Abstract
Digitalis drugs are selective inhibitors of the plasma membrane Na+/K+-ATPase. There are many studies on molecular mechanisms of digitalis interaction with purified pig kidney enzyme, with the tacit assumption that it is a good model of human kidney enzyme. However, previous studies on crude or recombinant human kidney enzymes are limited, and have not resulted in consistent findings on their digitalis sensitivities. Hence, we prepared comparably purified enzymes from human and pig kidneys and determined inhibitory constants of digoxin, ouabain, ouabagenin, bufalin, and marinobufagenin (MBG) on enzyme activity under optimal turnover conditions. We found that each compound had the same potency against the two enzymes, indicating that (i) the pig enzyme is an appropriate model of the human enzyme, and (ii) prior discrepant findings on human kidney enzymes were either due to structural differences between the natural and recombinant enzymes or because potencies were determined using binding constants of digitalis for enzymes under nonphysiological conditions. In conjunction with previous findings, our newly determined inhibitory constants of digitalis compounds for human kidney enzymes indicate that (i) of the compounds that have long been advocated to be endogenous hormones, only bufalin and MBG may act as such at kidney tubules, and (ii) beneficial effects of digoxin, the only digitalis with extensive clinical use, does not involve its inhibitory effect on renal tubular Na+/K+-ATPase.
Introduction
Na+/K+-ATPase (the sodium pump) is the energy-transducing enzyme of the plasma membrane of most eukaryotic cells that catalyzes the coupled active transport of Na+ and K+, maintains the resting membrane potential, regulates the cell volume, and allows Na+-coupled transport of many nutrients and other ions across the cell membrane.1,2 The enzyme has two subunits (α and β) that are necessary for ion pumping and a third subunit (a FXYD protein) that regulates functions in some cells.1,2 There are multiple isoforms of each of the subunits, with cell-type and species specificities.1−3
Digitalis compounds, such as digoxin, digitoxin, and ouabain, are highly specific inhibitors of all Na+/K+-ATPases; however, these enzymes from various sources exhibit significantly different digitalis sensitivities depending on the chemical structure of the specific digitalis and on the nature of the subunit isoforms of the enzyme used for assessing digitalis sensitivity.2−4
Na+/K+-ATPase from the mammalian kidneys has occupied a special place in the history for understanding the molecular mechanisms of digitalis interaction with the sodium pump. There are two main reasons for this: (i) since the early classical work on the purification of the Na+/K+-ATPase,5 it has been realized that the membrane-bound enzyme purified from the outer medulla of the mammalian kidneys are homogeneous in isoform composition, consisting of α1, β1, and FXYD2/γ;6 (ii) the convenience of the large-scale preparation of the purified enzyme from pig kidney has made the crystallization and analysis of the crystal structure in native and digitalis-bound forms possible.7−11 This and the tacit assumption that the pig kidney Na+/K+-ATPase (PKE) is a good model of the human kidney Na+/K+-ATPase (HKE) has led to a wealth of new information on the molecular mechanisms of digitalis interaction with the renal enzyme and on the potential functional consequences of the renal enzyme inhibition by different digitalis compounds.11 As is the case for all studies on experimental animals, however, the question arises as to whether the specific conclusions and interpretations of studies on the pig kidney enzyme also apply to the case of digitalis interaction with the human kidney enzyme. From this point of view, it is of considerable concern that the limited number of past studies that have been done on digitalis sensitivities of the HKE have not been consistent in results and interpretations.12−15 These studies have had several shortcomings owing to the legitimate difficulties of working with human tissues. First, nearly all of the previous work has been done on recombinant enzymes,12−14 creating a real possibility that the different membrane environments of the recombinant enzymes may have influenced their digitalis sensitivities. Second, examination of this limited literature shows that digitalis sensitivities of the preparations have been assessed by different means in different laboratories; for example, comparison of the different potencies of digitalis compounds as inhibitors of Na+/K+-ATPase activity have been done under different assay conditions14,15 and comparison of the binding constants of various digitalis compounds to those of recombinant enzymes have been done under different conditions.12−14 Therefore, the present study was initiated with two primary aims: (i) to use purified Na+/K+-ATPase prepared from healthy human kidneys and to assay inhibitory potencies of five structurally different digitalis compounds on their activity; (ii) to determine the inhibitory potencies of the same compounds on the purified pig kidney enzyme to see if the two enzymes respond differently. Our findings show identical sensitivities of the two enzymes to each of the tested compounds, indicating that the pig kidney enzyme is indeed an appropriate model of the human kidney enzyme. We discuss the causes of previous disagreements on the digitalis sensitivities of the human enzyme, and we consider the important implications of our findings for the suggested hormonal roles of some digitalis compounds and for their current clinical use in man.
