Abstract
In this study, we demonstrate the theranostic capability of actively targeted, site-specific multibranched gold nanoantennas (MGNs) in triple-negative breast cancer (TNBC) cells in vitro. By utilizing multiplexed surface-enhanced Raman scattering (SERS) imaging, enabled by the narrow peak widths of Raman signatures, we simultaneously targeted immune checkpoint receptor programmed death ligand 1 (PDL1) and the epidermal growth factor receptor (EGFR) overexpressed in TNBC cells. A 1:1 mixture of MGNs functionalized with anti-PDL1 antibodies and Raman tag 5,5-dithio-bis-(2-nitrobenzoic acid) (DTNB) and MGNs functionalized with anti-EGFR antibodies and Raman tag para-mercaptobenzoic acid (pMBA) were incubated with the cells. SERS imaging revealed a cellular traffic map of MGN localization by surface binding and receptor-mediated endocytosis, enabling targeted diagnosis of both biomarkers. Furthermore, cells incubated with anti-EGFR–pMBA–MGNs and illuminated with an 808 nm laser for 15 min at 4.7 W/cm2 exhibited photothermal cell death only within the laser spot (indicated by live/dead cell fluorescence assay). Therefore, this study not only provides an optical imaging platform that can track immunomarkers with spatiotemporal control but also demonstrates an externally controlled light-triggered therapeutic approach enabling receptor-specific treatment with biocompatible theranostic nanoprobes.
Introduction
Theranostic nanostructures that combine both diagnostic and therapeutic components within a single nanoscale platform have attracted significant interest in the past decade,1−4 as they enable both highly specific detection of disease markers and subsequent targeted, image-guided treatment all within a single clinical procedure. Gold nanostructures are particularly well-suited for multifunctional theranostics due to their high biocompatibility and straightforward surface chemistry, enabling functionalization of multiple molecules including targeting moieties, contrast agents, and charge-neutralizing polymers. The theranostic capabilities of several shape- and size-controlled gold nanostructures have been studied in the past decade,5−12 and some are already in clinical trials.13 However, the combination of multiplexed imaging enabling targeted detection of multiple receptors with a therapeutic modality has not been demonstrated successfully. In this work, we show the theranostic functionality of near-infrared (NIR) resonant multibranched gold nanoantennas (MGNs) in triple-negative breast cancer (TNBC) cells in vitro by combining surface-enhanced Raman scattering (SERS) imaging with photothermal therapy (PTT). TNBC is highly aggressive and one of the most lethal types of breast cancer due to the lack of response to endocrine treatment and other targeted therapies.14,15 We particularly show the multiplexed detection of immunomarker programmed death ligand 1 (PDL1) and epidermal growth factor receptor (EGFR) in TNBC cells. The upregulation of immune checkpoint receptor programmed death protein 1, PD-1, expressed on activated CD8+ T-cells, impedes effector T-cell function by binding to its ligand, PDL1.16−18 Upon binding, interferons (IFN-γ) produced by activated T-cells upregulate PDL1 expression, which contributes to immunosuppression in the tumor microenvironment. Cancer immunotherapy, particularly the blockade of inhibitory pathways such as PDL1, has recently driven a paradigm shift in the treatment of multiple cancers and shown significant advantages over chemotherapy and radiation.18 However, the patient response rate of PDL1 therapies remains low (<30%) in part due to both heterogeneous PDL1 expression among patients and within the same patients’ primary and metastatic disease sites. Although the histopathology of invasive biopsies remains the gold standard, they are grossly misinterpreted, necessitating a reliable, noninvasive immunoimaging platform that can accurately detect PDL1 and simultaneously identify other relevant biomarkers to enable combination therapies. PDL1 has also been shown to be overexpressed in breast cancer cells, which underscores the significance of PDL1 as a target receptor for immunoimaging.19−21 In addition to PDL1, EGFRs have been implicated in the pathogenesis and progression of up to 80% of TNBCs.22,23 EGFR is a transmembrane glycoprotein that is a member of the erbB family of tyrosine kinases. EGFR gene amplification and gene mutation both contribute to the overexpression of this receptor, and recently monoclonal antibodies and small-molecule inhibitors targeting EGFR have shown improved outcomes in TNBC.14 Therefore, the strengths of this work are the simultaneous detection of PDL1 and EGFR with high-resolution multiplexed SERS mapping achieved with MGNs, providing a qualitative distribution of both markers in vitro accompanied with light-controlled induction of cell death.
SERS imaging has rapidly progressed from a bench-scale spectroscopic tool to a preclinical diagnosis technique.24−27 Relative to other clinical imaging techniques, SERS offers high spatial resolution (cellular-level) and the capability to perform multiplexing, facilitated by the narrow spectral characteristics of the vibrational footprint of Raman molecules.28−30 This enables the detection of multiple receptors with high specificity, but unlike fluorescence imaging, SERS imaging is not limited by tissue autofluorescence and photobleaching.31 The combination of this preclinical imaging technique with therapeutic MGNs is highly promising, as it enables the detection of multiple biomarkers of cancer and, simultaneously, externally controlled light-mediated therapy.
MGNs consist of an “antenna-like” spherical core that absorbs incident light, and the light is then routed through the protrusions and concentrated at the tips.32 This unique geometry of MGNs gives rise to the “nanoantenna effect,” generating intense electromagnetic near fields in their vicinity, which is useful for efficiently converting resonant light to heat for PTT.32−34 Furthermore, the one-step synthesis of MGNs in the presence of a biological buffer, 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid (HEPES, pH 7.4), promotes their biocompatibility. HEPES binds weakly to gold surfaces, which facilitates straightforward surface modification with Raman-active molecules and antibodies for targeted detection. In addition, their sub-100 nm size is ideal for cellular uptake, enabling rapid surface binding and endocytosis in cells. Here, by employing a mixture of MGNs functionalized with Raman tag para-mercaptobenzoic acid (pMBA) and anti-EGFR antibodies and Raman tag 5,5-dithio-bis-(2-nitrobenzoic acid) (DTNB) and anti-PDL1 antibodies, we successfully demonstrated multiplexed SERS mapping as well as highly localized cell death via PTT. We anticipate the findings of this work will ultimately be translated to theranostics in vivo, contributing insights into the emerging field of immunoimaging and immunotherapy.
Results and Discussion
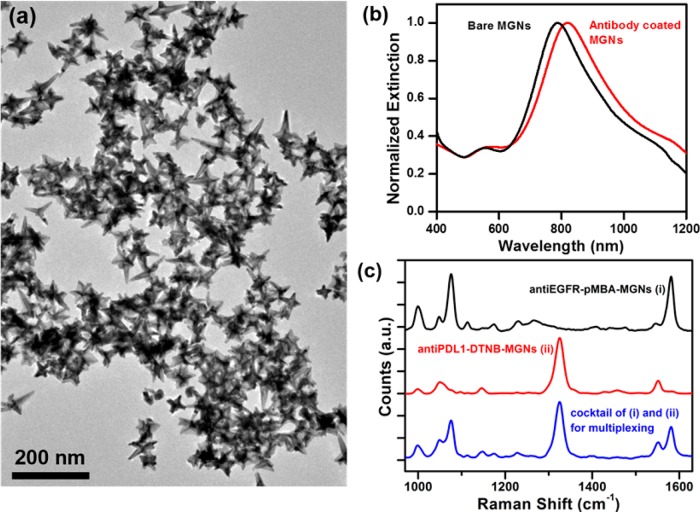
MGNs were synthesized via a one-step HEPES-mediated synthesis as described in our previously reported work.32 The average size of the MGNs, including the core and protrusions, was ∼65 ± 7 nm (branch-to-branch length) as visualized under transmission electron microscopy (TEM, Figure 1a). The heterogeneity in dimension is correlated to the variable protrusion length and protrusion density occurring during the synthesis process.32 The plasmon resonance of MGNs was tuned to ∼800 nm prior to functionalization (Figure 1b) to enable absorption of tissue-penetrating NIR light (690–900 nm). The NIR light absorption is advantageous for PTT, as an ∼800 nm laser can be utilized to activate the photothermal process. MGNs were further functionalized with antibodies to enable site-specific, targeted delivery of both diagnostic and therapeutic functions in a model TNBC cell line, MDA-MB-231. MGNs were therefore functionalized with anti-EGFR antibodies via a heterobifunctional poly(ethylene glycol) (PEG) linker, orthopyridyl disulfide (OPSS)–PEG–N-hydroxysuccinimide (NHS) ester. In this bioconjugation chemistry, the thiols on the OPSS group form a covalent link with the gold surface, whereas the NHS ester reacts with the amines of the antibodies to form an amide bond. Furthermore, the MGNs were tagged with a Raman molecule, pMBA, to enable SERS imaging and coated with an additional layer of PEG to neutralize any surface charge and minimize off-target interactions. The zeta potential of MGNs was −4.3 ± 1 mV after PEG coating, indicating their desirable near-neutral surface. The successful functionalization of antibodies, pMBA, and PEG was monitored with a spectrophotometer, and a ∼32 nm redshift in the plasmon resonance was observed due to an increase in the local refractive index of the MGNs (Figure 1b). The Raman footprint of the final functionalized MGNs, anti-EGFR–pMBA–MGNs, was confirmed by examining the SERs spectra (Figure 1c-i), which showed dominant peaks at 1074 and 1580 cm–1 corresponding to C–C stretching modes and ring stretching modes of pMBA.35 The weaker peaks between 1000 and 1200 cm–1 correspond to the antibody and PEG branching, as determined through control experiments.
Figure 1.
