliar with the conspecific under distress.
Discussion
Large-scale exome-sequencing studies strongly implicate genes involved in chromatin remodeling in the etiology of ASDs.3 Demonstrating this in a genetic mouse model is a critical step toward characterizing pathways, through which perturbations in epigenetic modifications may lead to neurodevelopmental disorders. Thus, it is significant that mice harboring a deletion of the chromatin remodeler gene Chd5 exhibit a number of the prototypical characteristics of ASDs. One defining feature of ASDs, impaired sociocommunicative interaction, was recapitulated in the mice as impoverished vocalizations and reduced interaction with conspecifics. A second defining feature of these disorders, a preference for sameness, was manifested in tests for exploratory behavior, social interaction and odor preference. The fact that this preference spanned several behavioral domains suggests that it reflects a pervasive, underlying endophenotype rather than a modality- or response-specific behavioral motif. The commonality between the behavior of the Chd5−/− mouse and the symptomatology of ASD was reinforced by our observation of impairments in emotional state-matching, a component of empathy in humans. In contrast, a variety of other behaviors, including locomotion, prepulse inhibition and Pavlovian fear conditioning, were normal. The selective pattern of behavioral deficits suggests that a disruption of chromatin remodeling can perturb the development of neurotypical behavior.
To begin to address the question of how the Chd5 chromatin remodeler may be involved in producing neurodevelopmental outcomes, we used RNA-Seq in tandem with qRT-PCR verification to identify genes whose expression in the frontal cortex is significantly affected by Chd5 deletion. Of those genes whose activity was upregulated or downregulated, the functions of a disproportionately large number were linked to transcription, cell development and signaling mechanisms. These canonical pathways are common to ASD etiology, and may therefore provide clues to the underlying pathophysiology of Chd5−/− mice. In addition, a significant proportion of differentially expressed genes (23 of 144) are ones implicated specifically in ASDs and/or socio-emotional function. Several such transcripts are especially noteworthy. For example, Magel2 is associated with the presentation of ASD symptoms in Prader–Willi Syndrome, a condition caused by absence of expression of paternal genes from chromosome 15q11-q13.38 Mice harboring the homologous copy number variation display reduced novelty-based exploration and feeding, features akin to the neophobic behaviors observed in the Chd5 mouse model.39 A second downregulated transcript was Npas4. Mice lacking this transcription factor show behavioral characteristics indicative of psychiatric diseases, including reduced social novelty preference and cognitive functions.40 Interestingly, Npas4 has not been linked to autism in sequencing studies. The present findings, therefore, raise the possibility that epigenetic dysregulation of Magel2 and/or Npas4 has a role in producing ASD endophenotypes.
We also identified upregulation of dopamine receptor-encoding transcripts (Drd1a and Drd2) in Chd5−/− cortical tissue. Dopamine signaling and specific polymorphisms in dopamine receptor genes have been linked to novelty-seeking behavior.41, 42, 43 Lastly, we saw upregulation of Foxp2, a transcription factor integral to speech/language development.44 Mice lacking the Foxp2 gene exhibit abnormal pup separation-induced USVs.45 Similar to these findings, we detected changes in the number and type of USVs elicited by Chd5−/− pups. In sum, known functions of a sizable proportion of the genes whose activity is affected by deletion of Chd5 relate to preference for sameness and socio-emotional cognition, and thus align with the behavioral impairments seen here in the Chd5−/− mouse.
Differential expression of transcripts related to transcriptional regulation, cell development and cell signaling might also be expected to have an impact on neuronal growth and structure. Consistent with this possibility, we discovered alterations in pyramidal neuron morphology within the cortex of Chd5−/− mice. Thus, neurons cultured from the frontal cortex displayed less branching of dendrites proximal to the soma. A similar reduction in dendritic complexity was one of the first documented neuropathological markers of ASDs,46 and has been replicated in mouse models following deletion of ASD risk genes, including MeCP2,47 SHANK3 (ref. 48) and various neurexins/neuroligins.49 It seems likely that such alterations in neuronal morphology affect functional signaling within the cortex so as to produce deleterious behavioral sequelae. Manipulation of expression of some of the genes dysregulated through Chd5 deletion may help us to determine whether indeed there is a causal relationship between these morphological and behavioral endophenotypes.
