Abstract
Pma1 is a plasma membrane H+-ATPase whose activity at the cell surface is essential for cell viability. In this paper we describe a temperature-sensitive pma1 allele, pma1-10 (with two point mutations in the first cytoplasmic loop of Pma1), in which the newly synthesized mutant protein fails to remain stable at the cell surface at 37°C. Instead, Pma1-10 appears to undergo internalization for vacuolar degradation in a manner dependent on End4, Vps27, Doa4, and Pep4. By contrast with wild-type Pma1, mutant Pma1-10 is hypophosphorylated and fails to associate with a Triton-insoluble fraction at 37°C, suggesting failure to enter lipid rafts. Kinetic analysis reveals that, at the permissive temperature, newly synthesized Pma1-10 acquires Triton-insolubility before becoming stabilized. We suggest that phosphorylation and lipid raft association may play important roles in maintaining protein stability at the plasma membrane.
The plasma membrane H+-ATPase, encoded by PMA1, is an electrogenic proton pump that serves to regulate intracellular pH and generate the membrane potential across the plasma membrane. Because of its critical physiological function, PMA1 is essential for cell viability (1). Pma1 is a member of the P-type ATPase family, the members of which include Ca2+-ATPases and the Na+,K+-ATPase in mammalian cells. Electron crystallography evidence supports predictions by hydropathy analysis that P-type enzymes are embedded in the membrane by 10 transmembrane segments: 4 at the amino terminus separated from 6 at the carboxyl terminus by a large cytoplasmic domain containing conserved ATP-binding and catalytic phosphorylation sites (2).
Because of its structural and mechanistic similarity with mammalian P-type ATPases, Pma1 of Saccharomyces cerevisiae has been the subject of extensive mutagenesis to analyze structure–function relationships (3). From these studies, it appears that many of the pma1 mutants exhibit defective transport of newly synthesized Pma1 through the secretory pathway. Defective trafficking of mutant Pma1 molecules appears to involve at least two different pathways. A large number of pma1 mutants have been described in which the cells cannot grow because newly synthesized Pma1 is retained in the endoplasmic reticulum (ER), and degraded by ER-associated degradation (4–6). The temperature-sensitive mutant pma1-7 represents a second class of mutants in which newly synthesized Pma1 is exported from the ER but fails to arrive at the plasma membrane and instead is targeted for vacuolar degradation (7, 8). Recognition of distinct conformational defects is likely the mechanism for both ER quality control and Golgi-to-vacuole delivery of different pma1 mutants.
At the plasma membrane, wild-type Pma1 is a paradigm of a stable membrane protein with a half-life of ≈11 h (9). Unlike other cell surface proteins, the endocytosis of which is signaled by ubiquitination, Pma1 is not ubiquitinated (10), and whether cell surface Pma1 undergoes recycling has not been established. Recent evidence has revealed that Pma1 is a major protein component of glycosphingolipid- and cholesterol-enriched microdomains in the plasma membrane, called lipid rafts (11). Although it has been proposed that entry into lipid rafts is a mechanism for regulating membrane traffic (12), the significance of lipid raft association for Pma1 trafficking, stability, and function remains unclear.
In this study, we describe a pma1 allele, pma1-10, that causes temperature-sensitive growth as newly synthesized mutant Pma1 fails to remain stable at the cell surface at 37°C and undergoes vacuolar degradation. Pma1-10 appears to have a misfolded conformation because it is more sensitive to proteolysis than wild-type Pma1. Unlike wild-type Pma1, Pma1-10 is hypophosphorylated, and it fails to enter a Triton-insoluble fraction, suggesting a failure to associate with lipid rafts. We suggest that these parameters contribute to maintaining membrane protein stability at the cell surface.
Materials and Methods
Strains and Media.
