Abstract
Premature chromatin condensation (PCC) is a hallmark of mammalian cells that begin mitosis before completing DNA replication. This lethal event is prevented by a highly conserved checkpoint involving an unknown, caffeine-sensitive mediator. Here, we have examined the possible involvement of the caffeine-sensitive ATM and ATR protein kinases in this checkpoint. We show that caffeine's ability to inhibit ATR (but not ATM) causes PCC, that ATR (but not ATM) prevents PCC, and that ATR prevents PCC via Chk-1 regulation. Moreover, mimicking cancer cell phenotypes by disrupting normal G1 checkpoints sensitizes cells to PCC by ATR inhibition plus low-dose DNA damage. Notably, loss of p53 function potently sensitizes cells to PCC caused by ATR inhibition by a small molecule. We present a molecular model for how ATR prevents PCC and suggest that ATR represents an attractive therapeutic target for selectively killing cancer cells by premature chromatin condensation.
Progression from one phase of the cell division cycle to the next is controlled by a set of sensors and arresting mechanisms called checkpoints (1, 2). At each checkpoint, the cell determines whether it is ready for progression to the next phase and halts progress if conditions are unfavorable; for example, if nutrients or nucleotides are insufficient or if DNA damage has not been repaired (1, 3). The replication (S/M) checkpoint determines whether DNA replication is complete and prevents the onset of mitosis, specifically the condensation of chromatin, if it is not. Schlegel and Pardee showed in 1986 that caffeine (at millimolar concentration) could “override” the replication checkpoint (4), causing chromosomes to condense despite incomplete DNA replication. However, the details of how caffeine blocks the replication checkpoint have remained unknown.
A family of large protein kinases related in sequence to phosphatidylinositol kinase is involved in sensing various stresses (1, 3). This family of kinases includes ATM (the gene mutated in ataxia telangiectasia), DNA-PK (required for DNA end-joining and antigen receptor gene rearrangement), FRAP (involved in nutrient sensing; modulated by FKBP12-rapamycin), and ATR (so named because it is related to ATM and Rad3) (1, 3, 5). Two lines of evidence suggest ATR as a possible target for caffeine in the replication checkpoint: (i) ATR and ATM were recently shown to be inhibited by caffeine in vitro (6, 7) (ii) and deletion of ATR in mouse leads to embryonic lethality with chromosomal fragmentation in cultured blastocyst cells (8). On the other hand, a prior report concluded that although ATR plays a role in the G2/M checkpoint, it was not involved in preventing the initiation of chromatin condensation if DNA replication is incomplete (9).
Studies to clarify the role of ATR have been hampered by the lack of a viable ATR-deficient animal or a specific chemical inhibitor. We have developed a set of stable cell lines derived from U2OS cells (human osteosarcoma) that are wild-type for p53, have an intact G1 DNA-damage checkpoint, and allow the inducible expression of either wild-type ATR or a dominant negative (kinase-dead) ATR point mutant (ATR-kd) by adding the small molecule doxycycline (see Materials and Methods). Similar to results from cells that are deficient in their p53 response (9), our p53-functional cells were sensitized to DNA damage if the dominant negative form of ATR was expressed. ATR has been shown in vitro and in vivo to phosphorylate p53 on Ser-15 and has been proposed to be upstream of p53 (10). Surprisingly, we found there was no loss of p21 up-regulation or of the p53-mediated G1 checkpoint when ATR function was inhibited, arguing that ATR is required for arrest elsewhere in the cell cycle (unpublished results). Here, we report that (i) ATR is in fact critical in preventing premature chromatin condensation in cells that have undergone DNA damage, (ii) ATR (but not the related caffeine-sensitive kinase ATM) is the relevant target of caffeine in the replication checkpoint, and (iii) several of the molecular hallmarks of cancer (including loss of p53 function) specifically sensitize cells to loss of ATR function.
Materials and Methods
Generation of Cell Lines.
U2OS (human osteosarcoma) cells, verified to have intact p53 and G1 checkpoint function, were used to generate doxycycline-inducible (11) stable cell lines. Amino-terminal FLAG epitope-tagged full-length constructs of ATR-wt or ATR-kd were inserted into the cytomegalovirus (CMV) promoter-based plasmid pcDNA4/TO (Invitrogen), which contains two tetracycline operator binding sites. These constructs were cotransfected with a 20-fold lower amount of pcDNA3.1, which contains the neomycin resistance gene. Beginning 2 days later, G418-resistant clones (400 μg/ml) were selected. Hygromycin was always present at 200 μg/ml to maintain expression of the tetracycline repressor as described (11). Approximately 120 G418-resistant clones were screened by FLAG-immunoprecipitation and FLAG-Western to obtain a pair of clones highly inducible for ATR-wt or ATR-kd and that had undetectable expression of the FLAG-tagged protein in the absence of doxycycline induction (unpublished data).