Chd5 is one of several Chd proteins that orchestrates gene expression through chromatin remodeling.50, 51 Mutations in a number of these genes—specifically, Chd1, Chd2, Chd3, Chd7 and Chd8—have been associated with ASDs.3, 52, 53, 54 In light of Chd5’s predominant localization in neurons (as well as the testes), it is noteworthy that instances of mutation-induced neurodevelopmental sequelae are sparse. This may be due to the relatively minor effects of heterozygous deletion. Similar to the limited reduction of protein from deletion of a single copy of Chd5 in our nestin-Cre mouse line, whole-body Chd5+/− mice do not show the deficits in spermatogenesis that are present in Chd5−/− mice.23 Moreover, although loss of a single copy of Chd5 is strongly implicated in various forms of cancer (primarily neuroblastomas), tumorigenesis typically occurs only when the remaining copy of the gene is silenced epigenetically.55 Thus, neurodevelopmental and pathogenic consequences of Chd5 mutations may result from genetic or epigenetic inactivation of both alleles, such as demonstrated in this study.
Recognition of the emotional states of others and exploration of novelty are evolutionarily conserved traits in humans, as in other primates.56, 57 Whereas the importance of these traits is evidenced by their disruption in psychiatric disease, their genetic and neural substrates remain largely unknown. The effects of deletion in mice of the neuron-specific Chd5 chromatin remodeler suggest that development of these facets of behavior is under epigenetic control. The Chd5−/− mouse promises to offer a window into the epigenetic programming of the brain structure and function that is necessary for adequate engagement in the outside world. Equally, it may help us to trace the etiology of neurodevelopmental diseases in which these capacities are impaired. Moreover, given this gene’s pivotal role in tumor suppression, advances in our understanding of Chd5 function in neurodevelopmental disorders may benefit the treatment of cancer, and vice versa.20
Acknowledgments
This research is supported by the Autism Initiative (University of Minnesota), the Gralnick Prize for Research on Severe Mental Disorders (American Psychological Foundation), Lieber Prize for Outstanding Schizophrenia Research (NARSAD), University of Louisville Grawemeyer Prize in Psychology, and University of Minnesota’s Center for Cognitive Sciences (5T32HD007151). We thank Britta Logdahl, Ruby Ouloch and Zofeyah McBrayer for assistance with data collection, Lorene Lanier and Thomas Bastian for guidance with cell-culturing experiments; Jon Vettervall and Robert Klink for PCR assistance; Emily Leathley Wozniak for western blot assistance; Michael Pazin for Chd5 protein antibodies; Michael Saxe and Yan-Ping Zhang for ultrasonic vocalization analysis code; Phu Tran for RNA-seq analysis assistance; and David Largespada for nestin-Cre transgenic mice. We also wish to express our humble gratitude to Irv Gottesman, our co-author and colleague, who passed away during the preparation of this manuscript.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
The authors declare no conflict of interest.
