Abstract
Alternate frame folding (AFF) and protein/fragment exchange (FREX) are related technologies for engineering allosteric conformational changes into proteins that have no pre-existing allosteric properties. One of their chief purposes is to turn an ordinary protein into a biomolecular switch capable of transforming an input event into an optical or functional readout. Here, we present a guide for converting an arbitrary binding protein into a fluorescent biosensor with Förster resonance energy transfer output. Because the AFF and FREX mechanisms are founded on general principles of protein structure and stability rather than a property that is idiosyncratic to the target protein, the basic design steps—choice of permutation/cleavage sites, molecular biology, and construct optimization—remain the same for any target protein. We highlight effective strategies as well as common pitfalls based on our experience with multiple AFF and FREX constructs.
Keywords: AFF, Biosensor, Fluorescence, FRET, FREX, Protein design, Protein engineering
1 Introduction
A biosensor is minimally composed of an input module, which interacts with the analyte, and an output module that reports on that interaction. Proteins excel at both roles. As input domains they are masters of molecular recognition, having the ability to bind targets tightly and specifically amidst a sea of similar-looking decoys. As output domains they possess a wide array of biological functions, among the most useful of which for biosensing are fluorescence and enzymatic activity. A major additional advantage of a protein-based biosensor is that it is genetically encodable for in vivo applications.
The main challenge in designing a protein-based biosensor is solving the problem of how to couple input and output domains, both physically and functionally, so that binding the analyte produces a detectable signal. Nature has given us some treasured, but rare clues in the form of proteins that undergo large-scale conformational changes in response to ligand binding (e.g., calmodulin, which has launched a family of fluorescent calcium sensors). The great majority of proteins, however, do not change their structure appreciably upon binding.
Calbindin D9k, fibronectin 3 (Fn3), and ribose binding protein (RBP) are three such examples in which the structures of the proteins in their ligand-free and ligand-bound states are similar. To address this challenge, we developed two methodologies for engineering a large, binding-dependent conformational change into each protein, which was then detected by placement of either chemical fluorophores or fluorescent proteins (FPs) [1–4]. The methods are known as alternate frame folding (AFF) and protein/fragment exchange (FREX).
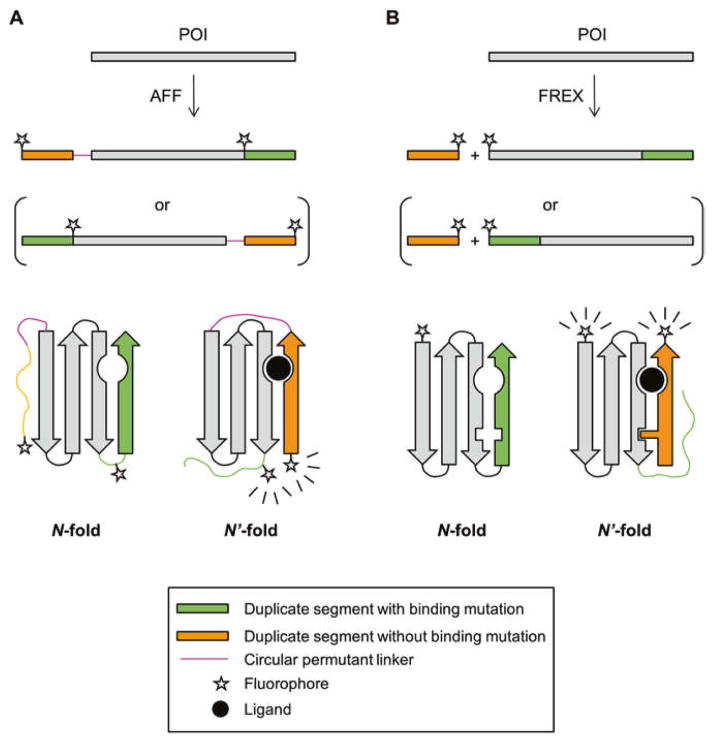
AFF and FREX both use partial sequence duplication to give a protein of interest (POI) a mutually exclusive choice between folding to its normal native state (N) or to an alternate form (N′) that possesses native-like structure and function (Fig. 1). Step 1 (see Subheading 3) of the protocol for creating an AFF-modified POI (POI-AFF) involves choosing an appropriate N-terminal or C-terminal segment of the POI to duplicate. One or more amino acids are identified in the segment that, when mutated, abrogate binding of the POI to its target ligand. Step 2 (see Subheading 4) entails attaching the duplicate copy of the N-terminal or C-terminal segment to the C- or N-terminus of the POI, respectively, by means of a peptide linker. Fig. 1a illustrates that POI-AFF can either fold by using the normal order of amino acids to yield N, or by using a rearranged order of amino acids to generate a circularly permuted structure (N′). The relative thermodynamic stabilities of N and N′ are tuned in Step 3 (see Subheading 5) of the protocol such that POI-AFF is predominantly in state N (or N′) in the absence of the target ligand, and chiefly in state N′ (or N) in its presence. Finally, fluorophores are incorporated at locations that consistently report on the N ⇆ N′ conformational change, independent of the choice of POI.