Mitotic Spreads.
Cells were harvested with trypsin 24 h (or at indicated times) after DNA damage and spun (300 × g for 10 min). All but about 50 μl of supernatant was discarded, and cells were resuspended with a pipettor. One milliliter of 75 mM KCl was added for 10 min at room temperature. Cells were spun, supernatant was discarded, and cells were resuspended in 300 μl of freshly prepared Carnoy's fixative (3 parts methanol, 1 part glacial acetic acid) for 10 min at room temperature. Cells were spun, supernatant was discarded, and cells were resuspended in 100 μl of Carnoy's fixative; 10 μl of this cell suspension was dropped from a height of 10 cm onto a glass slide and allowed to dry. Twelve microliters of DAPI solution (Vectashield with DAPI, Vector Laboratories) was spotted onto the slide, a coverslip was placed above it, and the edges were sealed with clear nail polish. A fluorescence microscope was used to count mitotic cells that had characteristic features of either a normal mitosis or PCC. Interphase cells and cells that were intermediate in morphology between normal and PCC were not counted. The following criteria were used to identify mitoses as PCC or normal. PCC characteristics include well-defined particles of DAPI staining material that were round, not oblong; space between the particles with no hazy chromatin material between particles; no chromatid-like pairs present; and borders of the cell's chromatin are jagged and composed of speckles, not smooth or with a creamy-hazy appearance (all characteristics must be met to be counted). Normal mitosis characteristics include well-formed oblong chromatids present in pairs and at least 20 such chromosome pairs found in a cluster.
Transient Transfection Experiments.
Transient transfection into 293T cells was performed with Fugene 6 (Roche Molecular Biochemicals) according to the manufacturer's specifications. ATR was detected with a rabbit polyclonal antibody we generated that was directed against amino acids 1–20 of ATR. ATM was detected with rabbit polyclonal anti-ATM Ab-3 (Calbiochem).
DNA Damage.
IR was delivered by Cesium-137 irradiation at a rate of 2.5 Gy/min. UV was delivered at a rate of 4 joules/m2 per second from a panel of 4 UV bulbs (8 watts/bulb, RPR-3000, Southern New England Ultraviolet, Hamden, CT), which had peak emission at 312 nm. For UV irradiation, phenol-red containing medium was removed for the 50 s during radiation. A 5-ml Kodacel filter (no. K6808, Eastman Kodak) was used to filter UV <295 nm, which is not encountered in the environment.
Double Thymidine Block.
Cells were plated into normal medium, and after adherence, 2.5 mM thymidine was added for 17 h. Cells were washed twice with PBS and placed in normal thymidine-free medium for 12 h. Thymidine (2.5 mM) was added for a second 17-h period, at which time cells were washed with PBS; this point was designated t = 0, at which 98% of cells were found to be at the G1/S border by propidium iodide–flow cytometry analysis.
Adenovirus Infections.
Adenoviral constructs and virus preparations were performed as described (12). Purified virus was added to the cell medium to a final titer of 1010 viral particles per ml 24 h before UV treatment. Expression constructs were driven by the CMV promoter, and protein expression was verified by Western blotting. Function of each protein was verified by flow cytometry for its expected cell cycle effects. Based on green fluorescent protein expression performed in parallel, this concentration of 1010 viral particles per ml yielded 100% infection and expression in these cells.
Results
ATR Prevents Premature Chromatin Condensation After DNA Damage or Replication Inhibitors.
