Abstract
Podocytes are morphologically complex cells, the junctions of which form critical elements of the final filtration barrier. Disruption of their foot processes and slit diaphragms occur early in the development of many glomerular diseases. Here, we biochemically purified fractions enriched with slit diaphragm proteins and performed a proteomic analysis to identify new components of this important structure. Several known slit diaphragm proteins were found, such as podocin and nephrin, confirming the validity of the purification scheme. However, proteins on the apical membrane such as podocalyxin were neither enriched nor identified in our analysis. The chloride intracellular channel protein 5 (CLIC5), predominantly expressed in podocytes, was enriched in these fractions and localized in the foot process apical and basal membranes. CLIC5 colocalized and associated with the ezrin/radixin/moesin complex and with podocalyxin in podocytes in vivo. It is important to note that CLIC5−/− mice were found to have significantly decreased foot process length, widespread foot process abnormalities, and developed proteinuria. The ezrin/radixin/moesin complex and podocalyxin were significantly decreased in podocytes from CLIC5−/− mice. Thus, our study identifies CLIC5 as a new component that is enriched in and necessary for foot process integrity and podocyte function in vivo.
Keywords: cytoskeleton, glomerular disease, podocyte, proteinuria
The podocyte is a postmitotic, structurally complex cell in the glomeruli of kidneys. Podocytes reside on the outer aspect of the glomerular basement membrane in the urinary space. Podocytes extend large and small processes that terminate in structures called foot processes.1 These foot processes interdigitate with neighboring foot processes that are further connected by ‘zipper-like’ protein complexes called slit diaphragm (SDs).2 In addition to endothelial cells and the glomerular basement membrane, podocytes, through their tight junction-like SDs, form the final filtration barrier that is essential for the filtering units of the kidneys.
Glomerular diseases are the most common cause of adult chronic and end-stage renal diseases, and current therapies are limited. There is a growing body of literature elucidating the importance of podocyte injury in the pathogenesis of many glomerular diseases.3–13 Two important studies using transgenic methods to make podocytes selectively susceptible to diphtheria toxin or anti-CD25 immunotoxin revealed that podocyte loss leads directly to glomerulosclerosis.8,9 Genetic evidence pointing to the importance of SD dysfunction in glomerular diseases comes from the existence of several human congenital and familial nephrotic syndromes that encode for mutations in SD proteins, such as nephrin, podocin, CD2AP, and TRPC6.14–20 In addition, studies in knockout mice implicate the loss of SDs and foot processes in these diseases.21 Given the importance of the SD complex in maintaining foot process integrity and function,22,23 we used a proteomics approach to identify molecules that are likely to make up the SD to ascertain their physiological function in podocytes. One such protein identified was chloride intracellular channel protein 5 (CLIC5).
CLIC5 was originally identified as a protein that associates with ezrin and is tightly associated with the cytoskeleton of placental microvilli.24,25 CLIC5 is a member of a family of intracellular chloride channels that are known to have a putative single transmembrane domain and have been shown to localize in the plasma membrane, cytosol, and intracellular organelles.25–28 CLIC proteins are not only capable of transporting chloride but can also function as nonselective ion channels.29–32 The topology of CLICs is highly divergent from other chloride channels and evidence of their function as ion channels in vivo is lacking, raising the possibility that they have other functions, such as cytoskeletal scaffolding.25,33,34 spontaneous CLC5−/− mouse, Jitterbug, has hearing loss and vestibular dysfunction from destruction of the stereocilia of cochlear and vestibular hair cells.35 Here, we identify CLIC5 as a protein enriched in podocyte processes that associates with the ezrin/radixin/moesin (ERM) complex and with podocalyxin. We further show the importance of CLIC5 for the integrity of foot processes and the filtration barrier in vivo.
RESULTS
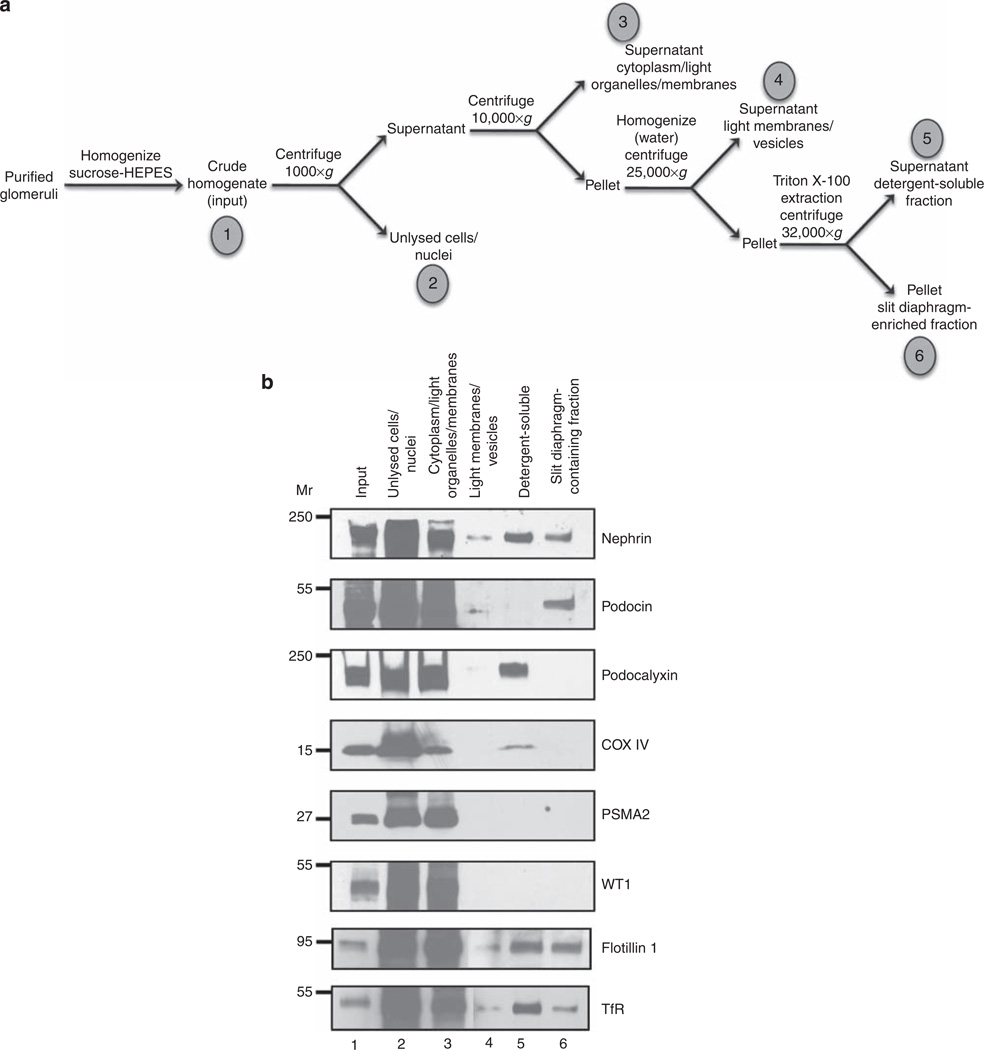
Biochemical isolation and enrichment of slit diaphragm proteins
The slit diaphragm is a protein complex that tightly interconnects two opposing membranes, similar to gap junctions and the synapses of neurons. This structure consists of a tightly associated complex of proteins involved in maintaining foot process integrity. Mutations of these known proteins have been linked to congenital and hereditary forms of nephrotic syndrome, such as α-actinin-4, nephrin, CD2AP, and podocin. We reasoned that because the slit diaphragm complex is similar to a tight junction, many of its constituents should be resistant to detergent and be bound or tightly associated with the plasma membrane. On the basis of this hypothesis, we have designed and validated a biochemical purification method to purify, or at least enrich for, proteins that make up the slit diaphragm complex. To this end, glomeruli that were purified from adult rats were homogenized in a sucrose-HEPES buffer, and nuclei and unlysed cells were removed from the extract by centrifugation. The supernatants were then centrifuged to sediment the dense membranes. This membrane pellet was then homogenized again in a hypotonic buffer and centrifuged. The pellet was detergent-extracted with Triton X-100 and centrifuged to isolate the insoluble protein fraction containing slit diaphragm proteins. A schematic diagram of the enrichment procedure is shown in Figure 1a. All of these fractions were analyzed by immunoblotting. Proteins known to be located in the slit diaphragm, such as nephrin and podocin, were present in the final insoluble pellet, indicating that this fraction did contain slit diaphragms (Figure 1b). Proteins located in the nucleus, mitochondria, and cytoplasm, WT1, COX IV, and PSMA2, respectively, were all located in earlier fractions of the purification protocol (Figure 1b). None of these proteins were detected in appreciable amounts in the final slit diaphragm-containing fraction. Podocalyxin, a protein located in the apical surface of foot processes, but not localized in the slit diaphragm itself, was not enriched in the final slit diaphragm-containing fraction (Figure 1b). This suggests that this protocol did enrich for proteins located specifically in the slit diaphragm. It is important to note that nephrin and podocin were present in the final protein fraction at considerably higher amounts than many other proteins. Membrane microdomains known as lipid rafts can be isolated from cells on the basis of their detergent insolubility. To ascertain to what extent the slit diaphragm-enriched fraction resembled a lipid raft, these fractions were immunoblotted with antibodies to flotillin 1 and transferrin receptor, markers of lipid rafts and nonraft domains, respectively. The final slit diaphragm-enriched fraction contained both flotillin 1 and transferrin receptor (Figure 1b), indicating that this purification procedure did not simply isolate lipid rafts.
Figure 1. Biochemical enrichment of slit diaphragm constituents.
Mouse glomeruli were isolated from kidney cortices, homogenized and fractionated to enrich for SD proteins. (a) The enrichment method is diagrammed schematically, indicating each fraction that was analyzed by immunoblotting (fractions 1–6). (b) An equal volume of extract was analyzed in lanes 2–6 to allow for a direct comparison between fractions. Replicate blots were probed with nephrin, podocin, podocalyxin, COX IV, PSMA2, WT1, flotillin 1, and transferrin receptor (TfR) antibodies (labeled to the right of each panel). As predicted, nephrin and podocin were located in the SD-enriched fraction, but podocalyxin, COX IV, PSMA2, and WT1 were absent from this fraction. Podocalyxin, COX IV, PSMA2, WT1, flotillin 1, and TfR receptor served as markers for apical membranes, mitochondria, cytoplasm, nuclei, lipid rafts, and nonraft domains, respectively.