Results
As the primary aim of this work was to compare the digitalis sensitivities of human kidney and PKEs and because only a small number of human kidneys were available, we chose to compare the inhibitory constants (Ki values) of five structurally different digitalis compounds on the Na+/K+-ATPase activities of the enzymes purified from single human and pig kidneys. Using the procedures described in Materials and Methods, purified preparations from the outer medulla of two pig kidneys (PKE-1 and PKE-2) and two human kidneys (HKE-1 and HKE-2) were obtained with the following characteristics: PKE-1, yield 30.7 mg of protein, specific activity (μmol of released inorganic phosphate (Pi) mg–1 h–1) 800 ± 40, n = 36; PKE-2, yield 47.6 mg of protein, specific activity (μmol of released Pi mg–1 h–1) 527 ± 32, n = 33; HKE-1, yield 39.3 mg of protein, specific activity (μmol of released Pi mg–1 h–1) 572 ± 30, n = 39; HKE-2, yield 48.8 mg of protein, specific activity (μmol of released Pi mg–1 h–1) 494 ± 50, n = 12. The indicated specific activity determinations were made during the 7 month duration of this work.
Comparison of Digitalis Sensitivities of the Two Enzymes
In previous studies on the human kidney enzyme,12−15 digitalis sensitivities have been assessed either by determining the inhibitory potencies on Na+/K+-ATPase activity under turnover conditions or by determining the digitalis binding constants of the enzyme that is inhibited by Mg2+ + Pi or Mg2+ + vanadate. We chose to use the former approach because the latter leads to generation of inexplicable data (see Discussion).
Using the above preparations and the procedures described in Materials and Methods, we first addressed two important preliminary issues using the more readily available PKEs.
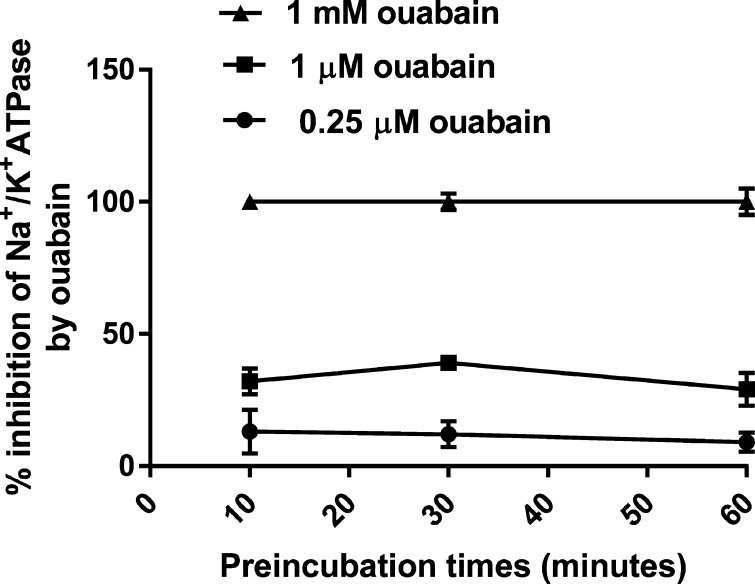
Some previous studies on the inhibitory effects of ouabain and other digitalis compounds on Na+/K+-ATPase activities had suggested the necessity of long contact times between the enzyme and the inhibitor for accurate determination of the Ki values.14−16 Therefore, we examined the possible effects of varying incubation times (10–60 min) of the enzyme with ouabain on inhibition of the PKE-1 activity. The results (Figure 1) clearly indicated that preincubation for 10 min was sufficient to obtain the maximal inhibitory effect of ouabain at any concentration in the range of 0.25–1000 μM.
Figure 1.
Effects of varying preincubation times of ouabain with PKE on the inhibitory effect of ouabain on enzyme activity. The indicated ouabain concentrations were preincubated with PKE-1 in the absence of ATP. The activity was then assayed after the addition of ATP, as described in Materials and Methods.
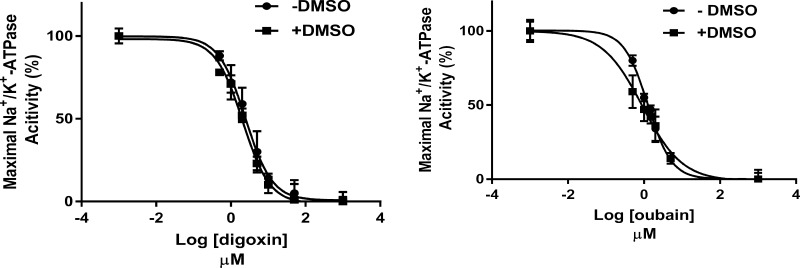
Because many digitalis compounds are highly water insoluble, for the study of their in vitro effects, it is often necessary to use solvents such as ethanol or dimethyl sulfoxide (DMSO).14,41 We chose to use DMSO in the present study, and hence we determined if 1% DMSO in the reaction mixtures affected the sensitivities of PKE to ouabain and digoxin. The results (Figure 2) showed no significant effects of DMSO on the Ki values of these representative digitalis drugs.