Fig. 1.
Schematic of AFF (a) and FREX (b) switching mechanisms. Primary amino acid sequences are indicated by horizontal bars with folded protein structures represented below the sequences. For the two sequences in parentheses an N-terminal segment (containing a critical binding residue) is duplicated. The other two sequences, and the structures that result from their folding, represent the analogous case in which a C-terminal segment is duplicated. Wavy lines indicate the copy of the duplicate segment that is orphaned in N and N′ conformations and is hence unfolded. The N-fold of POI-FREX is shown with a packing mutation in the green arrow that is swapped out by the wild-type residue from the orange arrow in the N′-fold
The FREX mechanism can be considered an intermolecular version of AFF, in which the duplicated segment is not covalently attached to the POI from which it was derived, but rather added in trans to the POI (Fig. 1b). The N′-state of a FREX-modified POI (POI-FREX) is the intermolecular complex of the POI and the fragment, which forms only in the presence of the target ligand and is detected by FRET between donor and acceptor fluorophores placed on the POI and fragment, respectively. The chief advantage of FREX and other two-component designs is that the ratiometric FRET change observed upon binding tends to be greater than that of single-component sensors, because FRET efficiency is typically reduced to near-zero values in the unbound state of the two-component sensors. The main limitation of FREX is that the POI and fragment should be present at close to equimolar concentrations to achieve maximum FRET response. The protocols for creating POI-AFF and POI-FREX sensors are very similar. We outline the protocol for AFF below, and enumerate the modifications for FREX after each step.
2 Gathering Ingredients: The POI
An available X-ray or other high-resolution structures of the POI or homolog thereof greatly facilitate the design process.
For AFF, the POI should ideally not contain any reduced Cys residues, as fluorophores are typically introduced by thiol-reactive chemistry. Cys in the POI may be tolerated vis-à-vis fluorescence labeling if they are buried and inaccessible to solvent. FREX can make use of fluorescent proteins for detection so reduced Cys residues are not inherently problematic. For both AFF and FREX, if oxidized Cys are present, the duplicate segment must be chosen (see Subheading 3) such that it is not crosslinked to the nonduplicated region of the POI by a disulfide bond.
Consider using the most stable variant of the POI available, e.g., one derived from a thermophilic organism. An axiom of protein folding is that it is far easier to destabilize a protein than to stabilize it. Accordingly, most of the modifications and tuning mutations employed herein either intentionally or unintentionally destabilize the POI. Starting with a stable template allows for greater design freedom.
3 Step 1 of AFF Protocol: Choosing the Segment of the POI to Duplicate
3.1 Identify a Binding Mutation
The only absolute requirement for the duplicate segment is that it contain at least one residue that, when mutated, greatly reduces affinity of the POI for its target ligand. Binding knockout mutations are often known from prior functional or genetic studies, and they may also be deduced from an existing crystal structure of the POI or homolog thereof. We have found that choosing a binding mutation close to the beginning or end of the amino acid sequence is advantageous, because this allows the duplicate segment to be short in length. Since the duplicated amino acids extend from POI-AFF as N- or C-terminal tails (Fig. 1a), shorter segments may present less of a risk for aggregation or degradation, although the opposite may be true. In any case, a binding mutation near one of the termini gives one the freedom to experiment with both short and long duplicate segments.
3.2 Identify a Circular Permutation Site
For a binding mutation near the C-terminus, the duplicated segment ends at the C-terminus (Fig. 1a). It begins at a position N-terminal to the binding mutation and this position is chosen to be a surface loop or turn. The reason is that this loop becomes the permutation site of the N′-fold, i.e., the location at which the polypeptide chain is broken and new N- and C-termini are generated. Interrupting an alpha helix, beta strand, or buried hydrophobic region is expected to be more destabilizing than disrupting a surface loop, although there are examples of successful permutation sites at the former locations [5–10]. The same considerations apply to the case where the binding mutation is near the N-terminus, except the duplicated segment begins with the N-terminus and ends at a surface loop C-terminal to the binding mutation (Fig. 1a).