Characterization of antibody-labeled MGNs. (a) TEM micrograph of MGNs. (b) Extinction spectra of MGNs showing a red shift after functionalization with antibodies, Raman molecule, and PEG. (c) SERS footprint of MGNs functionalized with (i) anti-EGFR/pMBA/PEG, (ii) anti-PDL1/DTNB/PEG, and (iii) MGN mixture (1:1) combining (i) and (ii) that was utilized for multiplexed Raman imaging, showing distinguishable peaks at 1074 and 1580 cm–1 corresponding to pMBA and at 1325 and 1551 cm–1 corresponding to DTNB.
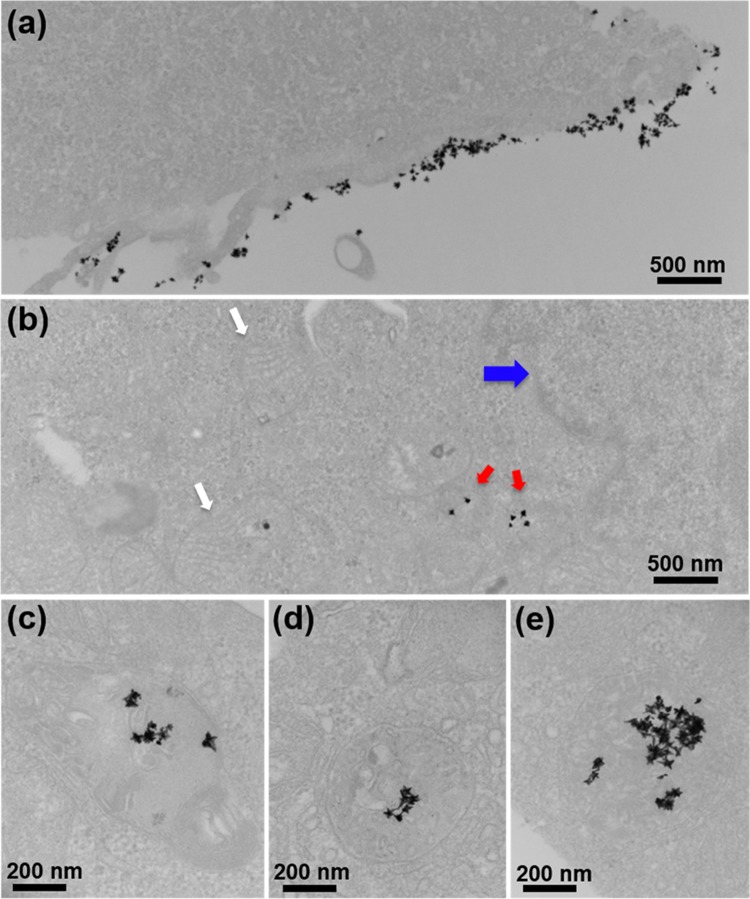
To further investigate the ability of MGNs for multiplexed SERS imaging, the PDL1 biomarker was targeted, as it has been found in numerous tumor types including breast cancer and is overexpressed in MDA-MB-231 cells.26,27 MGNs were functionalized with anti-PDL1 antibodies via the same heterobifunctional linker and Raman molecule DTNB via a gold–thiol bond. The MGNs were further coated with an additional layer of PEG for in vitro stability. The Raman footprint of the functionalized MGNs, anti-PDL1–DTNB–MGNs, was assessed with SERS (Figure 1c-ii), and dominant peaks at 1325 and 1551 cm–1, corresponding to the symmetric stretching mode of the nitro group of DTNB and the ring stretching mode, respectively, were observable.36 The SERS spectra of the mixture of MGNs utilized for multiplexing (anti-EGFR–pMBA–MGNs + anti-PDL1–DTNB–MGNs) at a 1:1 ratio demonstrated distinguishable peaks from both pMBA and DTNB with minimal overlap (Figure 1c-iii), which enabled us to successfully detect both EGFR and PDL1 receptors in vitro with high sensitivity and specificity. Prior to in vitro studies, we wanted to validate the utility of EGFR as a viable target and qualitatively demonstrate overexpression of EGFR in the MDA-MB-231 cells with a fluorescent secondary antibody assay. MDA-MB-231 cells were incubated with the anti-EGFR primary antibody followed by washing, incubation with blocking solution to minimize nonspecific binding, and addition of a secondary antibody complexed with phycoerythrin (PE). PE is a fluorescent protein with excitation at 488 nm and emission at 565–605 nm. Confocal fluorescence images (Figure S1) clearly indicate the high overexpression of EGFR in TNBC cells with fluorescent signal localized on the cellular edges, indicating EGFR expression on the cell surface. Following this assay, we performed SERS imaging targeting the EGFR biomarker on MDA-MB-231 cells. Cells were cultured on six-well plate dishes to ∼90% confluency, followed by incubation with anti-EGFR–pMBA–MGNs (170 μg/mL) for 16 h at 37 °C and repeated washing. After fixation, the cells were visualized under TEM to confirm surface binding and internalization of MGNs. The functionalized MGNs were both localized on the surface (Figure 2a) and internalized within (Figure 2b) the cells. MGNs were also found in cytoplasmic vesicles, demonstrating endocytosis (Figure 2c–e). We also note that MGNs maintained their structural integrity in cells and that they were not found within the nucleus or mitochondria, which is in agreement with previous literature.37,38
Figure 2.
Cellular binding and uptake of antibody-coated MGNs by MDA-MB-231 cells. (a) Transmission electron micrographs of cells incubated with anti-EGFR–pMBA–MGNs for 16 h show both (a) surface binding and (b) internalization by cells. The arrows denote organelles (blue, nucleus; white, mitochondria) with MGNs present in the cytoplasm (red). (c–e) High-magnification images show that internalization occurs via intracellular vesicles ranging ∼400–500 nm in size. Both on the surface and within these vesicles, the MGNs maintain their morphology.
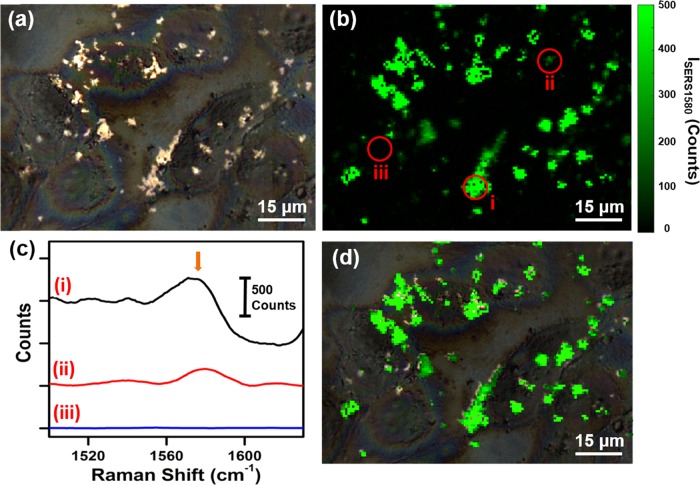
The targeted SERS mapping of MDA-MB-231 cells was performed by growing the cells on calcium fluoride (CaF2) disks to minimize autofluorescence, thus enabling higher-resolution SERS mapping. Cells were incubated with anti-EGFR–pMBA–MGNs (170 μg/mL) for 16 h at 37 °C followed by repeated washing, fixation, and imaging with a Raman microscope. The bright spots in the brightfield confocal image (Figure 3a) show scattering from MGN localization on the cell surface. The rectangular area of the brightfield image provides the x and y coordinates for SERS mapping. The Raman spectra of the cells were collected at 0.75 μm steps, providing a biochemical “traffic map” of the cells in the area. Static spectra were recorded with a 1200 cm–1 center with 10 s acquisition times at 2 mW power. The Raman intensity of the 1580 cm–1 peak, which corresponds to the C–C stretching modes and ring stretching modes of pMBA, was plotted as a function of position after analysis and background subtraction, creating a ∼100 μm × 100 μm SERS map (Figure 3b). The 1580 cm–1 peak of pMBA does not overlap with the Raman peaks of the intracellular lipid and provides an accurate means to demonstrate localization of anti-EGFR–pMBA–MGNs by examining the intensity. SERS point spectra are provided from the SERS map at selected locations, indicating high (Figure 3c-i), low (Figure 3c-ii), and no pMBA signals (Figure 3c-iii), which directly correspond to the amount of anti-EGFR–pMBA–MGNs localized in and on the surface of the cells. The intense SERS signal in areas with a high concentration of MGNs arises from the formation of SERS hot spots where strong electromagnetic and chemical enhancements occur.39−41 Furthermore, the brightfield image overlapped with the SERS map (Figure 3d) shows excellent correlation with the MGN distribution, demonstrating the efficacy of SERS in targeted imaging of the EGFR biomarker in TNBC cells. We also performed control studies where MGNs conjugated to pMBA without antibodies (pMBA–MGNs) under the same conditions and at the same concentrations were incubated in MDA-MB-231 cells on CaF2 disks, and SERS maps were obtained (Figure S2). The relatively weak Raman signal observable in these control experiments suggests that highly specific detection of EGFR is achieved only by MGNs labeled with the targeting antibodies. To further demonstrate the efficacy of MGNs in specifically targeting EGFR-overexpressing breast cancer cells, MCF7 was chosen as a control cell line, due to its low EGFR expression.42,43 MCF7 cells were seeded on CaF2 disks, anti-EGFR–pMBA–MGNs were incubated with cells, and SERS mapping was performed (Figure S3b). We observed minimal 1580 cm–1 intensity throughout the disk, indicating minimal MGNs bound to the MCF7 surface (Figure S3c).
Figure 3.