Figure 2.
Effect of 1% DMSO on the sensitivity of PKE to the inhibitory effects of ouabain and digoxin on enzyme activity. The Ki values of the two inhibitors were determined for PKE-1 or PKE-2 activities in the absence and presence of DMSO, as described in Materials and Methods. At each inhibitor concentration (n = 6), the activity was calculated relative to maximal activity. The curves are the fit of data to the four-parameter variable slope nonlinear model. R2 values (goodness fit): 0.96, digoxin (−DMSO); 0.98, digoxin (+DMSO); 0.97, ouabain (−DMSO); and 0.94, ouabain (+DMSO). Ki ± SE values: 2.4 ± 0.12 μM, digoxin (−DMSO); 2.1 ± 0.07 μM (+DMSO); 1.26 ± 0.04 μM, ouabain (−DMSO); and 0.90 ± 0.06 μM, ouabain (+DMSO).
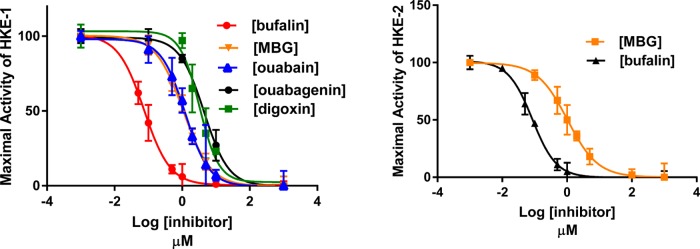
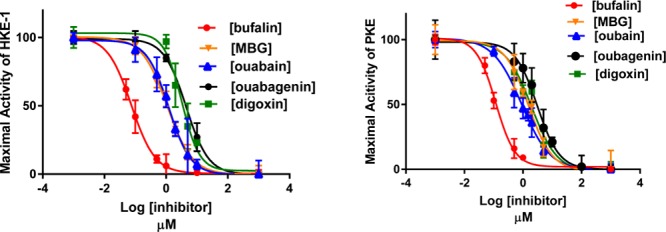
After ensuring that the preincubation time of 10 min was sufficient for ouabain to interact with the enzyme for obtaining maximal inhibition at any concentration of ouabain, and that DMSO does not interfere with inhibitor potencies, we determined the Ki values of ouabain, ouabagenin, bufalin, marinobufagenin (MBG), and digoxin on Na+/K+-ATPase activities of PKE and HKE in the presence of 1% DMSO. The results (Figures 3 and 4) showed that (a) the Ki value of each tested inhibitor was nearly the same for PKE and HKE; (b) of the tested inhibitors, bufalin was the most potent and ouabagenin the least potent; (c) the genins were significantly less potent than the corresponding glycosylated inhibitor; and (d) all tested inhibitors produced complete inhibition of Na+/K+-ATPase activity.
Figure 3.
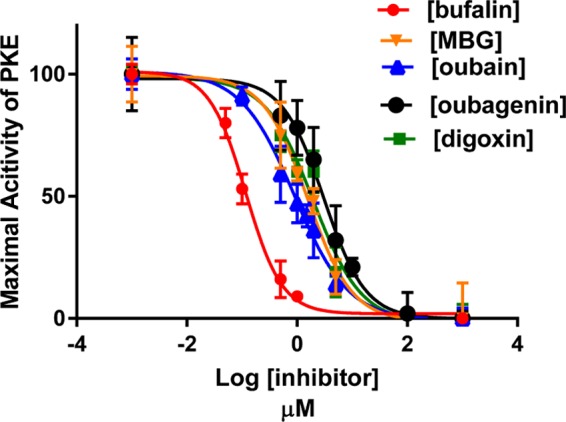
Inhibitory effects of varying concentrations of ouabain, ouabagenin, digoxin, bufalin, and MBG on Na+/K+-ATPase activity of PKE under optimal turnover conditions. Experiments were conducted, and the Ki values were determined, as described in Materials and Methods and in the legend to Figure 2 using PKE-1 or PKE-2 preparations. The calculated Ki values were: bufalin, 0.11 ± 0.005 μM; MBG, 1.5 ± 0.12 μM; ouabain, 0.9 ± 0.05 μM; ouabagenin, 3.07 ± 0.23 μM; and digoxin, 1.95 ± 0.15 μM.
Figure 4.
Inhibitory effects of varying concentrations of ouabain, ouabagenin, digoxin, bufalin, and MBG on Na+/K+-ATPase activity of HKE-1 (4A) and HKE-2 (4B) under optimal turnover conditions. Experiments were conducted, and the Ki values were determined, as described in Materials and Methods and in the legend to Figure 2, using the indicated HKE preparations. The calculated Ki values for A were: bufalin, 0.075 ± 0.003 μM; MBG, 1.07 ± 0.04 μM; ouabain, 1.22 ± 0.09 μM; ouabagenin, 4.4 ± 0.27 μM; and digoxin, 3.2 ± 0.22 μM. The Ki values for B were: bufalin, 0.085 ± 0.003 μM and MBG, 1.04 ± 0.06 μM.