Standard yeast media and genetic manipulations were as described (13). Strains used in this study are listed in Table 1. All strains except those marked with an asterisk are isogenic with L3852. XGY32 bearing the pma1-10 mutation was generated by pop-in pop-out gene replacement of PMA1 (14) after transformation of L3852 with pXG39. XGX28 is an isogenic cross between XGY32 and XGX19–2B (MATa his3Δ200 lys2Δ201 leu2-3,112 ura3-52 ade2 pep4Δ∷URA3). XGX47 is an isogenic cross between XGY32 and WLX4-2A (MATa his3Δ200 lys2Δ201 leu2-3,112 ura3-52 ade2 vps27∷LEU2). XGX42 is a cross between XGY32 and RH268–1C (MATa leu2 his4 ura3 bar1-1 end4-1) (15). XGX44 is a cross between XGX28-1C and ACX18-4B (MATα his3Δ200 leu2-3,112 ura3-52 ade2 sec6-4) (7). XGX55 is a cross between XGX28-1C and MHY623 (MATa his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 doa4-Δ1∷LEU2) from M. Hochstrasser (Yale University, New Haven, CT).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| L3852 | MATα his3Δ200 lys2Δ201 leu2-3,112 ura3-52 ade2 | Ref. 7 |
| XGY32 | MATα his3Δ200 lys2Δ201 leu2-3,112 ura3-52 ade2 pma1-10 | This study |
| XGX28-1A | MATα his3Δ200 lys2Δ201 leu2-3,112 ura3-52 ade2 pep4Δ∷URA3 pma1-10 | This study |
| XGX28-1C | MATa his3Δ200 lys2Δ201 leu2-3,112 ura3-52 ade2 pma1-10 | This study |
| XGX28-1D | MATα his3Δ200 lys2Δ201 leu2-3,112 ura3-52 ade2 | This study |
| XGX42-8B* | MATa his− leu2-3,112 ura3-52 ade2 pma1-10 end4-1 | This study |
| XGX44-1A* | MATa his3Δ200 lys2Δ201 leu2-3,112 ura3-52 ade2 pma1-10 sec6-4 | This study |
| XGX44-1C* | MATa his3Δ200 lys2Δ201 leu2-3,112 ura3-52 ade2 pma1-10 | This study |
| XGX47-1D | MATa his3Δ200 lys2Δ201 leu2-3,112 ura3-52 ade2 pma1-10 vps27∷LEU2 | This study |
| XGX55-5C* | MATα his3Δ200 lys2Δ201 leu2-3,112 ura3-52 ade2 pma1-10 doa4Δ∷LEU2 | This study |
Not isogenic with L3852.
Molecular Biology.
The temperature-sensitive pma1-10 allele was identified by plasmid shuffle after in vitro hydroxylamine mutagenesis of PMA1 (G. R. Fink laboratory collection). For pop-in pop-out gene replacement, pma1-10 was cloned into a URA3-marked YIp, pRS306 (16) as a 5-kb HindIII fragment to generate pXG39. pXG39 was linearized with PacI for transformation.
Metabolic Labeling, Cell Fractionation, and Immunoprecipitation.
For metabolic labeling, cultures were grown overnight in minimal medium supplemented with essential amino acids. For pulse–chase experiments with pma1-10, cells were resuspended at 1 OD600/ml and incubated at room temperature for 15 min before the start of the experiment. For labeling at the restrictive temperature, cells were shifted to 37°C for 5 min before adding Expre35S35S (NEN). After a 5-min pulse, an equal volume of synthetic complete medium containing 20 mM methionine and 20 mM cysteine was added to start the chase. Aliquots were removed at various times of chase and placed on ice in the presence of 10 mM sodium azide.
For immunoprecipitation, cell lysate was prepared by mixing with glass beads in a sorbitol buffer in a Vortex mixer, as described (17). For fractionation, lysates were prepared by mixing with glass beads in Tris/EDTA buffer in a Vortex mixer, as described (8). Renografin density gradients were prepared with the load at the bottom of the tube as described (8), and centrifuged in an SW50.1 rotor at 40,000 × g for >16 h. Fourteen fractions (350 μl) were collected from the top, every two fractions were pooled, and immunoprecipitations were in 0.15 M NaCl/0.05 M Tris⋅HCl, pH 7.5/1% Triton X-100/1% sodium deoxycholate/0.1% SDS (RIPA) buffer. A protease inhibitor mixture was included in lysis and RIPA buffers (17). Immunoprecipitations were normalized to acid-precipitable cpm and analyzed by SDS/PAGE and fluorography. Gas1 and yeast alkaline phosphatase (ALP) markers were assayed by Western blotting of membranes pelleted from gradient fractions. Anti-Gas1, anti-ALP, and anti-carboxypeptidase Y (CPY) antibodies were from Tamara Doering (Washington University, St. Louis), Greg Payne (University of California, Los Angeles), and Molecular Probes, respectively.
For alkaline phosphatase treatment, immunoprecipitated Pma1 was released from protein A-beads as described (17). Samples were divided and incubated for 1 h at 37°C in the presence and absence of 1 unit of calf intestinal alkaline phosphatase (Boehringer Mannheim). Samples were analyzed on 8% polyacrylamide gels, and electrophoresis time was extended to discern small mobility changes.
For limited trypsinolysis, cells were shifted to 37°C for 5 min before pulse-labeling for 2 min. After cell lysis, a total membrane fraction was generated by centrifugation at 100,000 × g for 1 h. Membranes were resuspended in 250 mM sucrose/5 mM MgCl2/20 mM Hepes (pH 7.5) buffer. Membranes were incubated at a protein concentration of 1 mg/ml at 30°C at a trypsin-to-protein ratio of 1:20. Diisopropyl fluorophosphate was added to terminate the reaction, and tryptic fragments were immunoprecipitated.