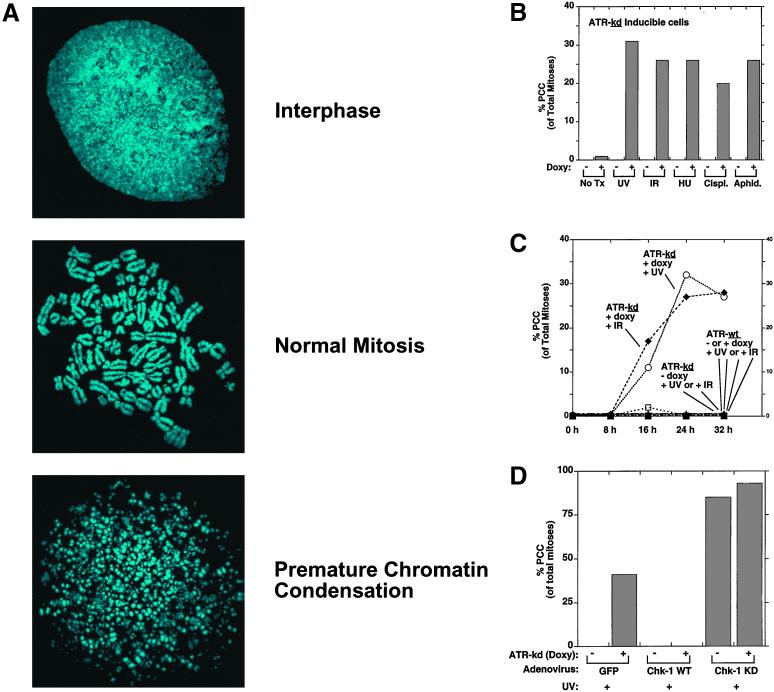
To test for a possible role for ATR in chromosomal integrity, we induced ATR-kd expression and performed mitotic spreads to observe details of chromosomal structure not visible by standard cell fixation and microscopy. No chromosomal fragmentation was seen after induction of ATR-kd alone. However, when hydroxyurea (which prolongs S phase by depleting deoxynucleotides) was added to ATR-kd expressing cells, significant PCC developed that was similar to what had been originally reported by Schlegel and Pardee (4) after treatment with hydroxyurea plus caffeine (Fig. 1 A and B). In addition, aphidicolin (a polymerase-alpha inhibitor) or low doses of DNA damage combined with expression of ATR-kd to cause a significant fraction (≈30%, depending on dose of damage) of mitotic cells to show massive chromosomal fragmentation. The onset of PCC was maximal ≈24 h after treatment of ATR-kd expressing cells (Fig. 1C). No PCC occurred in cells exposed to these treatments if ATR-wt was overexpressed (Fig. 1C). Also, if no doxycycline was added to induce ATR-kd expression, these treatments did not result in PCC (Fig. 1C). Thus, PCC occurred only in cells expressing ATR-kd and treated with DNA damage or an agent that directly causes S phase prolongation.
Figure 1.
Low-dose DNA damage or S phase-arresting agents induce PCC in cells in which ATR or Chk-1 has been inhibited. (A) Mitotic spreads showing three characteristic DNA-staining patterns were performed as described (see Materials and Methods). (B) Doxycycline-induced expression of a full-length kinase-inactive ATR point mutant (Asp-2475 → Ala, ATR-kd) caused asynchronous U2OS cells to undergo PCC by 24 h after treatment with UV (200 J/m2 of UV-B), ionizing radiation (IR, 10 Gy), hydroxyurea (HU, 1 mM), cisplatinum (cisplat., 600 nM), or aphidicolin (aphid., 2.5 μg/ml) (see Materials and Methods). (C) Cells that were not damaged or did not have ATR-kd induced did not undergo PCC. (D) Adenoviral expression of wild-type Chk-1 administered 24 h before UV irradiation completely rescued cells from PCC promoted by ATR-kd. Expression of a kinase-dead (kd) Chk-1 mutant under the same conditions augmented PCC and induced premature mitosis even if ATR-kd was not induced. In the indicated cases, doxycycline (1 mcg/ml) was added 2 days before cells were treated as specified. Nocodazole (100 ng/ml) was added at the time of damage (to block cells from exiting mitosis), and 24 h later (or at the times indicated in C) cells were harvested. The values represent the mean of two to five independent experiments or a representative experiment from within a dose-response curve. For each condition, over 2,000 cells were assessed, usually with >100 mitoses counted per condition. Agreement between experiments and three observers was excellent based on the criteria described (see Materials and Methods) to distinguish PCC and normal mitoses.
Because Chk-1 has been shown to be a target of ATR in Xenopus (13, 14) and mammalian cells (15), we tested whether expression of wild-type Chk-1 could rescue cells from PCC caused by ATR-kd induction. Indeed, overexpressing wild-type Chk-1 completely prevented PCC induced by UV plus ATR-kd, whereas overexpressing a dominant negative (kinase-dead) mutant of Chk-1 promoted PCC in UV-treated cells even if ATR-kd was not induced (Fig. 1D).
ATR (Not ATM) Is the Target of Caffeine in the Replication Checkpoint.
