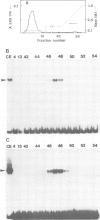
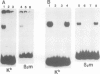
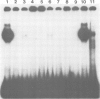
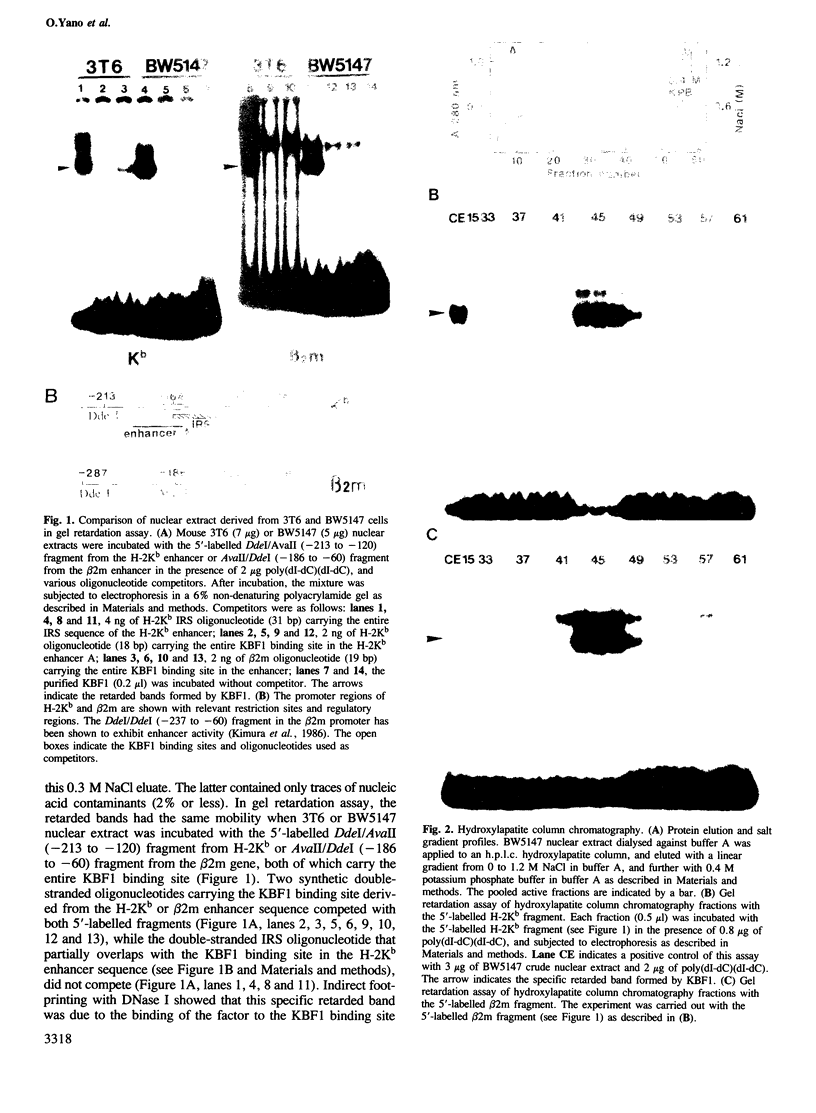
Abstract
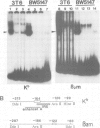
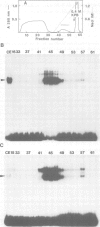
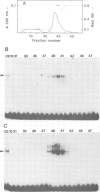
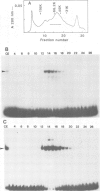
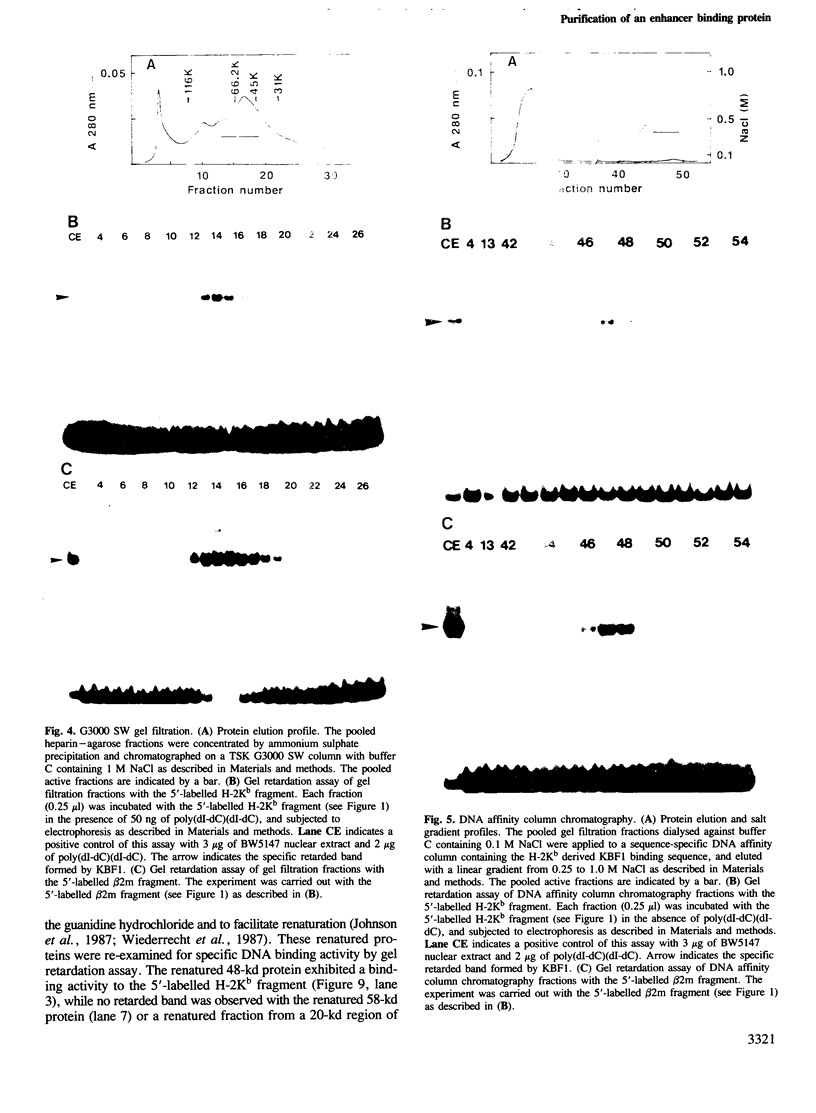
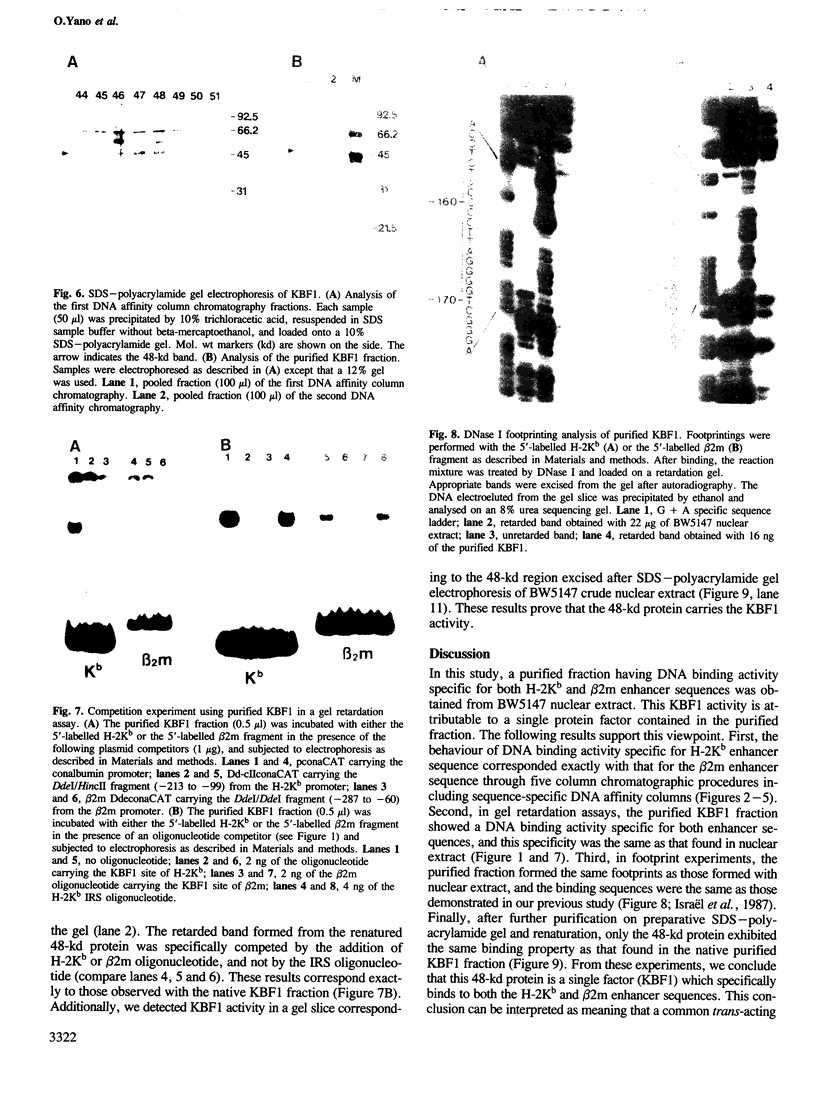
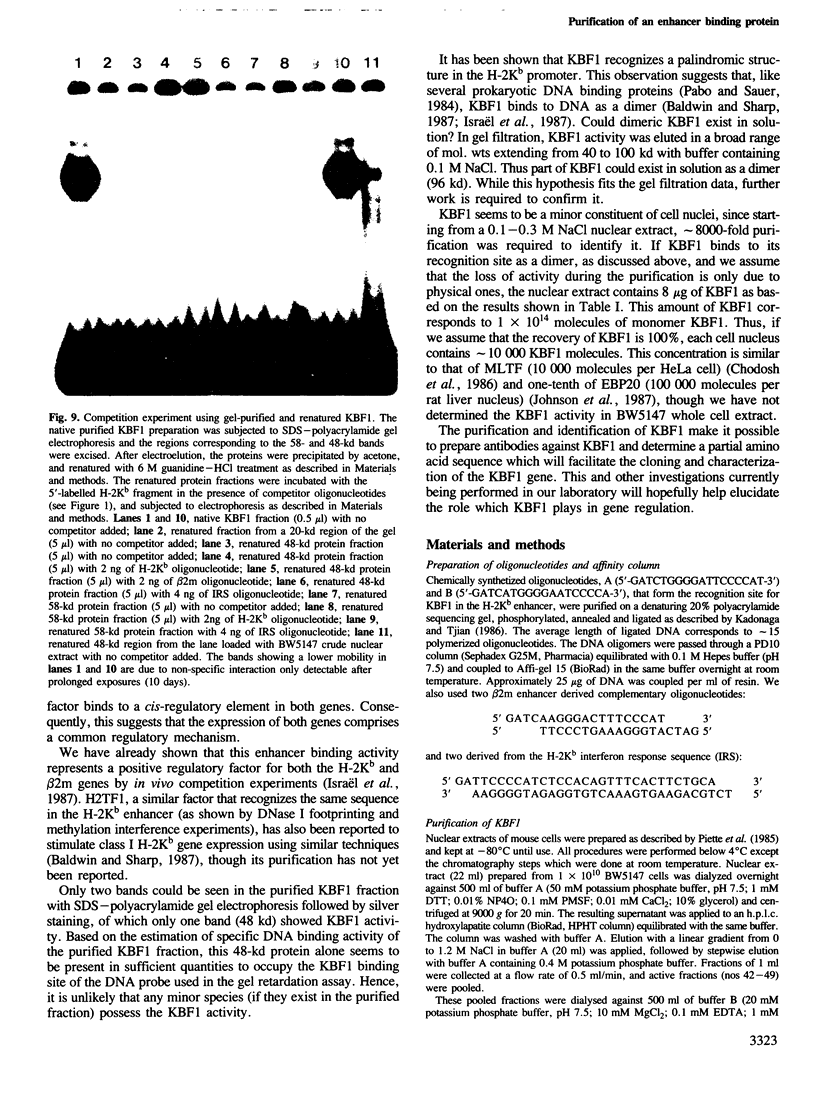
An enhancer binding factor, designated KBF1, has been purified from the nuclear extract of mouse BW5147 thymoma cells by five column chromatography steps including a sequence-specific DNA affinity column. Gel retardation and footprint analysis have shown that purified KBF1 has a binding activity specific for both H-2 and beta 2-microglobulin enhancer sequences. After SDS-polyacrylamide gel electrophoresis of the most purified preparation a 48-kd protein showed, after elution and renaturation, a binding activity to both enhancer sequences. These findings suggest that the expression of both H-2 and beta 2-microglobulin genes utilizes a common regulatory mechanism.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arcangioli B., Lescure B. Identification of proteins involved in the regulation of yeast iso- 1-cytochrome C expression by oxygen. EMBO J. 1985 Oct;4(10):2627–2633. doi: 10.1002/j.1460-2075.1985.tb03980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin A. S., Jr, Sharp P. A. Binding of a nuclear factor to a regulatory sequence in the promoter of the mouse H-2Kb class I major histocompatibility gene. Mol Cell Biol. 1987 Jan;7(1):305–313. doi: 10.1128/mcb.7.