Supplementary Material
References
- Association APDiagnostic and Statistical Manual of Mental Disorders. 5th edn, American Psychiatric Publishing: Arlington, VA, 2013. [Google Scholar]
- Sullivan PF, Daly MJ, O’Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet 2012; 13: 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Ercument Cicek A et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 2014; 3: 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet 2014; 94: 677–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. Chromatin remodelling during development. Nature 2010; 463: 474–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet 2013; 14: 347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier R, Golzio C, Xiong B, Stessman Ha, Coe BP, Penn O et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell 2014; 158: 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugathan A, Biagioli M, Golzio C, Erdin S, Blumenthal I, Manavalan P et al. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc Natl Acad Sci USA 2014; 111: 4468–4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotney J, Muhle Ra, Sanders SJ, Liu L, Willsey AJ, Niu W et al. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat Commun 2015; 6: 6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Gotoh T, Kok M, White PS, Brodeur GM. CHD5a new member of the chromodomain gene family, is preferentially expressed in the nervous system. Oncogene 2003; 22: 1002–1011. [DOI] [PubMed] [Google Scholar]
- Stanley FKT, Moore S, Goodarzi AA. CHD chromatin remodelling enzymes and the DNA damage response. Mutat Res 2013; 750: 31–44. [DOI] [PubMed] [Google Scholar]
- Eberl HC, Spruijt CG, Kelstrup CD, Vermeulen M, Mann M. A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol Cell 2013; 49: 368–378. [DOI] [PubMed] [Google Scholar]
- Vestin A, Mills Aa. The tumor suppressor Chd5 is induced during neuronal differentiation in the developing mouse brain. Gene Expr Patterns 2013; 13: 482–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan CM, Nyman U, Skotte J, Streubel G, Turner S, O’Connell DJ et al. CHD5 is required for neurogenesis and has a dual role in facilitating gene expression and polycomb gene repression. Dev Cell 2013; 26: 223–236. [DOI] [PubMed] [Google Scholar]
- Potts RC, Zhang P, Wurster AL, Precht P, Mughal MR, Wood WH et al. CHD5, a brain-specific paralog of Mi2 chromatin remodeling enzymes, regulates expression of neuronal genes. PLoS ONE 2011; 6: e24515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi A, Papazoglu C, Wu Y, Capurso D, Brodt M, Francis D et al. CHD5 is a tumor suppressor at human 1p36. Cell 2007; 128: 459–475. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009; 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012; 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira T, Blackman A, Oyrer J, Jayabal S, Chung A, Watt A et al. Neuronal morphometry directly from bitmap images. Nat Methods 2014; 11: 982–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley JN, Heyer WD, LaSalle JM. Autism and cancer share risk genes, pathways, and drug targets. Trends Genet 2016; 32: 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer M, Shair H, Brunelli S. Ultrasonic vocalizations in rat and mouse pups. Curr Protoc Neurosci 2001; 8: 1–16. [DOI] [PubMed] [Google Scholar]
- Crawley JN. Designing mouse behavioral tasks relevant to autistic-like behaviors. Ment Retard Dev Disabil Res Rev 2004; 10: 248–258. [DOI] [PubMed] [Google Scholar]
- Li W, Wu J, Kim S-Y, Zhao M, Hearn Sa, Zhang MQ et al. Chd5 orchestrates chromatin remodelling during sperm development. Nat Commun 2014; 5: 3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar M, Tasic B, Miyamichi K, Li L, Luo L. A global doublefluorescent Cre reporter mouse. Genesis 2007; 605: 593–605. [DOI] [PubMed] [Google Scholar]
- de la Torre-Ubieta L, Won H, Stein JL, Geschwind DH. Advancing the understanding of autism disease mechanisms through genetics. Nat Med 2016; 22: 345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Olave Ia, Qiu Z, Ghosh A, Graef Ia et al. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron 2007; 56: 94–108. [DOI] [PubMed] [Google Scholar]
- Goedert M, Crowther RA, Garner CC. Molecular characterization of microtubule-associated proteins tau and map2. Trends Neurosci 1991; 14: 193–199. [DOI] [PubMed] [Google Scholar]
- Gillott A, Furniss F, Walter A. Anxiety in high-functioning children with autism. Autism 2001; 5: 277–286. [DOI] [PubMed] [Google Scholar]
- Bevins Ra, Besheer J. Object recognition in rats and mice: a one-trial non-matching-to-sample learning task to study ‘recognition memory’. Nat Protoc 2006; 1: 1306–1311. [DOI] [PubMed] [Google Scholar]
- Lyon L, Saksida LM, Bussey TJ. Spontaneous object recognition and its relevance to schizophrenia: a review of findings from pharmacological, genetic, lesion and developmental rodent models. Psychopharmacology 2012; 220: 647–672. [DOI] [PubMed] [Google Scholar]