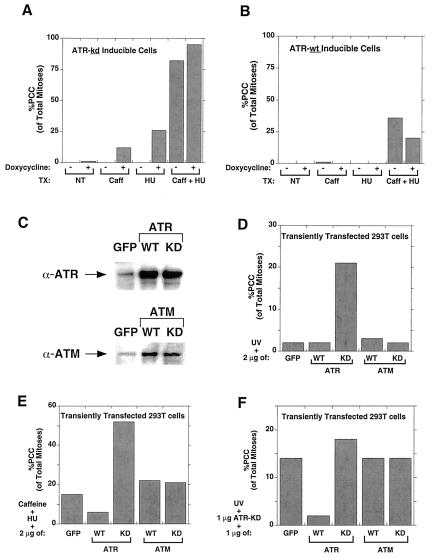
Caffeine inhibits many proteins, including phosphodiesterases and protein kinases (6, 7). Our PCC data led us to test whether ATR is the relevant target of caffeine in the PCC pathway. We tested the effect of expressing wild-type or kinase-dead ATR on PCC induced by treatment with caffeine and hydroxyurea (4). Indeed, we found that inducing ATR-kd augmented the effects of caffeine plus hydroxyurea, whereas ATR-wt overexpression diminished PCC after caffeine and hydroxyurea (Fig. 2 A and B). Moreover, the ability of caffeine to induce PCC together with UV was augmented by overexpression of Chk-1-kd and diminished by Chk-1-wt (data not shown). These results suggest that ATR is the relevant target of caffeine in promoting PCC and that Chk-1 is an essential downstream component of this pathway.
Figure 2.
Involvement of ATR, but not ATM, in PCC. (A) Induction of ATR-kd expression by doxycycline promoted PCC in U2OS cells treated with caffeine (1 mM) and/or hydroxyurea (HU, 1 mM). (B) ATR-wt expression diminished PCC caused by the combination of caffeine and hydroxyurea. (C) 293T cells were transiently transfected (see Materials and Methods), and expression of CMV promoter-driven constructs of ATR or ATM were compared by Western blot to endogenous levels present in control (green fluorescent protein-transfected) cells. (D) Transient transfection in 293T cells of ATR-kd synergized with low-level UV (200 J/m2) to cause PCC, but ATM expression had no effect. (E) Neither ATM-wt nor ATM-kd transfection had any effect on PCC induced by caffeine and hydroxyurea, whereas transfection of the ATR-wt and ATR-kd constructs into 293T cells had effects similar to those shown in A and B in which their expression had been induced in the U2OS cell lines by doxycycline. (F) Analogously, neither ATM-wt nor ATM-kd transfection had any effect on PCC induced by ATR-kd and UV, whereas additional ATR-kd augmented and ATR-wt diminished PCC. Cells were transfected with 2 μg of DNA (1 μg of ATR-kd and 1 μg of the indicated construct) and were treated with UV (200 J/m2) and nocodazole (10 ng/ml) 48 h later, then harvested for mitotic spreads 24 h later (see Materials and Methods).
We have used a kinase-dead ATR construct to study the role of ATR in cellular function because there is no viable animal model or specific small molecule inhibitor of ATR. Recently, Zhou and Elledge (1) raised an important concern with this approach, suggesting that ATR-kd overexpression may inhibit the function of endogenous ATM, and thus the observed effects of ATR-kd could be because of ATM inhibition. Indeed, ATR and ATM are similar in several ways: both are inhibited by millimolar caffeine (6, 7), there are similarities between the substrate specificities of ATR and ATM (16), both kinases phosphorylate p53 on serine 15 in vivo after DNA damage (1), and overexpression of wild-type ATR has been shown to rescue the radio-resistant DNA synthesis characteristic of ATM-deficient cells (9). These data suggest there may be an overlap of ATR and ATM function in certain processes. To clarify this issue, we investigated whether ATM plays a role in PCC and thus whether the ATR-kd construct could be working indirectly by inhibiting ATM function.
Transient transfections in human 293T cells were performed, and the effects of full-length, CMV promoter-driven constructs of ATM and ATR were compared. For both ATM and ATR, expression of recombinant protein was severalfold higher than the endogenous protein level (Fig. 2C). Lim et al. have shown that expression of this ATM-kd construct in this same 293T cell line potently blocks the ability of endogenous ATM to phosphorylate p95/nbs-1, an event that is required for S phase arrest (17). We found that transient transfection of ATR-kd promoted PCC when cells were damaged but that there was no effect of either ATM-kd or ATM-wt expression on PCC after damage (Fig. 2D). Similarly, when testing the ability of these constructs to rescue cells from PCC caused by caffeine and DNA damage, ATR-kd promoted PCC, ATR-wt blocked PCC, and neither ATM construct affected the incidence of PCC (Fig. 2E). Similar results were obtained with these constructs when PCC was caused by ATR-kd and DNA damage (Fig. 2F). These data suggest that ATR is the relevant target of caffeine in causing PCC, that ATM is not required for the checkpoint that prevents PCC, and that the ATR-kd construct causes PCC by inhibiting ATR and not ATM.