SERS mapping of MDA-MB-231 cells with anti-EGFR–pMBA–MGNs. (a) Brightfield image of cells after 16 h incubation with anti-EGFR–pMBA–MGNs provides coordinates for the Raman map. (b) Spatial Raman intensity map of the 1580 cm–1 peak corresponding to pMBA, with spectra recorded at 0.75 μm steps for the rectangular area in (a). (c) Selected SERS spectra corresponding to the indicated points on the SERS map in (b) representing a (i) high or (ii) low pMBA signal as well as (iii) no signal or cell response. The arrow denotes the 1580 cm–1 peak of interest for pMBA. (d) Overlap of (a) and (b) showing a direct correlation between the SERS map and the localization of the MGNs.
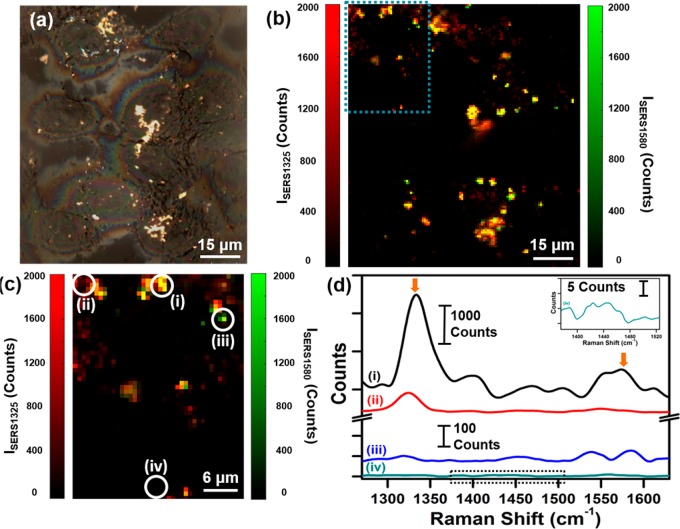
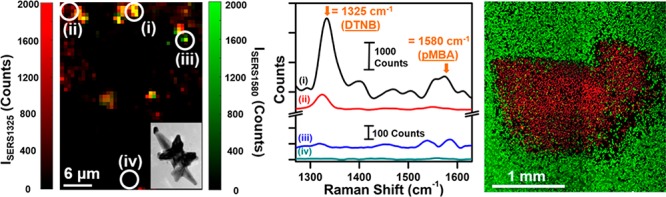
The immune checkpoint receptor PD-1 and its ligand PDL1 have been shown to be overexpressed in many subtypes of breast cancer, including in MDA-MB-231 TNBC cells.19−21 Immunotherapies with checkpoint blockade have already demonstrated tremendous success in multiple cancers and are now in clinical trials.44−46 Therefore, multiplexed targeted detection of PDL1 combined with EGFR offers a transformational approach to ultimately impact personalized immunotherapies. We performed multiplexed SERS imaging by simultaneously incubating cells with a mixture of anti-PDL1–DTNB–MGNs and anti-EGFR–pMBA–MGNs in a 1:1 ratio (see Materials and Methods Section for details). Because the vibrational signatures of DTNB and pMBA do not overlap, this approach allows us to detect both PDL1 and EGFR by mapping the signals of DTNB (1325 cm–1) and pMBA (1580 cm–1), respectively. Brightfield imaging (Figure 4a) shows an image of cells with light scattering from MGNS localized on the cell surface. A Raman spectral map of ∼100 μm × 100 μm was measured with 0.75 μm steps, and the intensities of the 1325–1 peak (ISERS1325) and 1580 cm–1 peak (ISERS1580) were plotted together as a function of position (Figure 4b). A strong signal is observable from both DTNB and pMBA, and the high spatiotemporal resolution of Raman allows us to qualitatively evaluate the localization of individual receptor bound MGNs. The intensities of the 1325 cm–1 peak and of the 1580 cm–1 peak are plotted individually (Figure S4a,c) and overlapped (Figure S4b) as a function of position in the SI. A magnified view of the local traffic map of the multiplexed SERS footprint is shown in Figure 4c, correlating the distributions of PDL1 and EGFR in the cells. Note that a strong signal is observed where multiple MGNs cluster, giving rise to SERS enhancements via hot spots. Specific locations on the magnified SERS map (Figure 4c) and corresponding SERS spectra (Figure 4d) indicate spatial localization from both probes (Figure 4d-i), DTNB alone (Figure 4d-ii), pMBA alone (Figure 4d-iii), or signal only from cellular lipids at 1440 cm–1 (Figure 4d-iv).47 We note that the intrinsic Raman peaks of biological lipids are typically very weak, necessitating the use of labeled MGNs for characterizing receptor expression.
Figure 4.
SERS multiplex mapping of MDA-MB-231 cells with (1:1) mixture of anti-EGFR–pMBA–MGNs and anti-PDL1–DTNB–MGNs. (a) Brightfield image of cells after 16 h incubation provides coordinates for a multiplex Raman map. (b) Overlap of spatial Raman intensity maps of both the 1325 cm–1 peak for DTNB and 1580 cm–1 peak for pMBA, recorded at 0.75 μm steps. (c) Magnified view of the blue rectangular area from (b) shows the Raman intensity plot of both ISERS1325 and ISERS1580, indicating a cellular traffic map with various points selected, (d) demonstrating (i) the signal from both probes, (ii) DTNB alone, (iii) pMBA alone, and (iv) no signal from probes or intracellular lipids (dashed box). The arrows denote peaks of interest, 1325 cm–1 peak for DTNB and 1580 cm–1 peak for pMBA.
Furthermore, to demonstrate that the observed Raman signal results from specific binding of antibody-conjugated MGNs rather than due to clustering via nonspecific binding, we performed a control study where 4 μg/mL monoclonal anti-PDL1 (1 h, 37 °C) was introduced in cells to block the PDL1 receptors prior to incubating with a 1:1 mixture of anti-PDL1–DTNB–MGNs and anti-EGFR–pMBA–MGNs (170 μg/mL). The brightfield image of the cells (Figure S5a) as well as SERS mapping (Figure S5b) at 0.75 μm steps indicated that the blocking of receptors results in significantly fewer MGNs binding to cells (Figure S5c). The multiplexed SERS traffic map presented for the experimental group (Figure 4b) and control group (Figure S5b) demonstrate that MGNs are both sensitive and specific in detecting biomarkers of cancer when conjugated with targeting moieties. In addition, SERS imaging provides a qualitative assessment of the distribution of both biomarkers in the cellular environment. We note that due to the possible colocalization of EGFR and PDL1 on the cell surfaces, a quantitative evaluation of the distribution of these receptors with SERS maps would be more challenging and would require sub-10 nm resolution.
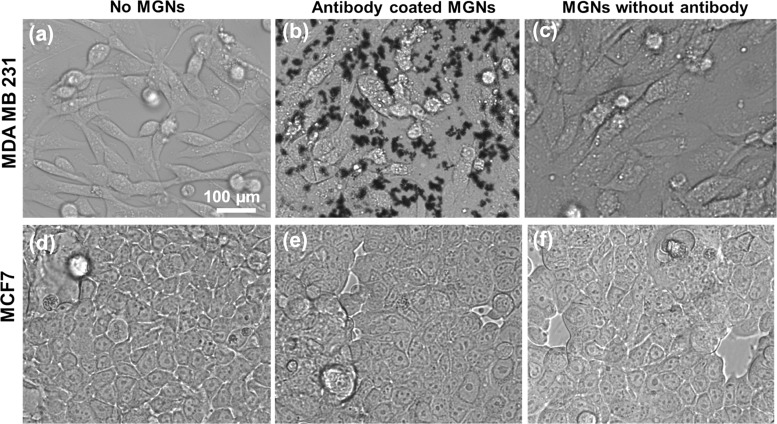
Antibody-conjugated theranostic MGNs not only enable high-resolution cellular mapping of cancer cells but are also ideal for inducing targeted cell death via PTT. We incubated MDA-MB-231 cells with anti-EGFR–pMBA–MGNs and performed control experiments where MGNs without antibodies (pMBA–MGNs) were utilized. Phase contrast images before (Figure 5a) and after incubating with MGNs demonstrated that cells incubated with anti-EGFR–pMBA–MGNs (Figure 5b) at 170 μg/mL for 16 h at 37 °C had a higher receptor-specific uptake relative to cells that were incubated with pMBA–MGNs without any antibodies (Figure 5c). We note that both antibody-conjugated MGNs and MGNs without antibodies were functionalized with a final layer of PEG to minimize cellular toxicity and nonspecific binding. This concentration of 170 μg/mL MGNs was measured by thermogravimetric analysis32,48 and was determined to be appropriate for enabling MGNs to be localized on the cell surface and achieve the necessary temperature increase to allow PTT.34 As a result, this MGN concentration was utilized throughout this study.
Figure 5.
Live phase contrast images of MDA-MB-231 breast cancer cells (a) without any MGNs, (b) after 16 h of incubation with anti-EGFR–pMBA–MGNs, and (c) after 16 h of incubation with pMBA–MGNs without antibodies. Phase contrast images of control breast cancer cell line MCF7 (d) without any MGNs, (e) after incubation with anti-EGFR–pMBA–MGNs, and (f) after incubation with pMBA–MGNs without antibodies, with incubation times also of 16 h duration.