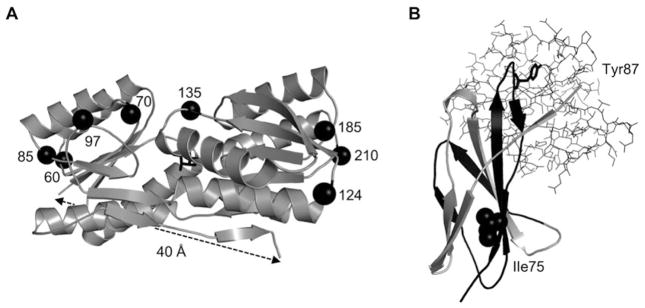
Inability to find a stable circular permutant (CP) is the most common failure point in the AFF protocol. Permutation almost always destabilizes a protein, and there is no reliable method for predicting the extent of destabilization for a given permutation site. The CP needs to be at least marginally stable (ΔGunfold ≥ 2–3 kcal/mol), with more stable CPs requiring less optimization (see Subheading 5). Our approach for selecting permutation sites is to choose the first three to four surface loops either N-terminal or C-terminal to the binding mutation, depending on whether the binding mutation is closer to the C-terminus or N-terminus, respectively. Loops that are close to the binding/active site should be avoided for functional reasons, although xylanase [9] and beta lactamase [11] were permuted at several loops proximal to their active sites without major loss of activity. Fortunately, all but the smallest POIs will have many loops from which to choose and at least one will usually be stable and functional enough for the AFF design. For example, RBP (277 amino acids) has 13 surface loops (Fig. 2a). We created CPs at eight of these loops and all were stable, soluble, and functional. All were destabilized compared to wild-type (WT) RBP, however, and this finding demonstrates the advantage of starting with the most stable variant of the POI available.
Fig. 2.
Examples of viable circular permutants for AFF and fragments for FREX. (a) Stable and functional CPs were generated by cleaving the RBP sequences at the eight surface loops centered around the positions indicated by black spheres, and joining the original termini by a Gly/Ala/Ser-based linker of 30 amino acids. The ribose ligand is shown as black sticks. (b) The Fn3-FREX sensor was created by duplicating residues 48 to the C-terminus (black segment). The binding mutation site (Tyr87) contacts the target ligand (SH2 domain, sticks) and the tuning mutation site (IIe75) packs against hydrophobic residues in the two gray beta strands shown
3.3 Design the Peptide Linker
The linker functions to physically bridge the original N- and C-termini of the POI. It effectively becomes a new surface loop of the CP. As such, the amino acid sequence should be hydrophilic and flexible enough to not impose any new constraints on the protein structure. We base our linkers on Gly/Ala/Ser repeats, although more advanced design criteria have been discussed [9, 11–15]. With regard to linker length, a rule of thumb is to measure the N-to-C distance (Cα–Cα) from the structure “as the crow flies,” and calculate length using ~2.5 Å per amino acid. For POIs in which the line-of-sight between termini is blocked by structure, we use a figure of <2.0 Å per amino acid to account for the arc that the linker must take over the curved surface of the protein. For example, RBP has an N-to-C distance of ~40 Å (Fig. 2a), which we spanned with a linker consisting of 30 amino acids [4]. If the N-to-C distance is not known precisely, it is best to err on the side of length, as we have found that using linkers longer than necessary does not dramatically destabilize the CP, in contrast to using linkers that are too short [16].
3.4 Characterize CPs
At this stage it is important to express and purify candidate CPs. CPs that exhibit degradation, aggregation, or loss of function should be rejected. The CPs are purified and their relative thermodynamic stabilities (as well as that of the POI containing binding mutation) are determined using chemical or thermal denaturation techniques [17] (see Subheadings 3.4.1 and 3.4.2). The most stable CP is then selected for AFF gene construction (Subheading 4). As a final note, fluorophores are typically introduced into the AFF protein by means of two Cys residues introduced at the N-terminus and in the surface loop of the N-fold selected as the permutation site. Although these Cys do not generally affect structure or stability of WT or CP forms of the POI, it is prudent to incorporate them into the constructs at this point to most accurately represent the N and N′ folds of POI-AFF.
3.4.1 Obtaining Stability Parameters from Chemical Denaturation Curves with Urea or Guanidine Hydrochlorie (GdnHCl)
Prepare solution A and solution B of the desired buffer, pH, salt, etc. The two solutions are made identically except solid, ultrapure urea (final concentration of 8 M) or GdnHCl (final concentration of 6 M) is added to solution B prior to the aliquots of stock buffer, salt, etc.
Add identical aliquots of concentrated protein to solution A and solution B. The final protein concentration depends on the instrumentation used to monitor unfolding, but typical concentrations for fluorescence and circular dichroism (CD) are 1–20 μM.
