Abstract
Eukaryotic genome is, not only linearly but also spatially, organized into non-random architecture. Though the linear organization of genes and their epigenetic descriptors are well characterized, the relevance of their spatial organization is beginning to unfold only recently. It is increasingly being recognized that physical interactions among distant genomic elements could serve as an important mean to eukaryotic genome regulation. With the advent of proximity ligation based techniques coupled with next generation sequencing, it is now possible to explore whole genome chromatin interactions at high resolution. Emerging data on genome-wide chromatin interactions suggest that distantly located genes are not independent entities and instead cross-talk with each other in an extensive manner, supporting the notion of “chromatin interaction networks”. Moreover, the data also advance the field to “3-dimensional (3D) chromatin structure and dynamics”, which would enable molecular biologists to explore the spatiotemporal regulation of genome. In this article, we introduce a stepwise topological transformation of genome from 1-dimension (1D, linear) to 2-dimension (2D, networks) to 3-dimension (3D, architecture) and discuss how such transformations could advance our understanding of genome biology.
Keywords: genome, chromatin interaction, interaction networks, chromatin 3D architecture
The availability of complete genome sequences and the rapid evolution of high throughput techniques had facilitated the genomic and epigenomic annotations of eukaryotic genomes. We now understand the non-random linear organization of genes and their epigenetic states in distinct species [Hurst et al., 2004; Davila Lopez et al., 2010]. Most of the available annotations are primarily 1D in the form of linear chromosomal coordinates of genes and associated regulatory elements. It was being assumed, for decades, that distantly located genes, especially those from different chromosomes, were autonomous transcriptional units on their own. However, the evidences accumulated in last decade nearly abandon this assumption and suggest that genes, like most other components in the real world, have intrinsic property to interact with each other and with other regulatory elements. Multiple distant enhancers (multipartite) could loop on to a single gene promoter and a single enhancer could dynamically interact with multiple gene promoters [Lomvardas et al., 2006; Deschenes et al., 2007]. Several genes from different chromosomes could converge at discrete foci in the nucleus [Osborne et al., 2004; Xu and Cook, 2008; Schoenfelder et al., 2009]. Interestingly, distant interactions could associate with alteration in transcriptional and other epigenetic states of the genes suggesting the functional nature of some, if not most, of these interactions [Lomvardas et al., 2006; Zhao et al., 2006; Sandhu et al., 2009]. It is also being speculated that chromatin interactions might play a crucial role in other essential nuclear processes like replication [Kitamura et al., 2006], DNA repair [Lisby et al., 2001; Lin et al., 2009], and chimeric transcription [Unneberg and Claverie, 2007]. Progressive innovations of proximity ligation assays, which include 3C [Dekker et al., 2002], 4C [Simonis et al., 2006; Zhao et al., 2006], 5C [Dostie et al., 2006], and more recently Hi-C [Lieberman-Aiden et al., 2009] and ChIA-PET [Fullwood et al., 2009], have enabled the identification of physical interactions among distant genomic loci from a local to global scale. As a result, large scale chromatin interaction data has started to populate and molecular biologists would soon require novel strategies and computational approaches, besides the present ones, to understand the epigenetic regulation of genome mediated through extensive chromatin interactions. To address such challenges, here we discuss a few strategies. First, we propose the concept of “chromatin interaction networks” as a mechanism to regulate genomic functions, and second, we outline a framework, which would be instrumental to analyze the chromatin interaction and associated epigenetic data in 3D conformation.