The increased uptake of anti-EGFR–pMBA–MGNs in cells clearly shows that (i) EGFR receptors are overexpressed in MDA-MB-231 cells, as supported by our fluorescence assay in Figure S1, (ii) active targeting of MGNs is enabled likely by both surface-bound receptors and receptor-mediated endocytosis,49−52 and (iii) MGNs without antibodies have a low passive uptake in cells within 16 h of incubation.49,53 Unlike the spindle morphology of the MDA-MB-231 cells, these MCF7 cells have more of a brick-shaped morphology and tend to grow in closer proximity to one another (Figure 5d). Phase contrast images clearly demonstrated that MGNs are not bound to cells either via active targeting (Figure 5e) or via passive uptake (Figure 5f). To further confirm the specificity of anti-EGFR–pMBA–MGNs, we incubated them with cocultured MCF7 and MDA-MB-231 cells (Figure S6). Phase contrast images clearly demonstrated that MGNs preferentially localized along the spindle-shaped MDA-MB-231 cells and minimal binding was observed for the brick-shaped MCF7 cells, indicating that anti-EGFR–pMBA–MGNs are highly specific in targeting EGFR high-expressing TNBC cells. We note that the morphology of both cell lines is unchanged in the presence of the MGNs and that no cytotoxic effects were observable.
PTT was performed in both MDA-MB-231 cells and control, MCF7, cells, with anti-EGFR–pMBA–MGNs and pMBA–MGNs (Figure 6). The plasmon resonance of the anti-EGFR–pMBA–MGNs was tuned to overlap with an 808 nm diode laser utilized for PTT, and the concentration of nanoparticles utilized was analogous to that used in SERS measurements (170 μg/mL). All culture dishes were illuminated for 15 min with the 808 nm laser at 4.7 W/cm2 with a spot size of ∼2 mm × 2 mm. A live/dead cell assay with calcein/propidium iodide (PI) stains clearly demonstrated that site-specific necrotic cell death was only observable when EGFR-overexpressing MDA-MB-231 cells were incubated with anti-EGFR-conjugated MGNs and illuminated with laser (Figure 6a). Cells incubated with pMBA–MGNs (no antibodies) showed no observable cell death due to the low passive uptake of MGNs in cells (Figure 6b). Control experiments with low-EGFR-expressing MCF7 cells also demonstrated no cell death with both anti-EGFR–pMBA–MGNs (Figure 6c) and pMBA–MGNs (Figure 6d). PTT with antibody-conjugated MGNs demonstrates that active targeting enables higher cellular accumulation, minimal nonspecific binding and off-site toxicities, and a highly specific and sensitive spatiotemporal control of cell death externally triggered with low NIR laser powers. Our control experiments also show that the 808 nm laser alone did not induce any cell death (Figure S7a) and in the absence of laser the anti-EGFR–pMBA–MGNs were not cytotoxic to the cells (Figure S7b).
Figure 6.
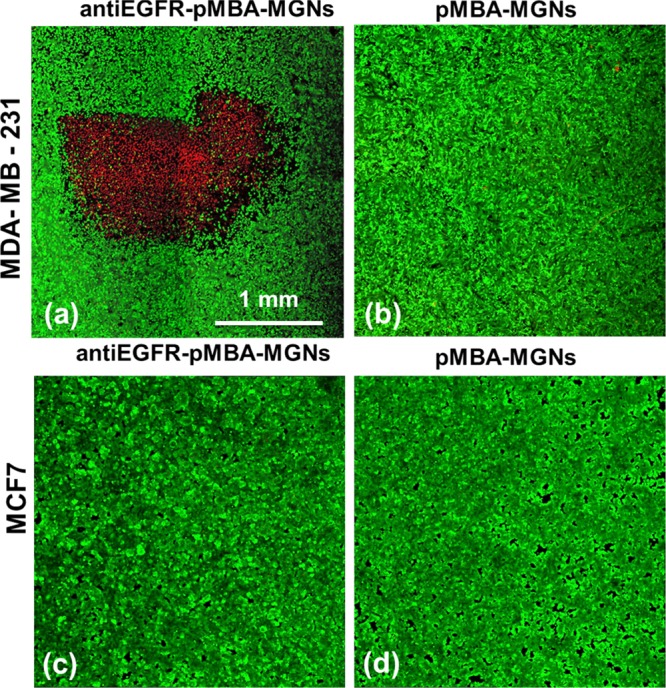
Confocal fluorescence images of live cells stained with calcein acetoxymethyl (AM) (green) and dead cells stained by PI (red) showing cell death via PTT 4 h post illumination with an 808 nm laser, 2 × 2 mm2 spot size, and 4.7 W/cm2 pump fluence. Localized cell death was observed only with (a) anti-EGFR–pMBA–MGNs, and no cell death was observed with (b) pMBA–MGNs. No cell death was observed for the control MCF7 cells with (c) anti-EGFR–pMBA–MGNs and (d) pMBA–MGNs. Note: the necrotic cells are found only within the laser spot.
Conclusions
In summary, we demonstrate the theranostic capabilities of MGNs conjugated with antibodies and Raman tags in vitro by combining multiplexed diagnosis of immunomarker PDL1 with EGFR in TNBC cells via SERS followed by PTT to treat the cells. The strong SERS signal enabled by both electromagnetic and chemical enhancement by MGNs, as well as the narrow vibrational signatures of Raman, allowed us to create cellular traffic maps, facilitating a highly sensitive and specific detection of both biomarkers. Furthermore, the nanoantenna effect with MGNs gives rise to enhanced light-to-heat conversion when illuminated with NIR light and subsequent photothermal cell death only within the laser spot, avoiding off-target toxicities. We envision that the successful detection of immune checkpoint ligand PDL1 will ultimately enable theranostic MGNs to provide a quantitative measure of immunomarkers and allow predictive, personalized image-guided immunotherapies with minimal toxicity.44,46,54 Furthermore, this work will impact the design of new theranostic nanoprobes capable of imaging multiple biomarkers of cancer and light-controlled therapeutic response, providing an avenue to diagnose TNBC tumors that typically do not respond to endocrine and targeted treatments.
Materials and Methods
MGN Synthesis
Reagents purchased from Sigma-Aldrich include both HEPES and gold(III) chloride trihydrate (HAuCl4) for MGN synthesis. For functionalization, Raman tags, pMBA and DTNB, were purchased from TCI America Inc.
MGNs were synthesized through the seedless, one-step HEPES growth protocol, as previously described.32,34,55 Briefly, 18 mL of Milli-Q (MQ) water (18 MΩ) was added to 12 mL 200 mM HEPES (pH 7.40 ± 0.1), mixing by gentle inversion. Quickly, 300 μL of 20 mM tetrachloroauric(III) acid (HAuCl4) was added, again mixing by inversion and leaving to react at room temperature for 75 min. Plasmon resonance of the MGNs, through the functionalization process, was monitored using a Varian Cary 5000 UV–vis NIR spectrophotometer (Agilent Technologies).
OPSS–PEG–NHS Ester Chemistry with Antibody
OPSS–PEG–Anti-EGFR
OPSS–PEG–NHS ester (Mw 2000 g/mol; JenKem Technology) and human EGF R/ErbB1 antibody (anti-EGFR; R&D Systems) were purchased in lyophilized forms. Chemistry to form the amide linkage from the ester group of OPSS–PEG–NHS with amine groups on the antibody structure was modified and troubleshot from previously reported methods.56−58 OPSS–PEG–NHS was resuspended in 100 mM pH 8.6 ± 0.1 sodium bicarbonate (NaHCO3) buffer to a concentration of 160 mg/mL, only immediately prior to the reaction with anti-EGFR, as the ester is sensitive to water hydrolysis. Also reconstituted in 100 mM NaHCO3 (pH 8.6 ± 0.1) buffer, the anti-EGFR protein was prepared at a concentration of 1 mg/mL. The higher pH bicarbonate buffer is used for the OPSS–antibody reaction, as the amide reaction is favored at a higher pH.59−61 Prepared on ice, the 160 mg/mL OPSS-PEGS–NHS solution was added at a (1:9) volumetric ratio57 to the 1 mg/mL anti-EGFR: 8 μL of 160 mg/mL OPSS–PEG–NHS was added to 72 μL of 1 mg/mL anti-EGFR. OPSS–PEG–antibody was reacted for 24 h on an inverter at 4 °C.
OPSS–PEG–Anti-PDL1
Human CD274 (B7-H1, PDL1) antibody (anti-PDL1; BioLegend) was already resuspended at 1 mg/mL stock in phosphate-buffered solution, pH 7.2. The OPSS–PEG–anti-PDL1 reaction was prepared in the same manner as the OPSS–PEG–anti-EGFR reaction but then further diluted fivefold with 100 mM NaHCO3 (pH 8.6 ± 0.1) buffer to ensure that the linker–antibody reaction occurred at the favorable pH 8.6.
Synthesis of Actively Targeted (Antibody) MGNs (Anti-EGFR–pMBA–MGNs and Anti-PDL1– DTNB–MGNs)
Quantification of MGNs
At points during the functionalization process, a known quantity of MGNs was required to allow for proper labeling efficiencies. To determine MGN concentration, an equation was developed from previous thermogravimetic analyzer32,48 and UV–vis spectroscopy correlative studies (refer to eq S1). The equation was utilized in calculations for all functionalized MGN chemistries.
Anti-EGFR–pMBA–MGNs
To begin, 60 mL MGNs at 170 μg/mL (ext. = 1.79) at max plasmon resonance ∼780 nm in the NIR region were synthesized. Ideal for nanomedicine work because of the weak absorption by biological tissues and proteins, MGN resonance in this regime allows for better penetration depth during therapy for future in vivo studies.62 MGNs were kept on ice through the functionalization process. pMBA, dissolved in 100% ethanol, was added to the MGNs in a small volume with high concentration, adapting from previous methods for SERS-tag labeling to gold nanoparticles.58,63−65 With slow stirring, 6 μL of 10 mM pMBA was injected to 60 mL of MGNs, mixing for 10 min at 4 °C and monitoring the shift in resonance due to the binding event via thiol linkage. Next, the 60 mL was centrifuged at 6000 rpm for 20 min.