Proteomic analysis of the slit diaphragm-enriched fraction
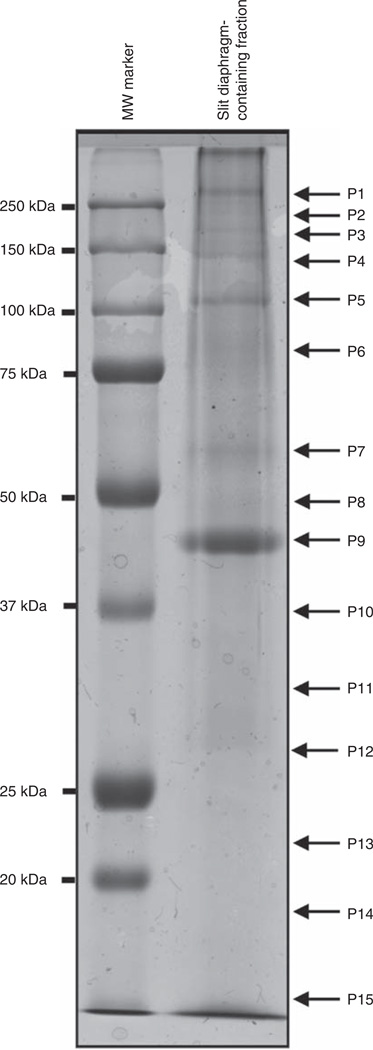
To identify proteins that are located in the slit diaphragm, we scaled up the purification procedure for proteomic analysis using glomeruli from 20 young adult rats. These fractions were then separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and the gel was stained with Coomassie blue (Figure 2). The lanes containing the slit diaphragm-enriched proteins were then cut and the proteins were in-gel digested with trypsin. The peptide fragments were then separated and analyzed by liquid chromatography-mass spectrometry/mass spectrometry (LC-MS). Two independent protein purifications and proteomic analyses were performed for protein identification, and these data were organized and analyzed according to their molecular weights. A partial list of identified proteins is given in Table 1.
Figure 2. Proteomic analysis of the SD-enriched fraction.
Protein fractions that were enriched in SD proteins were separated according to molecular weight using sodium dodecyl sulfate–polyacrylamide gel electrophoresis and stained with Coomassie blue. In all, 15 regions of the lane were excised, labeled from P1 to P15, and digested in gel with trypsin for further proteomic analysis.
Table 1.
List of proteins identified in the slit diaphragm-enriched fraction
| Fraction | Approximate MW range |
Entry | Description | No. of peptides |
Percentage coverage |
Mascot score |
|---|---|---|---|---|---|---|
| P1 | 300–250 | PLEC1_mouse | Plectin-1 | 16 | 5 | 270 |
| FINC_mouse | Fibronectin | 2 | 1 | 61 | ||
| FLNA_mouse | Filamin-A | 15 | 8 | 391 | ||
| FLNB_mouse | Filamin-B | 10 | 4 | 149 | ||
| P2 | 250–200 | NPHN_rat | Nephrin | 5 | 6 | 187 |
| SPTA2_rat | Spectrin-α chain, brain | 70 | 61 | 3005 | ||
| SPTB2_mouse | Spectrin-β chain, brain 1 | 61 | 52 | 2270 | ||
| MYH9_rat | Myosin-9 | 59 | 52 | 2277 | ||
| MYH10_mouse | Myosin-10 | 19 | 15 | 551 | ||
| TLN1_mouse | Talin-1 | 12 | 5 | 191 | ||
| TLN2_mouse | Talin-2 | 5 | 5 | 49 | ||
| ZO1_mouse | ZO-1 | 2 | 1 | 62 | ||
| P3 | 200–170 | MYH11_mouse | Myosin-11 | 18 | 10 | 457 |
| MYO6_mouse | Myosin-VI | 8 | 8 | 148 | ||
| CLH_mouse | Clathrin heavy chain | 4 | 4 | 60 | ||
| P4 | 170–120 | AT1A1_mouse | Sodium/potassium-transporting ATPase subunit α-1 | 12 | 22 | 389 |
| SYNPO_rat | Synaptopodin | 12 | 24 | 259 | ||
| KIRR1_rat | Kin of IRRE-like protein 1 | 2 | 5 | 65 | ||
| ARVC_mouse | Armadillo repeat protein deleted in velo-cardio-facial syndrome homolog |
1 | 1 | 56 | ||
| MYO1B_rat | Myosin-lb | 2 | 3 | 39 | ||
| MYO1C_mouse | Myosin-lc | 31 | 48 | 946 | ||
| VINC_mouse | Vinculin | 13 | 17 | 190 | ||
| CTND1_mouse | Catenin δ-1 | 10 | 12 | 186 | ||
| MYO1D_rat | Myosin-ld | 13 | 19 | 173 | ||
| MYO1E_rat | Myosin-le | 3 | 3 | 101 | ||
| NCKP1_mouse | Nck-associated protein 1 | 4 | 3 | 86 | ||
| P5 | 120–90 | ACTN4_mouse | α-Actinin-4 | 24 | 66 | 1533 |
| ACTN1_rat | α-Actinin-1 | 21 | 52 | 1265 | ||
| CTNA1_mouse | Catenin α-1 | 19 | 52 | 784 | ||
| CTNB1 | Catenin β-1 | 11 | 23 | 272 | ||
| AP2A2_rat | AP-2 complex subunit α-2 | 18 | 37 | 380 | ||
| AP2B1_mouse | AP-2 complex subunit β-1 | 7 | 17 | 111 | ||
| CTNA3_mouse | Catenin α-3 | 2 | 4 | 56 | ||
| GIT1_mouse | ARF GTPase-activating protein GIT-1 | 3 | 6 | 45 | ||
| P6 | 90–65 | GRP78_mouse | 78 kDa glucose-regulated protein precursor | 7 | 18 | 254 |
| MOES_mouse | Moesin | 15 | 42 | 418 | ||
| GELS_rat | Gelsolin | 6 | 12 | 72 | ||
| EZRLrat | Ezrin | 15 | 45 | 408 | ||
| RADI_mouse | Radixin | 12 | 32 | 335 | ||
| HS12B_mouse | Heat shock 70 kDa protein 12B | 9 | 23 | 168 | ||
| HS12A_mouse | Heat shock 70 kDa protein 12A | 3 | 5 | 59 | ||
| HSP7C_CRIGR | Heat shock cognate 71 kDa protein | 12 | 29 | 351 | ||
| DTNB_rat | Dystrobrevin-β | 9 | 17 | 223 | ||
| TCPA_CRIGR | T-complex protein-1 subunit-α | 1 | 5 | 53 | ||
| EFR3A_mouse | Protein EFR3 homolog A | 2 | 4 | 59 | ||
| PLAK_mouse | Junction plakoglobin | 10 | 23 | 205 | ||
| CALD1_rat | Non-muscle caldesmon | 3 | 6 | 38 | ||
| FXR1_mouse | Fragile X mental retardation syndrome-related protein 1 | 1 | 2 | 42 | ||
| P7 | 65–55 | VIME_rat | Vimentin | 10 | 45 | 511 |
| PLVAP_rat | Plasmalemma vesicle-associated protein | 12 | 46 | 438 | ||
| SNTB2_mouse | β2-syntrophin | 14 | 44 | 627 | ||
| SNTA1_mouse | α1-syntrophin | 4 | 12 | 53 | ||
| TCPD_mouse | T-complex protein 1 subunit-δ | 4 | 9 | 134 | ||
| 2AAA_mouse | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A-α isoform |
5 | 14 | 123 | ||
| TCPQ_mouse | T-complex protein 1 subunit-θ | 4 | 8 | 113 | ||
| TCPH_mouse | T-complex protein 1 subunit-φ | 4 | 11 | 100 | ||
| FYN_mouse | Tyrosine kinase Fyn | 3 | 7 | 97 | ||
| SRC_mouse | Tyrosine kinse Src | 3 | 7 | 96 | ||
| YES_mouse | Tyrosine kinase Yes | 7 | 20 | 83 | ||
| COR2B_mouse | Coronin-2B | 5 | 10 | 87 | ||
| TCPZ_mouse | T-complex protein 1 subunit ζ | 3 | 11 | 87 | ||
| TCPE_mouse | T-complex protein 1 subunit-ε | 1 | 3 | 66 | ||
| EHD1_mouse | EH domain-containing protein-1 | 2 | 5 | 61 | ||
| VATB2_mouse | Vacuolar ATP synthase subunit B, brain isoform | 10 | 30 | 363 | ||
| AP2M1 | AP-2 complex subunit μ-1 | 8 | 21 | 178 | ||
| DESM_rat | Desmin | 6 | 21 | 175 | ||
| TBB5_CRIGR | Tubulin β-5 chain | 5 | 30 | 117 | ||
| TBA1 B_CRIGR | Tubilin α-1B chain | 6 | 17 | 90 | ||
| SAM50_mouse | Sorting and assembly machinery component 50 homolog | 3 | 8 | 59 | ||
| TCPB_mouse | T-complex protein 1 subunit-β | 3 | 8 | 42 | ||
| P8 | 55–45 | PODO_rat | Podocin | 10 | 54 | 298 |
| SERPH | Serpin H1 | 5 | 23 | 242 | ||
| FLOT1_mouse | Flotillin-1 | 8 | 30 | 241 | ||
| FLOT2_mouse | Flotillin-2 | 7 | 21 | 146 | ||
| ARP3_mouse | Actin-related protein 3 | 6 | 41 | 142 | ||
| EF1G_rat | Elongation factor 1-γ | 3 | 10 | 76 | ||
| SNX8_mouse | Sorting nexin-8 | 1 | 2 | 73 | ||
| GNAS1_rat | Guanine nucleotide-binding protein Gs subunit-α isoform XLas | 3 | 4 | 73 | ||
| HA1 B_mouse | H-2 class I histocompatibility antigen, K-α chain | 3 | 8 | 64 | ||
| NHRF2_rat | Na+/H+ exchange regulatory cofactor NHE-RF2 | 3 | 14 | 49 | ||
| ILK_mouse | Integrin-linked protein kinase | 3 | 8 | 107 | ||
| P9 | 45–37 | ACTG_mouse | Actin, cytoplasmic 2 | 10 | 81 | 1743 |
| TMOD3_mouse | Tropomodulin-3 | 6 | 22 | 94 | ||
| PARVA_mouse | α-parvin | 3 | 10 | 149 | ||
| PARVB_mouse | β-parvin | 1 | 3 | 79 | ||
| PP1B_mouse | Serine/threonine protein phosphatase PP1-β catalytic subunit | 6 | 34 | 263 | ||
| LIMS2_mouse | LIM and senescent cell antigen-like-containing domain protein 2 | 5 | 16 | 83 | ||