The use of purified enzyme preparations made from single kidneys was imposed in these studies due to scarcity of human kidneys. This, however, raised the question of whether or not the above-described results could be due to unknown peculiarities of the enzymes made from single kidneys. Therefore, we prepared PKE on a large scale, as described in Materials and Methods with specific activity of 1035 μmol of released Pi mg–1 h–1, and assayed its ouabain sensitivity, as we did for the single kidney preparations. The results (not shown) were not significantly different from those presented in Figure 3 for PKEs prepared from single kidneys.
Discussion
Some of our findings and conclusions are in conflict with those of previously published studies; hence, it is necessary that we address these discrepancies.
The most significant of the disagreements between earlier findings and ours are on the sensitivity of HKE to digitalis compounds. This is best illustrated by comparing the ouabain and digoxin sensitivities of the human enzyme reported by others and those reported here. Katz et al.14 reported Ki values of 97 and 250 nM for ouabain and digoxin (their Table 2), whereas Touza et al.15 reported values of 63 and 280 nM for ouabain and digoxin (their Table 1), respectively, for Na+/K+-ATPase under turnover conditions. These values are about 10-fold lower than what we find for ouabain and digoxin (Figure 4). Why are their values so significantly different from ours? There are multiple possibilities.
Katz et al.14 used a recombinant human enzyme expressed in yeast, reconstituted it with a set of chosen lipids, and assayed the activity of the resulting detergent-solubilized enzyme for determining the Ki values. Recently, there have been elegant studies on crystal structures of P-ATPases and on the properties of recombinant enzymes, reviewed by Cornelius et al.,17 identifying both general and specific lipid interactions with Na+/K+-ATPase and related enzymes. There have been numerous older studies cited in Hegyvary et al.,18 suggesting the regulation of ouabain interaction with Na+/K+-ATPase by membrane lipids. Therefore, it is reasonable to suspect that the Ki values obtained by Katz et al.14 may have been distorted by differences between the lipids they chose for reconstitution and the authentic lipids of the membrane-bound human kidney enzyme that are yet to be identified.
Another possible explanation for the different Ki values found by us and those of Katz et al.14 is that they chose (their footnote 4) to use assay times equal to or longer than 40 min for their experiments because they felt such long reaction times were necessary for accurate Ki determination. We suggest that the need for long reaction times in such studies is a misconception introduced to the field by the limited data of a respected laboratory.16 From our findings for ouabain inhibition of PKE (Figure 1), and as others have shown for inhibitory effects of several digitalis drugs on the activity of purified lamb kidney enzymes,19 short preincubation and assay times of 10 min are clearly sufficient for accurate Ki determinations. On the whole, we consider the Ki values reported here for the HKE to be more reliable than those of Katz et al.,14 obtained with a recombinant human enzyme.
The second set of Ki values reported earlier by Touza et al.15 was not obtained with a recombinant enzyme but with the use of samples taken from the unaffected poles of diseased human kidneys, preparation of crude unfractionated membranes from sample homogenates, and assay of Na+/K+-ATPase activities for 2 h durations, most likely due to low specific activities of the crude preparations. We suggest that the unusually long assay duration, potential for enzyme inactivation during such long reaction times, and the unreported specific activities of the crude preparations make the reported Ki values of these investigators also less reliable than our values reported here.