Prepare at least 25 samples consisting of evenly spaced mixtures (in denaturant concentration) of solution A and solution B. For example, sample #1 is 100% solution A, sample #25 is 100% solution B, and samples #2–24 contain linearly increasing concentrations of denaturant. Use of a two-syringe Hamilton dilutor or a manual repeating pipet is recommended to minimize denaturant concentration error. Incubate samples at the desired temperature until equilibrium is reached (typically ≥2 h).
-
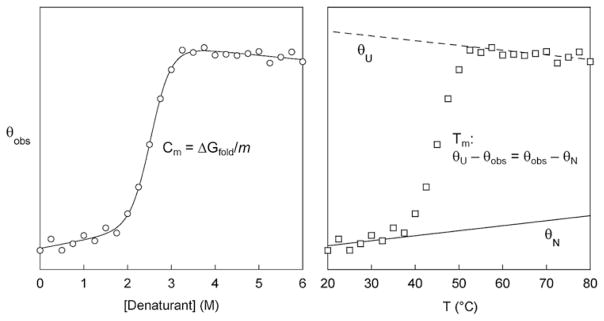
Scan samples using the instrument of choice (UV/Vis spectrophotometer, fluorimeter, CD spectrophotometer) at the desired wavelength. The observed signal (θobs) follows a sigmoidal curve, as shown in Fig. 3 (left). Fit θobs to the linear extrapolation equation (Eq. 1):
(1) where [D] is denaturant concentration and parameters θN and θU are the signals of the native and unfolded forms of the protein at zero denaturant concentration, sN and sU are the slopes of the native and unfolded baselines, ΔGfold is the stability of the protein at zero denaturant concentration, and m is the cooperativity parameter. The midpoint of chemical denaturation (Cm; equal to ΔGunfold/m) is a particularly useful parameter for ranking the relative stabilities of related protein variants with similar m-values (e.g., the POI and mutants thereof), because it is more accurate and reproducible than ΔGunfold.
Fig. 3.
Simulated chemical (left) and thermal (right) denaturation curves illustrating how Cm and Tm values are calculated and interpolated, respectively. Lines are best fits of the data to the linear extrapolation equation (left) and to linear portions of the native and unfolded baselines (right)
3.4.2 Obtaining Stability Parameters from Thermal Denaturation Curves
Prepare a solution of protein in the desired buffer. For CD-monitored denaturation, one should generally use dilute protein solutions (1–5 μM) and a long path length cuvette (1 cm) to minimize aggregation at higher temperatures.
To establish native and unfolded baselines, start the melt at a temperature at least 10 °C below the beginning of the unfolding transition, and continue the melt at least 10 °C after the transition is 90% complete. After the melt is finished, let the sample equilibrate at the starting temperature and record the CD signal. Thermal denaturation is considered reversible if the signal returns to ≥90% of its original value.
One can obtain ΔGfold and melting temperature (Tm) parameters by fitting θobs to the v’ant Hoff equation (17) if denaturation is reversible. However, since thermal transitions are often irreversible, and because we are mostly interested in the relative stabilities of closely related protein variants, it is sufficient to report apparent Tm values by interpolating the midpoints of melting curves obtained under identical solution and heating conditions. To do so, fit the linear portions of the native and unfolded baselines as shown in Fig. 3 (right) and calculate θN and θU at each experimental temperature. Interpolate the temperature at which θU−θobs= θobs−θN.
3.5 Modifications to Step 1 for FREX
The steps described in Subheadings 3.1 and 3.2 are identical in the FREX protocol. Just as there is no method for predicting CP stability as a function of permutation site, there is no means for predicting the affinity of two complementary protein fragments based on the location of the cleavage site. We therefore apply the same criteria in choosing the binding mutation and duplicate segment for FREX. The steps described in Subheadings 3.3 and 3.4 do not apply to FREX. At this design stage, however, one should identify a site in the POI for making tuning mutations. A tuning mutation is often needed for AFF and will always be necessary for FREX, since the duplicate fragment will not form a complex with the POI (at reasonable fragment concentrations) unless a structural defect is introduced into the POI which is then “swapped out” by fragment binding. A reliable tuning mutation site consists of a large, buried hydrophobic residue (Leu, Ile, Phe, or Tyr) that is in the duplicated portion of the POI and packs against hydrophobic residues in the unduplicated portion (Fig. 2b). A single tuning site should (and usually can) be selected that will service all fragment lengths to be tested. Tuning consists of substituting progressively smaller hydrophobic residues (Val, Ala, Gly, and for extreme destabilization, a charged residue) into that site to progressively destabilize the POI.