| PP1A_rat | Serine/threonine protein phosphatase PP1-α catalytic subunit | 7 | 41 | 261 | ||
| PP1G_mouse | Serine/threonine protein phosphatase PP1-γ catalytic subunit | 6 | 45 | 231 | ||
| ANXA2_rat | Annexin A2 | 7 | 36 | 178 | ||
| CAZA2_mouse | F-actin-capping protein subunit α-2 | 5 | 26 | 175 | ||
| CAZA1_mouse | F-actin-capping protein subunit α-1 | 6 | 41 | 120 | ||
| GNAI2_rat | Guanine nucleotide-binding protein Gi, α-2 subunit | 4 | 21 | 80 | ||
| AAKG1_mouse | 5′-AMP-activated protein kinase subunit γ-1 | 1 | 5 | 59 | ||
| P10 | 37–32 | GBB2_mouse | Guanine nucleotide-binding protein Gi/Gs/Gt subunit β-2 | 9 | 62 | 391 |
| GBB1_mouse | Guanine nucleotide-binding protein Gi/Gs/Gt subunit β-1 | 6 | 40 | 349 | ||
| AQP1_mouse | Aquaporin-1 | 3 | 13 | 84 | ||
| KC1A_mouse | Casein kinase I isoform-α | 2 | 5 | 55 | ||
| PP2AA_mouse | Serine/threonine-protein phosphatase 2A catalytic subunit-α | 1 | 3 | 54 | ||
| LPP1_rat | Lipid phosphate phosphohydrolase 1 | 1 | 7 | 53 | ||
| LPP3_rat | Lipid phosphate phosphohydrolase 3 | 2 | 9 | 38 | ||
| STX4_mouse | Syntaxin-4 | 3 | 13 | 48 | ||
| ARPC2 | Actin-related protein 2/3 complex subunit 2 | 7 | 24 | 108 | ||
| C1O9A_mouse | Coiled-coil domain-containing protein 109A | 2 | 12 | 90 | ||
| TPM4_rat | Tropomyosin α-4 | 3 | 14 | 75 | ||
| K0152_mouse | Uncharacterized protein KIAA0152 | 2 | 8 | 80 | ||
| P11 | 32–28 | CLIC5_mouse | Chloride intracellular channel protein 5 | 7 | 50 | 216 |
| VAPB_mouse | Vesicle-associated membrane protein-associated protein B | 2 | 13 | 123 | ||
| VAPA_mouse | Vesicle-associated membrane protein-associated protein A | 3 | 23 | 111 | ||
| RSU1_mouse | Ras suppressor protein 1 | 6 | 46 | 86 | ||
| RAB5C_mouse | Ras-related protein Rab-5c | 2 | 10 | 57 | ||
| TM111_mouse | Transmembrane protein 111 | 3 | 14 | 94 | ||
| P12 | 28–23 | RAB5B_mouse | Ras-related protein Rab-5B | 3 | 17 | 81 |
| RAB5C_mouse | Ras-related protein Rab-5C | 5 | 42 | 238 | ||
| RAB5A_mouse | Ras-related protein Rab-5A | 5 | 33 | 161 | ||
| HSPB1 | Heat shock protein β-1 | 5 | 38 | 131 | ||
| RAB21_mouse | Ras-related protein Rab-21 | 3 | 29 | 106 | ||
| SNP23_mouse | Synaptosomal-associated protein 23 | 3 | 25 | 91 | ||
| RRAS_mouse | Ras-related protein R-ras | 3 | 17 | 133 | ||
| RRAS2_mouse | Ras-related protein R-ras2 | 5 | 27 | 106 | ||
| MLRA_rat | Myosin regulatory light chain 2-A | 3 | 23 | 120 | ||
| THY1_rat | Thy-1 membrane glycoprotein | 3 | 17 | 68 | ||
| RAP1A_mouse | Ras-related protein Rap-1A | 1 | 6 | 54 | ||
| CAV1_mouse | Caveolin-1 | 4 | 24 | 102 | ||
| RAB18_mouse | Ras-related protein Rab-18 | 3 | 22 | 97 | ||
| P13 | 23–18 | RAP1B_rat | Ras-related protein Rap-1b | 4 | 36 | 215 |
| CDC42_rat | Cell division control protein 42 homolog | 5 | 36 | 171 | ||
| RAC1_mouse | Ras-related C3 botulinum toxin substrate 1 | 6 | 52 | 123 | ||
| RASH_mouse | GTPase HRas precursor | 3 | 28 | 99 | ||
| RASN_CAVPO | GTPase NRas | 3 | 26 | 76 | ||
| RHOA_mouse | Transforming protein ρA | 4 | 48 | 71 | ||
| TP4A1_mouse | Protein tyrosine phosphatase type IVa | 2 | 12 | 48 | ||
| RASK_mouse | GTPase KRas | 4 | 31 | 48 | ||
| MLRN_mouse | Myosin regulatory light chain 2 | 3 | 27 | 44 | ||
| MLRA_rat | Myosin regulatory light chain 2-A | 3 | 62 | 337 | ||
| RAP2B_mouse | Ras-related protein Rap-2b | 1 | 6 | 88 | ||
| MLRB_mouse | Myosin regulatory light chain 2-B | 3 | 62 | 367 | ||
| ARPC3_mouse | Actin-related protein 2/3 complex subunit 3 | 4 | 30 | 165 | ||
| ARPC4_mouse | Actin-related protein 2/3 complex subunit 4 | 4 | 46 | 94 | ||
| COF1_mouse | Cofilin-1 | 2 | 30 | 152 | ||
| P14 | 18–14 | MYL6_mouse | Myosin light chain polypeptide 6 | 3 | 66 | 422 |
| PDZ11_mouse | PDZ domain-containing protein 11 | 3 | 35 | 99 | ||
| AP2S1_mouse | AP-2 complex subunit σ-1 | 2 | 10 | 47 | ||
| P15 | 14–8 | DYL1_mouse | Dynein light chain 1 | 2 | 49 | 109 |
| UBIQ_CAVPO | Ubiquitin | 2 | 32 | 50 |
A mascot score of 32 or greater was considered significant.
The bold text highlights CLIC5 proteins.
Several known slit diaphragm proteins were identified, such as nephrin, α-actinin-4, and podocin (Table 1). Thus, this fraction consists of proteins that are tightly associated with the slit diaphragm, or with membrane complexes of similar density and detergent resistance. Many identified proteins are members or regulators of the cytoskeleton. Interestingly, the presence of myosins and dynein components suggests that vesicular trafficking or migration takes place at slit diaphragms.36,37 Consistent with this idea, several members of the endocytic machinery were identified, including clathrin heavy chain and adaptor protein-2 (AP-2) complex subunits (Table 1).38 Several molecules that are located in lipid rafts were also identified, including flotillin, caveolin, and Src family kinases, indicating that the membrane complexes that were identified contained some lipid raft components (Table 1).39 Finally, although not indicated in the table, almost every band that was excised and analyzed contained ubiquitin, indicating that many of the proteins in this complex were ubiquitinated. This observation suggests that this post-translational modification may have a role in slit diaphragm homeostasis and function.
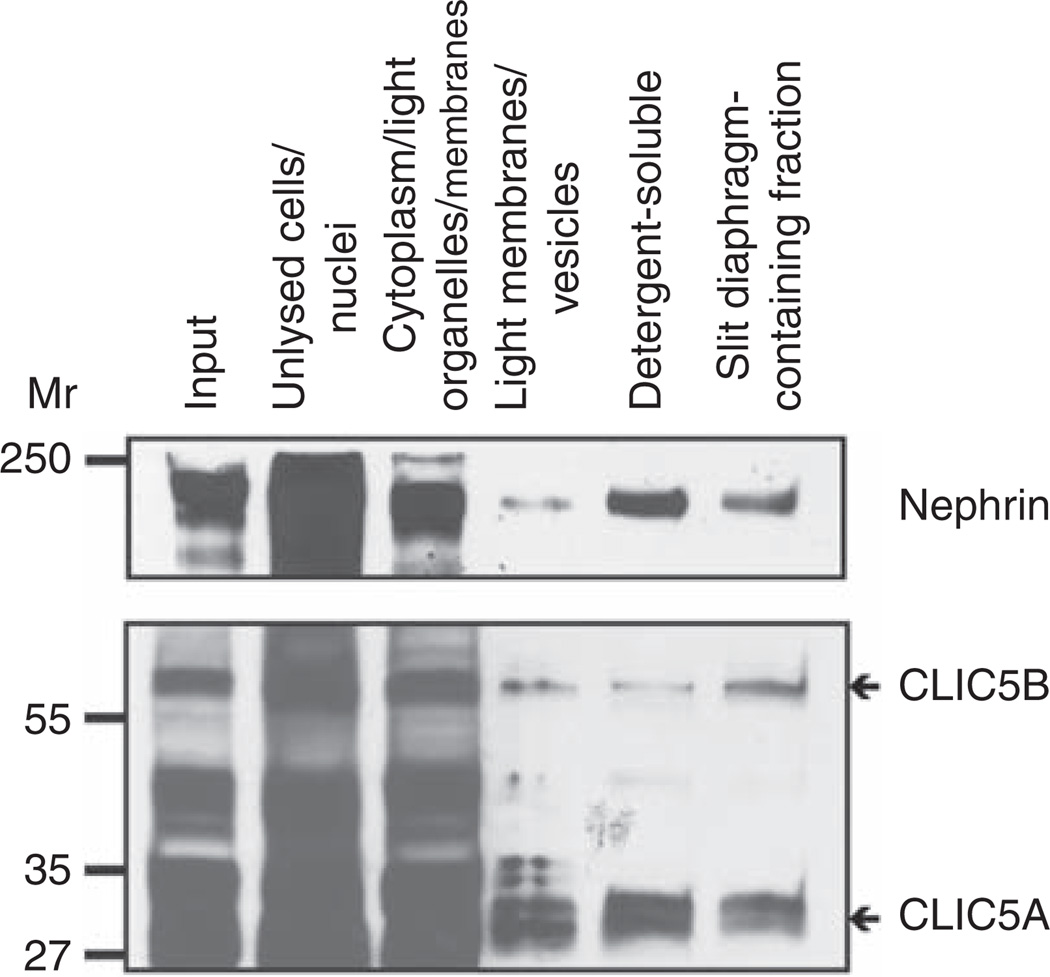
CLIC5 is present in slit diaphragm-enriched fractions
One of the proteins identified from our analysis was CLIC5 (band 11; Table 1). CLIC5 is a member of the CLIC family of putative chloride channels. To confirm that CLIC5 was indeed located in this biochemical fraction, immunoblotting was performed. Both isoforms of CLIC5, CLIC5A and CLIC5B were located in the final SD-enriched fraction (Figure 3). CLIC5B seemed to be more enriched in the SD-enriched fraction as compared with CLIC5A (Figure 3). These data verified the proteomic identification of CLIC5 in this protein mixture, suggesting that CLIC5 is either located in SDs or is present in membrane regions with similar biochemical properties. The immunoblotting also suggested that CLIC5 is located in other subcellular locations as well, such as in the detergent-soluble fraction in which podocalyxin was enriched.
Figure 3. Chloride intracellular channel protein 5 (CLIC5) is present in the slit diaphragm-containing fraction.
Protein fractions produced during the biochemical isolation of SDs were immunoblotted with antibodies to nephrin and CLIC5. Nephrin and both isoforms of CLIC5 were identified in the final SD-enriched fraction, as well as in other fractions (antibodies are labeled to the right of each panel).