Leaving aside the disagreements on the Ki values, another important issue regarding digitalis sensitivities of Na+/K+-ATPases requires further discussion. In a number of studies either on recombinant enzymes12−14 or with purified kidney enzymes,8,11,19 comparison of sensitivities have been made by measuring binding constants of various digitalis compounds to a phosphorylated form of the enzyme (E2-Pi) obtained in the presence of Mg2+ and Pi or to the equivalent of E2-Pi that is obtained in the presence of Mg2+ and vanadate. Although these forms of the enzymes are known to have the highest affinities for digitalis inhibitors, it is debatable if they provide useful information about the relative potencies of the inhibitors. Paula et al.19 correctly pointed out the lack of correlation between binding affinities for E2-Pi and inhibitory potencies on Na+/K+-ATPase activities. Examination of the work done since their observations adds to the doubts about the use of such binding affinities for comparison of digitalis potencies. Consider, for example, that with the use of relative binding constants and a recombinant human kidney enzyme, Katz et al.14 noted that MBG was about 200-fold less potent than ouabain (their Table 1), whereas our results show that in Na+/K+-ATPase activity assays ouabain and MBG are equally potent (Figure 4A). Also consider that with the use of relative binding constants and the purified PKE, Laursen et al.11 concluded that ouabagenin was about 800-fold less potent than ouabain and that ouabagenin was only a partial inhibitor of the enzyme (their Figure 3), whereas our results show that in Na+/K+-ATPase activity assays, ouabagenin is about 3-fold less potent than ouabain and that it produces complete inhibition (Figure 3). Why are there such dramatic and strange differences between the binding affinities and inhibitory potencies of digitalis compounds? We suggest that it is due to the forgotten fact that there is no solid evidence to indicate that the phosphoenzyme obtained from Pi (E2-Pi) is related to the phosphoenzyme obtained from ATP or that E2-Pi can be converted to the physiologically relevant phosphoenzymes.20,21 It may be regrettable that the uncertainties about E2-Pi and digitalis interaction with it that were identified years ago22 still remain unresolved, but the same misgivings also emphasize that for comparison of relative potencies of digitalis compounds it is prudent not to rely on the binding data obtained in the presence of Pi or vanadate and rather rely on Ki values measured under turnover conditions. Another unfortunate consequence of the use of Pi and Mg2+ for the binding of digitalis compounds to the purified enzyme is the uncertainty that it has introduced into the elegant structural studies that have been done on the crystals of various digitalis-bound PKE preparations.10,11 Because all such crystals were obtained in the presence of Pi and Mg2+, with the dubious assumption that the complexes have the same structures as those obtained from the phosphoenzyme generated from ATP, we suggest the necessity of reexamination of some of the conclusions derived from these studies.
To summarize the main point of the present study, we conclude that our findings on Ki values are the best representations of the potencies of the tested digitalis compounds as inhibitors of HKE and PKE and that there are no significant differences between sensitivities of HKE and PKE.
Physiological and Therapeutic Implications
Perhaps the most important physiological implication of this work is related to the more than half a century of interest of the field in the hypothesis that sodium pump inhibitors may be naturetic hormones.23,24 This hypothesis has always been controversial since its proposal and still remains so.25−30
For proponents of the hypothesis, such as some coauthors of this report,31−33 our findings of the potencies of ouabain, digoxin, MBG, and bufalin as inhibitors of the HKE (Figure 4) become a guide for deciding if any measured change in the blood level of the advocated hormones in human beings may exert a meaningful renal effect. Considering the Ki values of the four putative hormones (Figure 4) and recalling that the highest reported blood level value of an “ouabain-like compound” is ≈130 nM,34 it seems that bufalin, ouabain, and MBG are the only compounds with sufficient potency to have some renal effect. As it has been aptly pointed out, however, whether or not any sodium pump inhibitor may exert a hormonal effect at any locus depends not only on its concentration but also on its circulating half-life.29 Taking into account the limited amount of information about the clearance of these compounds in human beings29 along with their potencies (Figure 4), we suggest that only bufalin and MBG remain as potential hormones with renal effects. Needless to say, however, our findings on the HKE do not rule out the possibility of hormonal effects of the sodium pump inhibitors at loci other than those of human kidney tubules.
For those who seriously doubt the existence of endogenous digitalis compounds and consider these sodium pump inhibitors simply as drugs, are there any therapeutic implications of our findings? Of the tested compounds, only digoxin is an approved drug and still used in the United States. When used clinically, blood levels are controlled by appropriate dosage regimens to achieve nearly constant levels in the low nM range (e.g., 0.64–1.15 nM).35 The Ki value of HKE for digoxin being ≈3000 nM (Figure 4), one would expect little or no renal effects of the clinically used digoxin. This, however, does not rule out significant and beneficial therapeutic effects of low nM blood levels of digoxin. As discussed elsewhere,36 any such effects need to be due to digoxin interactions with human Na+/K+-ATPase isoforms other than α1 at loci other than kidneys, most likely in the heart.
Materials and Methods
Kidneys
Pig kidneys were obtained from a local slaughterhouse and kept frozen at −80 °C until use. Healthy human kidneys were those that were intended for transplantation but did not match with any available recipients at the time of donation, hence becoming available for research instead of being discarded. We received a limited number of such kidneys through Life Connection of Ohio, following this organization’s policies and our institutional policies for research on organs and tissue samples from a human source. These kidneys had been kept on ice for a few hours before being frozen and delivered to our laboratory. They were then kept at −80 °C until use.