ATR Function Is Most Critical During S Phase.
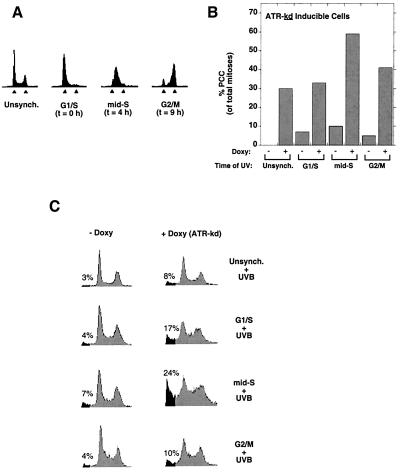
At what point in the cell cycle is ATR function most important in the response to DNA damage? To address this question, we performed cell cycle synchronization experiments by using a double thymidine block protocol that effectively arrested 98% of the cells at the G1/S border (2C DNA content) of the cell cycle (Fig. 3A). Synchronized cells were irradiated at various points in the cell cycle, and 24 h later the extent of PCC was assessed for cells expressing ATR-kd or ATR-wt. Compared with unsynchronized, G1, or M phase cells, S phase cells were most sensitive to irradiation if ATR-kd had been induced before UV treatment (Fig. 3B). Induction of ATR-wt had no effect on the extent of PCC after UV damage in synchronized cells (data not shown). The increased PCC in ATR-kd expressing cells irradiated during S phase corresponded with increased death among these cells 36 h after damage, as demonstrated by the fraction of cells with <2C DNA content (Fig. 3C). Inhibition of ATR function thus promotes death selectively in S phase, a phase disproportionately represented in frequently dividing cells such as cancer cells.
Figure 3.
ATR is required after damage in mid-S phase. (A) U2OS cells were synchronized by using double thymidine block (see Materials and Methods) and harvested for propidium-iodide flow cytometry at the indicated times after release into normal medium. At the time of release (t = 0 h), 98% of cells were at the G1/S border, with progression into G2/M by 9 h later, as indicated. Arrowheads indicate 2C and 4C DNA content. (B) The effect of ATR-kd expression on the incidence of PCC in cells irradiated (UVB 200 J/m2) at different cell cycle phases. Cells irradiated in mid-S phase were most susceptible to undergoing PCC. Cells were harvested 24 h after UV. (C) When ATR-kd was induced, cells irradiated (UVB 200 J/m2) in mid-S phase underwent high levels of cell death, as evidenced by the increase in the fraction of cells with a sub-2C DNA content (shaded cell population, with percent of sub2C DNA content cells indicated).
G1 Checkpoint Deficiency Sensitizes Cells to ATR Inhibition.
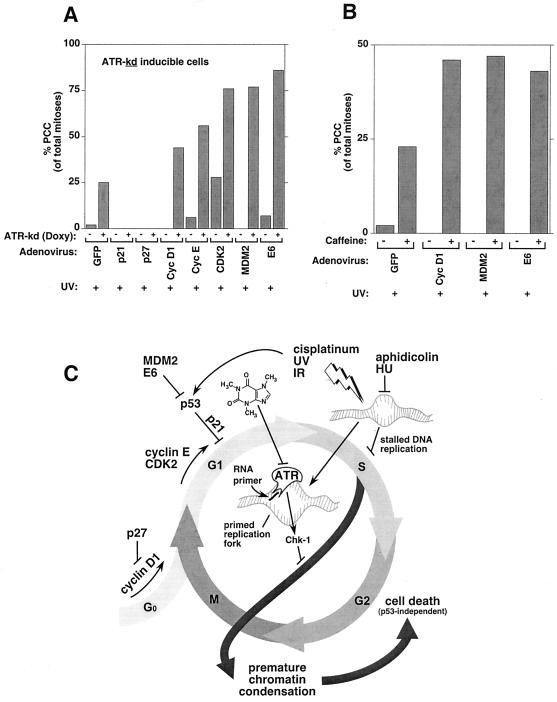
Cancer cells often progress rapidly through the cell cycle with minimal delay in G0 or G1 because of two factors: (i) mutations in the Rb/p16 pathway make cancer cells growth factor-independent and (ii) mutations in the p53 pathway render them unable to arrest in G1 after DNA damage (18). These alterations result in cancer cells spending proportionately more time in S phase and in progressing from G0/G1 into S phase with more DNA damage than normal cells (which would arrest in G1 and repair damage before beginning DNA synthesis). Based on our cell synchronization data, we predicted that loss of the G1 checkpoint would sensitize cells to ATR inhibition. By targeting processes that are often defective in cancer cells, we sought to mimic G1 checkpoint deficiencies in our U2OS cell lines (which have a functional G1 arrest pathway following DNA damage and an impaired Rb/p16 pathway because of p16 deletion). To inhibit the Rb pathway further, we overexpressed cyclin D1, CDK2, or cyclin E, each of which promotes G1–S progression and are commonly up-regulated in cancer. In cells expressing ATR-wt, adenoviral expression of cyclin E or CDK2 followed by damage caused no PCC (data not shown). In cells expressing ATR-kd, however, adenovirus-mediated expression of cyclin D1, cyclin E, or CDK2 followed by damage markedly promoted PCC (Fig. 4A). Because our ATR-inducible U2OS cell lines have an intact p53 pathway, we were able to assess the role of p53 in preventing