Abstract
On phosphorylation of Cbl, the c-Cbl-associated protein (CAP)/Cbl complex dissociates from the insulin receptor and translocates to a lipid raft membrane fraction to form a ternary complex with flotillin. Deletion analyses of the CAP gene identified a 115-aa region responsible for flotillin binding. This region is homologous to the peptide sorbin and is referred to as the sorbin homology (SoHo) domain. This domain is present in two other proteins, vinexin and ArgBP2. Vinexin also interacted with flotillin, and deletion of its SoHo domain similarly blocked flotillin binding. The overexpression of a CAP mutant in which the SoHo domain had been deleted (CAPΔSoHo) prevented the translocation of Cbl to lipid rafts and subsequently blocked the recruitment of CrkII and C3G. Moreover, overexpression of CAPΔSoHo prevented the stimulation of glucose transport and GLUT4 translocation by insulin. These results suggest a mechanism for localization of signaling proteins to the lipid raft that mediates the compartmentalization of crucial signal transduction pathways.
Lipid rafts are microdomains of the plasma membrane enriched in cholesterol and sphingolipids (1, 2). These regions concentrate certain signaling molecules (3, 4), including heterotrimeric and small G proteins (5–7), Src-family tyrosine kinases (8, 9), endothelial nitric oxide synthase (10, 11), G-protein-coupled receptors (12), and certain tyrosine kinase receptors (13). This concentration of signaling molecules suggests that these microdomains might function as a site for compartmentalization of signaling events. We observed that insulin stimulated the tyrosine phosphorylation of c-Cbl (14) and its subsequent translocation to a caveolin-enriched, lipid raft membrane fraction (15, 16). The c-Cbl-associated protein (CAP)/Cbl complex interacts with the insulin receptor and dissociates after insulin stimulation, subsequently migrating to rafts because of the interaction of CAP with the lipid raft-associated protein flotillin (17–19). Moreover, the localization of the Cbl/CAP complex to this microdomain recruits the guanyl nucleotide exchange protein C3G, resulting in the activation of the G protein TC10. This activation of TC10 generates a PI 3-kinase-independent signal that is crucial to the regulation of glucose uptake by insulin (20, 21).
CAP is a multifunctional adapter protein with three SH3 domains in its C terminus and a region of homology to the gut peptide sorbin in its N terminus. The overall sorbin homology (SoHo)/SH3 organization of CAP is also found in other proteins, including vinexinα and ArgBP2A. The similar organization of these proteins suggests that they may similarly function as adapters, linking signaling or cytoskeletal proteins to the lipid raft. We demonstrate here that the SoHo domain does indeed mediate the interaction of CAP and vinexin with flotillin. Moreover, this interaction is crucial for the localization of SH3-binding proteins such as Cbl to the lipid raft and propagation of the downstream signal. These data suggest a signaling paradigm in which the SoHo domain of these adapter proteins can mediate interactions with the lipid raft that are crucial to intracellular communication.
Materials and Methods
Expression Plasmids and Antibodies.
FLAG epitope-tagged CAP and CAPΔSH3 expression constructs were made as described (15, 17). A FLAG epitope-tagged CAPΔSoHo (deletion of amino acid residues 219 to 263) in a eukaryotic expression vector was constructed as follows. The cDNA of the amino-terminal region of CAPΔSoHo was amplified by PCR with CAP cDNA in pCI-neo (15) as the template. The 5′ primer was designed to have an EcoRI restriction site followed by coding sequences for a FLAG epitope in frame and the start codon. The 3′ primer was designed to have a XbaI restriction site followed by the codon for amino acid residue 219. The 5′ primer of the carboxyl-terminal region of CAPΔSoHo was designed to have an XbaI restriction site followed by the codon for amino acid residue 263. The 3′ primer was designed to have a NotI restriction site followed by the stop codon for amino acid residue 685. These PCR products were cloned into pCR2.1-TOPO (Invitrogen); digested with EcoRI and XbaI, and XbaI and NotI, respectively; and ligated into pCI-neo digested with EcoRI and NotI. FLAG epitope-tagged vinexinα, vinexinαΔSoHo, and vinexinα ΔSH3 in a eukaryotic expression vector were constructed as follows. The cDNA of vinexinα was amplified by PCR with a 3T3-L1 adipocyte cDNA library as the template. The primer 5′-ATCGAATTCAGCATGGACTACAAGGACGACGATGATAAGGCATGGTATCAGACCTGGCCAGGCCCT-3′ for vinexinα and vinexinαΔSH3, 5′-ATCGAATTCAGCATGGACTACAAGCAGGTGCCTAGACATCGAGAGAAAGTA-3′ for vinexinαΔSoHo were designed to have a EcoRI restriction site followed by a coding sequence for a FLAG epitope in frame with amino acid 125 or 286, respectively. The primers 5′-ATCGCGGCCGCTCACACTGGGGCTACATAATTTCCAGGGAATGT-3′ for vinexinα and vinexinΔSoHo and 5′-ATCGCGGCCGCTTACAGTTCTCTCTCTAGCAGGACCTCAAT-3′ for vinexinΔSH3 correspond to amino acids 724 to 734 and 330 to 340, respectively.