Preparation of Purified Na+/K+-ATPase
The enzyme was purified from a single pig or human kidney by the widely used procedure of Jorgensen,5 involving the preparation of microsomes from dissected outer red medulla, sodium dodecyl sulfate treatment of the microsomal suspension to achieve selective extraction of proteins other than Na+/K+-ATPase, and fractionation of the remaining crude membranes by sucrose density gradient centrifugation in an angle rotor5,37 to obtain pellets of the purified enzyme. These were suspended in 25 mM imidazole-HCl, 1 mM EDTA (pH 7.5) and stored at −80 °C until use. The specific activities of the preparations from the individual kidneys are indicated in Results. For a limited number of experiments, the pig kidney enzyme prepared by the large-scale purification procedure using the combined outer medullas of a large number of kidneys was also needed. This was made by the same general procedure,5 as described before.38
Assay of Na+/K+-ATPase Activity
The steady-state ouabain-sensitive activity was assayed at 37 °C by measuring the initial rate of release of Pi from ATP in a reaction mixture (1 mL), containing 100 mM NaCl, 25 mM KCl, 2 mM ATP, 3 mM MgCl2, 1 mM EGTA, and 20 mM Tris–HCl (pH 7.4) in the presence and absence of 1 mM ouabain. This medium composition ensures that optimal activity of Na+/K+-ATPase is achieved.39 To conduct the assay under standard conditions, the enzyme was preincubated at 37 °C for 10 min in the above medium without ATP and MgCl2. The reaction was then started by the simultaneous addition of ATP and MgCl2 and stopped after 10 min by the addition of 0.6 ml of 8% trichloroacetic acid. The enzyme amount (2 μg or less of protein) was chosen to obtain less than 10% hydrolysis of ATP. To determine the inhibitory constant (Ki) of a digitalis compound, the effects of varying concentrations of the inhibitor relative to those of 1 mM ouabain were determined. Ki values were calculated from curves fit to the data by a nonlinear regression model using GraphPad Prism software, version 7.
Other Assays
Pi was measured with Malachite Green,40 using a commercially available kit (Biomol Green; Enzo Life Sciences, Farmingdale, NY). Protein was determined by the BioRad DC colorimetric assay.
Digitalis Drugs
Ouabain, ouabagenin, bufalin, and digoxin were purchased from Sigma-Aldrich. MBG was purified from the parotid glands of Bufomarinus toads, as previously described.41
Data Analysis
Data are presented as mean ± standard error. Analyses were performed using GraphPad Prism 7 software. Differences were considered significant at p < 0.05.
Acknowledgments
We are grateful to Life Connection of Ohio for arranging the use of donated human kidneys for this research. We also thank Dr. Lijun Liu for advice on data analysis and Jenifer Zak for capable assistance in the preparation of the article.
Glossary
Abbreviations
- DMSO
dimethyl sulfoxide
- E2-Pi
phosphorylated Na+/K+-ATPase formed from inorganic phosphate
- HKE
human kidney Na+/K+-ATPase
- Ki
inhibitory constant or the inhibitor concentration causing half-maximal inhibition
- MBG
marinobufagenin
- Pi
inorganic phosphate
- PKE
pig kidney Na+/K+-ATPase
The authors declare no competing financial interest.
Notes
This study was supported by the National Heart, Lung, and Blood Institute Grant P01-HL36573 and Intramural Research Program, National Institute of Aging, NIH.
References
- Skou J. C.; Esmann M. The Na,K-ATPase. J. Bioenerg. Biomembr. 1992, 24, 249–261. 10.1007/BF00768846. [DOI] [PubMed] [Google Scholar]
- Kaplan J. H. Biochemistry of Na,K ATPase. Annu. Rev. Biochem. 2002, 71, 511–535. 10.1146/annurev.biochem.71.102201.141218. [DOI] [PubMed] [Google Scholar]
- Blanco G.; Mercer R. W. Isozymes of the Na-K-ATPase: heterogeneity in structure, diversity in function. Am. J. Physiol. 1998, 275, F633–F650. [DOI] [PubMed] [Google Scholar]
- McDonough A. A.; Velotta J. B.; Schwinger R. H.; Philipson K. D.; Farley R. A. The cardiac sodium pump: structure and function. Basic Res. Cardiol. 2002, 97, I19–I24. 10.1007/s003950200024. [DOI] [PubMed] [Google Scholar]
- Jørgensen P. L. Purification of Na+,K+-ATPase: enzyme sources, preparative problems, and preparation from mammalian kidney. Methods Enzymol. 1988, 156, 29–43. [DOI] [PubMed] [Google Scholar]