4 Step 2 of AFF Protocol: Gene Construction
AFF genes are generated by constructing two half-genes corresponding to the N- and N′-frames, mutating them as needed, and then fusing them together. Keeping the frames separate is necessary because mutagenic PCR primers will bind to two locations in the full-length AFF gene due to sequence duplication. This redundancy also makes it difficult to obtain fully synthetic AFF genes from commercial sources.
A His6–8 sequence or other purification/expression tag can be introduced at this point. The tag should be expressed at the terminus of the protein that comprises the N′-fold, i.e., at the N-terminus of POI-AFF if a C-terminal fragment was duplicated, or at the C-terminus if an N-terminal fragment was duplicated. This arrangement will help select against purifying degradation products, because if a cellular protease cleaves POI-AFF it will likely do so by attacking the less stable N′-fold. The following steps describe construction of an AFF-POI in which a C-terminal fragment is duplicated and appended to the N-terminus; analogous steps are taken for an AFF-POI with a duplicated N-terminal fragment.
4.1 N-frame Half-Gene
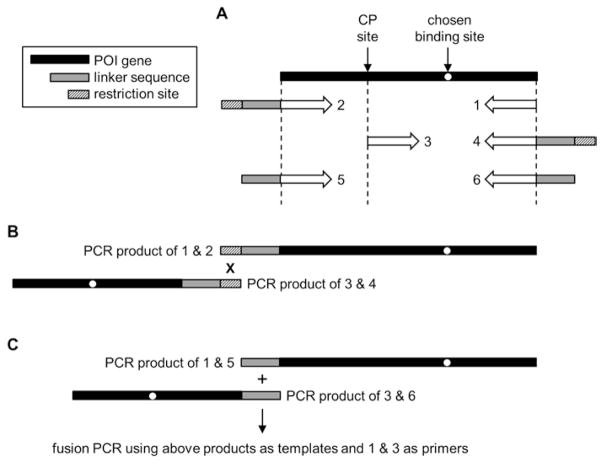
Introduce binding, Cys, and tuning mutations into the WT POI gene as dictated in Subheading 3. Amplify the N-frame half-gene by annealing primer 1 (containing a stop codon) to the 3′-end of the above-modified POI gene, and primer 2 to the 5′-end (Fig. 4a). Primer 2 encodes for the C-terminal half of the desired linker and bears a restriction site of choice at its 5′-end. The restriction site necessarily becomes incorporated into the linker and for this reason we favor the Not I restriction site because it encodes for Ala-Ala-Ala.
Fig. 4.
Cloning strategy for constructing AFF genes, (a) Annealing sites and compositions of primers 1–6 are indicated below the POI gene. The restriction enzyme digestion/ligation and fusion PCR methods for connecting the N and N′ half-genes are shown in (b, c), respectively
4.2 N′-frame Half-Gene
Introduce an N′-frame tuning mutation into the WT POI gene if necessary. Design primer 3 such that it binds the POI gene at a position that defines the start of the duplicate polypeptide segment (and the N-terminus of POI-AFF) (Fig. 4a). Primer 3 begins with a codon for Cys. Primer 4 anneals to the 3′-end of the POI gene, encodes for the N-terminal half of the linker, and ends with the same restriction site as in Subheading 4.1. PCR amplify to generate the N′-frame half-gene. Digest the two half-genes with the restriction enzyme. The products are now ready to be ligated and subcloned into an expression vector of choice (Fig. 4b).
4.3 Alternate Method: Fusion PCR
The amino acids imposed by the restriction site usually have little effect on the properties of the linker, especially when the linker is long. For short linkers or those that must be composed of a particular sequence (e.g., protease site), these leftover residues may be undesirable. The alternate method allows one to eliminate the restriction site signature altogether. Construct the N-frame PCR template as in Subheading 4.1 except use primer 5 instead of primer 2 (Fig. 4a). Primer 5 contains enough of the linker sequence to overlap with primer 6 by at least 20 base pairs; this can correspond to the full or partial linker sequence depending on linker length. Primer 5 does not contain a restriction site. Create the N′-frame template as in Subheading 4.2 except use primer 6 instead of primer 4. Primer 6 binds to the 3′-end of the POI gene and ends with a full or partial linker sequence, complementary to that in primer 5 with at least a 20 base pair overlap, again without a restriction site. PCR amplify the N′-frame template by annealing primer 3 and primer 6 to the POI gene. Mix the N-frame template, N′-frame template, primer 1, and primer 3, and generate the full-length AFF gene using the overlap extension PCR method (Fig. 4c) [18]. Primer 1 and primer 3 will each anneal to a second, undesirable site within the N′-frame and N-frame template, respectively, and this results in shorter PCR products. Purify the longest PCR product by agarose gel and subclone into the expression vector.