The chemistry ratio for OPSS–PEG–antibody to SERS-tag-MGNs was modified and troubleshot from other literature methods.56,57,61,66 Briefly, 80 μL of OPSS–PEG–anti-EGFR ([anti-EGFR]Final = 0.5 mg/mL) was added to 5.95 mL of pMBA-labeled-MGNs at 1.14 mg/mL, pipetting to mix. The antibody with the MGN reaction was then left to react for 24 h on an inverter at 4 C. Post 24 h incubation, PEGylation chemistry34,67,68 was performed to passivate any free surface area to negate charge effects by the anti-EGFR–pMBA–MGNs. To the aqueous MGN mixture, 700 μL of 5 μM PEG–thiol (mPEG-SH, Mw 5000 g/mol; JenKem Technology) was added and reacted for 10 min at room temperature. After the reaction, MGNs were then centrifuged at 4000 rpm for 10 min. Pellets were resuspended in 200 μL of sterile 8.6 mM pH 7.40 ± 0.1 monophosphate buffer at 5 mg/mL, rechecking concentration. For cellular assays, functionalized MGNs were further diluted to 170 μg/mL with normal media [no fetal bovine serum (FBS)] as this concentration was previously shown to have strong SERS and photothermal activity.32,34
Anti-PDL1–DTNB–MGNs
To synthesize the complementary MGNs particles, the functionalization method was repeated with similar steps but substituting anti-PDL1 as the targeting moiety and DTNB as the SERS tag.
Synthesis of MGNs Lacking the Targeting Moiety (pMBA–MGNs)
MGNs without a targeting moiety, but SERS-tagged and PEG-bound, pMBA–MGNs, were synthesized via a modified version of previous procedures.34,67,68 pMBA labeling was performed as with anti-EGFR–pMBA–MGN chemistry. For the pegylation step, 50 μM mPEG-SH was added to 60 mL of pMBA-labeled MGNs, incubating for 5 min, and centrifuging at 6000 rpm for 20 min. To be certain of the surface neutrality of the pMBA–MGNs, a second addition of mPEG-SH was performed. To the aqueous 5.95 mL pMBA-labeled-MGNs at 1.14 mg/mL (ext. = 12.0), 200 μL of 5 μM mPEG-SH was added in a cold vessel for 10 min with constant stirring at room temperature. The particles were then centrifuged at 4000 rpm for 10 min and resuspended in 200 μL of sterile 8.6 mM pH 7.40 ± 0.1 monophosphate buffer.
Characterization of Antibody-Labeled MGNs
Antibody-tag-MGNs were imaged with an Osiris TEM at 200 keV to ensure the proper morphology and functionalization of the particles. Raman spectra of anti-EGFR–pMBA–MGNs, anti-PDL1–DTNB–MGNs, and a (1:1) mixture were obtained using an inVia Raman microscope (Renishaw) with a 785 nm wavelength laser source with 1200 lines/mm grating in high confocal mode.
Fluorescent Secondary Antibody Assay Testing EGFR Overexpression in MDA-MB-231 Cells
MDA-MB-231 cells, purchased from ATCC (HTB-26), were cultured in Dulbecco’s Modified Eagle’s medium (DMEM; Gibco) supplemented with 10% FBS (ATCC) and 1% penicillin/streptomycin (Gibco) and maintained at 37 °C and 5% CO2. MDA-MB-231 cells were seeded on a 96-well plate and allowed to grow to ∼90% confluency. Cells were then fixed with cytofix/cytoperm (BD Biosciences), incubating for 20 min at room temperature. The fixative was removed, and the cells were washed with MQ water (×2). Next, the cells were blocked to prevent any nonspecific binding by incubating with a 1× animal-free blocking solution (Cell Signaling Technology) for 1 h at 4 °C. The blocking solution was removed, followed by washing with MQ water (×2). The cells were then incubated with human EGF R/ErbB1 antibody (monoclonal mouse anti-EGFR, 100 μg/mL; R&D Systems) diluted in 8.6 mM pH 7.4 ± 0.1 monophosphate buffer for 24 h at 4 °C. After 24 h, the cells were washed with monophosphate buffer (×2) and next incubated with secondary antibody (mouse IgG2B PE-conjugated, 2 μg/mL; R&D Systems) for 2 h at 4 °C. The cells were washed with monophosphate buffer (×2) and then visualized under fluorescent microscopy using a Zeiss LSM 710 confocal microscope.
TEM Imaging of Surface Binding and Uptake of Antibody-Labeled MGNs for MDA-MB-231 Cells
Anti-EGFR–pMBA–MGNs (170 μg/mL) were added to cells at ∼90% confluency. The cells were further incubated for another 16 h at 37 °C. Next, the cells were washed with cacodylate buffer before fixing in 2.5% gluteraldehyde in 0.1 M cacodylate buffer. Sample processing and imaging were handled by the Vanderbilt Cell Imaging Shared Resource. The samples were postfixed in 1% osmium tetraoxide at room temperature and then washed with 0.1 M cacodylate buffer (×3). Postfixation, the samples were dehydrated through a graded ethanol series followed by incubation in 100% ethanol and propylene oxide (PO) with two exchanges of pure PO. Finally, cell samples were embedded in epoxy resin and polymerized at 60 °C for 48 h. For thin section preparation, ultrathin slices (70–80 nm) were cut from the block and added to copper grids for imaging. The grids were further stained with 2% uranyl acetate and lead citrate. Imaging was performed using a Philips/FEI Tecnai T12 electron microscope operating at 200 keV.
SERS Singleplex Mapping of MDA-MB-231 Cells (Active vs Passive) and Control MCF7 Cells
For SERS mapping experiments, MDA-MB-231 or MCF7 cells were seeded on CaF2 disks (20 mm × 2 mm Raman grade; Crystran Ltd.). Cells at ∼90% confluency were incubated with media (no FBS, nDMEM) supplemented with either 170 μg/mL anti-EGFR–pMBA–MGNs or 170 μg/mL pMBA–MGNs for 16 h at 37 °C. Next, the cells were washed with nDMEM (×2) and then placed back in complete media (with FBS, cDMEM) to equilibrate for 2 h at 37 °C. The cells were then fixed with cytofix/cytoperm (BD Biosciences), incubating for 20 min at room temperature. The fixative was removed, and the cells were washed with MQ water (×2).69
Control SERS mapping experiments, MGNs coated with antibody (anti-EGFR–pMBA–MGNs, 170 μg/mL) and nontargeted MGNs (pMBA–MGNs, 170 μg/mL) experiments, as well as the MCF7 control assay, were treated the same for SERS mapping and data collection, as well as processing. After allowing the samples to dry overnight at 4 °C, the cells on the CaF2 disks were visualized using a Raman microscope. Using an 100× objective, a video montage of the cells was captured, and the resulting brightfield image provided x, y coordinates for the Raman instrument to record SERS chemical footprints of a rectangular area of ∼100 μm × 100 μm. Recording spectra with 0.75 μm steps, the Raman microscope took measurements of the cells with a 785 nm wavelength laser source with 1200 lines/mm grating in high confocal mode. Static spectra (1200 cm–1 center) were recorded using a 100× objective with a ∼1 μm spot size at a power of 2 mW with a 10 s acquisition time.
SERS Multiplex Imaging of MDA-MB-231 Cells (No Block and Blocking PDL1 Receptor)
Multiplex with (1:1) Cocktail with MDA-MB-231 Cells
Again, CaF2 disks were seeded with MDA-MB-231 cells. Cells were incubated with media (no FBS, nDMEM) supplemented with a 170 μg/mL (1:1) cocktail of anti-EGFR–pMBA–MGNs to anti-PDL1–DTNB–MGNs for 16 h at 37 °C. Next, the cells were washed with nDMEM (×2) and equilibrated for 2 h at 37 °C. After 2 h in cDMEM, the cells were fixed, washed, and dried overnight at 4 °C.
Measurement conditions for the multiplex cell studies were the same as those for singleplex except the use of a lower laser power here at 1 mW.
Multiplex with (1:1) Cocktail with 231 Cells after Blocking the PDL1 Receptor
To test the fidelity of the multiplex technology, a blocking PDL1 receptor control experiment was performed to determine the specificity and mapping capabilities of this (1:1) cocktail of functionalized antibody-specific MGNs. MDA-MB-231s were seeded on CaF2 disks and preblocked by incubating with nDMEM supplemented with 4 μg/mL human CD274 (B7-H1, PDL1) antibody (monoclonal anti-PDL1) for 1 h at 37 °C.70−72 After washing and MGN incubation step for 16 h at 37 °C, the cells were again washed, equilibrated in cDMEM, fixed, and dried.
Analysis of SERS Imaging Maps (Singleplex, Multiplex, PDL1 Block)
Using wire 3.4 software, cosmic ray removal was performed with the nearest-neighbor method implemented to ensure that the scaling of the spectra was not jeopardized. A custom code was built using MATLAB software to perform smoothing and background fluorescent subtraction. Truncation was first performed, and the data underwent smoothing and data differentiation using the Savitzsky and Golay method.73−75 The S and G filter used an order of 5 and a coefficient value of 61. Following the modified polyfit method,76 an automated approach was applied to perform background subtraction of fluorescence using polynomial fitting to uncover SERS peaks even at low-intensity regimes.76 A seventh-order polynomial was utilized for fluorescent background subtraction through an iterative process with a threshold of 0.0001. Finally, overlays of SERS intensity maps and brightfield were performed using Adobe Photoshop graphic software.