- Arystarkhova E.; Sweadner K. J. Splice variants of the gamma subunit (FXYD2) and their significance in regulation of the Na, K-ATPase in kidney. J. Bioenerg. Biomembr. 2005, 37, 381–386. 10.1007/s10863-005-9475-y. [DOI] [PubMed] [Google Scholar]
- Morth J. P.; Pedersen B. P.; Toustrup-Jensen M. S.; Sørensen T. L.; Petersen J.; Andersen J. P.; Vilsen B.; Nissen P. Crystal structure of the sodium-potassium pump. Nature 2007, 450, 1043–1049. 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- Yatime L.; Laursen M.; Morth J. P.; Esmann M.; Nissen P.; Fedosova N. U. Structural insights into the high affinity binding of cardiotonic steroids to the Na+,K+-ATPase. J. Struct. Biol. 2011, 174, 296–306. 10.1016/j.jsb.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Nyblom M.; Poulsen H.; Gourdon P.; Reinhard L.; Andersson M.; Lindahl E.; Fedosova N.; Nissen P. Crystal structure of Na+,K(+)-ATPase in the Na(+)-bound state. Science 2013, 342, 123–127. 10.1126/science.1243352. [DOI] [PubMed] [Google Scholar]
- Laursen M.; Yatime L.; Nissen P.; Fedosova N. U. Crystal structure of the high-affinity Na+K+-ATPase-ouabain complex with Mg2+ bound in the cation binding site. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 10958–10963. 10.1073/pnas.1222308110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen M.; Gregersen J. L.; Yatime L.; Nissen P.; Fedosova N. U. Structures and characterization of digoxin- and bufalin-bound Na+,K+-ATPase compared with the ouabain-bound complex. Proc. Natl. Acad. Sci. U.S.A. 2015, 112, 1755–1760. 10.1073/pnas.1422997112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crambert G.; Hasler U.; Beggah A. T.; Yu C.; Modyanov N. N.; Horisberger J. D.; Lelièvre L.; Geering K. Transport and pharmacological properties of nine different human Na,K-ATPase isozymes. J. Biol. Chem. 2000, 275, 1976–1986. 10.1074/jbc.275.3.1976. [DOI] [PubMed] [Google Scholar]
- Hauck C.; Potter T.; Bartz M.; Wittwer T.; Whalers T.; Mehlhorn U.; Scheiner-Bobis G.; McDonough A. A.; Bloch W.; Schwinger R. H.; Müller-Ehmsen J. Isoform specificity of cardiac glycosides binding to human Na+,K+-ATPase alpha1beta1, alpha2beta1 and alpha3beta1. Eur. J. Pharmacol. 2009, 622, 7–14. 10.1016/j.ejphar.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz A.; Lifshtiz Y.; Bab-Dinitz E.; Kapri-Pardes E.; Goldshleger R.; Tai D. M.; Karlish S. J. Selectivity of digitalis glycosides for isoforms of human Na,K-ATPase. J. Biol. Chem. 2010, 285, 19582–19592. 10.1074/jbc.M110.119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touza N. A.; Pôcas E. S.; Quintas L. E.; Cunha-Filho G.; Santos M. L.; Noël F. Inhibitory effect of combinations of digoxin and endogenous cardiotonic steroids on Na+/K+-ATPase activity in human kidney membrane preparation. Life Sci. 2011, 88, 39–42. 10.1016/j.lfs.2010.10.027. [DOI] [PubMed] [Google Scholar]
- Wallick E. T.; Schwartz A. Interaction of cardiac glycosides with Na+,K+-ATPase. Methods Enzymol. 1988, 156, 201–213. [DOI] [PubMed] [Google Scholar]
- Cornelius F.; Habeck M.; Kanai R.; Toyoshima C.; Karlish S. J. General and specific lipid-protein interactions in Na,K-ATPase. Biochim. Biophys. Acta 2015, 1848, 1729–1743. 10.1016/j.bbamem.2015.03.012. [DOI] [PubMed] [Google Scholar]
- Hegyvary C.; Chigurupati R.; Kang K.; Mahoney D. Reversible alterations in the kinetics of cardiac sodium- and potassium-activated adenosine triphosphatase after partial removal of membrane lipids. J. Biol. Chem. 1980, 255, 3068–3074. [PubMed] [Google Scholar]
- Paula S.; Tabet M. R.; Ball W. J. Jr. Interactions between cardiac glycosides and sodium potassium-ATPase: three-dimensional structure-activity relationship models for ligand binding to the E2-Pi form of the enzyme versus activity inhibition. Biochemistry 2005, 44, 498–510. 10.1021/bi048680w. [DOI] [PubMed] [Google Scholar]
- Hansen O. Interaction of cardiac glycosides with (Na+ + K+)-activated ATPase. A biochemical link to digitalis-induced inotropy. Pharmacol. Rev. 1984, 36, 143–163. [PubMed] [Google Scholar]
- Askari A.; Kakar S. S.; Huang W. H. Ligand binding sites of the ouabain-complexed (Na+ + K+)-ATPase. J. Biol. Chem. 1988, 263, 235–242. [PubMed] [Google Scholar]
- Kakar S. S.; Huang W. H.; Askari A. Properties of the Na+, K+ and ATP binding sites of the ouabain-complexed (Na+ + K+)-ATPase: implications of the mechanism of ouabain action. Prog. Clin. Biol. Res. 1988, 268A, 211–218. [PubMed] [Google Scholar]
- de Wardener H. E.; Clarkson E. M. Concept of natriuretic hormone. Physiol. Rev. 1985, 65, 658–759. [DOI] [PubMed] [Google Scholar]