Cell Culture.
Human embryo kidney (HEK) 293T cells were maintained in DMEM containing 10% FBS at 37°C with 5% CO2. 3T3-L1 adipocytes were cultured in DMEM containing 25 mM glucose and 10% calf serum. Confluent cultures were induced to differentiate into adipocytes as described (22). HEK293T cells were transfected with a Lipofectamine Plus transfection kit as described by the manufacturer (Life Technologies, Rockville, MD). 3T3-L1 adipocytes were transfected by electroporation as described (23).
In Vitro Protein Associations.
FLAG epitope-tagged CAP, CAPΔSoHo, and CAPΔSH3 expression constructs were overexpressed into HEK293T cells. Whole-cell detergent extracts from HEK293T cells were prepared by HNTG buffer containing 50 mM Hepes (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10% glycerol, 1% Triton X-100, 1 mM Na3VO4, 100 mM NaF, 1 mM benzamidine and phenylmethylsulfonylfluoride, and 10 μg/ml of aprotinin and leupeptin. Cell lysates were incubated with either glutathione S-transferase (GST) alone or with GST-flotillin immobilized on glutathione-agarose beads for 1 h at 4°C. The beads were washed extensively three times with 1 ml of HNTG buffer, and the samples were then resuspended in Laemmli sample buffer, heated for 5 min at 100°C, separated by SDS/PAGE, and immunoblotted with FLAG M2 monoclonal antibody (Stratagene).
Immunoprecipitations and Immunoblotting.
After insulin treatment, electroporated adipocytes were washed with PBS and lysed in MES buffer containing protease inhibitors and sodium orthovanadate. The Triton-insoluble pellet fraction was generated as described (14). Lysates and immunoprecipitations were resolved by SDS/PAGE and transferred to nitrocellulose for immunoblot analysis. Immunoprecipitations and immunoblots were performed with the use of polyclonal c-Cbl antisera (Santa Cruz Biotechnology), monoclonal 4G10 anti-phosphotyrosine antibodies (Upstate Biotechnology, Lake Placid, NY), monoclonal phospho-Akt and phospho-mitogen-activated protein kinase antibodies (New England Biolabs), polyclonal C3G antibodies (Santa Cruz Biotechnology), and monoclonal caveolin I antibodies (Transduction Laboratories, Lexington, KY). We used horseradish peroxidase-conjugated secondary antibodies (Bio-Rad) and enhanced chemiluminescence (Amersham Pharmacia) for immunoblot detection.
Metabolic Assays.
Determination of 2-deoxyglucose uptake was performed as described (24). All enhanced green fluorescence protein (EGFP) fusion proteins and translocation assays were constructed by the method of Thurmond et al. (25).
Results
Identification of the SoHo Domain, a Region That Directs Interaction of CAP with the Lipid Raft Protein Flotillin.
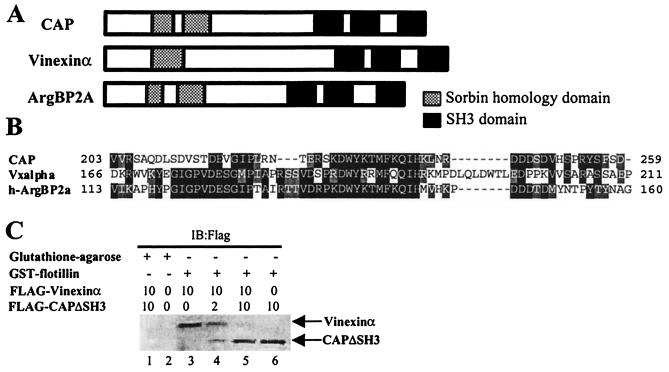
CAP is a bifunctional adapter protein with three SH3 domains and a region of similarity to the peptide sorbin. A similar overall organization is found in two other proteins, vinexinα and ArgBP2 (Fig. 1A). Fig. 1B shows the amino acid alignment in the first SoHo domain of the three proteins. To determine whether a similar region in these proteins might direct their interaction with flotillin, we cloned flotillin into a bacterial vector for expression as a GST fusion protein and assayed interaction by precipitation (Fig. 1C). As was described for CAP (17), full-length FLAG epitope-tagged vinexinα was quantitatively precipitated with the immobilized GST-flotillin fusion protein, but not with control beads. This interaction was blocked by addition to the assay of a fusion protein lacking the carboxyl-terminal sequences of CAP (CAPΔSH3). Thus, vinexinα can interact directly with flotillin and can be displaced with CAP, suggesting that the two proteins interact with flotillin in a similar manner.
Figure 1.
Structure of the sorbin homology (SoHo) family of adapter proteins. (A) Schematic description of mouse CAP, mouse vinexinα, and human ArgBP2A. The sorbin homology domain (divided into two at CAP and ArgBP2A; shaded) and the SH3 domains (SH3A, SH3B, and SH3C; solid) are represented. (B) Sequence alignments of SoHo domains. Identity and similarity are highlighted. (C) Various amounts of FLAG-vinexinα or FLAG-CAPΔSH3 overexpressing lysates from HEK293T cells were premixed before the addition of glutathione-agarose beads or GST-flotillin for 1 h at 4°C. The samples were then washed, resuspended in Laemmli sample buffer, and subjected to SDS/PAGE followed by immunoblotting with an anti-FLAG antibody.