Triton insolubility was determined by adding an equal volume of ice-cold 2% Triton X-100 to lysate (3–4 OD600 equivalents), and incubating for 30 min on ice. Samples were then centrifuged at 100,000 × g for 1 h. Pellets were resuspended in 1% SDS. Detergent concentrations in aliquots of total, supernatant, and pellet samples were adjusted for immunoprecipitation. Immunoprecipitates were analyzed by SDS/PAGE and quantitated by phosphorimaging (Molecular Dynamics).
Indirect Immunofluorescence.
Indirect immunofluorescence staining was done essentially as described (18). Exponentially growing cultures were fixed overnight with 4.4% formaldehyde in 0.1 M potassium phosphate (pH 6.5). Cells were spheroplasted and permeabilized with methanol and acetone before staining with affinity-purified rabbit anti-Pma1 and mouse monoclonal antibody against the 60-kDa subunit of V-ATPase (Molecular Probes). Primary antibody staining was detected with Cy3- and Cy2-conjugated secondary antibodies (Jackson ImmunoResearch). Images were visualized with an Olympus IX70 microscope (Lake Success, NY), and collected digitally and adjusted with ADOBE PHOTOSHOP 4.0 (Adobe Systems, Mountain View, CA).
Results
Newly Synthesized Pma1-10 Is Delivered for Vacuolar Degradation After Arriving at the Cell Surface.
A temperature-sensitive allele of pma1, pma1-10, was isolated by plasmid shuffle (14). DNA sequencing of the pma1-10 allele revealed two nucleotide changes resulting in A165G and V197I changes. The residues are both predicted to reside in the first cytoplasmic loop between transmembrane segments 2 and 3 (3). Previous work suggests that this region may interact with other domains of the protein (19).
A strain bearing the pma1-10 allele as its sole copy of pma1 was constructed by pop-in, pop-out gene replacement (see Materials and Methods). The strain grows well at 24°C and 30°C, but cannot grow at 37°C, as shown in Fig. 1A. Previously, we have characterized a class of temperature-sensitive pma1 mutants, represented by pma1–7, in which newly synthesized mutant Pma1 is degraded on shift to 37°C (7). To determine whether pma1-10 belongs to a similar class of mutants, pulse–chase experiments were performed to examine the stability of newly synthesized Pma1. Cells were pulse-labeled for 5 min with Expre35S35S, and chased for various times. Pma1 was then analyzed by immunoprecipitation, SDS/PAGE, and fluorography. At room temperature, Pma1-10 remains stable (see Fig. 6). At 37°C, however, newly synthesized Pma1-10 is largely degraded after 30-min chase (Fig. 1B). Pma1-10 is stabilized in pep4Δ cells, indicating that degradation requires PEP4 and vacuolar proteolytic activity (20). Nevertheless, pma1-10 pep4 cells cannot grow at 37°C (Fig. 1A), reflecting the requirement for Pma1 activity at the plasma membrane for growth (3). Strikingly, Pma1-10 degradation is prevented by a block in the late secretory pathway in sec6–4 cells (21), or by blocking internalization from the cell surface in end4–1 cells (15) (Fig. 1B). These observations suggest that degradation of Pma1-10 occurs after its delivery to the plasma membrane.
Figure 1.
Newly synthesized Pma1-10 is degraded in a Pep4-dependent manner. (A) Growth of cells on plates containing synthetic complete medium at 30°C and 37°C. Strains shown are wild-type (L3852), pma1-10 (XGY32), pma1-10 pep4Δ (XGX28–1A), and pma1-10 vps27Δ (XGX47–1D). (B) Pulse–chase analysis of Pma1-10. Cells were shifted to 37°C, labeled for 5 min with Expre35S35S, and chased for various times. Pma1 immunoprecipitation from cell lysate was normalized to acid-precipitable radiolabeled material, and analyzed by SDS/PAGE and fluorography. Strains analyzed are XGX28–1D, XGX44–1C, XGX28–1A, XGX44–1A, and XGX42–8B.
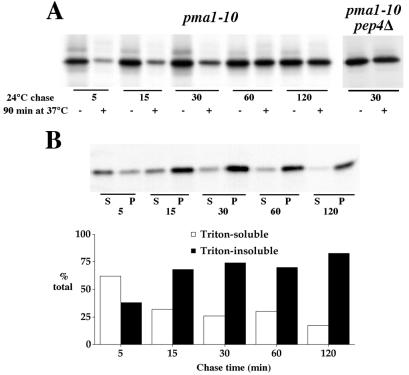
Figure 6.