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs M. R., Kadonaga J. T., Bell S. P., Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986 Oct 3;234(4772):47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- Buc-Caron M. H., Condamine H., Jacob F. The presence of F9 antigen on the surface of mouse embryonic cells until day 8 of embryogenesis. J Embryol Exp Morphol. 1978 Oct;47:149–160. [PubMed] [Google Scholar]
- Camier S., Gabrielsen O., Baker R., Sentenac A. A split binding site for transcription factor tau on the tRNA3Glu gene. EMBO J. 1985 Feb;4(2):491–500. doi: 10.1002/j.1460-2075.1985.tb03655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodosh L. A., Carthew R. W., Sharp P. A. A single polypeptide possesses the binding and transcription activities of the adenovirus major late transcription factor. Mol Cell Biol. 1986 Dec;6(12):4723–4733. doi: 10.1128/mcb.6.12.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen R. B., Sheffery M., Kim C. G. Partial purification of a nuclear protein that binds to the CCAAT box of the mouse alpha 1-globin gene. Mol Cell Biol. 1986 Mar;6(3):821–832. doi: 10.1128/mcb.6.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coligan J. E., Kindt T. J., Uehara H., Martinko J., Nathenson S. G. Primary structure of a murine transplantation antigen. Nature. 1981 May 7;291(5810):35–39. doi: 10.1038/291035a0. [DOI] [PubMed] [Google Scholar]