4.4 Modifications to Step 2 for FREX
The cloning steps for creating FREX constructs are simpler, since the duplicated DNA sequences are never physically joined and no linker is present. At this stage, we find it useful to fuse the gene of a donor and acceptor FP to the either end of the POI and fragment genes. In addition to providing a direct binding assay for sensor tuning (Subheading 5), an FP serves as a carrier protein to help the fragment express well, resist degradation, and stay soluble in cells.
5 Step 3 of AFF Protocol: Optimization
The objective of this step is to tune the thermodynamics of the sensor so that it is mainly in the N-form in the absence of ligand, and switches to the N′-form upon ligand binding. For the sake of discussion, we assume the most common outcome of Subheading 3, i.e., that the CP form of the POI was found to be less stable than the WT POI, and that the binding mutation was introduced into the N-frame. The ideal distribution of N:N′ populations in the absence of ligand is ~10:1. This scenario, which corresponds to the N-fold being ~1.4 kcal/mol more stable than the N′-fold, achieves an optimal balance of near-maximum fluorescence signal change and minimal reduction of observed ligand-binding affinity1. Which tuning mutation (if any) needs to be made to achieve this balance, and into which frame it should be placed, can be predicted from the results of Subheading 3.4. Introducing this mutation into POI-AFF during Subheading 4 can save time by requiring less subsequent optimization in Subheading 5.
5.1 Binding Positive Control
A common problem with POI-AFFs prior to their thermodynamic balancing is that the N-fold is so much more stable than the N′-fold that the ligand binds weakly or not at all. This snag is likely to be encountered if the POI was found in Subheading 3.4 to be much more stable than the CP (e.g., ΔTm > 10 °C or ΔΔGunfold > 3 kcal/mol; see Subheadings 3.4.1 and 3.4.2). We therefore recommend that one initially perform a positive binding test using saturating levels of ligand by means of a rapid and not necessarily quantitative assay including size exclusion chromatography, affinity pull-down, or shift in Tm or Cm. If no binding is observed, then one or more destabilizing mutations are introduced into the N-fold, e.g., substituting Gly, Ala, or Val at one or more hydrophobic packing sites identified in Subheading 3. A positive binding result indicates that the protein has adopted the N′-fold in the presence of ligand but does not reveal whether it had switched to the N′-fold from the N-fold, or was already in the N′-fold in the absence of ligand.
5.2 Stability Tuning
Once a binding interaction has been established, the final step is to optimize the sensor for maximum fluorescence response and binding affinity. Fluorescent donor and acceptor groups are attached to POI-AFF at its N-terminus and in the permutation loop of the N-frame. We favor attaching maleimide dyes to engineered Cys residues, although there are a variety of alternate chemistries and labeling strategies from which to choose2. The important considerations are that one obtains close to a 1:1 donor:acceptor ratio and that the chosen dyes are sensitive to short-range distance changes (short Förster radii for FRET), as they will be close enough to be in contact in the N′-fold and of variable distance apart in the N-fold, depending on the length and residual structure of the duplicated segment. To achieve 1:1 donor:acceptor labeling one may have to experiment with different ratios of dyes in the labeling step. We have also obtained satisfactory results by labeling both positions with a single fluorophore (pyrene excimer formation and BODIPY-FL self-quenching).
To evaluate sensor performance, increasing amounts of ligand are added to a fixed concentration of fluorescently labeled sensor. Fitting the observed fluorescence (θobs) to the one-site binding equation (Eq. 2) yields the dissociation constant (Kd) and fluorescence change (Δθ=θbound−θfree) as the parameters. LT and PT are total concentrations of ligand and protein.
| (2) |
The three possible outcomes are that a binding curve: (i) is obtained with a fitted Kd value close to that of WT POI; (ii) is obtained with a fitted Kd significantly larger than that of WT POI; and (iii) cannot be generated because no fluorescence change is observed. Outcome (i) indicates that no further optimization is necessary and the sensor is ready for use. Result (ii) suggests that the N-fold is too stable and that a moderately destabilizing mutation should be introduced into the N-frame. If Δθ is close to zero (outcome (iii)) then one can conclude that POI-AFF either never switched conformation (i.e., it was in the N′-fold even in the absence of ligand), or that it changed conformation and the fluorophores did not report on the change. In the former scenario the solution is to introduce tuning mutations into the N′-frame instead of the N-frame. The expectation is that Δθ will initially increase and then approach a maximum value as the N′-fold is progressively destabilized by packing mutations of increasing severity [3]. Any one of these variants can be used for sensing, although for the typical case in which a balance between high Δθ and low Kd is desired, the mutant that yields outcome (i) is selected as the final optimized product.