Kinetics of stabilization and acquisition of Triton-insolubility. (A) pma1-10 (XGY32) and pma1-10 pep4Δ (XGX28–1A) cells were pulse-labeled for 5 min at 24°C with Expre35S35S and chased at 24°C. At various times of chase, aliquots were removed and either placed on ice with sodium azide or shifted to 37°C for an additional 90-min incubation. Lysates were prepared, and Pma1 was immunoprecipitated and analyzed by SDS/PAGE and fluorography. (B) Lysates from pma1-10 cells (XGY32) pulse-labeled and chased at 24°C were extracted with ice-cold Triton X-100 and separated into Triton-soluble and Triton-insoluble fractions. Pma1 was immunoprecipitated and analyzed by SDS/PAGE and phosphorimaging. Both gel and quantitation are shown. Newly synthesized Pma1-10 is predominantly found in a Triton-insoluble fraction by 15-min chase, whereas resistance to degradation at 37°C is achieved by 1- to 2-h chase.
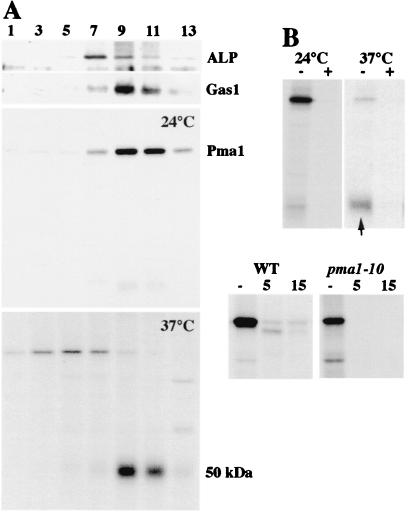
To observe directly Pma1-10 delivery to the plasma membrane, cells were fractionated on Renografin density gradients at various times of chase after a 5-min pulse-labeling period. Previous work has shown that Renografin density gradients resolve plasma membrane from intracellular membranes (8, 22). Fig. 2A confirms the separation of the plasma membrane marker protein Gas1, maximal in fractions 9–10, from the vacuolar marker ALP, maximal in fractions 7–8. Like wild-type Pma1 (data not shown), Pma1-10, pulse-labeled for 5 min and chased for 30 min at 24°C, is predominantly localized in fractions 9–10 representing plasma membrane; a faint, ≈50-kDa fragment was also observed (Fig. 2A). After pulse and chase at 37°C, however, an ≈50-kDa band was immunoprecipitated from plasma membrane-enriched fractions instead of intact 100-kDa Pma1 polypeptide (Fig. 2A). To determine whether the 50-kDa band is derived from Pma1 or represents a nonspecifically immunoprecipitated protein, purified Pma1 protein was included in immunoprecipitations. Purified Pma1 competed with both 100- and 50-kDa bands, suggesting the 50-kDa band is a proteolytic fragment of Pma1 (Fig. 2B). The 50-kDa band was also the predominant form observed in end4 pma1-10 cells at 37°C (not shown).
Figure 2.
Cell surface delivery of Pma1-10. (A) Density gradient fractionation. Cells (XGY32) were pulse-labeled for 5 min and chased for 30 min at 24°C or 37°C. Lysates were prepared and separated on Renografin density gradients as described in Materials and Methods. Pma1 was immunoprecipitated from gradient fractions. Distribution of ALP and Gas1 was determined by Western blotting gradient fractions. (B) Specificity of immunoprecipitation. Anti-Pma1 antibody was incubated in the presence (+) or absence (−) of 5 μg of partially purified Neurospora Pma1 (gift of Carolyn Slayman, Yale University) before adding to RIPA buffer containing peak plasma membrane fractions (9 and 10) from pulse–chase experiments in A. Arrow indicates 50-kDa fragment. (C) Limited trypsinolysis. Total membranes were prepared from cells pulse-labeled for 2 min at 37°C and incubated in the absence (−) and presence of trypsin at 30°C for 5 and 15 min, as described in Materials and Methods. Immunoprecipitated Pma1 was analyzed by SDS/PAGE and fluorography.
Because the 50-kDa fragment was obvious only after fractionation, it appears to reflect increased susceptibility of Pma1-10 to proteolysis in vitro. To support this idea, a total membrane fraction generated after pulse-labeling at 37°C was subjected to limited digestion with trypsin. Limited trypsinolysis has been used extensively to examine Pma1 folding (3, 23). As shown in Fig. 2B, newly synthesized Pma1-10 has extreme sensitivity to trypsin by comparison with wild-type Pma1. These data indicate that at 37°C Pma1-10 has a conformation that is more accessible to proteolytic attack than that of wild-type Pma1. Nevertheless, Pma1-10 reaches the plasma membrane.
Localization of Pma1-10 to the Vacuole and Prevacuolar Compartment.