- Silverman J, Yang M, Lord C, Crawley J. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci 2011; 11: 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsley JMS, Monaghan JJM, Wenstrup JJ. Development of social vocalizations in mice. PLoS ONE 2011; 6: e17460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucksmith E, Allison C, Baron-Cohen S, Chakrabarti B, Hoekstra Ra. Empathy and emotion recognition in people with autism, first-degree relatives, and controls. Neuropsychologia 2013; 51: 98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolphs R, Spezio ML, Parlier M, Piven J. Distinct face-processing strategies in parents of autistic children. Curr Biol 2008; 18: 1090–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo MV, Barnes JL, Wheelwright SJ, Baron-Cohen S. Self-referential cognition and empathy in autism. PLoS ONE 2007; 2: e883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon D, Kim S, Chetana M, Jo D, Ruley HE, Lin S-Y et al. Observational fear learning involves affective pain system and Cav1.2 Ca2+ channels in ACC. Nat Neurosci 2010; 13: 482–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman P. Nepotism and the evolution of alarm calls. Science 1977; 197: 11–18. [DOI] [PubMed] [Google Scholar]
- Schaaf C, Gonzalez-Garay M, Xia F. Truncating mutations of MAGEL2 cause Prader-Willi phenotypes and autism. Nat Genet 2013; 45: 1405–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada K, Tomonaga S, Hatanaka F, Nakai N, Takao K, Miyakawa T et al. Decreased exploratory activity in a mouse model of 15q duplication syndrome; implications for disturbance of serotonin signaling. PLoS ONE 2010; 5: e15126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutellier L, Beraki S, Ardestani PM, Saw NL, Shamloo M. Npas4: a neuronal transcription factor with a key role in social and cognitive functions relevant to developmental disorders. PLoS ONE 2012; 7: e46604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhara T, Yasuno F, Sudo Y, Yamamoto M. Dopamine D2 receptors in the insular cortex and the personality trait of novelty seeking. Neuroimage 2001; 13: 891–895. [DOI] [PubMed] [Google Scholar]
- Gjedde A, Kumakura Y, Cumming P, Linnet J, Møller A. Inverted-U-shaped correlation between dopamine receptor availability in striatum and sensation seeking. Proc Natl Acad Sci USA 2010; 107: 3870–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalley JW, Fryer TD, Brichard L, Robinson ESJ, Theobald DEH, Laane K et al. Nucleus accumbens D2/3 receptors predict trait impulsivity and cocaine reinforcement. Science 2007; 315: 1267–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C, Fisher S, Hurst J. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 2001; 413: 519–523. [DOI] [PubMed] [Google Scholar]
- Shu W, Cho JY, Jiang Y, Zhang M, Weisz D, Elder Ga et al. Altered ultrasonic vocalization in mice with a disruption in the Foxp2 gene. Proc Natl Acad Sci USA 2005; 102: 9643–9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond GV, Bauman ML, Kemper TL. Hippocampus in autism: a Golgi analysis. Acta Neuropathol 1995; 91: 117–119. [DOI] [PubMed] [Google Scholar]
- Kishi N, Macklis JD. MeCP2 functions largely cell-autonomously, but also non-cell-autonomously, in neuronal maturation and dendritic arborization of cortical pyramidal neurons. Exp Neurol 2010; 222: 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Perroy J, Loll F, Perrais D, Fagni L, Bourgeron T et al. SHANK3 mutations identified in autism lead to modification of dendritic spine morphology via an actin-dependent mechanism. Mol Psychiatry 2012; 17: 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SX, Tari PK, She K, Haas K. Neurexin-neuroligin cell adhesion complexes contribute to synaptotropic dendritogenesis via growth stabilization mechanisms in vivo. Neuron 2010; 67: 967–983. [DOI] [PubMed] [Google Scholar]
- Marfella CGa, Imbalzano AN. The Chd family of chromatin remodelers. Mutat Res 2007; 618: 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JA, Georgel PT. CHD proteins: a diverse family with strong ties. Biochem Cell Biol 2007; 85: 463–476. [DOI] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 2012; 485: 5–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 2012; 485: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014; 515: 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolla V, Zhuang T, Higashi M, Naraparaju K, Brodeur GM. Role of CHD5 in human cancers: 10 years later. Cancer Res 2014; 74: 652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Waal FBM. Putting the altruism back into altruism: the evolution of empathy. Annu Rev Psychol 2008; 59: 279–300. [DOI] [PubMed] [Google Scholar]
- Russell PAPsychological studies of exploration in animals: a reappraisal. In: Archer J, Birke, L (eds). Exploration in Animals and Humans. Van Nostrand Reinhold Co., London, 1983, pp. 22–54. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.