To identify the precise regions in CAP or vinexin that mediate their interaction with flotillin, we designed a series of FLAG epitope-tagged deletion mutants of both proteins (Fig. 2A). We expressed in HEK293T cells full-length CAP, a mutant in which sorbin homology domain 1 was deleted (CAPΔSoHo), a mutant that lacks all three SH3 domains (CAPΔSH3), and a mutant that contains only the first sorbin homology domain in residues 152–269 (CAPSoHo). We also constructed FLAG epitope-tagged mutants of vinexinα, including full-length vinexinα (Vxα); VxΔSoHo, which lacked the N-terminal sorbin homology domain of vinexinα; and VxΔSH3, which lacked the C terminus of vinexinα. Immunoblotting of whole-cell detergent extracts demonstrated similar levels of expression of all mutants (Fig. 2B). To determine whether these constructs bound to GST-flotillin, the immobilized fusion protein was incubated with lysates, and the precipitates were immunoblotted with anti-FLAG antibodies. Whereas GST-flotillin bound to FLAG-CAP, FLAG-CAPΔSH3, and FLAG-CAPSoHo, it did not interact with FLAG-CAPΔSoHo. Moreover, GST-flotillin precipitated FLAG-Vxα and FLAG-VxΔSH3 but did not interact with FLAG-VxΔSoHo. These data indicate that the interaction of flotillin with both CAP and vinexinα requires the region of homology to sorbin. Moreover, the direct interaction of the isolated SoHo domain of CAP with flotillin suggests that this domain is both necessary and sufficient for this interaction and thus represents a bonafide protein interaction domain.
Figure 2.
The SoHo domain is necessary and sufficient for flotillin binding. (A) Schematic diagram of FLAG-epitope-tagged expression constructs of CAP, CAP mutants, vinexinα, vinexinα mutants, and chimeras of CAP-vinexinα. (B) LacZ, FLAG-epitope-tagged CAP, CAPΔSoHo, CAPΔSH3 CAPSoHo, Vxα, VxΔSoHo and VxΔSH3 were overexpressed in HEK293T cells. Lysates were subject to SDS/PAGE and immunoblotted with monoclonal FLAG antibody to evaluate expression level (Left, arrowheads). Lysates were incubated with glutathione-agarose-bound GST-flotillin for 1 h at 4°C. Precipitates were subjected to SDS/PAGE and blotted with monoclonal FLAG antibody or with Anti-Cbl antibody (Right, arrowheads). (C) LacZ, FLAG-epitope-tagged CAP, Vxα, Vx-CAP, and CAP-Vx were overexpressed in HEK293T cells, and lysates were subject to SDS/PAGE and immunoblotted with monoclonal FLAG antibody to evaluate the expression level (Left). Lysates were incubated with glutathione-agarose-bound GST-flotillin for 1 h at 4°C. Precipitates were subjected to SDS/PAGE and blotted with monoclonal FLAG antibody or with anti-Cbl antibody (Right).
To confirm the role of the SoHo domain, we designed chimeras between the two proteins (Fig. 2a). FLAG epitope-tagged Vx-CAP, which has the N terminus of vinexin and the C terminus of CAP, and CAP-Vx, which has the N terminus of CAP and the C terminus of vinexin, were expressed in HEK293T cells, and whole-cell detergent extracts from these cells were immunoblotted with anti-FLAG antibody for evaluation of the expression level (Fig. 2C). GST-flotillin fusion proteins were incubated with these lysates, and the precipitates were immunoblotted with anti-FLAG antibodies. Interestingly, both Vx-CAP and CAP-Vx chimeras interacted with flotillin in vitro.
Overexpression of a CAPΔSoHo Mutant Blocks the Translocation of Phosphorylated Cbl into Triton-Insoluble Lipid Rafts.