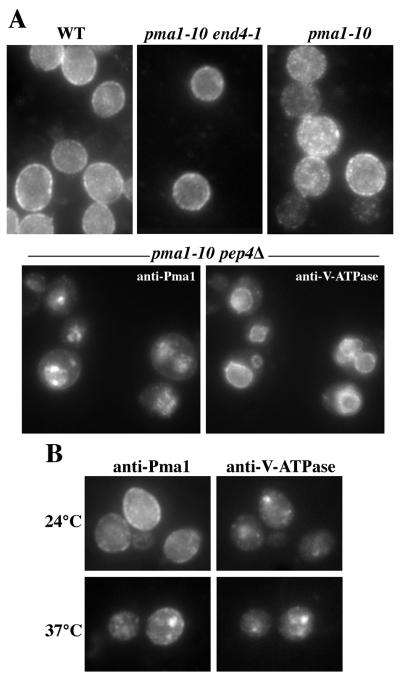
Because Pma1-10 is stabilized in pep4Δ cells (Fig. 1B), indirect immunofluorescence was carried out to visualize vacuolar localization of Pma1-10 after shifting cells to 37°C. Staining of wild-type Pma1 appears exclusively at the perimeter and surface of cells (Fig. 3A, Upper Left). Although some surface staining of Pma1-10 was observed, some punctate staining was also apparent (Fig. 3A, Upper Right), possibly reflecting Pma1-10 in endocytic intermediates. When internalization from the cell surface is blocked in end4–1 pma1-10 cells, Pma1-10 staining resembles that of wild-type Pma1 (Fig. 3A, Upper Middle). Pma1-10 staining coincident with vacuoles, indicated by the vacuolar membrane marker V-ATPase, was readily observed only in pep4Δ cells defective in vacuolar protease activity (Fig. 3A, Lower). Presumably, Pma1-10 molecules delivered to the vacuole after shift to 37°C are rapidly degraded in PEP4+ cells and, thus, escape detection by indirect immunofluorescence.
Figure 3.
Indirect immunofluorescence localization of Pma1-10. Cells were shifted to 37°C for 3 h, and then fixed, permeabilized, and stained with polyclonal anti-Pma1 and monoclonal anti-V-ATPase antibodies. (A) Pma1 localization in wild-type (L3852), pma1-10 end4–1 (XGX42–8B), pma1-10 (XGX28–1C), and pma1-10 pep4Δ (XGX28–1A) cells. Colocalization of Pma1 and V-ATPase at the vacuolar membrane is seen in pma1-10 pep4 cells. (B) Double labeling with anti-Pma1 and anti-V-ATPase in pma1-10 vps27Δ (XGX47–1D). Pma1 colocalizes in a large punctate spot with V-ATPase.
Interestingly, intracellular localization of Pma1-10 was detected in vps27 cells, a class-E vps mutant, in which delivery from the prevacuolar compartment to the vacuole is inhibited (24). Fig. 3B shows that V-ATPase accumulates in the prevacuolar compartment visualized as large perivacuolar spots in vps27 cells (25). After 3 h at 37°C, Pma1-10 staining also appeared as spots coincident with V-ATPase staining, indicating accumulation in the prevacuolar compartment. vps27 mutation does not, however, suppress temperature-sensitive growth of pma1-10 cells (Fig. 1A; although vps27 is a suppressor of pma1-7; see ref. 26). These data are consistent with delivery of Pma1-10 from the surface to the vacuole via the prevacuolar compartment.
A role has been suggested for the Doa4 deubiquitinating enzyme in trafficking of membrane proteins to the vacuole (27). Thus, we were prompted to test the effect of doa4 mutation on trafficking of Pma1-10. Pulse–chase analysis of pma1-10 doa4Δ double mutants was performed at 37°C. Fig. 4A shows that at 37°C newly synthesized Pma1-10 is stabilized in doa4Δ cells, in contrast with rapid degradation seen in DOA4+ cells (compare with Fig. 1B). Indirect immunofluorescence localization confirms Pma1-10 accumulation in doa4 cells at the vacuolar membrane (Fig. 4B). One possible explanation for inhibition of Pma1-10 degradation is that doa4Δ causes inactivation of the vacuolar proteases. To examine this possibility, intracellular transport of the vacuolar enzyme CPY was analyzed from the same lysates used to analyze Pma1-10 (Fig. 4A). In doa4Δ cells, mature CPY is produced rapidly, appearing by 30-min chase, indicating normal proteolytic cleavage. Thus, Pma1-10 is stable at the vacuole of doa4Δ cells, despite the presence of active proteases.
Figure 4.