CLIC5 is predominantly expressed in podocytes of the glomeruli
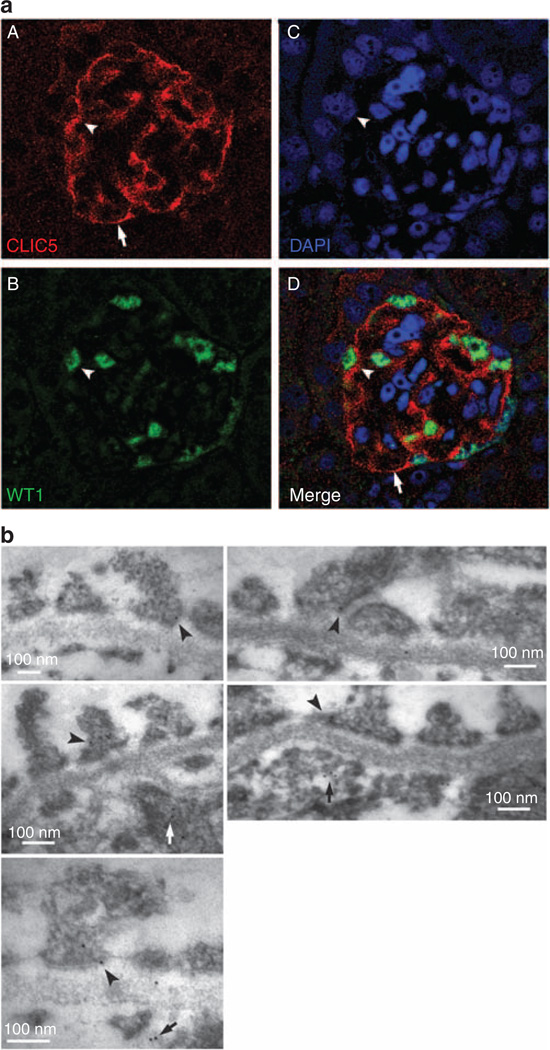
To determine which cell type in the glomeruli expresses CLIC5, immunofluorescent labeling of CLIC5 in kidney tissues was initially performed. The immunofluorescent labeling of CLIC5 revealed that CLIC5 mainly localized in cells with foot processes (Figure 4a). The merged image indicated that CLIC5 labeled predominantly WT1+ cells, a marker of podocytes (Figure 4aD). Therefore, CLIC5 is expressed in podocytes and is localized to both cell body and foot processes. Immunoelectron microscopy of mouse kidneys revealed CLIC5 labeling in foot processes of podocytes and glomerular endothelial cells (Figure 4b). In foot processes, CLIC5 localized to both the apical and basal membranes, and to areas adjacent to slit diaphragms. Control labeling with the secondary gold-conjugated antibody did not label any of these areas (data not shown).
Figure 4. Chloride intracellular channel protein 5 (CLIC5) is predominantly expressed in podocytes.
(a) Adult mouse kidney sections were double fluorescently labeled with (A) CLIC5 (red), (B) WT1 (green), a marker for podocytes, and (C) 4′,6-diamidino-2-phenylindole (DAPI) (blue), a nuclear marker. (D) A merged image that shows WT1 + podocytes colabeled with CLIC5 (arrowheads). CLIC5 was noted to localize to the cell body and cell processes (arrows) of podocytes. This is a representative fluorescence confocal image of a mouse glomerulus. (b) Immunoelectron microscopic localization of CLIC5 in adult mouse kidney. CLIC5 labeling was seen in foot processes with predominant localization in the apical and basal membranes and in regions proximal to slit diaphragms (arrowheads). CLIC5 also localized to glomerular endothelial cells (arrows). These are representative images taken at × 64,000–180,000 magnification.
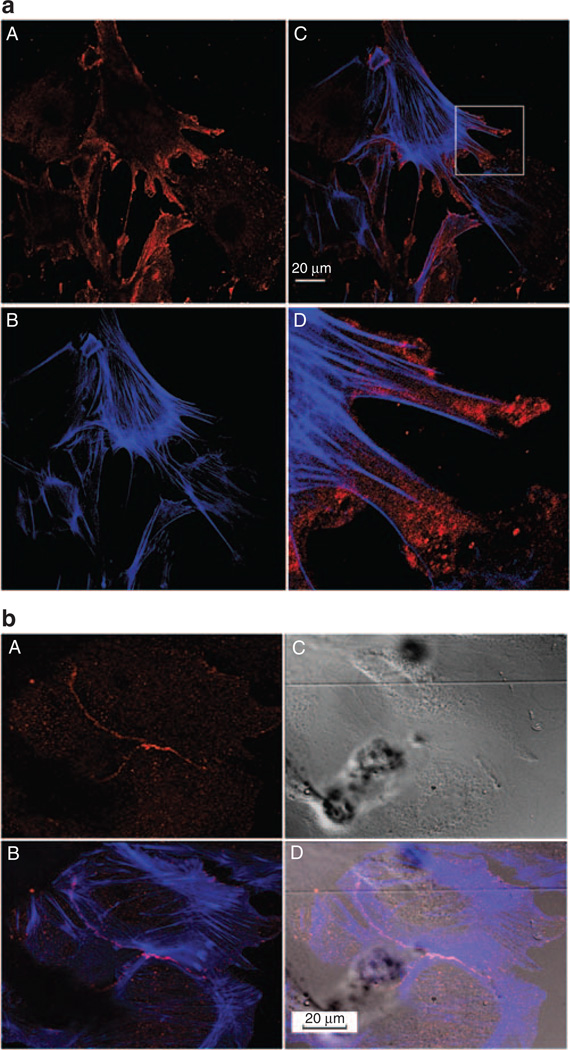
CLIC5 is localized at actin filament tips and is enriched in regions of cell–cell contact
The subcellular distribution of CLIC5 was determined using primary podocytes, which made elaborate extensions and developed a complex morphology. Immunofluorescent labeling of CLIC5 and actin filaments revealed that CLIC5 has a punctate subcellular distribution that is predominantly located at actin filament tips, characteristic of filopodia (Figure 5aA and D). Interestingly, CLIC5 was concentrated in regions where podocytes contacted each other (Figure 5bD), suggesting that CLIC5 is important in subcellular regions of cell–cell contact and possibly cell–substratum contact.
Figure 5. Chloride intracellular channel protein 5 (CLIC5) is localized at the tips of actin filaments and is enriched in regions of cell–cell contact.
(a) Primary podocyte cultures were labeled with (A) CLIC5 (red) and (B) actin (phalloidin, blue). (C) A merged image and (D) a magnified inset from the merged image showing that CLIC5 localized at the tips of actin filaments. This is a representative fluorescence confocal image of primary mouse podocytes after 14 days in vitro. (b) Three adjacent primary podocytes were fluorescently labeled and imaged with (A) CLIC5 (red), (B) merged with phalloidin (blue), (C) imaged with differential interference contrast, and (D) merged with differential interference contrast. CLIC5 had a punctate labeling pattern enriched at regions of cell–cell contact. Images were taken at × 60 magnification for (a) and at × 40 magnification for (b).
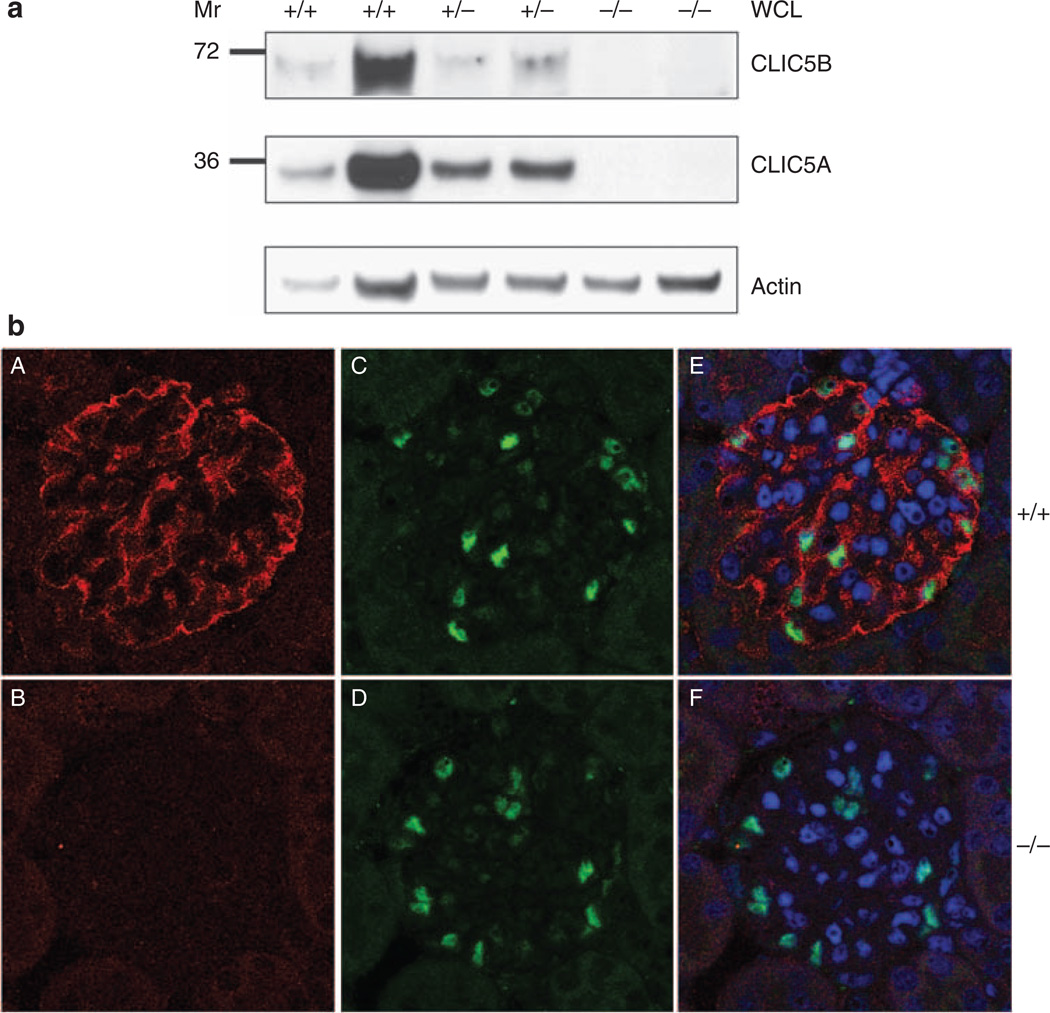
CLIC5 expression in podocytes is absent in CLIC5−/− mice
To verify whether Jitterbug mice lacked CLIC5 expression in podocytes, whole cellular lysates prepared from CLIC5+/+ , CLIC5+/−, and CLIC5−/− glomeruli were immunoblotted with CLIC5 and actin antibodies (Figure 6a). Both isoforms of CLIC5 were present in the highest abundance in CLIC5+/+ glomeruli, at a relatively lower abundance in CLIC5+/− glomeruli, and were not detectable in CLIC5−/− glomeruli. In addition, CLIC5B expression was lower than that of CLIC5A in glomeruli. These findings were further supported by immunofluorescent labeling of CLIC5 in glomerular sections (Figure 6b). CLIC5 expression in CLIC5−/− kidney was not evident as compared with the CLIC5+/+ kidney. It is important to note that CLIC5 and WT1 double immunofluorescent labeling revealed that WT1+ cells expressed CLIC5 (Figure 6b). These data confirmed the findings in Figure 4 that CLIC5 was expressed in podocytes, and that both CLIC5A and CLIC5B were absent in CLIC5−/− mice.
Figure 6. Chloride intracellular channel protein 5 (CLIC5) expression in podocytes is absent in CLIC5−/− mice.