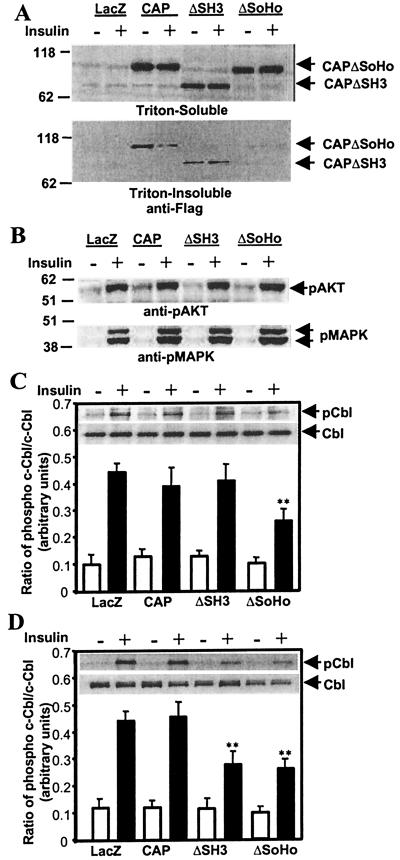
To explore the role of CAP/Cbl association with flotillin in insulin action, we overexpressed CAP or CAPΔSoHo in 3T3-L1 adipocytes and evaluated the subcellular localization of the wild-type and mutant proteins. Whereas wild-type CAP was detected at the plasma membrane by confocal microscopy, CAPΔSoHo exhibited a clear perinuclear staining, with little if any of the protein found in the plasma membrane (data not shown). This difference in cellular localization allowed us to evaluate the role of CAP/flotillin binding with these mutants. Wild-type CAP, CAPΔSH3, and CAPΔSoHo were expressed in 3T3-L1 adipocytes, and insulin-stimulated tyrosine phosphorylation was evaluated. Each of the constructs was expressed at similar levels in the cells, as detected in the Triton-soluble fraction by immunoblotting with anti-FLAG antibodies (Fig. 3A). Whereas the wild-type and CAPΔSH3 mutants were detected in the Triton-insoluble fraction, CAPΔSoHo appeared to be absent, presumably because the interaction of CAP with flotillin requires its SoHo domain. Expression of CAP, CAPΔSH3, or CAPΔSoHo had no effect on the ability of insulin to stimulate the phosphorylation of two downstream targets, Akt and MAP kinase, that are activated as a consequence of well-characterized phosphorylation cascades (Fig. 3b). Because Akt activation occurs as a direct consequence of PI 3-kinase stimulation (26, 27), these data also demonstrate that the CAP pathway did not affect the stimulation of PI 3-kinase activity by insulin, as described (17). Overexpression of CAPΔSH3 or CAPΔSoHo also had no effect on the tyrosine phosphorylation of the insulin receptor β-subunit or its substrate IRS-1 (data not shown).
Figure 3.
CAPΔSoHo blocks the translocation of phosphorylated Cbl into Triton-insoluble complexes. (A) A representative FLAG immunoblot of the Triton-soluble fraction (Upper) and Triton-insoluble pellet fraction (Lower) of 3T3-L1 adipocytes expressing LacZ, CAP, CAPΔSH3, and CAPΔSoHo. (B) Lysates from basal and insulin-treated adipocytes expressing LacZ, CAP, CAPΔSH3, and CAPΔSoHo were immunoblotted with phospho-Akt (Upper) and phospho-mitogen-activated protein kinase (Lower) antibodies. (C) Cbl was immunoprecipitated from the Triton-soluble fraction of lysates from basal and insulin-treated adipocytes expressing LacZ, CAP, CAPΔSH3, and CAPΔSoHo. Immunoprecipitates were resolved by SDS/PAGE, immunoblotted with monoclonal phosphotyrosine antibodies, and reprobed with the Cbl antibody. A representative blot is shown within the graph. Films were scanned and quantified, and the ratio of total Cbl to phospho-Cbl was calculated. (D) Cbl was immunoprecipitated from the Triton-insoluble pellet fraction of lysates from basal and insulin-treated adipocytes expressing LacZ, CAP, CAPΔSH3, and CAPΔSoHo. Immunoprecipitates were resolved and analyzed as described in Fig. 2C. **, Significant difference, P < 0.01, n = 3.
The phosphorylation of c-Cbl by insulin is associated with the interaction of the protein with CAP and the insulin receptor (15). As observed, overexpression of CAP or CAPΔSH3 had no effect on insulin-stimulated Cbl phosphorylation in the Triton-soluble fraction (Fig. 3C). In contrast, CAPΔSoHo overexpression caused a significant reduction in the appearance of phosphorylated Cbl in this fraction. Overexpression of CAPΔSH3 or CAPΔSoHo inhibited the appearance of phospho-Cbl in the Triton-insoluble fraction, presumably because of the blockade of recruitment of the CAP/Cbl complex to flotillin-enriched lipid rafts (Fig. 3D). Considering that the average efficiency of transfection in these experiments was 50%, the observed reduction in Cbl phosphorylation in both Triton-soluble and insoluble fractions suggests that the expression of the CAPΔSoHo mutant almost completely blocked insulin-stimulated Cbl phosphorylation. These data reflect the likelihood that overexpressed CAPΔSoHo can associate with endogenous Cbl and sequester the protein to a perinuclear site. This mutant can thus block both the initial phosphorylation of Cbl and its translocation to the lipid raft, consequently preventing the appearance of phosphorylated Cbl in both Triton-soluble and insoluble plasma membrane fractions.
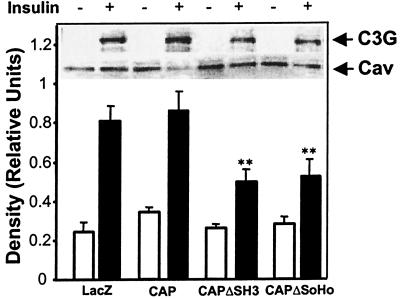
CAPΔSoHo Blocks the Translocation of C3G into the Triton-Insoluble Lipid Raft.