5.3 Optional: Kinetic Tuning
The AFF sensing mechanism is reversible and the speed at which the forward and reverse conformational changes occur determines how rapidly the sensor can detect fluctuations in analyte concentration. It may be desirable in some applications to increase this response rate. For calbindin D9k and RBP, the rate limiting step of the N→N′ and N′→N reactions appears to involve a local but not global unfolding event, most likely unfolding/dissociation of the copy of the duplicate segment that is docked to the shared segment of the protein. If this result is general, any mutation that destabilizes the interaction between the shared and duplicate regions has the potential to accelerate the conformational switch. If one wishes to attempt to engineer a faster AFF sensor, we recommend first following all of the thermodynamic balancing procedures described above, then introducing kinetic tuning mutation(s) in a manner that does not perturb the existing N/N′ balance or ligand-binding affinity. For example, consider a hypothetical 200-amino acid POI from which residues 150–200 were duplicated to generate POI-AFF. Further suppose that residues Phe130 and Leu170 pack tightly against each other. Kinetic tuning could consist of a single Phe130→Ala substitution in the shared region, or Leu170→Ala + Leu170′→Ala mutations in the respective duplicate segments. In either case the N-fold and N′-fold are in principle destabilized equally but the kinetic barrier to their interconversion may be lowered, depending on the extent to which the Phe130-Leu170/Leu170′ interaction is formed in the transition state ensemble.
5.4 Modifications to Step 3 for FREX
As noted, it is simplest to fuse donor and acceptor FPs to the full-length POI and duplicate fragment at the cloning stage; however, if chemical fluorophores are desired then the labeling procedure is identical to that of AFF. One difference of FREX is that the two components of the sensor are on average very far apart in the absence of ligand (at reasonable protein concentrations), so donor/acceptor pairs with a wide range of Förster distances can be considered.
Thermodynamic tuning for FREX consists of binding experiments in which ligand is added to equimolar concentrations of fluorescently tagged full-length POI and fragment. For example, we optimized the Fn3-FREX sensor by screening the I75V, I75A, and I75G tuning variants of the POI in the background of the Y87A binding mutation (Fig. 2b). Fluorescence data are fit to Eq. 2. Our simulations have shown that it is possible, although highly unlikely, that a ternary complex will form in the absence of a packing mutation, as the entropic barrier for intermolecular folding is usually too large to be overcome by ligand-binding energy alone [3]. A more realistic concern is that the binary complex of full-length POI and fragment will form without ligand present. This occurs when the full-length POI is “overbalanced,” i.e., so destabilized by the tuning mutation that restoration of the native packing interaction by the fragment drives folding in the absence of ligand binding. It is not uncommon to span these extremes of stability by placing a set of packing mutations at a single well-chosen site. A nicely balanced FREX construct is characterized by unusually high Δθ (achieved by reducing FRET in the free state to near zero) and Kd similar to that of the WT POI. This condition was met in the Fn3-FREX sensor by the I75A tuning mutation.
6 Concluding Remarks
Both AFF and FREX involve a folding competition akin to a two-person game of molecular musical chairs: one copy of the duplicate segment is left standing and is at least partially unfolded at any given time. One must be alert to degradation and/or aggregation originating from the orphaned copy. Computational methods for predicting the stability and solubility of CPs, protein fragments, and complexes thereof will greatly facilitate the design of FREX and AFF sensors, as well as that of conformational switches based on other mechanisms.
Acknowledgments
This work was supported by NTH grant R01 GM115762 to S.N.L.
Abbreviations
- AFF
Alternate frame folding
- CP
Circular permutant
- Fn3
Fibronectin 3
- FP
Fluorescent protein
- FRET
Förster resonance energy transfer
- FREX
Protein/fragment exchange
- N
Normal fold of protein
- N′
Alternate fold of protein
- POI
Protein of interest
- RBP
Ribose-binding protein
- WT
Wild-type
Footnotes
Because a portion of the binding energy is used to drive the N → N′ conformational change, the observed Kd will be greater than the intrinsic Kd of the POI by a factor of (1 + KN+KN′)/KN, where K= exp (−ΔGunfold/R.T) for the respective N- and N′-folds [3].