Photothermal Ablation of Breast Cancer Cells Utilizing Actively Targeted MGNs
The MCF7 (HTB-22) breast cancer cell line was purchased from ATCC. MCF7 cells were cultured in Eagle’s Minimum Essential medium (EMEM; ATCC) supplemented with 10% FBS (ATCC), 1% penicillin/streptomycin (Gibco), and 0.01 mg/mL human recombinant insulin (Invitrogen). Culturing on 35 mm culture dishes for the PTT experiment, both MDA-MB-231 cells and MCF7s were seeded to reach ∼90% confluent layers on the day of MGN addition.
The two cell lines were incubated with new media (no FBS, nDMEM, or nEMEM) supplemented with either 170 μg/mL anti-EGFR–pMBA–MGNs or 170 μg/mL pMBA–MGNs for 16 h at 37 °C. Next, the cells (231s and MCF7s) were washed with nDMEM or nEMEM (×2) and then placed back in complete media (with FBS, cDMEM, or cEMEM) prior to irradiation with laser. The localization of anti-EGFR–pMBA–MGNs and pMBA–MGNs was compared for the two cell types through visualization with a Zeiss Observer Z1 microscope. In addition, anti-EGFR–pMBA–MGNs uptake and localization in the coculture of MCF7 and MDA-MB-231 cells were observed with a Zeiss Observer Z1 microscope.
Custom PTT laser optics were purchased from Thorlabs, Inc. and built to provide controlled light to heat conversion. The optics included an 808 nm laser diode (L808P1WJ), a current controller (LDC240C), a thermoelectric controller (TEDC200C), a collimating lens (C230TME-B), and a silver polished mirror (PF-10-03-P01). The photothermal setup was also completed with an aluminum block wrapped with antireflecting black films and a 37 °C heating mantle to simulate biological conditions. Calipers and a power meter (PM130D; Thorlabs, Inc.) were used to measure the laser spot size and power.
After allowing to equilibrate at 37 °C for 2 h, the cells were illuminated for 15 min at 4.7 W/cm2 pump fluence with an 808 nm laser and a spot size of 2 mm × 2 mm. The cells were then placed back in the 37 °C incubator. After 4 h, fresh complete media was added with calcein AM (5 μM) and PI (1 μM) (live/dead stain) and incubated for 20 min at 37 °C. A Zeiss LSM 710 confocal microscope was utilized for fluorescent imaging of the cells. Additional controls were performed on the MDA-MB-231 cells to show that the 808 nm laser alone did not induce any cell death and that the anti-EGFR–pMBA–MGNs were not cytotoxic to the cells.
Acknowledgments
J.A.W. acknowledges support from the National Science Foundation Graduate Research Fellowship Program under Grant Number 1445197. Y.-C.O. acknowledges support from the American Cancer Society Institutional Research Grant (IRG-58-009-56). S.F. and L.M.B. acknowledge support from the National Institutes of Health under Grant Number 4R00EB013630 (NIBIB). TEM micrographs of MGNs were acquired with an instrument supported by NSF EPS 1004083. TEM images of surface binding and cellular uptake of anti-EGFR–pMBA–MGNs were acquired using the VUMC Cell Imaging Shared Resource (supported by NIH grants CA68485, DK20593, DK58404, DK59637, and EY08126).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.7b00527.
Fluorescence assay to validate overexpression of EGFR in MDA-MB-231 cells; SERS imaging controls including mapping of MDA-MB-231 cells with pMBA–MGNs; singleplex mapping of MCF7 cells with anti-EGFR–pMBA–MGNs; multiplex mapping of cells; SERS mapping of cells with PDL1 receptors blocked; phase contrast imaging of MCF7 and MDA-MB-231 coculture incubated with anti-EGFR–pMBA–MGNs; PTT control assays with laser alone and anti-EGFR–pMBA–MGNs alone (PDF)
Author Contributions
# J.A.W. and Y.-C.O. are equally contributing first authors.
The authors declare no competing financial interest.
Supplementary Material
References
- Chen W.; Zhang S.; Zu Y.; Zhang H.; He Q. Structural-Engineering Rationales of Gold Nanoparticles for Cancer Theranostics. Adv. Mater. 2016, 28, 8567–8585. 10.1002/adma.201602080. [DOI] [PubMed] [Google Scholar]
- Webb J. A.; Bardhan R. Emerging Advances in Nanomedicine with Engineered Gold Nanostructures. Nanoscale 2014, 6, 2502–2530. 10.1039/c3nr05112a. [DOI] [PubMed] [Google Scholar]
- Bardhan R.; Lal S.; Joshi A.; Halas N. J. Theranostic Nanoshells: From Probe Design to Imaging and Treatment for Cancer. Acc. Chem. Res. 2011, 44, 936–946. 10.1021/ar200023x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K. Y.; Liu G.; Lee S.; Chen X. Theranostic Nanoplatforms for Simultaneous Cancer Imaging and Therapy: Current Approaches and Future Perspectives. Nanoscale 2012, 4, 330–342. 10.1039/C1NR11277E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardhan R.; Chen W.; Bartels M.; Perez-Torres C.; Botero M. F.; McAninch R. W.; Contreras A.; Schiff R.; Pautler R. G.; Halas N. J.; Joshi A. Tracking of Multimodal Therapeutic Nanocomplexes Targeting Breast Cancer in Vivo. Nano Lett. 2010, 10, 4920–4928. 10.1021/nl102889y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardhan R.; Chen W.; Perez-Torres C.; Bartels M.; Huschka R. M.; Zhao L. L.; Morosan E.; Pautler R. G.; Joshi A.; Halas N. J. Nanoshells with Targeted Simultaneous Enhancement of Magnetic and Optical Imaging and Photothermal Therapeutic Response. Adv. Funct. Mater. 2009, 19, 3901–3909. 10.1002/adfm.200901235. [DOI] [Google Scholar]
- Chen W.; Ayala-Orozco C.; Biswal N. C.; Perez-Torres C.; Bartels M.; Bardhan R.; Stinnet G.; Liu X.-D.; Ji B.; Deorukhkar A.; Brown L. V.; Guha S.; Pautler R. G.; Krishnan S.; Halas N. J.; Joshi A. Targeting of Pancreatic Cancer with Magneto-Fluorescent Theranostic Gold Nanoshells. Nanomedicine 2014, 9, 1209–1222. 10.2217/nnm.13.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.; Bardhan R.; Bartels M.; Perez-Torres C.; Pautler R. G.; Halas N. J.; Joshi A. A Molecularly Targeted Theranostic Probe for Ovarian Cancer. Mol. Cancer Ther. 2010, 9, 1028–1038. 10.1158/1535-7163.MCT-09-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Maltzahn G.; Centrone A.; Park J.-H.; Ramanathan R.; Sailor M. J.; Hatton T. A.; Bhatia S. N. SERS-Coded Gold Nanorods as a Multifunctional Platform for Densely Multiplexed Near-Infrared Imaging and Photothermal Heating. Adv. Mater. 2009, 21, 3175–3180. 10.1002/adma.200803464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Yang M.; Zhang J.; Zhi X.; Li C.; Zhang C.; Pan F.; Wang K.; Yang Y.; Fuentea J. M. d. l.; Cui D. Human Induced Pluripotent Stem Cells for Tumor Targeted Delivery of Gold Nanorods and Enhanced Photothermal Therapy. ACS Nano 2016, 10, 2375–2385. 10.1021/acsnano.5b07172. [DOI] [PubMed] [Google Scholar]
- Gao Y.; Li Y.; Chen J.; Zhu S.; Liu X.; Zhou L.; Shi P.; Niu D.; Gu J.; Shi J. Multifunctional Gold Nanostar-Based Nanocomposite: Synthesis and Application for Noninvasive MR-SERS Imaging-Guided Photothermal Ablation. Biomaterials 2015, 60, 31–41. 10.1016/j.biomaterials.2015.05.004. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Ashton J. R.; Moding E. J.; Yuan H.; Register J. K.; Fales A. M.; Choi J.; Whitley M. J.; Zhao X.; Qi Y.; Ma Y.; Vaidyanathan G.; Zalutksy M. R.; Kirsch D. G.; Badea C. T.; Vo-Dinh T. A Plasmonic Gold Nanostar Theranostic Probe for In Vivo Tumor Imaging and Photothermal Therapy. Theranostics 2015, 5, 946–960. 10.7150/thno.11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakor A. S.; Gambhir S. S. Nanooncology: The Future of Cancer Diagnosis and Therapy. CA-Cancer J. Clin. 2013, 63, 395–418. 10.3322/caac.21199. [DOI] [PubMed] [Google Scholar]
- Abramson V. G.; Lehmann B. D.; Ballinger T. J.; Pietenpol J. A. Subtyping of Triple-Negative Breast Cancer: Implications for Therapy. Cancer 2015, 121, 8–16. 10.1002/cncr.28914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peddi P. F.; Ellis M. J.; Ma C. Molecular Basis of Triple Negative Breast Cancer and Implications for Therapy. Int. J. Breast Cancer 2011, 2012, 217185 10.1155/2012/217185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardoll D. M. The Blockade of Immune Checkpoints in Cancer Immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips G. K.; Atkins M. Therapeutic Uses of anti-PD-1 and anti-PD-L1 Antibodies. Int. Immunol. 2015, 27, 39–46. 10.1093/intimm/dxu095. [DOI] [PubMed] [Google Scholar]