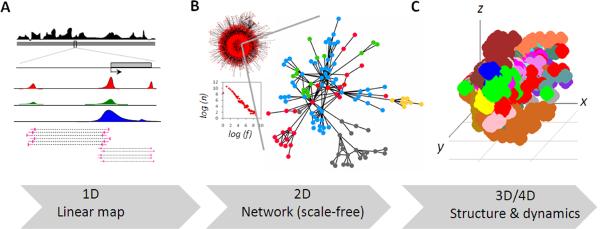
Networks provide a 2D understanding of 1D genomic information in the form of nodes and their inter-connections (edges) [fig. 1]. Ever since the discovery of scale freeness of real world networks, which states that there would be very few nodes with large number of connections, i.e., hubs, while most others would have fewer connections [Albert et al., 2000; Barabási and Bonabeau, 2003; Barabasi and Oltvai, 2004], molecular biologists are fascinated to gain systems insight to cellular regulations. Metabolic networks, protein-protein interaction networks, gene regulatory networks are extensively being studied; however, networks in the context of chromatin interactions have been lacking largely due to the lack of genome scale datasets. Now, with the availability of such datasets [Fullwood et al., 2009; Lieberman-Aiden et al., 2009; Duan et al., 2010], we could apply the concepts of network science to chromatin interactions. A brief analysis of RNAPII bound chromatin interactome recently solved by ChIA-PET technique suggests that chromatin interaction networks also follow scale free distribution of number of interactions per locus, which suggests the presence of “genomic hubs” having disproportionately large number of chromatin interactions. The data also hints at another interesting aspect, the fractal hierarchy of chromatin structures starting from gene loops at very local level, to distant enhancer-promoter interactions at middle level and long range cis and trans enhancer-promoter and promoter-promoter interactions at top level, suggesting the possibility of “hierarchical networks” of chromatin interactions. As more data accumulate in future, these observations could be scrutinized. Though very preliminary, these observations might open up new prospects in the field. For example, “hubs” in the chromatin interaction networks would suggest a possible emergence of “pleiotropy” mediated by chromatin interaction networks. In fact, there are certain reports which hint at such pleiotropy in the system [Steidl et al., 2007; Sandhu et al., 2009]. Analysis of network descriptors (betweenness, closeness, transitivity, modularity, etc) and dynamics (date/party interactions, perturbation analysis, evolution, etc) under distinct normal and diseased conditions could serve as one of the future directions in the field. Moreover, with the realization that real world networks are often interdependent i.e., “network of networks” [Buldyrev et al., 2010; Parshani et al., 2010], it would be interesting to explore the cross-talk of chromatin interaction networks with other cellular networks like that of protein-protein interactions bridging the distant chromatin loci or neighboring signaling networks altering the chromatin interactions remotely. Similarly, chromatin interaction networks could determine gene co-regulatory network downstream [Schoenfelder et al., 2009], suggesting that genetic or epigenetic errors in one network could propagate to other “interacting” networks. Multiple errors at hub loci, thus, could create mayhem of dysregulations and compromise with robustness of cellular systems. Therefore, the networks approach would not only provide a hawk eye view of the widespread chromatin interactions, but would also enrich naïve systems knowledge on genome regulation and perturbations therein mediated by chromatin interactions. Tracking the error propagation in the “coupled” networks could also help designing the strategies to keep the cellular networks healthy and robust.
Figure 1.
Progressive transformation of 1D genomic information to 2D interaction networks and to 3D chromatin architecutres. (a) Linear map of genomic and epigenomic data. (b) Chromatin interaction network of Human RNAPII mediated chromatin interaction data and corresponding log-log plot, which is a trademark of scale-free networks. `n' is number of interactions and `f' is fraction of nodes with `n' interactions. An enlarged sub-network, where colors of the nodes represent genomic loci from distinct chromosomes, is shown. (c) A crude 3D space-filled model of human genome reverse engineered from RNAPII mediated chromatin interaction data. Color depicts different chromosomes. (All the data shown here is unpublished data from authors' laboratory)
Though transformation 1D genomic information into 2D network interactions is a significant leap forward, it is still largely an artificial abstraction of real in situ scenario. To achieve higher resolution visual insights to genomic conformations and their regulation, an extension of 2D to 3D would be desirable. This could be achieved by reconstructing the 3D model of chromatin fiber or even simulating the dynamics in the 4th dimension (4D) of time [fig. 1]. Presently, the 3D conformations are, in general, being represented by 2D graphics. Though it does provide a rough approximation of chromatin looping, it is far from representing real in situ conformations. Reverse modeling and visually navigating the 3D conformation of chromatin fiber under the spatial and biophysical constraints in the nucleus could advance our understanding of condition specific or general architectural regulation of essential genomic functions like transcription. Since chromatin states also attribute to chromatin mobility, incorporating open, closed and other chromatin states in the model would be instrumental in determining the dynamics of a locus of interest. Several hypotheses could be tested by analyzing 3D structure and dynamics of genome and novel principles of chromatin folding and gene regulation may surface. For example, Lieberman-Aiden et al. (2009) revealed how fractal organization of human genome packs the chromatin in highly dense but knot-free conformation, which could allow the rapid storage and retrieval of information in the genome. More recently, the role of transcription factories in determining the global chromatin organization was investigated using Monte Carlo simulation of polymer chains in confined nuclear space. It was demonstrated that contacts mediated by transcription factories could explain several global features of 3D genome organization [Dorier and Stasiak, 2010]. It would also be interesting to address, partly if not thoroughly, the long standing questions concerning nuclear reprogramming during cellular differentiation like that of rod nuclei in nocturnal animals [Solovei et al., 2009], disease progression, chromatin mobility versus transcriptional stochasticity/bursts, mechanism of allelic exclusion, etc. by analyzing 3D conformation of genome and simulating chromatin dynamics. The limitation presently is the lack of a unified framework for modeling and simulating the dynamics of chromatin and the long-range interactions. An integrated platform, which incorporates reverse engineering of Cartesian coordinates from chromatin interaction frequency data, the dynamic loop construction, visual representation at varying resolutions, overlay of other epigenomics data, 3D BLAST search, cross-comparison of conformations in differential conditions, and simulating global and local dynamics of chromatin conformations would help the molecular biologists to explore the genome architecture and regulation in a way previously unanticipated.