Existing AFF sensors have thus far made exclusive use of chemical fluorophores rather than genetically encoded FPs. The reason is that inserting an FP into the permutation loop of the N-frame would likely destabilize the N-fold and necessitate extensive thermodynamic rebalancing. It may be possible, however, to insert a CP form of the FP (in which the close proximity of its N- and C-termini would be less perturbing to the POI), or to move the FP from the permutant loop to the C-terminus of POI-AFF.
References
- 1.Stratton MM, Mitrea DM, Loh SN. A Ca2+-sensing molecular switch based on alternate frame protein folding. ACS Chem Biol. 2008;3:723–732. doi: 10.1021/cb800177f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stratton MM, Loh SN. On the mechanism of protein fold-switching by a molecular sensor. Proteins. 2010;78:3260–3269. doi: 10.1002/prot.22833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng H, Bi J, Krendel M, Loh SN. Converting a binding protein into a biosensing conformational switch using protein fragment exchange. Biochemistry. 2014;53(34):5505–5514. doi: 10.1021/bi500758u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ha JH, Shinsky SA, Loh SN. Stepwise conversion of a binding protein to a fluorescents witch: application to Thermoanaerobacter tengcongensis ribose binding protein. Biochemistry. 2013;52(4):600–612. doi: 10.1021/bi301105u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graf R, Schachman HK. Random circular permutation of genes and expressed polypeptide chains: application of the method to the catalytic chains of aspartate transcarbamoylase. Proc Natl Acad Sci U S A. 1996;93(21):11591–11596. doi: 10.1073/pnas.93.21.11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hennecke J, Sebbel P, Glockshuber R. Random circular permutation of DsbA reveals segments that are essential for protein folding and stability. J Mol Biol. 1999;286(4):1197–1215. doi: 10.1006/jmbi.1998.2531. [DOI] [PubMed] [Google Scholar]
- 7.Iwakura M, Nakamura T, Yamane C, Maki K. Systematic circular permutation of an entire protein reveals essential folding elements. Nat Struct Biol. 2000;7(7):580–585. doi: 10.1038/76811. [DOI] [PubMed] [Google Scholar]
- 8.Qian Z, Lutz S. Improving the catalytic activity of Candida antarctica lipase B by circular permutation. J Am Chem Soc. 2005;127(39):13466–13467. doi: 10.1021/ja053932h. [DOI] [PubMed] [Google Scholar]
- 9.Reitinger S, Yu Y, Wicki J, Ludwiczek M, D’Angelo I, Baturin S, Okon M, Strynadka NC, Lutz S, Withers SG, McIntosh LP. Circular permutation of Bacillus circulans xylanase: a kinetic and structural study. Biochemistry. 2010;49(11):2464–2474. doi: 10.1021/bi100036f. [DOI] [PubMed] [Google Scholar]
- 10.Carlson HJ, Cotton DW, Campbell RE. Circularly permuted monomeric red fluorescent proteins with new termini in the beta-sheet. Protein Sci. 2010;19(8):1490–1499. doi: 10.1002/pro.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guntas G, Mansell TJ, Kim JR, Ostermeier M. Directed evolution of protein switches and their application to the creation of ligand-binding proteins. Proc Natl Acad Sci U S A. 2005;102(32):11224–11229. doi: 10.1073/pnas.0502673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwakura M, Nakamura T. Effects of the length of a glycine linker connecting the N-and C-termini of a circularly permuted dihydrofolate reductase. Protein Eng. 1998;11(8):707–713. doi: 10.1093/protein/11.8.707. [DOI] [PubMed] [Google Scholar]
- 13.Flores G, Soberon X, Osuna J. Production of a fully functional, permuted single-chain penicillin G acylase. Protein Sci. 2004;13(6):1677–1683. doi: 10.1110/ps.03436604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian Z, Horton JR, Cheng X, Lutz S. Structural redesign of lipase B from Candida antarctica by circular permutation and incremental truncation. J Mol Biol. 2009;393(1):191–201. doi: 10.1016/j.jmb.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Correia BE, Holmes MA, Huang PS, Strong RK, Schief WR. High-resolution structure prediction of a circular permutation loop. Protein Sci. 2011;20(11):1929–1934. doi: 10.1002/pro.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butler JS, Mitrea DM, Mitrousis G, Cingolani G, Loh SN. Structural and thermodynamic analysis of a conformationally strained circular permutant of barnase. Biochemistry. 2009;48(15):3497–3507. doi: 10.1021/bi900039e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimsley GR, Trevino SR, Thurlkill RL, Scholtz JM. Determining the conformational stability of a protein from urea and thermal unfolding curves. Curr Protoc Protein Sci. 2013;Chapter 28(Unit28):24. doi: 10.1002/0471140864.ps2804s71. [DOI] [PubMed] [Google Scholar]
- 18.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]