Degradation of Pma1-10 depends on Doa4. (A) Pulse–chase analysis of Pma1-10. pma1-10 doa4Δ cells (XGX55–5C) were shifted to 37°C, pulse-labeled for 5 min with Expre35S35S, and chased for various times. Pma1 and CPY were immunoprecipitated and analyzed by SDS/PAGE and fluorography. Arrow indicates mature CPY. (B) Indirect immunofluorescence localization of Pma1 in pma1-10 doa4Δ cells. Cells were grown at 25°C or shifted to 37°C for 3 h, and fixed, permeabilized, and stained with anti-Pma1 antibody followed by a Cy3-conjugated secondary antibody. Cells were also stained with DAPI, 4′,6-diamidino-2-phenylindole, to reveal nuclear localization.
Pma1-10 Is Hypophosphorylated and Does Not Enter a Triton-Insoluble Membrane Fraction.
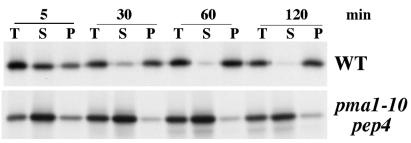
Recent work shows that wild-type Pma1 is a major protein component of specific lipid microdomains in yeast that, like those in mammalian cells, have the property of being insoluble in cold, nonionic detergents such as Triton X-100 (11). Because these lipid rafts have been suggested to act as platforms for protein assembly and membrane traffic, we examined the ability of newly synthesized Pma1-10 to associate with a Triton X-100-insoluble fraction. Cells were pulse-labeled and chased at 37°C. After extraction of lysates with ice-cold Triton X-100, samples were centrifuged to generate detergent-soluble and detergent-insoluble fractions, and Pma1 was immunoprecipitated and analyzed by SDS/PAGE and fluorography. Fig. 5 Upper shows that newly synthesized wild-type Pma1 is distributed nearly equally in Triton-soluble and -insoluble fractions after a 5-min chase, and by 30 min, the major fraction is Triton-insoluble. Triton X-100 fractionation of Pma1-10 was then examined. A pep4Δ strain background was used to stabilize newly synthesized Pma1-10. Unlike wild-type Pma1, Pma1-10 remains predominantly Triton-soluble even after 2-h chase at 37°C, suggesting defective association with lipid rafts (Fig. 5 Lower).
Figure 5.
At 37°C, Pma1-10 does not associate with a Triton-insoluble fraction. Wild-type (XGX28–1D) and pma1-10 pep4Δ (XGX28–1A) cells were pulse-labeled for 5 min at 37°C with Expre35S35S and chased for various times. Lysates were prepared, extracted with ice-cold Triton X-100, and separated into Triton-soluble and Triton-insoluble fractions, as described in Materials and Methods. T, total lysate; S, Triton-soluble; P, Triton-insoluble. Pma1 was immunoprecipitated and analyzed by SDS/PAGE and fluorography.
Because Pma1-10 fails to remain stable at the plasma membrane at 37°C, the possibility of a link is suggested between lipid raft association and plasma membrane stability. To examine further the relationship between Triton-insolubility and stability of Pma1-10, temperature shift experiments were carried out. At the permissive temperature, Pma1-10 is stable like wild-type Pma1 (Fig. 6A), and it associates with a Triton-insoluble fraction (Fig. 6B). When cells are shifted to 37°C, Pma1-10 is degraded, even after pulse-labeling and 5-min chase at the permissive temperature. However, as the time of chase at the permissive temperature increases, Pma1-10 becomes progressively resistant to degradation on shift to 37°C. At 30-min chase at 24°C, Pma1-10 has arrived at the plasma membrane (Fig. 2), and yet it remains sensitive to degradation at 37°C; degradation is Pep4-dependent (Fig. 6A). Maximal stabilization, i.e., maximal resistance to degradation at 37°C, is not achieved until 1- to 2-h chase (Fig. 6A). By contrast with its stabilization kinetics, Pma1-10 achieves near-maximal Triton insolubility by 15-min chase at room temperature (Fig. 6B). These observations indicate that Triton insolubility is a prelude to stabilization.
It has been demonstrated that association of some proteins with lipid rafts is affected by their phosphorylation state (28). Because wild-type Pma1 undergoes progressive kinase-mediated phosphorylation during transport through the secretory pathway (17), we examined Pma1-10 phosphorylation. Previously, we established that the extent of phosphorylation is reflected by the electrophoretic mobility of Pma1 on SDS/polyacrylamide gels, and newly synthesized Pma1 is maximally phosphorylated after 2-h chase (17). Fig. 7 shows Pma1 immunoprecipitates from wild-type and pma1-10 cells pulse-labeled with Expre35S 35S and chased for 2 h at 37°C. An end4–1 strain background was used to stabilize mutant Pma1-10 at the cell surface. Strikingly, there is a slight, but reproducible, increase in the electrophoretic mobility of Pma1-10 by comparison with that of wild-type Pma1. After digestion of the immunoprecipitates with alkaline phosphatase, Pma1-10, and wild-type Pma1 migrated with the same mobility, indicating that hypophosphorylation accounts for the faster migration of Pma1-10 at 37°C. At room temperature, no mobility difference was detected between mutant and wild-type Pma1 (not shown).