Stimulation of the tyrosine phosphorylation of Cbl by insulin generates a specific docking site for the amino-terminal SH2 domain of the adapter protein CrkII (14). The SH3 domain of CrkII directly interacts with a proline-rich domain in the guanyl nucleotide exchange factor C3G (28). We examined the insulin-stimulated translocation of C3G into the lipid raft in cells overexpressing CAPΔSoHo. 3T3-L1 adipocytes were transfected with FLAG-tagged constructs encoding CAP, CAPΔSH3, or CAPΔSoHo. Caveolin was primarily found in the Triton-insoluble fraction and was unaffected by insulin or by expression of CAP, CAPΔSH3, or CAPΔSoHo (Fig. 4). Insulin recruited C3G into the triton-insoluble fraction in both LacZ- and CAP-expressing cells. However, expression of CAPΔSH3 or CAPΔSoHo decreased the insulin-stimulated recruitment of C3G by ≈40%. Taking into account a 50% transfection efficiency, this decrease in recruitment of C3G corresponds to a nearly complete blockade of C3G recruitment in the transfected cell population. Thus, insulin-stimulated translocation of C3G into caveolin-enriched compartments requires both the SH3 domains and the SoHo domain of CAP.
Figure 4.
CAPΔSoHo blocks the translocation of C3G into Triton-insoluble complexes. Triton-insoluble fraction of lysates from basal and insulin-treated adipocytes expressing LacZ, CAP, CAPΔSH3, and CAPΔSoHo were immunoblotted with anti-C3G and anti-caveolin antibodies. A representative blot is shown within the graph. Films were scanned and quantified, and the ratio of caveolin to C3G was calculated. **, Significant difference, P < 0.01, n = 3.
Overexpression of CAPΔSoHo in 3T3-L1 Adipocytes Specifically Attenuates Insulin-Stimulated Glucose Uptake.
To investigate the role of the association of the CAP/Cbl complex with flotillin in insulin action, we evaluated the effect of CAPΔSoHo expression on insulin-stimulated [14C]2-deoxyglucose uptake in 3T3-L1 adipocytes (Table 1). In LacZ-transfected cells, insulin produced an 8- to 10-fold increase in [14C]2-deoxyglucose uptake. Expression of CAP had no effect on this process. However, the expression of either CAPΔSH3 or CAPΔSoHo led to a 50% reduction in insulin-stimulated glucose uptake, without effecting basal activity.
Table 1.
Overexpression of CAPΔSoHo attenuates insulin-stimulated glucose uptake
| LacZ | CAP | CAPΔSH3 | CAPΔSoHo | |
|---|---|---|---|---|
| Basal | 1 | 1.19 ± 0.2 | 0.78 ± 0.11 | 0.86 ± 0.09 |
| Insulin | 8.87 ± 0.77 | 8.51 ± 0.61 | 4.76 ± 0.46** | 4.88 ± 0.44** |
Differentiated 3T3-L1 adipocytes were transfected with LacZ, CAP, CAPΔSH3, and CAPΔSoHo. The cells were then either left untreated or stimulated with 100 nM insulin for 30 min. The rate of [14C]2-deoxyglucose uptake was determined. Results are expressed as fold stimulation over the LacZ basal control, and are the mean of three independent experiments ± standard error from individual experiments performed in triplicate with transfection efficiency about 50%.
, Significant difference, P < 0.01.
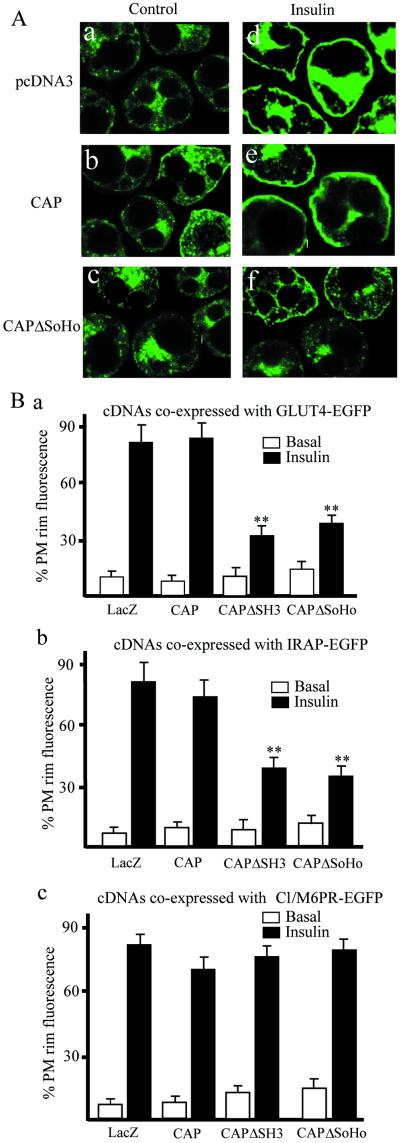
To examine the mechanism by which CAPΔSoHo blocks glucose transport, the translocation of GLUT4 was directly examined. 3T3-L1 adipocytes were electroporated with pcDNA3-LacZ, CAP, or CAPΔSoHo together with a construct encoding an enhanced green fluorescence protein fusion of GLUT4 (GLUT4-EGFP) (Fig. 5A). Insulin stimulated the translocation of GLUT4-EGFP into the plasma membrane in cells transfected with pcDNA3-LacZ or CAP. In contrast, expression of CAPΔSoHo markedly inhibited GLUT4-EGFP translocation to the cell surface.