- Croce C. M., Linnenbach A., Huebner K., Parnes J. R., Margulies D. H., Appella E., Seidman J. G. Control of expression of histocompatibility antigens (H-2) and beta 2-microglobulin in F9 teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5754–5758. doi: 10.1073/pnas.78.9.5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell R. A., Allen H., Burkly L. C., Sherman D. H., Waneck G. L., Widera G. Molecular biology of the H-2 histocompatibility complex. Science. 1986 Jul 25;233(4762):437–443. doi: 10.1126/science.3726537. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager D. A., Burgess R. R. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal Biochem. 1980 Nov 15;109(1):76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- Hood L., Steinmetz M., Malissen B. Genes of the major histocompatibility complex of the mouse. Annu Rev Immunol. 1983;1:529–568. doi: 10.1146/annurev.iy.01.040183.002525. [DOI] [PubMed] [Google Scholar]
- Israel A., Kimura A., Fournier A., Fellous M., Kourilsky P. Interferon response sequence potentiates activity of an enhancer in the promoter region of a mouse H-2 gene. Nature. 1986 Aug 21;322(6081):743–746. doi: 10.1038/322743a0. [DOI] [PubMed] [Google Scholar]
- Israël A., Kimura A., Kieran M., Yano O., Kanellopoulos J., Le Bail O., Kourilsky P. A common positive trans-acting factor binds to enhancer sequences in the promoters of mouse H-2 and beta 2-microglobulin genes. Proc Natl Acad Sci U S A. 1987 May;84(9):2653–2657. doi: 10.1073/pnas.84.9.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. F., Landschulz W. H., Graves B. J., McKnight S. L. Identification of a rat liver nuclear protein that binds to the enhancer core element of three animal viruses. Genes Dev. 1987 Apr;1(2):133–146. doi: 10.1101/gad.1.2.133. [DOI] [PubMed] [Google Scholar]