(a) Whole cellular lysates (WCLs) were produced from isolated glomeruli, and CLIC5A and CLIC5B expression levels were determined by western blot analysis. Actin immunoblotting served as a loading control to enable protein expression comparisons between samples. Expression of CLIC5A and CLIC5B decreased between CLIC5+/+, CLIC5+/−, and CLIC5−/− mice. There was no detectable expression of either CLIC5 isoform in CLIC5−/− mice. (b) Mouse kidney sections were double immunofluorescently labeled with CLIC5 (A and B) and WT1 (C and D) antibodies, and nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, merge, E and F). CLIC5+/+ glomeruli exhibited CLIC5 expression, whereas no CLIC5 was evident in kidney sections from CLIC5−/− mice. Immunofluorescence for CLIC5, WT1, and DAPI was merged, indicating that CLIC5 expression was absent in podocytes of CLIC5−/− mice. Images were taken at × 60 magnification.
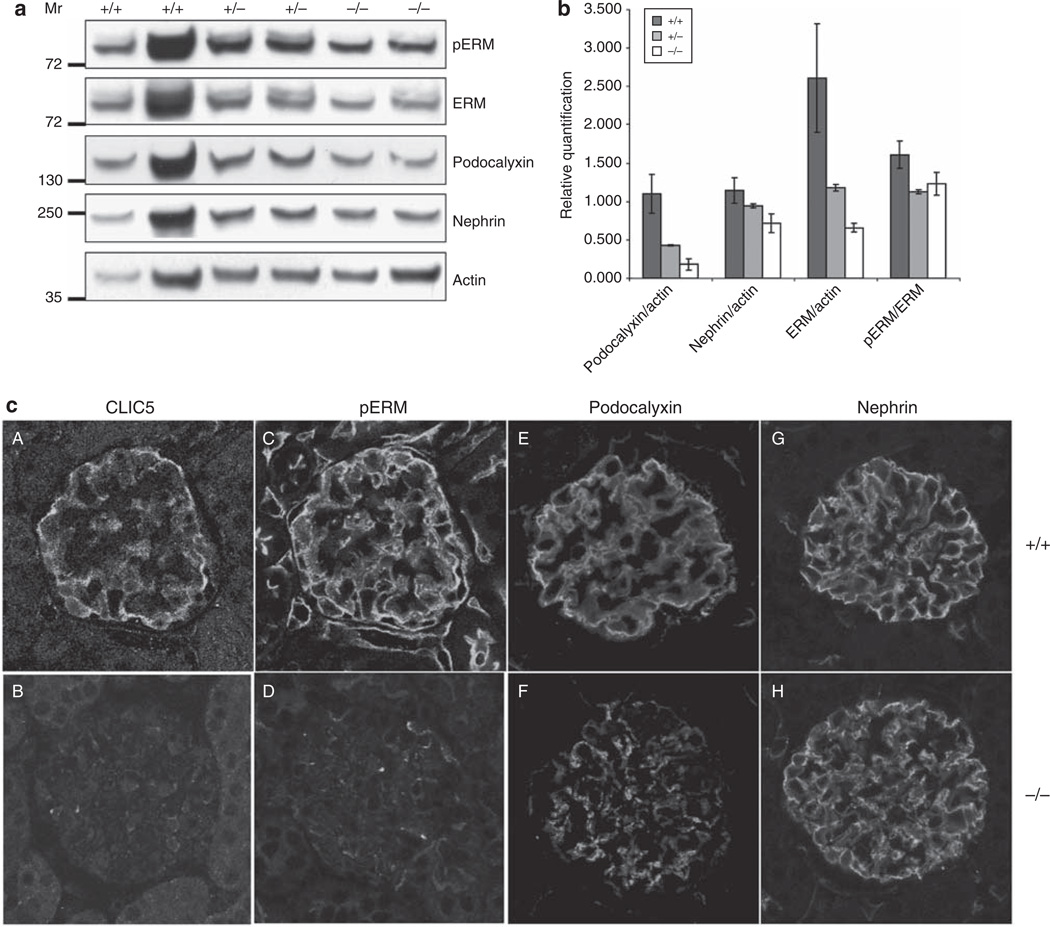
ERM and podocalyxin expressions are dependent on CLIC5
To determine whether CLIC5 modifies the expression of other podocyte proteins, western blot analysis was performed on whole cellular lysates derived from CLIC5+/+, CLIC5+/−, and CLIC5−/− glomeruli (Figure 7a). ERM,40 podocalyxin, and to a lesser extent, nephrin levels were reduced in CLIC5−/− podocytes. The expression levels of podocalyxin, nephrin, and ERM were normalized against actin (Figure 7b). A graded reduction of podocalyxin and ERM was observed in CLIC5+/− and CLIC5−/− glomeruli, which suggests that the genetic dosage of CLIC5 affects ERM and podocalyxin levels. Although not as pronounced, nephrin also exhibited a decrease in expression between CLIC5+/+ and CLIC5−/− glomeruli (Figure 7a and b). There were no differences between the levels of phosphorylated-ERM (pERM) and ERM among genotypes (Figure 7b), indicating that CLIC5 regulated the level of ERM, but not its phosphorylation. These data were reinforced by pERM, podocalyxin, and nephrin immunofluorescent labeling in CLIC5+/+ mice compared with CLIC5−/− mice (Figure 7c). Total ERM antibodies did not reliably immunofluorescently label glomerular tissues. Taken together, these results indicated that CLIC5 expression directly influenced the abundance of podocalyxin and ERM in podocytes in vivo.
Figure 7. ERM and podocalyxin expression levels are dependent on chloride intracellular channel protein 5 (CLIC5).
(a) Western blot analysis was performed on whole cellular lysates (WCLs) derived from CLIC5+/+, CLIC5+/−, and CLIC5−/− glomeruli. pERM, total ERM, podocalyxin, and nephrin (labeled to the right of each blot) were detected using the appropriate antibodies, and immunoblotting for actin was performed to allow protein expression comparisons between each genotype. (b) To determine the relative protein expression levels, the signal intensities for ERM, podocalyxin, and nephrin were normalized to actin by densitometric analysis. To investigate whether the phosphorylation of the ERM protein complex differed between genotypes, a ratio of phosphorylated ERM to total ERM expression was calculated. Data were graphed as the mean ± range. No difference between genotypes was detected for the level of pERM compared with total ERM. However, there was more than a 1.5- and 2.5-fold decrease in the expression of total ERM in CLIC5+/− and CLIC5−/− mice, respectively, when compared with total ERM expression in CLIC5+/+ mice. Podocalyxin expression diminished by almost 1.5- and 3-fold for CLIC5+/− and CLIC5−/− mice, respectively. There was also a decrease in nephrin expression when CLIC5+/+ mice were compared with CLIC5−/− mice. (c) Images of glomeruli were obtained from kidney sections that were immunofluorescently labeled for CLIC5, pERM, podocalyxin, and nephrin. CLIC5+/+ glomeruli (A, C, E and G) and CLIC5−/− glomeruli (B, D, F and H).
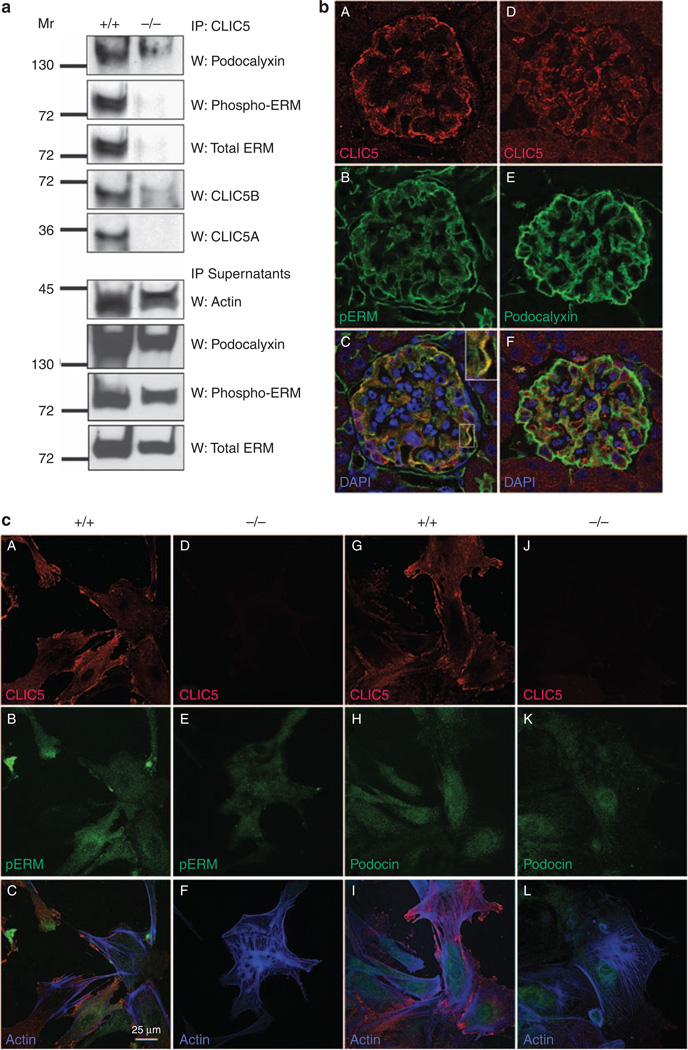
Podocalyxin and ERM associate with CLIC5
Because CLIC5 deletion resulted in a decrease of ERM and podocalyxin expression, the association of these proteins with CLIC5 was examined by coimmunoprecipitation. Detergent extracts from CLIC5+/+ and CLIC5−/− glomeruli were subjected to CLIC5 immunoprecipitation, followed by immunoblotting using pERM, ERM, podocalyxin, and CLIC5 antibodies. pERM, ERM, and podocalyxin associated with CLIC5 (Figure 8a). As expected, CLIC5 was not detected in immunoprecipitates from CLIC5−/− glomeruli, and the immunoprecipitations of pERM, ERM, and podocalyxin were lost as well (Figure 8a). The lack of coimmunoprecipitation of ERM and podocalyxin was not due to their reduced expression in CLIC5−/− glomeruli, as pERM, ERM, and podocalyxin were all detected in the supernatants from the immunoprecipitations (Figure 8a). Neither nephrin nor podocin coimmunoprecipitated with CLIC5 (data not shown), indicating that CLIC5 did not associate with these proteins.
Figure 8. Chloride intracellular channel protein 5 (CLIC5) associates with pERM, total ERM, and podocalyxin.
(a) Glomeruli were isolated from CLIC5+/+ and CLIC5−/− kidneys, detergent extracts were produced from them, and CLIC5 was immunoprecipitated. Immunoblotting of podocalyxin (top panel), pERM (second panel), and total ERM (third panel) showed coimmunoprecipitation of CLIC5 with these proteins. Blots were reprobed with anti-CLIC5 to confirm immunoprecipitation of CLIC5 (fourth and fifth panels). Immunoblotting of supernatants with actin controlled for equal amounts of lysates analyzed, and immunoblotting the supernatants for podocalyxin, pERM, and ERM confirmed their presence in extracts from CLIC5+/+ and CLIC5−/− glomeruli. (b) Images of mouse whole glomeruli fixed and labeled with CLIC5 (A and D, red), pERM (B, green), podocalyxin (E, green), and merged images (C and F). (C) The pERM-CLIC5 merged image shows colocalization of these proteins that appeared as yellow labeling in the podocyte processes (magnified inset). Colocalization of CLIC5 with podocalyxin was also noted as yellow labeling in the merged image (F). Fluorescence confocal images were taken at × 60 magnification. (c) Primary podocyte cultures from CLIC5+/+ (A–C and G–I) and CLIC5−/− (D–F and J–L) mice were grown for 14 days in vitro before they were fluorescently labeled with CLIC5 (A, D, G, and J, red), pERM (B and E, green), podocin (H and K, green), and phalloidin-labeled actin filaments (blue); merged images (C, F, I, and L). CLIC5 labeling was only detected in CLIC5+/+ podocytes and was absent from CLIC5−/− podocytes. The merged image (C) indicated that CLIC5 colocalized with pERM and was most evident near edges and cell-cell contact regions of the podocytes. The merged image of CLIC5 with podocin (I) showed no colocalization.