Figure 5.
CAPΔSoHo blocks insulin-stimulated GLUT4 and IRAP translocation to the plasma membrane, without affecting translocation of Cl/M6PR. (A) Differentiated 3T3-L1 adipocytes were transfected with GLUT4-EGFP plus vector (a and d), FLAG-CAPΔSH3 (b and e), or FLAG-CAPΔSoHo (c and f), and allowed to recover for 18 h. The cells were treated without (a–c) or with (d–f) 100 nM insulin for 30 min. Cells were fixed and fluorescence visualized by confocal microscopy. This image is a representative collection of three to six cell images per field. (B) Number of GLUT4-EGFP transfected cells displaying visually detectable plasma membrane (PM) rim fluorescence. The numbers of IRAP-EGFP and Cl/M6PR-EGFP-transfected cells displaying visually detectable rim fluorescence are plotted. These data were obtained by blind counting of more than 60 cells from three independent experiments. **, Significant difference, P < 0.01, n =3.
To confirm the specificity of this effect, we examined the translocation of another cargo protein known to reside in the GLUT4 vesicles, the insulin-responsive aminopeptidase (IRAP) (29, 30). IRAP-EGFP was cotransfected with pcDNA3-LacZ, CAP, or CAPΔSoHo. The translocation of IRAP to the plasma membrane was examined by immunofluorescence and quantitated as the percentage of electroporated cells in a representative field displaying rim or plasma membrane fluorescence. As seen with GLUT4-EGFP, CAPΔSoHo blocked the insulin-stimulated translocation of IRAP-EGFP, whereas CAP and empty vector were without effect (Fig. 5B).
Insulin also stimulates the net exocytosis of the cation-independent mannose-6-phosphate receptor (CI/M6PR), which is located in a distinct vesicular compartment (31, 32). CI/M6PR traffics between the trans-Golgi network, late endosomes, and lysosomes, distinct from the GLUT4/IRAP-containing compartments (33). In contrast to GLUT4 and IRAP, expression of CAPΔSoHo did not attenuate the insulin-stimulated translocation of CI/M6PR-EGFP fusion (Fig. 5B). Taken together, these data indicate that the CAPΔSoHo mutant has a dominant interfering effect on insulin-stimulated translocation of GLUT4 and IRAP-containing vesicles into the plasma membrane, without exerting an impact on other insulin-regulated vesicle trafficking processes.
Discussion
There is abundant evidence that the dynamic changes in plasma membranes produced by extracellular stimuli occur in restricted compartments. One such segment that may be crucial in segregating receptor signals is a microdomain defined by its lipid and protein composition, referred to as lipid rafts (3, 4). These structures are highly enriched in cholesterol and sphingolipids and often contain phospholipids with saturated fatty acids that pack well in a highly ordered lipid environment (34). Lipid rafts may also represent sites for the sequestered localization of certain membrane proteins. Among these are proteins with lipid modifications, such as glycosylphosphatidylinositol-anchored cell surface proteins and cytoplasmically oriented proteins with closely spaced myristoylation and palmitoylation, as well as other hydrophobic integral membrane proteins such as caveolin and flotillin (17). We report here the identification of a protein motif responsible for the targeting of a family of adapter proteins to lipid raft microdomains in the plasma membrane. The proteins of the SoHo family, CAP, ArgBP2, and vinexin, each contain three C-terminal SH3 domains and an N-terminal region with similarity to the peptide sorbin, termed the sorbin homology (SoHo) domain. Whereas the SH3 domains of these proteins can bind to different signaling or cytoskeletal molecules, the SoHo domain interacts specifically with the lipid raft protein flotillin. Thus these proteins serve as adapters that link signaling or cytoskeletal proteins to the lipid raft.
The critical role of lipid rafts in permitting the generation of segregated signals is typified by the hormonal control of glucose transport. Although there is little doubt that the IRS-mediated activation of PI 3-kinase represents a critical step in this process (35), insulin action also requires the tyrosine phosphorylation of Cbl, which is recruited to the insulin receptor by the adapter protein CAP (14, 15) and translocates to a lipid raft membrane fraction, because of the interaction of CAP with flotillin (17). The critical role of this translocation event led us to focus on the region of CAP responsible for its interaction with flotillin. A series of deletion mutants of CAP revealed that the SoHo domain is both necessary and sufficient for this interaction. Furthermore, this domain appears to serve the same function in vinexin, inasmuch as deletion of the corresponding sequences also blocked flotillin binding, and a CAP/vinexin chimera generated the expected binding pattern.