- Jones K. A., Kadonaga J. T., Rosenfeld P. J., Kelly T. J., Tjian R. A cellular DNA-binding protein that activates eukaryotic transcription and DNA replication. Cell. 1987 Jan 16;48(1):79–89. doi: 10.1016/0092-8674(87)90358-8. [DOI] [PubMed] [Google Scholar]
- Kadonaga J. T., Tjian R. Affinity purification of sequence-specific DNA binding proteins. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A., Israël A., Le Bail O., Kourilsky P. Detailed analysis of the mouse H-2Kb promoter: enhancer-like sequences and their role in the regulation of class I gene expression. Cell. 1986 Jan 31;44(2):261–272. doi: 10.1016/0092-8674(86)90760-9. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Morello D., Daniel F., Baldacci P., Cayre Y., Gachelin G., Kourilsky P. Absence of significant H-2 and beta 2-microglobulin mRNA expression by mouse embryonal carcinoma cells. Nature. 1982 Mar 18;296(5854):260–262. doi: 10.1038/296260a0. [DOI] [PubMed] [Google Scholar]
- Morello D., Duprey P., Israel A., Babinet C. Asynchronous regulation of mouse H-2D and beta-2 microglobulin RNA transcripts. Immunogenetics. 1985;22(5):441–452. doi: 10.1007/BF00418090. [DOI] [PubMed] [Google Scholar]
- Morello D., Gachelin G., Dubois P., Tanigaki N., Pressman D., Jacob F. Absence of reaction of a xenogenic anti-H-2 serum with mouse embryonal carcinoma cells. Transplantation. 1978 Aug;26(2):119–125. doi: 10.1097/00007890-197808000-00012. [DOI] [PubMed] [Google Scholar]