Figure 7.

Pma1-10 is hypophosphorylated. Cells were shifted to 37°C, pulse-labeled for 5 min, and chased for 2 h. Pma1 was immunoprecipitated from cell lysate. Immunoprecipitates were divided, incubated in the presence (+) or absence (−) of alkaline phosphatase, and analyzed by extended electrophoresis on an 8% polyacrylamide gel followed by fluorography.
Discussion
In this study we describe a temperature-sensitive pma1-10 mutant in which newly synthesized Pma1 is degraded at 37°C (Fig. 1B). Unlike the Pma1-7 mutant, which we have previously characterized as defective for targeting to the plasma membrane (7), Pma1-10 appears to move correctly to the cell surface (Fig. 2). At 37°C, newly synthesized Pma1-10 is proteolytically cleaved during cell fractionation on Renografin density gradients (Fig. 2). The susceptibility to cleavage is likely a consequence of Pma1-10 misfolding. Indeed, Pma1-10 has extreme sensitivity to limited trypsinolysis (Fig. 2B). Because Pma1-10 is stabilized when internalization from the cell surface is prevented in end4–1 cells (Figs. 1B and 3A), we suggest that Pma1-10 degradation in vivo occurs after internalization from the plasma membrane. Degradation occurs on delivery to the vacuole because Pma1-10 is stabilized by pep4Δ (Figs. 1B and 3). Thus, newly synthesized Pma1-10 at 37°C fails to remain stable at the plasma membrane.
pma1-10 represents a valuable tool for future work to address how membrane protein stability is maintained. A genetic screen to isolate suppressors of pma1-10 may yield answers. In this regard, it is of interest that stabilization of Pma1-10, per se, without correct localization, is not sufficient to suppress the temperature-sensitive growth defect of pma1-10 cells: pep4Δ and vps27Δ stabilize Pma1-10 at the vacuole and prevacuolar compartment, respectively (Fig. 3), without allowing the cells to grow at 37°C (Fig. 1A). Similarly, Pma1-10 is stabilized in doa4Δ cells, as are other membrane protein substrates for vacuolar proteolysis (27, 29). Although Doa4 is a deubiquitinating enzyme required for degradation of substrates of the ubiquitin–proteasome pathway, it remains unclear whether Pma1-10 is ubiquitinated, or whether the effect of doa4Δ on Pma1-10 degradation is indirect.
Cell fractionation shows that Pma1-10, like wild-type Pma1, is delivered to the plasma membrane by 30-min chase (Fig. 2). Pma1 arrival at the cell surface occurs faster than previously suggested based on the time required for wild-type Pma1 to achieve maximal phosphorylation (17). Like wild-type Pma1, Pma1-10 at the permissive temperature is stable (Fig. 6A), and its steady-state distribution appears exclusively plasma membrane (X.G., unpublished result). However, temperature-shift experiments reveal that newly synthesized Pma1-10 is susceptible to degradation when shifted to 37°C after 30-min chase at 24°C (Fig. 6A). Degradation is Pep4-dependent, suggesting vacuolar delivery after internalization. Only after 1- to 2-h chase at the permissive temperature does Pma1-10 become resistant to degradation at 37°C. This suggests that Pma1-10 stabilization is achieved after arrival at the plasma membrane, perhaps by association with one or more plasma membrane components. Indeed, stabilization of the Na+,K+-ATPase of epithelial cells appears to depend on association with spectrin at the plasma membrane (30, 31).
Triton insolubility of Pma1-10 was measured to assess association with lipid rafts. Although lipid rafts were not isolated on density gradients in this study, it has been shown that Pma1 within the detergent-insoluble fraction is entirely contained in rafts (11). The failure to enter a Triton-insoluble fraction and hypophosphorylation of Pma1-10 at 37°C correlate with instability of Pma1-10 (Figs. 5 and 7). A model in which cell surface stability is maintained by association with lipid rafts is consistent with evidence from several cell types that lipid rafts are restricted from entering degradative compartments (32). Indeed, ergosterol and sphingolipid, the major lipid components of rafts, are excluded from the yeast vacuole (33). Nevertheless, kinetic experiments reveal that newly synthesized Pma1-10 becomes Triton-insoluble at 24°C before it becomes fully resistant to degradation on shift to 37°C (Fig. 6). Therefore, although association with lipid rafts may promote Pma1 stabilization, it is not sufficient for stabilization.
Acknowledgments
We thank Peter Arvan, Michel Bagnat, and Kai Simons for helpful comments. This work was supported by National Institutes of Health Grant GM 58212.