The expression patterns of CLIC5, pERM, and podocalyxin were examined in mouse glomeruli. As seen previously (Figures 4a and 6), CLIC5 was highly expressed in podocytes that extended processes along the outer aspect of glomeruli. pERM also had a similar pattern of expression and, on overlaying the coimmunofluorescent labels, a significant amount of colocalization was observed by confocal microscopy (Figure 8bC). Double immunofluorescent labeling of CLIC5 and podocalyxin also showed regions of colocalization within glomeruli, but to a lesser extent than pERM (Figure 8b). The lower level of colocalization between podocalyxin and CLIC5 was likely to be partly due to the lower sensitivity of the rabbit CLIC5 antibody that was necessary to colabel with the goat podocalyxin antibody (compare Figure 8bA versus D). To examine the subcellular distribution of colocalization between pERM and CLIC5, primary podocytes were examined using coimmunofluorescent labeling. There was some colocalization between pERM and CLIC5, in contrast to podocin and CLIC5, which did not overlap (Figure 8cC and I respectively). CLIC5−/− podocytes were also examined for pERM and podocin localization. CLIC5 labeling was absent in primary podocytes from CLIC5−/− mice, and pERM labeling decreased dramatically, suggesting that a considerable amount of its expression requires CLIC5. Interestingly, the highest level of colocalization between CLIC5 and pERM occurred at the cellular edges and at regions of cell–cell contact, where actin filaments terminate. We were unable to obtain consistent labeling with the available total ERM antibodies, but immunoblotting analysis indicated that both pERM and total ERM decline coordinately (Figure 7), suggesting that CLIC5 is required to maintain the level of ERM complexes. In contrast, there was no change in podocin expression or localization in CLIC5+/+ and CLIC5−/− primary podocytes. Taken together, these data indicate that CLIC5 associated with podocalyxin and pERM in vivo, and is most highly colocalized at the regions of cell–cell contact and at the cellular edges of actin filaments.
CLIC5 is critical for podocyte morphology and the glomerular filtration barrier
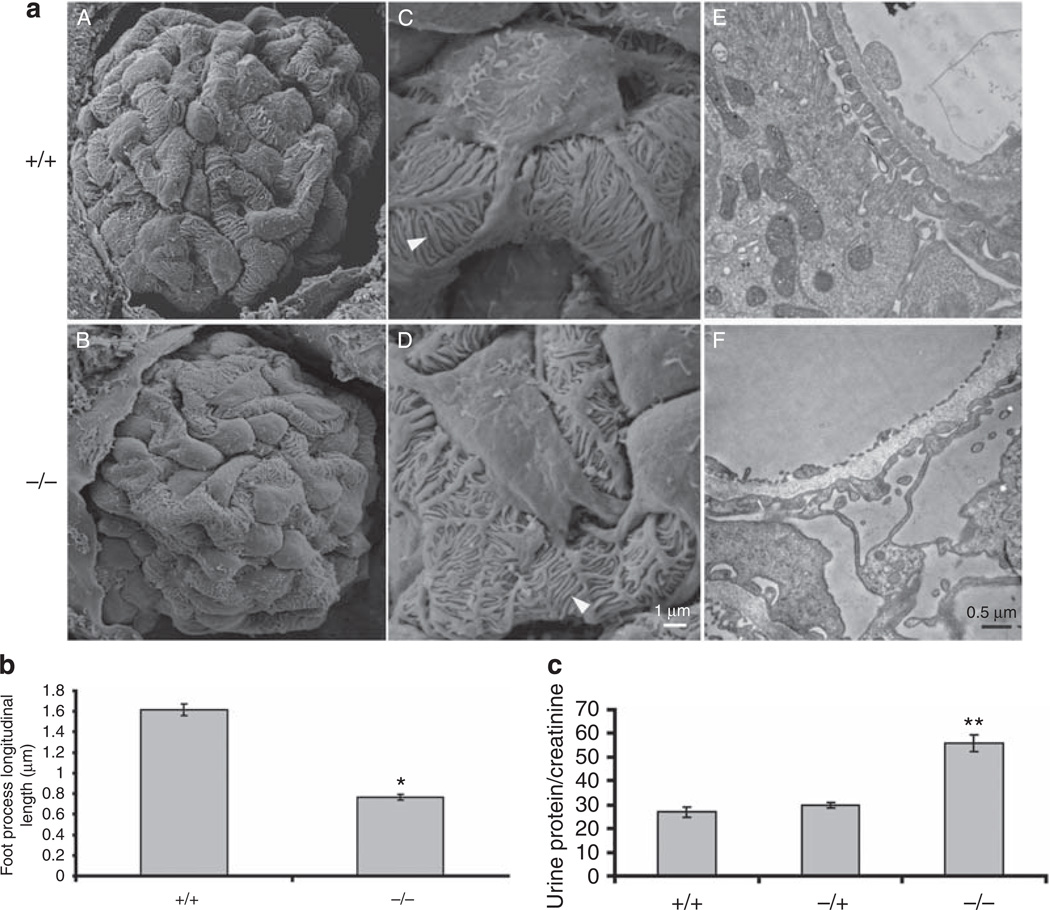
Given the dependence of expression and association of podocalyxin, ERM, and, to some extent, nephrin on CLIC5, and its unique subcellular localization, we examined the ultrastructural morphology of CLIC5+/+ and CLIC5−/− podocytes using s.e.m. and transmission electron microscope. Most of the CLIC5−/− podocyte cell bodies (Figure 9aB and D) seemed to have a ‘smoother’ cell surface than CLIC5+/+ podocytes (Figure 9aA and C), which were found to have microvilli-like structures extension on their cell surfaces using s.e.m. analysis. At higher magnifications, many foot processes were observed to be shorter in length in CLIC5−/− versus CLIC5+/+ podocytes (Figure 9aC–D), and measurements of these processes revealed a significant reduction in longitudinal length by >50% in CLIC5−/− versus CLIC5+/+ mice (Figure 9b). These foot processes under a transmission electron microscope were also noted to have significant disorganization, but not complete effacement (Figure 9aE and F), because at higher magnification, the SDs were intact (data not shown). The extent of proteinuria was measured and expressed as urine protein/creatinine ratios from adult CLIC5+/+, CLIC5+/−, and CLIC5−/− mice. There was an increase in proteinuria with CLIC5 deletion (Figure 9c). By light microscopy, we did not observe glomerulosclerosis using periodic acid-Schiff and tricrome staining, and examination of a smaller group of animals to 6 months did not show further worsening of proteinuria (data not shown). Taken together, these data showed that a lack of CLIC5 significantly altered the podocyte morphology at the foot process level and these changes resulted in a partial loss of the glomerular filtration barrier.
Figure 9. Chloride intracellular channel protein 5 (CLIC5) is critical for podocyte morphology and the glomerular filtration barrier.
(a) Representative s.e.m. images of whole glomeruli from CLIC5+/+ mice (A) versus CLIC5−/− mice (B) at 4 weeks of age. These images were taken at × 2000 magnification. (C and D) Representative images of podocyte cell bodies and processes were taken at higher magnification, × 10,000 (C, CLIC5+/+ and D, CLIC5−/−). White arrowheads show the ‘longitudinal’ length of foot processes that were measured using ImageJ in b below. Images of transmission electron microscopy were taken at × 19,000 magnification. (b) Longitudinal lengths of foot processes in micrometers. Data are represented as mean ± s.e.m. from three to four mice of each genotype. A total of 140–180 processes were measured from each mouse (see methods for details.) *P-value <0.001 as compared with CLIC5+/+ mice. (c) Urine protein/creatinine ratios (mg/mg) were determined from male, 3-month-old CLIC5+/+, CLIC5+/−, and CLIC5−/− mice. Data were represented as mean ± s.e.m., n=4 per data group. **P-value <0.001 as compared with the CLIC5+/+ group.
DISCUSSION
A proteomic analysis of biochemical fractions enriched in SD proteins led to the identification of CLIC5. Immunofluorescence microscopy of glomeruli indicated that CLIC5 was expressed in podocytes and was present throughout their processes. By immunoelectron microscopy, CLIC5 was seen in foot processes on both the apical and basal membranes and within close proximity to SDs. CLIC5 was also observed in glomerular endothelial cells. At the subcellular level, CLIC5 was concentrated at the tips of actin filaments and was enriched at points of contact between podocytes. Consistent with a potential role as a linker between actin filaments and the plasma membrane, CLIC5 associated with the ERM complex and podocalyxin. In CLIC5−/− mice, the expression levels of ERM, podocalyxin, and, to some extent, nephrin were reduced in glomeruli. Ultrastructurally, CLIC5−/− podocytes exhibited significantly reduced longitudinal foot process length. It is important to note that CLIC5−/− mice had significant proteinuria within 3 months of age, consistent with the morphological changes observed in the foot processes by transmission electron microscope in summary. CLIC5 is a protein necessary for the maintenance of foot process morphology, which likely serves as a linkage point between foot process membranes and the actin cytoskeleton.
The purification procedure that we used to enrich for SD proteins did not make use of stronger detergents, such as sarcosyl. Although these more stringent conditions would isolate tightly associated proteins and are more likely to enrich specifically for the SD complex, these conditions would exclude proteins that are loosely associated with this complex, as well as with closely located regions of the plasma membrane. Our intention in using the Triton X-100 fraction as the starting material for proteomic analysis was to ensure that we did not remove potentially important regulators of foot process structure and function that are not tightly associated with the SD. CLIC5 seems to be such a protein. It cannot be concluded with certainty from this study that CLIC5 is in SD, but the immunofluorescence and immunoEM analyses suggest that CLIC5 is located proximal to SDs, and is more broadly present in both apical and basal membranes throughout the podocyte processes. In addition, although CLIC5B expression was lower than that of CLIC5A in glomeruli, CLIC5B was more highly abundant in the SD-enriched fraction compared with CLIC5A, which was more abundant in the Triton-soluble fraction. The identification of CLIC5 from this proteomic analysis, and its characterization as being critical for foot process morphology and podocyte function, serves to validate the utility of this proteomic procedure for the identification of proteins that are important for podocytes. Using sarcosyl extraction and density gradients to further isolate proteins that are solely present in the SD complex will reveal which proteins are tightly associated components of the SD and which proteins are peripherally associated regulators and closely localized membrane constituents. These data may unravel how the SD is regulated, turned over, and modified by the local foot process milieu. These studies will also form a basis for examining how diseases alter these structures in adaptive and pathophysiological manners.