- Tumeh P. C.; Harview C. L.; Yearley J. H.; Shintaku I. P.; Taylor E. J. M.; Robert L.; Chmielowski B.; Spasic M.; Henry G.; Ciobanu V.; West A. N.; Carmona M.; Kivork C.; Seja E.; Cherry G.; Gutierrrez A. J.; Grogan T. R.; Mateus C.; Tomasic G.; Glaspy J. A.; Emerson R. O.; Robins H.; Pierce R. H.; Elashoff D. A.; Robert C.; Ribas A. PD-1 Blockade Induces Responses by Inhibiting Adaptive Immune Resistance. Nature 2014, 515, 568–571. 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista M. Z.; Sarian L. O.; Derchain S. F. M.; Pinto G. A.; Vassallo J. Prognostic Significance of PD-L1 and PD-L2 in Breast Cancer. Hum. Pathol. 2016, 47, 78–84. 10.1016/j.humpath.2015.09.006. [DOI] [PubMed] [Google Scholar]
- Cimino-Mathews A.; Thompson E.; Taube J. M.; Ye X.; Lu Y.; Meeker A.; Xu H.; Sharma R.; Lecksell K.; Cornish T. C.; Cuka N.; Argani P.; Emens L. A. PD-L1 (B7-H1) Expression and the Immune Tumor Microenvironment in Primary and Metastatic Breast Carcinomas. Hum. Pathol. 2016, 47, 52–63. 10.1016/j.humpath.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno B. H.; Ribas A. Anti-Programmed Cell Death Protein-1/Ligand-1 Therapy in Different Cancers. Br. J. Cancer 2015, 112, 1421–1427. 10.1038/bjc.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balko J. M.; Giltnane J. M.; Wang K.; Schwarz L. J.; Young C. D.; Cook R. S.; Owens P.; Sanders M. E.; Kuba M. G.; Sánchez V.; Kurupi R.; Moore P. D.; Pinto J. A.; Doimi F. D.; Gómez H.; Horiuchi D.; Goga A.; Lehmann B. D.; Bauer J. A.; Pietenpol J. A.; Ross J. S.; Palmer G. A.; Yelensky R.; Cronin M.; Miller V. A.; Stephens P. J.; Arteaga C. L. Molecular Profiling of the Residual Disease of Triple-Negative Breast Cancers after Neoadjuvant Chemotherapy Identifies Actionable Therapeutic Targets. Cancer Discovery 2014, 4, 232–245. 10.1158/2159-8290.CD-13-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balko J. M.; Stricker T. P.; Arteaga C. L. The Genomic Map of Breast Cancer: Which Roads Lead to Better Targeted Therapies?. Breast Cancer Res. 2013, 15, 209. 10.1186/bcr3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreou C.; Kishore S. A.; Kircher M. F. Surface-Enhanced Raman Spectoscopy: A New Modality for Cancer Imaging. J. Nucl. Med. 2015, 56, 1295–1299. 10.2967/jnumed.115.158196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jermyn M.; Mok K.; Mercier J.; Desroches J.; Pichette J.; Saint-Arnaud K.; Bernstein L.; Guiot M.-C.; Petrecca K.; Leblond F. Intraoperative Brain Cancer Detection with Raman Spectroscopy in Humans. Sci. Transl. Med. 2015, 7, 274ra19. 10.1126/scitranslmed.aaa2384. [DOI] [PubMed] [Google Scholar]
- Karabeber H.; Huang R.; Iacono P.; Samii J. M.; Pitter K.; Holland E. C.; Kircher M. F. Guiding Brain Tumor Resection Using Surface-Enhanced Raman Scattering Nanoparticles and a Hand-Held Scanner. ACS Nano 2014, 8, 9755–9766. 10.1021/nn503948b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M. F.; Zerda A. d. l.; Jokerst J. V.; Zavaleta C. L.; Kempen P. J.; Mittra E.; Pitter K.; Huang R.; Campos C.; Habte F.; Sinclair R.; Brennan C. W.; Mellinghoff I. K.; Holland E. C.; Gambhir S. S. A Brain Tumor Molecular Imaging Strategy Using a New Triple-Modality MRI-Photoacoustic-Raman Nanoparticle. Nat. Med. 2012, 18, 829–835. 10.1038/nm.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matschulat A.; Drescher D.; Kneipp J. Surface-Enhanced Raman Scattering Hybrid Nanoprobe Multiplexing and Imaging in Biological Systems. ACS Nano 2010, 4, 3259–3269. 10.1021/nn100280z. [DOI] [PubMed] [Google Scholar]
- Zavaleta C. L.; Smith B. R.; Walton I.; Doering W.; Davis G.; Shojaei B.; Natan M. J.; Gambhir S. S. Multiplexed Imaging of Surface Enhanced Raman Scattering Nanotags in Living Mice Using Noninvasive Raman Spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 13511–13516. 10.1073/pnas.0813327106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez de Aberasturi D.; Serrano-Montes A. B.; Langer J.; Henriksen-Lacey M.; Parak W. J.; Liz-Marzán L. M. Surface Enhanced Raman Scattering Encoded Gold Nanostars for Multiplexed Cell Discrimination. Chem. Mater. 2016, 28, 6779–6790. 10.1021/acs.chemmater.6b03349. [DOI] [Google Scholar]
- Zhang Y.; Hong H.; Myklejord D. V.; Cai W. Molecular Imaging with SERS-Active Nanoparticles. Small 2011, 7, 3261–3269. 10.1002/smll.201100597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb J. A.; Erwin W. R.; Zarick H. F.; Aufrecht J.; Manning H. W.; Lang M. J.; Pint C. L.; Bardhan R. Geometry-Dependent Plasmonic Tunability and Photothermal Characteristics of Multibranched Gold Nanoantennas. J. Phys. Chem. C 2014, 118, 3696–3707. 10.1021/jp408793a. [DOI] [Google Scholar]
- Webb J. A.; Aufrecht J.; Hungerford C.; Bardhan R. Ultrasensitive Analyte Detection with Plasmonic Paper Dipsticks and Swabs Integrated with Branched Nanoantennas. J. Mater. Chem. C 2014, 2, 10446–10454. 10.1039/C4TC01634C. [DOI] [Google Scholar]
- Ou Y.-C.; Webb J. A.; Faley S.; Shae D.; Talbert E. M.; Lin S.; Cutright C. C.; Wilson J. T.; Bellan L. M.; Bardhan R. Gold Nanoantenna-Mediated Photothermal Drug Delivery from Thermosensitive Liposomes in Breast Cancer. ACS Omega 2016, 1, 234–243. 10.1021/acsomega.6b00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P.; Jeon S.-H.; Mack N. H.; Doorn S. K.; Williams D. J.; Han X.; Wang H.-L. Field-Assisted Synthesis of SERS-Active Silver Nanoparticles Using Conducting Polymers. Nanoscale 2010, 2, 1436–1440. 10.1039/c0nr00106f. [DOI] [PubMed] [Google Scholar]
- Wang C.; Xu J.; Wang J.; Rong Z.; Li P.; Xiao R.; Wang S. Polyethylenimine-Interlayered Silver-Shell Magnetic-Core Microspheres as Multifunctional SERS Substrates. J. Mater. Chem. C 2015, 3, 8684–8693. 10.1039/C5TC01839K. [DOI] [Google Scholar]
- Chithrani B. D.; Ghazani A. A.; Chan W. C. W. Determining the Size and Shape Dependence of Gold Nanopartilce Uptake into Mammalian Cells. Nano Lett. 2006, 6, 662–668. 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- Lévy R.; Shaheen U.; Cesbron Y.; Sée V. Gold Nanoparticles Delivery in Mammalian Live Cells: A Critical Review. Nano Rev. 2010, 1, 4889. 10.3402/nano.v1i0.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willets K. A. Super-Resolution Imaging of SERS Hot Spots. Chem. Soc. Rev. 2014, 43, 3854–3864. 10.1039/C3CS60334B. [DOI] [PubMed] [Google Scholar]
- Ameer F. S.; Zhou Y.; Zou S.; Zhang D. Wavelength-Dependent Correlations between Ultraviolet–Visible Intensities and Surface Enhanced Raman Spectroscopic Enhancement Factors of Aggregated Gold and Silver Nanoparticles. J. Phys. Chem. C 2014, 118, 22234–22242. 10.1021/jp5073395. [DOI] [Google Scholar]
- Tian L.; Jiang Q.; Liu K. K.; Luan J.; Naik R. R.; Singamaneni S. Bacterial Nanocellulose-Based Flexible Surface Enhanced Raman Scattering Substrate. Adv. Mater. Interfaces 2016, 3, 1600214 10.1002/admi.201600214. [DOI] [Google Scholar]
- Kirk J.; Carmichael J.; Stratford I. J.; Harris A. L. Selective Toxicity of TGF-α-PE40 to EGFR-positive Cell Lines: Selective Protection of Low EGFR-expressing Cell Lines by EGF. Br. J. Cancer 1994, 69, 988–994. 10.1038/bjc.1994.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.-H.; Richert N.; Ito S.; Merlino G. T.; Pastan I. Characterization of Epidermal Growth Factor Receptor Gene Expression in Malignant and Normal Human Cell Lines. Proc. Natl. Acad. Sci. U.S.A. 1984, 81, 7308–7312. 10.1073/pnas.81.23.7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L. T.; Ohashi P. S. Clinical Blockade of PD1 and LAG3 - Potential Mechanisms of Action. Nat. Rev. Immunol. 2015, 15, 45–56. 10.1038/nri3790. [DOI] [PubMed] [Google Scholar]