Abbreviations
- ALP
yeast alkaline phosphatase
- CPY
carboxypeptidase Y
References
- 1.Serrano R, Kielland-Brandt M C, Fink G R. Nature (London) 1986;319:689–693. doi: 10.1038/319689a0. [DOI] [PubMed] [Google Scholar]
- 2.Auer M, Scarborough G A, Kuhlbrandt W. Nature (London) 1998;392:840–843. doi: 10.1038/33967. [DOI] [PubMed] [Google Scholar]
- 3.Morsomme P, Slayman C W, Goffeau A. Biochim Biophys Acta. 2000;1469:133–157. doi: 10.1016/s0304-4157(00)00015-0. [DOI] [PubMed] [Google Scholar]
- 4.Harris S L, Na S, Zhu X, Seto-Young D, Perlin D, Teem J H, Haber J E. Proc Natl Acad Sci USA. 1994;91:10531–10535. doi: 10.1073/pnas.91.22.10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeWitt N D, Tourinho dos Santos C F, Allen K E, Slayman C W. J Biol Chem. 1998;273:21744–21751. doi: 10.1074/jbc.273.34.21744. [DOI] [PubMed] [Google Scholar]
- 6.Wang Q, Chang A. EMBO J. 1999;18:5972–5982. doi: 10.1093/emboj/18.21.5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang A, Fink G R. J Cell Biol. 1995;128:39–49. doi: 10.1083/jcb.128.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo W-j, Chang A. Mol Biol Cell. 2000;11:579–592. doi: 10.1091/mbc.11.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benito B, Moreno E, Lagunas R. Biochim Biophys Acta. 1991;1063:265–268. doi: 10.1016/0005-2736(91)90381-h. [DOI] [PubMed] [Google Scholar]
- 10.Kolling R, Losko S. EMBO J. 1997;16:2251–2261. doi: 10.1093/emboj/16.9.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bagnat M, Keranen S, Shevchenko A, Simons K. Proc Natl Acad Sci USA. 2000;97:3254–3259. doi: 10.1073/pnas.060034697. . (First Published March 14, 2000; 10.1073/pnas.060034697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simons K, Ikonen E. Nature (London) 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 13.Sherman F, Hicks J B, Fink G R. Methods in Yeast Genetics: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1986. [Google Scholar]
- 14.Boeke J D, Trueheart J, Natsoulis G, Fink G R. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 15.Raths S, Rohrer J, Crausaz F, Riezman H. J Cell Biol. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang A, Slayman C W. J Cell Biol. 1991;115:289–295. doi: 10.1083/jcb.115.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rose M D, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 19.Maldonado A M, Portillo F. J Biol Chem. 1995;270:8655–8659. doi: 10.1074/jbc.270.15.8655. [DOI] [PubMed] [Google Scholar]
- 20.Jones E. J Biol Chem. 1991;266:7963–7966. [PubMed] [Google Scholar]
- 21.Novick P J, Ferro S, Schekman R. Cell. 1981;25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- 22.Schandel K A, Jenness D D. Mol Cell Biol. 1994;14:7245–7255. doi: 10.1128/mcb.14.11.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang A, Rose M D, Slayman C W. Proc Natl Acad Sci USA. 1993;90:5805–5812. doi: 10.1073/pnas.90.12.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piper R C, Cooper A A, Yang H, Stevens T H. J Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raymond C K, Howald-Stevenson I, Vater C A, Stevens T H. J Cell Biol. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo W-j, Chang A. J Cell Biol. 1997;138:731–746. doi: 10.1083/jcb.138.4.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amerik A Y, Nowak J, Swaminathan S, Hochstrasser M. Mol Biol Cell. 2000;11:3365–3380. doi: 10.1091/mbc.11.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harder T, Simons K. Curr Opin Cell Biol. 1997;9:534–542. doi: 10.1016/s0955-0674(97)80030-0. [DOI] [PubMed] [Google Scholar]
- 29.Loayza D, Michaelis S. Mol Cell Biol. 1998;18:779–789. doi: 10.1128/mcb.18.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubreuil R R, Wang P, Dahl S, Lee J, Goldstein L S B. J Cell Biol. 2000;149:647–656. doi: 10.1083/jcb.149.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammerton R W, Krzeminski K A, Mays R W, Ryan T A, Wollner D A, Nelson W J. Science. 1991;254:847–850. doi: 10.1126/science.1658934. [DOI] [PubMed] [Google Scholar]
- 32.Simons K, Gruenberg J. Trends Cell Biol. 2000;10:459–462. doi: 10.1016/s0962-8924(00)01847-x. [DOI] [PubMed] [Google Scholar]
- 33.Zinser E, Daum G. Yeast. 1995;11:493–536. doi: 10.1002/yea.320110602. [DOI] [PubMed] [Google Scholar]