Although CLIC5 is a member of a family of putative intracellular chloride channels, current evidence to support this function in intact cells is limited. Furthermore, CLIC5 is not homologous to known chloride channels, suggesting that there may be other cellular functions for CLIC5. The results presented here indicate that CLIC5 likely functions as an adapter between the plasma membrane of podocytes and the actin cytoskeleton. This is consistent with previous studies identifying CLIC5 as a protein associated with the ERM complex.24 Furthermore, the localization of CLIC5 to the base of stereocilia in hair cells suggests that it has a linkage function between the cytoskeleton and membrane structures.35 CLIC5 seems to be a critical regulator of podocyte morphology, which is necessary for maintenance of the filtration barrier. Future studies will examine the mechanism of this effect and map the portions of CLIC5 that interact with podocalyxin and other associated proteins. Furthermore, whether CLIC5A and CLIC5B have similar or unique functions and subcellular targeting motifs in podocytes is an important unanswered question. The results reported here raise the possibility that CLIC5 regulates or contributes to the pathogenesis of glomerular diseases that results in proteinuria. Examining tissues of human and animal models of glomerular diseases for alterations in CLIC5 expression will be a critical first step.
MATERIALS AND METHODS
Purification and analysis of slit diaphragm-containing protein fractions
Given the similarities between synapses of neurons and the slit diaphragm of podocytes, we used a modified synaptic junctional complex purification protocol41–43 to produce protein fractions that were enriched in slit diaphragms. Briefly, glomeruli were purified from either rats or mice using a differential sieving technique. These glomeruli were then homogenized in a sucrose-HEPES buffer (0.32 mol/l sucrose, 4 mmol/l HEPES, pH 7.4, with protease and phosphatase inhibitors). The crude homogenates obtained were centrifuged at 1000 × g for 15 min to remove nuclei and unlysed cells, and this supernatant was next centrifuged at 10,000 × g for 10 min to isolate mitochondria and dense cellular compartments. These pellets were further homogenized in water and then quickly adjusted to pH 7.4 with HEPES. The homogenate was centrifuged at 25,000 × g for 20 min and the pellet was detergent-extracted with 0.1% Triton X-100. Finally, the resulting extracts were centrifuged at 32,000 × g for 20 min and this final pellet was analyzed by immunoblotting for the presence of slit diaphragm proteins. This final, detergent-insoluble pellet was used as the protein source for the proteomic analysis.
To identify the proteins in the slit diaphragm-enriched fraction, this fraction was subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis. The gels were stained with Coomassie blue and the bands (and gel regions) were cut out using a robotic spot cutter. The proteins were digested in gel with trypsin and the peptide fragments were eluted and subjected to LC-MSMS. Protein identifications were based on a Mascot search of the peptide analysis and proteins were considered truly identified only if their Mascot score was >32. A Mascot probability score of 200, for example, represents a 10−20 chance of the protein match being a random event. Two independent proteomic analyses were performed and the identified proteins were compared. The second preparation contained several additional proteins because it had five-fold more proteins. These analyses were conducted by the Roswell Park Proteomics Core Facility (Buffalo, NY, USA).
Immunoblotting and immunoprecipitations
Protein extracts were separated using sodium dodecyl sulfate– polyacrylamide gel electrophoresis and electroblotted onto polyvinylidene fluoride membranes. These blots were washed with Tris-buffered saline with Tween 20 and blocked with 3% bovine serum albumin in Tris-buffered saline with Tween 20. The blots were probed with primary antibodies, followed by use of the appropriate horseradish peroxidase-conjugated secondary antibody. The immmunoblots were developed using a chemiluminescent substrate (Supersignal, Pierce, Rockford, IL, USA) and quantified using ImageJ software (National Institute of Health, Bethesda, MD, USA). The primary antibodies used were as follows: goat polyclonal anti-nephrin and goat polyclonal anti-podocalyxin (R&D Systems, Minneapolis, MN, USA); goat polyclonal anti-CLIC5 (Santa Cruz Biotechnology, Santa Cruz, CA, USA); rabbit polyclonal anti-CLIC5 and anti-transferrin receptor (Sigma, Saint Louis, MO, USA); anti-COX IV, anti-PSMA2, rabbit polyclonal anti-ERM, rabbit polyclonal anti-pERM and anti-flotillin 1 (Cell Signaling Technology, Danvers, MA, USA); rabbit polyclonal anti-podocin (Sigma); and goat anti-actin and rabbit anti-WT1 (Santa Cruz). Immunoblotting and immunoprecipitations were performed as described previously.
Mouse podocyte primary cell cultures
Kidneys were harvested from postnatal day-7 CLIC5+/+ and CLIC5−/− mice (Jitterbug, Jackson Laboratory, Bar Harbor, ME, USA). The cortices were removed and minced into 1 mm sections that were then sieved successively through cell strainers (BD Biosciences, San Jose, CA, USA). Pelleted glomeruli were resuspended and plated in 10% fetal bovine serum and RPMI (Invitrogen, Carlsbad, CA, USA). Glomeruli were seeded on collagen-coated chamber slides. Podocytes were allowed to mature in vitro for at least 14 days.
Immunofluorescent analysis of cells
Cells were fixed in 4% paraformaldehyde, and 0.1% Triton X-100 was used to permeabilize the cells. Cells were incubated with the following primary antibodies: goat anti-CLIC5 (Santa Cruz Biotechnology), goat anti-podocalyxin (R&D Biosciences), rabbit anti-podocin (Santa Cruz Biotechnology), rabbit anti-pERM (Cell Signaling Technology), and rabbit anti-ERM (Cell Signaling Technology). This was followed by incubation with appropriate secondary antibodies (Invitrogen). Actin filaments were stained with Phalloidin (Invitrogen).
Immunofluorescent analysis of kidney tissues
Kidneys from mice were perfused with phosphate-buffered saline, pH 7.4, before fixation with either buffered formalin or Methyl Carnoy’s solution before sectioning into 4 µm thickness. Immunofluorescence labeling was performed as previously described.44 Antibodies used were as follows: goat anti-CLIC5, rabbit anti-WT1, goat anti-podocalyxin, rabbit anti-phospho-ERM (Cell Signaling Technology), goat anti-nephrin (R&D Systems), and 4′,6-diamidino-2-phenylindole (DAPI, Vector Labs, Burlingame, CA, USA). Images were taken using a Nikon Eclipse C1 Plus confocal microscope (Melville, NY, USA) and analyzed using ImageJ (NIH, Bethesda, MD, USA).
Immunoelectron microscopy
Mice were cardiac perfused with phosphate-buffered saline, followed by perfusion fixation in 3% paraformaldehyde in 0.1 mol/l phosphate buffer, pH 7.4. Tissues were dehydrated through a graded series of ethanol, and embedded in LR white (Electron Microscopy Sciences, Hatfield, PA). Ultrathin kidney cortical sections (70 nm) were mounted onto Formvar/carbon-coated nickel grids (Electron Microscopy Sciences). Duplicate sections were analyzed for each condition. Aldehyde quenching with 0.05 mol/l glycine and antigen retrieval with citrate buffer (Covance, Princeton, NJ, USA) were performed before blocking with normal donkey serum, bovine serum albumin, Cold Water Fish Gelatin, and Tween20 (Electron Microscopy Sciences). Grids were placed in goat anti-CLIC5 (Santa Cruz) at 1:10 and 1:5 overnight at 4°C in blocking buffer plus 0.2% bovine serum albumin-c (Aurion; Electron Microscopy Sciences). After washing, sections were incubated with a donkey anti-goat antibody conjugated to 10 nmol/l gold particles at 1:20. After rinsing, grids were fixed in 2.5% glutaraldehyde in 0.1 mol/l phosphate buffer, and poststained with uranyl acetate and lead citrate. The sections were examined using a Philips CM100 electron microscope at 60 kV.
Transmission and scanning electron microscopy
Mice were cardiac perfused with phosphate-buffered saline, followed by perfusion with 2.5% glutaraldehyde in 0.1 mol/l Sorensen’s buffer (pH 7.4). For transmission electron microscopy, kidney cortices were dissected and minced into 1–2 mm pieces. Further processing was performed by the Microscopy & Image-analysis Laboratory at the University of Michigan, Ann Arbor, MI, USA. This processing included postfixation in 1% osmium tetroxide in the same buffer, rinsing in double-distilled water to remove phosphate salt, and then en bloc staining with aqueous 3% uranyl acetate for 1 h. The tissues were then dehydrated in ascending concentrations of ethanol, rinsed twice in propolynoxide, and embedded in epoxy resin. The samples were ultrathin sectioned (70 nm) and stained with uranyl acetate and lead citrate. The sections were examined using a Philips CM100 electron microscope (FEI, Hillsboro, OR, USA) at 60 kV. Images were recorded digitally using a Hamamatsu ORCA-HR digital camera system operated using AMT software (Advanced Microscopy Techniques, Danvers, MA, USA). For s.e.m., samples were postfixed similarly. They were then immersed in hexamethyldisilazane before mounting and sputter coating with gold using a ‘Polaron’ sputter coater (POLARON SEM Coating System, Hatfield, PA, USA). Images were examined using an Amray 1910FE scanning electron microscope (SEMTech Solutions, North Billerica, MA, USA) and digitally imaged using X-stream Imaging Software (SEMTech Solutions). All samples were imaged by a naïve observer.
Foot process longitudinal length measurements
The s.e.m. images that were acquired at × 10,000 magnification were assessed. Longitudinal length was traced from the beginning of each final branch point to the end of a foot process from only areas that are perpendicular to the plane of the camera image. Each measurement was analyzed using ImageJ. ‘Foot process length’ from each animal was averaged from a total of 140–180 measured processes. Briefly, 15–18 individual foot processes were traced from each image taken from 5–6 different glomeruli of at least 3–4 adult mice (4 months old) of each genotype by a naïve observer. Data are represented as mean ± s.e.m. from three to four mice of each genotype.
Urine protein/creatinine ratio measurements
Urine samples from CLIC5+/+, CLIC5+/−, and CLIC5−/− mice were collected by placing them into urine collection containers for 4–6 h. A physical barrier prevented the mixing of solid and liquid wastes, and urine samples free of fecal contamination were collected. Samples were diluted to 1:5, 1:10, or 1:50 with distilled water before total protein was measured using a bicinchoninic acid protein assay kit (Pierce). Urine creatinine measurements were taken using the Creatinine Companion kit (Exocell, Philadelphia, PA, USA). Absorbances were measured using an EL × 800 microplate reader (BioTek Instruments, Winooski, VT, USA).
ACKNOWLEDGMENTS
This research is supported by a Pilot Research Grant from the University of Michigan George M O’Brien Renal Core Center, P30 DK081943, by the National Institutes of Health Career Development Award K08, DK084210 (CCT), and by National Institutes of Health R01, NS058510 (BAP). We thank Paul Krebsbach and the Department of Biologic and Materials Sciences for confocal microscopy support, and William Couser and Roger Wiggins for critical reading and comments. We also appreciate the Microscopy Image Laboratory at the University of Michigan, especially Dorothy Sorenson, and the Roswell Park Proteomics Core Facility in Buffalo, NY for their technical expertise.