Thus, the transformation from 1D linear genome to 2D and to 3D topologies would not only provide additional means to analyze the chromatin interactions, but might also radically change our perspectives on genetic and epigenetic control of eukaryotic genome regulation. We anticipate that the application of these approaches to the upcoming large-scale chromatin interaction data would uncover several interesting insights and an integrated systems paradigm might emerge.
ACKNOWLEDGMENTS
The authors are supported by A*STAR of Singapore. In addition, YR is supported by NIH ENCODE grants R01HG004456-01 and U54 HG004557-01.
References
- Albert R, Jeong H, Barabasi AL. Error and attack tolerance of complex networks. Nature. 2000;406:378–82. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- Barabási A-L, Bonabeau E. Scale-Free Networks. Scientific American. 2003;288:60–69. doi: 10.1038/scientificamerican0503-60. [DOI] [PubMed] [Google Scholar]
- Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–13. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Buldyrev SV, Parshani R, Paul G, Stanley HE, Havlin S. Catastrophic cascade of failures in interdependent networks. Nature. 2010;464:1025–8. doi: 10.1038/nature08932. [DOI] [PubMed] [Google Scholar]
- Davila Lopez M, Martinez Guerra JJ, Samuelsson T. Analysis of gene order conservation in eukaryotes identifies transcriptionally and functionally linked genes. PLoS One. 2010;5:e10654. doi: 10.1371/journal.pone.0010654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Deschenes J, Bourdeau V, White JH, Mader S. Regulation of GREB1 transcription by estrogen receptor alpha through a multipartite enhancer spread over 20 kb of upstream flanking sequences. J Biol Chem. 2007;282:17335–9. doi: 10.1074/jbc.C700030200. [DOI] [PubMed] [Google Scholar]
- Dorier J, Stasiak A. The role of transcription factories-mediated interchromosomal contacts in the organization of nuclear architecture. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS. A three-dimensional model of the yeast genome. Nature. 2010;465:363–7. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EG, Huang PY, Welboren WJ, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY, Wansa KD, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li H, Desai KV, Thomsen JS, Lee YK, Karuturi RK, Herve T, Bourque G, Stunnenberg HG, Ruan X, Cacheux-Rataboul V, Sung WK, Liu ET, Wei CL, Cheung E, Ruan Y. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Pal C, Lercher MJ. The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet. 2004;5:299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- Kitamura E, Blow JJ, Tanaka TU. Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell. 2006;125:1297–308. doi: 10.1016/j.cell.2006.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, Rosenfeld MG. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–83. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci U S A. 2001;98:8276–82. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–13. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–71. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Parshani R, Buldyrev SV, Havlin S. Interdependent networks: reducing the coupling strength leads to a change from a first to second order percolation transition. Phys Rev Lett. 2010;105:048701. doi: 10.1103/PhysRevLett.105.048701. [DOI] [PubMed] [Google Scholar]
- Sandhu KS, Shi C, Sjolinder M, Zhao Z, Gondor A, Liu L, Tiwari VK, Guibert S, Emilsson L, Imreh MP, Ohlsson R. Nonallelic transvection of multiple imprinted loci is organized by the H19 imprinting control region during germline development. Genes Dev. 2009;23:2598–603. doi: 10.1101/gad.552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, Eskiw CH, Luo Y, Wei CL, Ruan Y, Bieker JJ, Fraser P. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2009;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–54. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Steidl U, Steidl C, Ebralidze A, Chapuy B, Han HJ, Will B, Rosenbauer F, Becker A, Wagner K, Koschmieder S, Kobayashi S, Costa DB, Schulz T, O'Brien KB, Verhaak RG, Delwel R, Haase D, Trumper L, Krauter J, Kohwi-Shigematsu T, Griesinger F, Tenen DG. A distal single nucleotide polymorphism alters long-range regulation of the PU.1 gene in acute myeloid leukemia. J Clin Invest. 2007;117:2611–20. doi: 10.1172/JCI30525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unneberg P, Claverie JM. Tentative mapping of transcription-induced interchromosomal interaction using chimeric EST and mRNA data. PLoS One. 2007;2:e254. doi: 10.1371/journal.pone.0000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Cook PR. Similar active genes cluster in specialized transcription factories. J Cell Biol. 2008;181:615–23. doi: 10.1083/jcb.200710053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–7. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]