Because the SoHo domain mediates the interaction of CAP with flotillin, we sought to determine whether this domain was singularly responsible for the insulin-stimulated translocation of the CAP/Cbl complex into lipid rafts. Whereas expression of CAPΔSoHo had no effect on the IRS-1/PI 3-kinase/Akt pathway, there were profound effects on the Cbl/CAP/C3G pathway. CAPΔSoHo overexpression blocked Cbl phosphorylation and translocation by CAPΔSoHo, in the process inhibiting the translocation of the guanyl nucleotide exchange factor C3G into lipid rafts. As was seen with CAPΔSH3, the SoHo domain mutant inhibited glucose uptake and GLUT4 translocation by insulin. These data support previous studies with CAP (17) and downstream players in the pathway (20), demonstrating that the CAP-dependent translocation of Cbl to lipid rafts generates a necessary step in the stimulation of glucose transport by insulin.
Do SoHo family proteins mediate similar interactions in other cell types that play critical roles in signaling? As shown in Fig. 1, the structural organization of this family is highly conserved, implicating its members as bifunctional proteins that localize their partners to discrete plasma membrane sites. Vinexinα has 32% identity and 38% similarity in its SoHo domain with CAP, whereas ArgBP2 exhibits 42% identity and 53% similarity. Vinexin was identified as a vinculin-binding protein through its first and second SH3 domains and plays a key role in cell spreading and cytoskeletal organization (36). Vinexinα has SoHo domains in its N terminus and three SH3 domains in its C terminus, whereas vinexinβ lacks the SoHo domains, suggesting that these two forms of the protein might have different functions. Expression of vinexinα (36) and CAP (37) increased focal adhesion size in fibroblasts and promoted actin stress fiber formation. Interestingly, when only the N-terminal sequences of vinexinα were expressed, the mutant protein was localized in focal adhesions but had no impact on the actin cytoskeleton. These data strongly suggest that like CAP, vinexin specifically targets proteins to the lipid raft for proper function. It is likely that the SoHo family of proteins carries out the targeting of signaling or cytoskeletal proteins to this important microdomain of the plasma membrane and thus represents a paradigm in the spatial compartmentalization of signal transduction. Further biochemical and structural analyses of the SoHo domain and its role in cellular signaling have yet to be made.
Abbreviations
- CAP
c-Cbl-associated protein
- SoHo
sorbin homology
- HEK
human embryo kidney
- GST
glutathione S-transferase
- EGFP
enhanced green fluorescence protein
- IRAP
insulin-responsive aminopeptidase
- CI/M6PR
cation-independent mannose-6-phosphate receptor
References
- 1.Song K S, Scherer P E, Tang Z, Okamoto T, Li S, Chafel M, Chu C, Kohtz D S, Lisanti M P. J Biol Chem. 1996;271:15160–15165. doi: 10.1074/jbc.271.25.15160. [DOI] [PubMed] [Google Scholar]
- 2.Scherer P E, Lisanti M P, Baldini G, Sargiacomo M, Mastick C C, Lodish H F. J Cell Biol. 1994;127:1233–1243. doi: 10.1083/jcb.127.5.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson R G. Annu Rev Biochem. 1998;67:199–225. doi: 10.1146/annurev.biochem.67.1.199. [DOI] [PubMed] [Google Scholar]
- 4.Harder T, Simons K. Curr Opin Cell Biol. 1997;9:534–542. doi: 10.1016/s0955-0674(97)80030-0. [DOI] [PubMed] [Google Scholar]
- 5.Sargiacomo M, Sudol M, Tang Z, Lisanti M P. J Cell Biol. 1993;122:789–807. doi: 10.1083/jcb.122.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li S, Okamoto T, Chun M, Sargiacomo M, Casanova J E, Hansen S H, Nishimoto I, Lisanti M P. J Biol Chem. 1995;270:15693–15701. doi: 10.1074/jbc.270.26.15693. [DOI] [PubMed] [Google Scholar]
- 7.Roy S, Luetterforst R, Harding A, Apolloni A, Etheridge M, Stang E, Rolls B, Hancock J F, Parton R G. Nat Cell Biol. 1999;1:98–105. doi: 10.1038/10067. [DOI] [PubMed] [Google Scholar]
- 8.Li S, Couet J, Lisanti M P. J Biol Chem. 1996;271:29182–29190. doi: 10.1074/jbc.271.46.29182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robbins S M, Quintrell N A, Bishop J M. Mol Cell Biol. 1995;15:3507–3515. doi: 10.1128/mcb.15.7.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaul P W, Smart E J, Robinson L J, German Z, Yuhanna I S, Ying Y, Anderson R G, Michel T. J Biol Chem. 1996;271:6518–6522. doi: 10.1074/jbc.271.11.6518. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Cardena G, Fan R, Stern D F, Liu J, Sessa W C. J Biol Chem. 1996;271:27237–27240. doi: 10.1074/jbc.271.44.27237. [DOI] [PubMed] [Google Scholar]