PCC by ATR, an experiment that would not have been possible in any other system known to us. Importantly, targeted loss of p53 function by either of two mechanisms that are relevant in cancer (expression of MDM2 or human papilloma virus E6) roughly tripled the amount of PCC in UV-damaged cells in which ATR-kd was expressed, with no PCC in identically treated cells that did not express ATR-kd (Fig. 4A). This pronounced effect of MDM2 or E6 in cells already expressing ATR-kd suggests that p53 is active in these cells and thus that ATR is not required for p53 activation. In contrast, artificially augmenting G1 arrest by expressing p21 or p27 completely prevented PCC (Fig. 4A).
Figure 4.
Loss of G1 arrest promotes PCC induced by ATR inhibition and treatments that prolong DNA synthesis. (A) Promotion of G1 checkpoint function by overexpressing p21 or p27 completely rescued PCC induced by ATR inhibition and UV, compared with control adenoviral expression of green fluorescent protein (GFP). Interference with G1 checkpoint function via adenoviral expression of cyclin D1, cyclin E, CDK2, MDM2, or E6 markedly increased susceptibility to PCC following UV exposure and ATR-kd induction. (B) Similarly, a small molecule inhibitor of ATR, caffeine (here used at 300 μM), sensitized cells with G1 checkpoint deficiencies to PCC induction by UV. Values shown are means of three independent experiments, and standard errors of the mean were <5% in all cases. (C) A model of the role of ATR in preventing PCC. UV, IR, cisplatinum, hydroxyurea, and aphidicolin have in common the ability to increase the number of stalled replication forks and prolong DNA synthesis. The signal that DNA synthesis is not complete (and that chromatin condensation should not start) is sent by ATR after it is recruited to replication forks that have been primed by addition of an RNA primer via polymerase-α (13, 19). ATR-mediated phosphorylation of Chk-1 causes delay of mitotic entry and prevention of PCC. G1 checkpoints are lost in cancer cells by overexpression of cyclin D1, CDK2, cyclin E, MDM2, or E6 because of mutations in the p53 or Rb/p16 pathways. Because G1 checkpoint loss causes cells to enter S phase with more unrepaired damage, such cells would have a greater requirement for functional ATR to delay mitotic entry until all damage has been repaired and the DNA has been replicated. The chemical structure of caffeine, a small molecule inhibitor of ATR, is depicted. An animated interactive model for ATR function is available as Movie 1, which is published as supplemental data on the PNAS web site, www.pnas.org.
To evaluate whether a small molecule inhibitor of ATR could also selectively induce PCC in G1 checkpoint-deficient cells, we expressed cyclin D1, MDM2, or E6 and assessed the effect of caffeine on PCC. Indeed, ATR inhibition via a small molecule also markedly augmented PCC selectively in cells made deficient in G1 checkpoints (Rb/p16 or p53) by each of these perturbations. These results demonstrate that DNA-damaged cells that no longer arrest at G1 before S phase entry are especially sensitive to loss of the replication checkpoint that requires ATR.
Discussion
Our data demonstrate that ATR functions to prevent premature chromatin condensation after DNA damage or replication inhibitors. This finding is in fact in disagreement with a prior report (9) that concluded “Therefore, in ATR-kd expressing cells, chromosome condensation does not occur in the absence of genome replication.” The most likely explanation for this prior conclusion is that the techniques used in that study were not well suited to visualize chromatin structure and that cells undergoing PCC were deleted from analysis.