- Nesher M.; Shpolansky U.; Rosen H.; Lichtstein D. The digitalis-like steroid hormones: new mechanisms of action and biological significance. Life Sci. 2007, 80, 2093–2107. 10.1016/j.lfs.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Nicholls M. G.; Lewis L. K.; Yandle T. G.; Lord G.; McKinnon W.; Hilton P. J. Ouabain, a circulating hormone secreted by the adrenals, is pivotal in cardiovascular disease. Fact or fantasy?. J. Hypertens. 2009, 27, 3–8. 10.1097/HJH.0b013e32831101d1. [DOI] [PubMed] [Google Scholar]
- Lewis L. K.; Yandle T. G.; Hilton P. J.; Jensen B. P.; Begg E. J.; Nicholls M. G. Endogenous ouabain is not ouabain. Hypertension 2014, 64, 680–683. 10.1161/HYPERTENSIONAHA.114.03919. [DOI] [PubMed] [Google Scholar]
- Baecher S.; Kroiss M.; Fassnacht M.; Vogeser M. No endogenous ouabain is detectable in human plasma by ultra-sensitive UPLC-MS/MS. Clin. Chim. Acta 2014, 431, 87–92. 10.1016/j.cca.2014.01.038. [DOI] [PubMed] [Google Scholar]
- Blaustein M. P. Why isn’t endogenous ouabain more widely accepted?. Am. J. Physiol. Heart Circ. Physiol. 2014, 307, H635–H639. 10.1152/ajpheart.00404.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlyn J. M. Natriuretic hormones, endogenous ouabain, and related sodium transport inhibitors. Front. Endocrinol. 2014, 5, 199. 10.3389/fendo.2014.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlyn J. M.; Blaustein M. P. Endogenous ouabain: Recent advances and controversies. Hypertension 2016, 68, 526–532. 10.1161/HYPERTENSIONAHA.116.06599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagrov A. Y.; Shapiro J. I.; Fedorova O. V. Endogenous cardiotonic steroids: physiology, pharmacology and novel therapeutic targets. Pharmacol. Rev. 2009, 61, 9–38. 10.1124/pr.108.000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova O. V.; Shapiro J. I.; Bagrov A. Y. Endogenous cardiotonic steroids and salt-sensitive hypertension. Biochim. Biophys. Acta 2010, 1802, 1230–1236. 10.1016/j.bbadis.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AlGhatrif M.; Wang M.; Fedorova O. V.; Bagrov A. Y.; Lakatta E. G. The pressure of aging. Med. Clin. North Am. 2017, 101, 81–101. 10.1016/j.mcna.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer N.; Müller-Ehmsen J.; Krämer U.; Hambarchian N.; Zobel C.; Schwinger R. H.; Neu H.; Kirch U.; Grünbaum E. G.; Schoner W. Ouabain-like compound changes rapidly on physical exercise in humans and dogs: effects of beta-blockade and angiotensin-converting enzyme inhibition. Hypertension 2005, 45, 1024–1028. 10.1161/01.HYP.0000165024.47728.f7. [DOI] [PubMed] [Google Scholar]
- Ahmed A.; Pitt B.; Rahimtoola S. H.; Waagstein F.; White M.; Love T. E.; Braunwald E. Effects of digoxin at low serum concentrations on mortality and hospitalization in heart failure: a propensity-matched study of the DIG trial. Int. J. Cardiol. 2008, 123, 138–146. 10.1016/j.ijcard.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.; Li D.; Du L.; Baldawi M.; Gable M. E.; Askari A.; Liu L. Ouabain prevents pathological cardiac hypertrophy and heart failure through activation of phosphoinositide 3-kinase α in mouse. Cell Biosci. 2015, 5, 64. 10.1186/s13578-015-0053-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson K. B. Light-dependent inactivation of (Na+ + K+)-ATPase with a new photoaffinity reagent, chromium arylazido-beta-alanyl ATP. J. Biol. Chem. 1981, 256, 3223–3230. [PubMed] [Google Scholar]
- Liu L.; Ivanov A. V.; Gable M. E.; Jolivel F.; Morrill G. A.; Askari A. A. Comparative properties of caveolar and noncaveolar preparations of kidney Na+/K+-ATPase. Biochemistry 2011, 50, 8664–8663. 10.1021/bi2009008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmann M. ATPase and phosphatase activity of Na+,K+-ATPase: molar and specific activity, protein determination. Methods Enzymol. 1988, 156, 105–115. 10.1016/0076-6879(88)56013-5. [DOI] [PubMed] [Google Scholar]
- Lanzetta P. A.; Alvarez L. J.; Reinach P. S.; Candia O. A. An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 1979, 100, 95–97. 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- Bagrov A. Y.; Roukoyatkina N. I.; Pinaev A. G.; Dmitrieva R. I.; Fedorova O. V. Effects of two endogenous Na+,K(+)-ATPase inhibitors, marinobufagenin and ouabain, on isolated rat aorta. Eur. J. Pharmacol. 1995, 274, 151–158. 10.1016/0014-2999(94)00735-P. [DOI] [PubMed] [Google Scholar]