- 12.Liu P, Ying Y, Ko Y G, Anderson R G. J Biol Chem. 1996;271:10299–10303. doi: 10.1074/jbc.271.17.10299. [DOI] [PubMed] [Google Scholar]
- 13.Gustavsson J, Parpal S, Karlsson M, Ramsing C, Thorn H, Borg M, Lindroth M, Peterson K H, Magnusson K E, Stralfors P. FASEB J. 1999;13:1961–1971. [PubMed] [Google Scholar]
- 14.Ribon V, Saltiel A R. Biochem J. 1997;324:839–845. doi: 10.1042/bj3240839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ribon V, Printen J A, Hoffman N G, Kay B K, Saltiel A R. Mol Cell Biol. 1998;18:872–879. doi: 10.1128/mcb.18.2.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mastick C C, Saltiel A R. J Biol Chem. 1997;272:20706–20714. doi: 10.1074/jbc.272.33.20706. [DOI] [PubMed] [Google Scholar]
- 17.Baumann C A, Ribon V, Kanzaki M, Thurmond D C, Mora S, Shigematsu S, Bickel P E, Pessin J E, Saltiel A R. Nature (London) 2000;407:202–207. doi: 10.1038/35025089. [DOI] [PubMed] [Google Scholar]
- 18.Bickel P E, Scherer P E, Schnitzer J E, Oh P, Lisanti M P, Lodish H F. J Biol Chem. 1997;272:13793–13802. doi: 10.1074/jbc.272.21.13793. [DOI] [PubMed] [Google Scholar]
- 19.Lang D M, Lommel S, Jung M, Ankerhold R, Petrausch B, Laessing U, Wiechers M F, Plattner H, Stuermer C A. J Neurobiol. 1998;37:502–523. doi: 10.1002/(sici)1097-4695(199812)37:4<502::aid-neu2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 20.Chiang S H, Baumann C A, Kanzaki M, Thurmond D C, Watson R T, Neudauer C L, Macara I G, Pessin J E, Saltiel A R. Nature (London) 2001;410:944–948. doi: 10.1038/35073608. [DOI] [PubMed] [Google Scholar]
- 21.Baumann C A, Brady M J, Saltiel A R. J Biol Chem. 2001;276:6065–6068. doi: 10.1074/jbc.C000856200. [DOI] [PubMed] [Google Scholar]
- 22.Olson A L, Knight J B, Pessin J E. Mol Cell Biol. 1997;17:2425–2435. doi: 10.1128/mcb.17.5.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamauchi K, Pessin J E. J Biol Chem. 1994;269:31107–31114. [PubMed] [Google Scholar]
- 24.Martin S S, Haruta T, Morris A J, Klippel A, Williams L T, Olefsky J M. J Biol Chem. 1996;271:17605–17608. doi: 10.1074/jbc.271.30.17605. [DOI] [PubMed] [Google Scholar]
- 25.Thurmond D C, Ceresa B P, Okada S, Elmendorf J S, Coker K, Pessin J E. J Biol Chem. 1998;273:33876–33883. doi: 10.1074/jbc.273.50.33876. [DOI] [PubMed] [Google Scholar]
- 26.Alessi D R, Downes C P. Biochim Biophys Acta. 1998;1436:151–164. doi: 10.1016/s0005-2760(98)00133-7. [DOI] [PubMed] [Google Scholar]
- 27.Corvera S, Czech M P. Trends Cell Biol. 1998;8:442–446. doi: 10.1016/s0962-8924(98)01366-x. [DOI] [PubMed] [Google Scholar]
- 28.Knudsen B S, Feller S M, Hanafusa H. J Biol Chem. 1994;269:32781–32787. [PubMed] [Google Scholar]
- 29.Keller S R, Scott H M, Mastick C C, Aebersold R, Lienhard G E. J Biol Chem. 1995;270:23612–23618. doi: 10.1074/jbc.270.40.23612. [DOI] [PubMed] [Google Scholar]
- 30.Kandror K V, Pilch P F. Proc Natl Acad Sci USA. 1994;91:8017–8021. doi: 10.1073/pnas.91.17.8017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oka Y, Mottola C, Oppenheimer C L, Czech M P. Proc Natl Acad Sci USA. 1984;81:4028–4032. doi: 10.1073/pnas.81.13.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wardzala L J, Simpson I A, Rechler M M, Cushman S W. J Biol Chem. 1984;259:8378–8383. [PubMed] [Google Scholar]
- 33.Malide D, Cushman S W. J Cell Sci. 1997;110:2795–2806. doi: 10.1242/jcs.110.22.2795. [DOI] [PubMed] [Google Scholar]
- 34.Brown D A, London E. J Biol Chem. 2000;275:17221–17224. doi: 10.1074/jbc.R000005200. [DOI] [PubMed] [Google Scholar]
- 35.Isakoff S J, Taha C, Rose E, Marcusohn J, Klip A, Skolnik E Y. Proc Natl Acad Sci USA. 1995;92:10247–10251. doi: 10.1073/pnas.92.22.10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kioka N, Sakata S, Kawauchi T, Amachi T, Akiyama S K, Okazaki K, Yaen C, Yamada K M, Aota S. J Cell Biol. 1999;144:59–69. doi: 10.1083/jcb.144.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ribon V, Herrera R, Kay B K, Saltiel A R. J Biol Chem. 1998;273:4073–4080. doi: 10.1074/jbc.273.7.4073. [DOI] [PubMed] [Google Scholar]