- Soliman H.; Khalil F.; Antonia S. PD-L1 Expression is Increased in a Subset of Basal Type Breast Cancer Cells. PLoS One 2014, 9, e88557 10.1371/journal.pone.0088557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taube J. M.; Klein A.; Brahmer J. R.; Xu H.; Pan X.; Kim J. H.; Chen L.; Pardoll D. M.; Topalian S. L.; Anders R. A. Association of PD-1, PD-1 Ligands, and Other Features of the Tumor Immune Microenvironment with Response to Anti-PD-1 Therapy. Clin. Cancer Res. 2014, 20, 5064–5074. 10.1158/1078-0432.CCR-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movasaghi Z.; Rehman S.; Rehman I. U. Raman Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. 10.1080/05704920701551530. [DOI] [Google Scholar]
- Soetan N.; Zarick H. F.; Banks C.; Webb J. A.; Libson G.; Coppola A.; Bardhan R. Morphology-Directed Catalysis with Branched Gold Nanoantennas. J. Phys. Chem. C 2016, 120, 10320–10327. 10.1021/acs.jpcc.6b01238. [DOI] [Google Scholar]
- Petros R. A.; DeSimone J. M. Strategies in the Design of Nanoparticles for Therapeutic Applications. Nat. Rev. Drug Discovery 2010, 9, 615–627. 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- Kang B.; Kukreja A.; Song D.; Huh Y.-M.; Haam S. Strategies for Using Nanoprobes to Perceive and Treat Cancer Activity: A Review. J. Biol. Eng. 2017, 11, 1–12. 10.1186/s13036-016-0044-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejman J.; Oberle V.; Zuhorn I. S.; Hoekstra D. Size-Dependent Internalization of Particles via the Pathways of Clathrin- and Caveolae-Mediated Endocytosis. Biochem. J. 2004, 377, 159–169. 10.1042/bj20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. Z.; Langer R.; Farokhzad O. C. Nanoparticle delivery of cancer drugs. Annu. Rev. Med. 2012, 63, 185–198. 10.1146/annurev-med-040210-162544. [DOI] [PubMed] [Google Scholar]
- Mitra S.; Gaur U.; Ghosh P. C.; Maitra A. N. Tumour Targeted Delivery of Encapsulated Dextran–Doxorubicin Conjugate Using Chitosan Nanoparticles as Carrier. J. Controlled Release 2001, 74, 317–323. 10.1016/S0168-3659(01)00342-X. [DOI] [PubMed] [Google Scholar]
- Blau R.; Krivitsky A.; Epshtein Y.; Satchi-Fainaro R. Are Nanotheranostics and Nanodiagnostics-Guided Drug Delivery Stepping Stones Towards Precision Medicine?. Drug Resist. Updates 2016, 27, 39–58. 10.1016/j.drup.2016.06.003. [DOI] [PubMed] [Google Scholar]
- Xie J.; Lee J. Y.; Wang D. I. C. Seedless, Surfactantless, High-Yield Synthesis of Branched Gold Nanocrystals in HEPES Buffer Solution. Chem. Mater. 2007, 19, 2823–2830. 10.1021/cm0700100. [DOI] [Google Scholar]
- Bishnoi S. W.; Lin Y.-j.; Tibudan M.; Huang Y.; Nakaema M.; Swarup V.; Keiderling T. A. SERS Biodetection Using Gold-Silica Nanoshells and Nitrocellulose Membranes. Anal. Chem. 2011, 83, 4053–4060. 10.1021/ac103195e. [DOI] [PubMed] [Google Scholar]
- Lowery A. R.; Gobin A. M.; Day E. S.; Halas N. J.; West J. L. Immunonanoshells for Targeted Photothermal Ablation of Tumor Cells. Int. J. Nanomed. 2006, 1, 149–154. 10.2147/nano.2006.1.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.; Wang Z.; Zong S.; Chen H.; Wang C.; Xu S.; Cui Y. Simultaneous Evaluation of p53 and p21 Expression Level for Early Cancer Diagnosis Using SERS Technique. Analyst 2013, 138, 3450–3456. 10.1039/c3an00181d. [DOI] [PubMed] [Google Scholar]
- Gobin A. M.; Moon J. J.; West J. L. EphrinA1-Targeted Nanoshells for Photothermal Ablation of Prostate Cancer Cells. Int. J. Nanomed. 2008, 3, 351–358. 10.2147/ijn.s2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson G. T.Bioconjugate Techniques, 2nd ed.; Academic Press: Cambridge, MA, 2008. [Google Scholar]
- Hirsch L. R.; Jackson J. B.; Lee A.; Halas N. J.; West J. L. A Whole Blood Immunoassay Using Gold Nanoshells. Anal. Chem. 2003, 75, 2377–2381. 10.1021/ac0262210. [DOI] [PubMed] [Google Scholar]
- Weissleder R. A Clearer Vision for In Vivo Imaging. Nat. Biotechnol. 2001, 19, 316–317. 10.1038/86684. [DOI] [PubMed] [Google Scholar]
- Chen K.; Han H.; Luo Z. Streptococcus suis II Immunoassay Based on Thorny Gold Nanoparticles and Surface Enhanced Raman Scattering. Analyst 2012, 137, 1259–1264. 10.1039/c2an15997j. [DOI] [PubMed] [Google Scholar]
- Kang H.; Jeong S.; Park Y.; Yim J.; Jun B.-H.; Kyeong S.; Yang J.-K.; Kim G.; Hong S.; Lee L. P.; Kim J.-H.; Lee H.-Y.; Jeong D. H.; Lee Y.-S. Near-Infrared SERS Nanoprobes with Plasmonic Au/Ag Hollow-Shell Assemblies for In Vivo Multiplex Detection. Adv. Funct. Mater. 2013, 23, 3719–3727. 10.1002/adfm.201203726. [DOI] [Google Scholar]
- Zou X.; Ying E.; Dong S. Seed-Mediated Synthesis of Branched Gold Nanoparticles with the Assistance of Citrate and Their Surface-Enhanced Raman Scattering Properties. Nanotechnology 2006, 17, 4758–4764. 10.1088/0957-4484/17/18/038. [DOI] [PubMed] [Google Scholar]
- Singh A. K.; Senapati D.; Wang S.; Griffin J.; Neely A.; Candice P.; Naylor K. M.; Varisli B.; Kalluri J. R.; Ray P. C. Gold Nanorod Based Selective Identification of Escherichia coli Bacteria using Two-Photon Rayleigh Scattering Spectroscopy. ACS Nano 2009, 3, 1906–1912. 10.1021/nn9005494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manson J.; Kumar D.; Meenan B. J.; Dixon D. Polyethylene Glycol Functionalized Gold Nanoparticles: The Influence of Capping Density on Stability in Various Media. Gold Bull. 2011, 44, 99–105. 10.1007/s13404-011-0015-8. [DOI] [Google Scholar]
- Rahme K.; Chen L.; Hobbs R. G.; Morris M. A.; O’Driscoll C.; Holmes J. D. PEGylated Gold Nanoparticles: Polymer Quantification as a Function of PEG Lengths and Nanoparticle Dimensions. RSC Adv. 2013, 3, 6085–6094. 10.1039/C3RA22739A. [DOI] [Google Scholar]
- Nima Z. A.; Mahmood M.; Xu Y.; Mustafa T.; Watanabe F.; Nedosekin D. A.; Juratli M. A.; Fahmi T.; Galanzha E. I.; Nolan J. P.; Basnakian A. G.; Zharov V. P.; Biris A. S. Circulating Tumor Cell Identification by Functionalized Silver-Gold Nanorods with Multicolor, Super-Enhanced SERS and Photothermal Resonances. Sci. Rep. 2014, 4, 4752 10.1038/srep04752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.-L.; Wang L.; Liu X.-Y.; Zhang Z.-P.; Guo H.-C.; Liu W.-M.; Tang S.-H. In Vitro Cancer Cell Imaging and Therapy Using Transferrin-Conjugated Gold Nanoparticles. Cancer Lett. 2009, 274, 319–326. 10.1016/j.canlet.2008.09.024. [DOI] [PubMed] [Google Scholar]
- Melancon M. P.; Lu W.; Yang Z.; Zhang R.; Cheng Z.; Elliot A. M.; Stafford J.; Olson T.; Zhang J. Z.; Li C. In Vitro and In Vivo Targeting of Hollow Gold Nanoshells Directed at Epidermal Growth Factor Receptor for Photothermal Ablation Therapy. Mol. Cancer Ther. 2008, 7, 1730–1739. 10.1158/1535-7163.MCT-08-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla R.; Chanda N.; Zambre A.; Upendran A.; Katti K.; Kulkarni R. R.; Nune S. K.; Casteel S. W.; Smith C. J.; Vimal J.; Boote E.; Robertson J. D.; Kan P.; Engelbrecht H.; Watkinson L. D.; Carmack T. L.; Lever J. R.; Cutler C. S.; Caldwell C.; Kannan R.; Katti K. V. Laminin Receptor Specific Therapeutic Gold Nanoparticles (198AuNP-EGCg) Show Efficacy in Treating Prostate Cancer. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 12426–12431. 10.1073/pnas.1121174109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorry P. A. General Least-Squares Smoothing and Differentiation by the Convolution (Savitzky-Golay) Method. Anal. Chem. 1990, 62, 570–573. 10.1021/ac00205a007. [DOI] [Google Scholar]
- Luo J.; Ying K.; Bai J. Savitzky–Golay Smoothing and Differentiation Filter for Even Number Data. Signal Process. 2005, 85, 1429–1434. 10.1016/j.sigpro.2005.02.002. [DOI] [Google Scholar]
- Savitzky A.; Golay M. J. E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. 10.1021/ac60214a047. [DOI] [Google Scholar]
- Lieber C. A.; Mahadevan-Jansen A. Automated Method for Subtraction of Fluorescence from Biological Raman Spectra. Appl. Spectrosc. 2003, 57, 1363–1367. 10.1366/000370203322554518. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.