A model of the role of ATR in the cell division cycle (Fig. 4C) emerges if our data are combined with recent studies on the Xenopus replication checkpoint (19), on ATR-chromatin binding in Xenopus (13, 14), and on the subcellular localization of ATR after damage in human cells (20). In our studies, ATR inhibition by caffeine or ATR-kd expression promoted PCC when combined with diverse cellular stresses including hydroxyurea (deoxynucleotide synthesis inhibitor), aphidicolin (polymerase-alpha inhibitor), or DNA damage via radiation or cisplatinum. We believe the essential feature common to these agents is the prolongation of S phase and a resulting increase in the number of stalled replication forks present in a given cell. These replication forks are then modified by the addition of an RNA primer that has been shown to have two effects: (i) RNA priming allows the replication checkpoint to be activated (19) and (ii) it promotes binding of ATR to the replication fork (13). ATR activity may then increase because ATR-DNA binding has been shown in vitro to increase ATR activity about 3-fold (6, 21), and ATR appears to colocalize on damage with at least one relevant substrate, BRCA-1, promoting their interaction by proximity (20).
Intriguingly, ATR appears to recognize and bind only “activated” replication forks that have already been primed with RNA (13, 19). The latter finding suggests that a common feature of the phosphatidylinositol kinase-related kinase family may be the recognition of specifically modified ends of nucleic acids (22). For example, DNA-PK has been shown to bind and be activated in a manner dependent on the length of single-stranded DNA at the end of a double-stranded segment (23). Likewise, the family members FRAP and the Tor proteins, which are key elements of the nutrient-sensing network (24), may function by recognizing the ends of tRNA molecules (22).
Frequent cell division and defective G1 checkpoints are hallmarks of cancer cells. We have shown that these features sensitize cells to dying after ATR inhibition and propose the following model as to why this occurs. G1 checkpoint deficiencies result in cancer cells spending a greater proportion of time in S phase and, if damaged, having a greater number of stalled replication forks relative to cells with intact G1 checkpoints. In particular, the loss of p53 function is a common feature of cancer cells that causes increased resistance to radiation or chemotherapy through loss of p53-dependent apoptosis and damage-induced anti-growth signals. Our results with ATR inhibition in cells made p53-deficient (by MDM2 or E6 expression) are therefore striking as p53 loss markedly sensitizes cells to lethal PCC (Fig. 4 A and B). This effect of p53 inhibition in ATR-kd expressing cells demonstrates that p53 is activated by UV in these cells and thus that ATR function is not required for p53 activation. The cell death that follows PCC has been shown to be independent of p53 function (8) (deletion of p53 in mouse did not rescue the ATR−/− phenotype), making this an appealing pathway for use in killing p53-deficient cancer cells. The mechanism of this cell death may involve the forces of microtubule action at kinetochore attachment sites that tear apart partially replicated, prematurely condensed chromosomes irreparably (25).
Our data show that features common to cancer cells make them more sensitive than normal cells to inhibition of ATR function as their increased number of stalled replication forks requires an increased supply of functional ATR (Fig. 4C). Here, we have identified that ATR is the relevant target of caffeine in causing PCC and suggest that a more potent and selective ATR inhibitor than caffeine is therefore an important goal in cancer research. Caffeine has been shown in vitro to selectively radiosensitize cancer cells (26) and, in fact, has been investigated in humans with incurable cancer. Unfortunately, because of its lack of specificity, caffeine causes seizures in humans at doses less than are required for inhibition of the replication checkpoint (27). A further implication of these studies is that an ideal ATR inhibitor would not affect ATM function, as this would have the undesirable effect of diminishing ATM-dependent p53/G1 arrest in normal cells, making them more sensitive to ATR inhibition. An ATR inhibitor may selectively sensitize cancer cells both by exploiting traits intrinsic to the malignant phenotype and by enhancing extrinsic treatments such as low-dose DNA damage, causing cancer cells to die by prematurely condensing their chromatin.
Supplementary Material
Acknowledgments
We thank members of the Schreiber group for valuable discussions and assistance. Full-length ATM-wt and ATM-kd constructs were a gift from M. Kastan. P.N. is supported by National Institutes of Health Grant K08-AR0208703, and this research was supported by GM-52067. P.K.P. was supported by the Harvard College Research Program. Y.S.K. is supported by the Mildred Scheel Cancer Foundation Fellowship. S.L.S. is an Investigator at the Howard Hughes Medical Institute.