- Ozato K., Wan Y. J., Orrison B. M. Mouse major histocompatibility class I gene expression begins at midsomite stage and is inducible in earlier-stage embryos by interferon. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2427–2431. doi: 10.1073/pnas.82.8.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Piette J., Kryszke M. H., Yaniv M. Specific interaction of cellular factors with the B enhancer of polyoma virus. EMBO J. 1985 Oct;4(10):2675–2685. doi: 10.1002/j.1460-2075.1985.tb03987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa F., Le Bouteiller P. P., Abadie A., Mishal Z., Lemonnier F. A., Bourrel D., Lamotte M., Kalil J., Jordan B., Fellous M. HLA class I genes integrated into murine cells are inducible by interferon. Eur J Immunol. 1983 Jun;13(6):495–499. doi: 10.1002/eji.1830130612. [DOI] [PubMed] [Google Scholar]
- Rosenfeld P. J., Kelly T. J. Purification of nuclear factor I by DNA recognition site affinity chromatography. J Biol Chem. 1986 Jan 25;261(3):1398–1408. [PubMed] [Google Scholar]
- Simon R. H., Felsenfeld G. A new procedure for purifying histone pairs H2A + H2B and H3 + H4 from chromatin using hydroxylapatite. Nucleic Acids Res. 1979 Feb;6(2):689–696. doi: 10.1093/nar/6.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederrecht G., Shuey D. J., Kibbe W. A., Parker C. S. The Saccharomyces and Drosophila heat shock transcription factors are identical in size and DNA binding properties. Cell. 1987 Feb 13;48(3):507–515. doi: 10.1016/0092-8674(87)90201-7. [DOI] [PubMed] [Google Scholar]
- Zinkernagel R. M., Doherty P. C. MHC-restricted cytotoxic T cells: studies on the biological role of polymorphic major transplantation antigens determining T-cell restriction-specificity, function, and responsiveness. Adv Immunol. 1979;27:51–177. doi: 10.1016/s0065-2776(08)60262-x. [DOI] [PubMed] [Google Scholar]