Abbreviations
- PCC
premature chromatin condensation
- CMV
cytomegalovirus
References
- 1.Zhou B B, Elledge S J. Nature (London) 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 2.Weinert T A, Kiser G L, Hartwell L H. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 3.Keith C T, Schreiber S L. Science. 1995;270:50–51. doi: 10.1126/science.270.5233.50. [DOI] [PubMed] [Google Scholar]
- 4.Schlegel R, Pardee A B. Science. 1986;232:1264–1266. doi: 10.1126/science.2422760. [DOI] [PubMed] [Google Scholar]
- 5.Cimprich K A, Shin T B, Keith C T, Schreiber S L. Proc Natl Acad Sci USA. 1996;93:2850–2855. doi: 10.1073/pnas.93.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall-Jackson C A, Cross D A, Morrice N, Smythe C. Oncogene. 1999;18:6707–6713. doi: 10.1038/sj.onc.1203077. [DOI] [PubMed] [Google Scholar]
- 7.Sarkaria J N, Busby E C, Tibbetts R S, Roos P, Taya Y, Karnitz L M, Abraham R T. Cancer Res. 1999;59:4375–4382. [PubMed] [Google Scholar]
- 8.Brown E J, Baltimore D. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 9.Cliby W A, Roberts C J, Cimprich K A, Stringer C M, Lamb J R, Schreiber S L, Friend S H. EMBO J. 1998;17:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tibbetts R S, Brumbaugh K M, Williams J M, Sarkaria J N, Cliby W A, Shieh S Y, Taya Y, Prives C, Abraham R T. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao F, Svensjo T, Winkler T, Lu M, Eriksson C, Eriksson E. Hum Gene Ther. 1998;9:1939–1950. doi: 10.1089/hum.1998.9.13-1939. [DOI] [PubMed] [Google Scholar]
- 12.Guo N, Faller D V, Vaziri C. J Biol Chem. 2000;275:1715–1722. doi: 10.1074/jbc.275.3.1715. [DOI] [PubMed] [Google Scholar]
- 13.Hekmat-Nejad M, You Z, Yee M, Newport J W, Cimprich K A. Curr Biol. 2000;10:1565–1573. doi: 10.1016/s0960-9822(00)00855-1. [DOI] [PubMed] [Google Scholar]
- 14.Guo Z, Kumagai A, Wang S X, Dunphy W G. Genes Dev. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q, Guntuku S, Cui X S, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, et al. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S T, Lim D S, Canman C E, Kastan M B. J Biol Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- 17.Lim D S, Kim S T, Xu B, Maser R S, Lin J, Petrini J H, Kastan M B. Nature (London) 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan D, Weinberg R A. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 19.Michael W M, Ott R, Fanning E, Newport J. Science. 2000;289:2133–2137. doi: 10.1126/science.289.5487.2133. [DOI] [PubMed] [Google Scholar]
- 20.Tibbetts R S, Cortez D, Brumbaugh K M, Scully R, Livingston D, Elledge S J, Abraham R T. Genes Dev. 2000;14:2989–3002. doi: 10.1101/gad.851000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lakin N D, Hann B C, Jackson S P. Oncogene. 1999;18:3989–3995. doi: 10.1038/sj.onc.1202973. [DOI] [PubMed] [Google Scholar]
- 22.Kuruvilla F G, Schreiber S L. Chem Biol. 1999;6:R129–R136. doi: 10.1016/S1074-5521(99)80070-2. [DOI] [PubMed] [Google Scholar]
- 23.Leuther K K, Hammarsten O, Kornberg R D, Chu G. EMBO J. 1999;18:1114–1123. doi: 10.1093/emboj/18.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shamji A F, Kuruvilla F G, Schreiber S L. Curr Biol. 2000;10:1574–1581. doi: 10.1016/s0960-9822(00)00866-6. [DOI] [PubMed] [Google Scholar]
- 25.Brinkley B R, Zinkowski R P, Mollon W L, Davis F M, Pisegna M A, Pershouse M, Rao P N. Nature (London) 1988;336:251–254. doi: 10.1038/336251a0. [DOI] [PubMed] [Google Scholar]
- 26.Russell K J, Wiens L W, Demers G W, Galloway D A, Plon S E, Groudine M. Cancer Res. 1995;55:1639–1642. [PubMed] [Google Scholar]
- 27.Stewart D J, Hugenholtz H, DaSilva V, Benoit B, Richard M, Russell N, Maroun J, Verma S. Semin Oncol. 1987;14:110–115. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.