Footnotes
DISCLOSURE
All the authors declared no competing interests.
REFERENCES
- 1.Mundel P, Kriz W. Structure and function of podocytes: an update. Anat Embryol (Berl) 1995;192:385–397. doi: 10.1007/BF00240371. [DOI] [PubMed] [Google Scholar]
- 2.Reiser J, Kriz W, Kretzler M, et al. The glomerular slit diaphragm is a modified adherens junction. J Am Soc Nephrol. 2000;11:1–8. doi: 10.1681/ASN.V1111. [DOI] [PubMed] [Google Scholar]
- 3.Kim YH, Goyal M, Kurnit D, et al. Podocyte depletion and glomerulosclerosis have a direct relationship in the PAN-treated rat. Kidney Int. 2001;60:957–968. doi: 10.1046/j.1523-1755.2001.060003957.x. [DOI] [PubMed] [Google Scholar]
- 4.Kriz W, Kretzler M, Nagata M, et al. A frequent pathway to glomerulosclerosis:deterioration of tuft architecture-podocyte damage-segmental sclerosis. Kidney Blood Press Res. 1996;19:245–253. doi: 10.1159/000174083. [DOI] [PubMed] [Google Scholar]
- 5.Kriz W, Elger M, Nagata M, et al. The role of podocytes in the development of glomerular sclerosis. Kidney Int Suppl. 1994;45:S64–S72. [PubMed] [Google Scholar]
- 6.Kriz W, Gretz N, Lemley KV. Progression of glomerular diseases: is the podocyte the culprit? Kidney Int. 1998;54:687–697. doi: 10.1046/j.1523-1755.1998.00044.x. [DOI] [PubMed] [Google Scholar]
- 7.Kriz W, Lemley KV. The role of the podocyte in glomerulosclerosis. Curr Opin Nephrol Hypertens. 1999;8:489–497. doi: 10.1097/00041552-199907000-00014. [DOI] [PubMed] [Google Scholar]
- 8.Matsusaka T, Xin J, Niwa S, et al. Genetic engineering of glomerular sclerosis in the mouse via control of onset and severity of podocyte-specific injury. J Am Soc Nephrol. 2005;16:1013–1023. doi: 10.1681/ASN.2004080720. [DOI] [PubMed] [Google Scholar]
- 9.Wharram BL, Goyal M, Wiggins JE, et al. Podocyte depletion causes glomerulosclerosis: diphtheria toxin-induced podocyte depletion in rats expressing human diphtheria toxin receptor transgene. J Am Soc Nephrol. 2005;16:2941–2952. doi: 10.1681/ASN.2005010055. [DOI] [PubMed] [Google Scholar]
- 10.Steffes MW, Schmidt D, Mccrery R, et al. Glomerular cell number in normal subjects and type I diabetic patients. Kidney Int. 2001;59:2104–2113. doi: 10.1046/j.1523-1755.2001.00725.x. [DOI] [PubMed] [Google Scholar]
- 11.Pagtalunan ME, Miller PL, Jumping-Eagle S, et al. Podocyte loss and progressive glomerular injury in type II diabetes. J Clini Invest. 1997;99:342–348. doi: 10.1172/JCI119163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer TW, Bennett PH, Nelson RG. Podocyte number predicts long-term urinary albumin excretion in Pima Indians with type II diabetes and microalbuminuria. Diabetologia. 1999;42:1341–1344. doi: 10.1007/s001250051447. [DOI] [PubMed] [Google Scholar]
- 13.Lemley KV, Lafayette RA, Safai M, et al. Podocytopenia and disease severity in IgA nephropathy. Kidney Int. 2002;61:1475–1485. doi: 10.1046/j.1523-1755.2002.00269.x. [DOI] [PubMed] [Google Scholar]
- 14.Kestila M, Lenkkeri U, Mannikko M, et al. Positionally cloned gene for a novel glomerular protein–nephrin–is mutated in congenital nephrotic syndrome. Mol Cell. 1998;1:575–582. doi: 10.1016/s1097-2765(00)80057-x. [DOI] [PubMed] [Google Scholar]
- 15.Lenkkeri U, Mannikko M, McCready P, et al. Structure of the gene for congenital nephrotic syndrome of the finnish type (NPHS1) and characterization of mutations. Am J Hum Genet. 1999;64:51–61. doi: 10.1086/302182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boute N, Gribouval O, Roselli S, et al. NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat Genet. 2000;24:349–354. doi: 10.1038/74166. [DOI] [PubMed] [Google Scholar]
- 17.Kim JM, Wu H, Green G, et al. CD2-associated protein haploinsufficiency is linked to glomerular disease susceptibility. Science. 2003;300:1298–1300. doi: 10.1126/science.1081068. [DOI] [PubMed] [Google Scholar]
- 18.Shih NY, Li J, Karpitskii V, et al. Congenital nephrotic syndrome in mice lacking CD2-associated protein. Science. 1999;286:312–315. doi: 10.1126/science.286.5438.312. [DOI] [PubMed] [Google Scholar]
- 19.Winn MP, Conlon PJ, Lynn KL, et al. A mutation in the TRPC6 cation channel causes familial focal segmental glomerulosclerosis. Science. 2005;308:1801–1804. doi: 10.1126/science.1106215. [DOI] [PubMed] [Google Scholar]
- 20.Reiser J, Polu KR, Moller CC, et al. TRPC6 is a glomerular slit diaphragm-associated channel required for normal renal function. Nat Genet. 2005;37:739–744. doi: 10.1038/ng1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moller CC, Pollak MR, Reiser J. The genetic basis of human glomerular disease. Adv Chronic Kidney Dis. 2006;13:166–173. doi: 10.1053/j.ackd.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 22.Huber TB, Benzing T. The slit diaphragm: a signaling platform to regulate podocyte function. Curr Opin Nephrol Hypertens. 2005;14:211–216. doi: 10.1097/01.mnh.0000165885.85803.a8. [DOI] [PubMed] [Google Scholar]
- 23.Patari-Sampo A, Ihalmo P, Holthofer H. Molecular basis of the glomerular filtration: nephrin and the emerging protein complex at the podocyte slit diaphragm. Ann Med. 2006;38:483–492. doi: 10.1080/07853890600978149. [DOI] [PubMed] [Google Scholar]
- 24.Berryman M, Bretscher A. Identification of a novel member of the chloride intracellular channel gene family (CLIC5) that associates with the actin cytoskeleton of placental microvilli. Mol Biol Cell. 2000;11:1509–1521. doi: 10.1091/mbc.11.5.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berryman M, Bruno J, Price J, et al. CLIC-5A functions as a chloride channel in vitro and associates with the cortical actin cytoskeleton in vitro and in vivo . J Biol Chem. 2004;279:34794–34801. doi: 10.1074/jbc.M402835200. [DOI] [PubMed] [Google Scholar]
- 26.Nishizawa T, Nagao T, Iwatsubo T, et al. Molecular cloning and characterization of a novel chloride intracellular channel-related protein, parchorin, expressed in water-secreting cells. J Biol Chem. 2000;275:11164–11173. doi: 10.1074/jbc.275.15.11164. [DOI] [PubMed] [Google Scholar]
- 27.Berry KL, Hobert O. Mapping functional domains of chloride intracellular channel (CLIC) proteins in vivo . J Mol Biol. 2006;359:1316–1333. doi: 10.1016/j.jmb.2006.04.046. [DOI] [PubMed] [Google Scholar]
- 28.Edwards JC. A novel p64-related Cl— channel: subcellular distribution and nephron segment-specific expression. Am J Physiol. 1999;276:F398–F408. doi: 10.1152/ajprenal.1999.276.3.F398. [DOI] [PubMed] [Google Scholar]
- 29.Tulk BM, Kapadia S, Edwards JC. CLIC1 inserts from the aqueous phase into phospholipid membranes, where it functions as an anion channel. Am J Physiol Cell Physiol. 2002;282:C1103–C1112. doi: 10.1152/ajpcell.00402.2001. [DOI] [PubMed] [Google Scholar]
- 30.Warton K, Tonini R, Fairlie WD, et al. Recombinant CLIC1 (NCC27) assembles in lipid bilayers via a pH-dependent two-state process to form chloride ion channels with identical characteristics to those observed in Chinese hamster ovary cells expressing CLIC1. J Biol Chem. 2002;277:26003–26011. doi: 10.1074/jbc.M203666200. [DOI] [PubMed] [Google Scholar]
- 31.Littler DR, Assaad NN, Harrop SJ, et al. Crystal structure of the soluble form of the redox-regulated chloride ion channel protein CLIC4. FEBS J. 2005;272:4996–5007. doi: 10.1111/j.1742-4658.2005.04909.x. [DOI] [PubMed] [Google Scholar]
- 32.Singh H, Cousin MA, Ashley RH. Functional reconstitution of mammalian ′chloride intracellular channels′ CLIC1, CLIC4 and CLIC5 reveals differential regulation by cytoskeletal actin. FEBS J. 2007;274:6306–6316. doi: 10.1111/j.1742-4658.2007.06145.x. [DOI] [PubMed] [Google Scholar]
- 33.Griffon N, Jeanneteau F, Prieur F, et al. CLIC6, a member of the intracellular chloride channel family, interacts with dopamine D(2)-like receptors. Brain Res Mol Brain Res. 2003;117:47–57. doi: 10.1016/s0169-328x(03)00283-3. [DOI] [PubMed] [Google Scholar]
- 34.Friedli M, Guipponi M, Bertrand S, et al. Identification of a novel member of the CLIC family, CLIC6, mapping to 21q22.12. Gene. 2003;320:31–40. doi: 10.1016/s0378-1119(03)00830-8. [DOI] [PubMed] [Google Scholar]
- 35.Gagnon LH, Longo-Guess CM, Berryman M, et al. The chloride intracellular channel protein CLIC5 is expressed at high levels in hair cell stereocilia and is essential for normal inner ear function. J Neurosci. 2006;26:10188–10198. doi: 10.1523/JNEUROSCI.2166-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vallee RB, Seale GE, Tsai JW. Emerging roles for myosin II and cytoplasmic dynein in migrating neurons and growth cones. Trends Cell Biol. 2009;19:347–355. doi: 10.1016/j.tcb.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hwang W, Lang MJ. Mechanical design of translocating motor proteins. Cell Biochem Biophys. 2009;54:11–22. doi: 10.1007/s12013-009-9049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin XS, Tsukaguchi H, Shono A, et al. Phosphorylation of nephrin triggers its internalization by raft-mediated endocytosis. J Am Soc Nephrol. 2009;20:2534–2545. doi: 10.1681/ASN.2009010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsui-Pierchala BA, Encinas M, Milbrandt J, et al. Lipid rafts in neuronal signaling and function. Trends Neurosci. 2002;25:412–417. doi: 10.1016/s0166-2236(02)02215-4. [DOI] [PubMed] [Google Scholar]
- 40.Hugo C, Nangaku M, Shankland SJ, et al. The plasma membrane-actin linking protein, ezrin, is a glomerular epithelial cell marker in glomerulogenesis, in the adult kidney and in glomerular injury. Kidney Int. 1998;54:1934–1944. doi: 10.1046/j.1523-1755.1998.00195.x. [DOI] [PubMed] [Google Scholar]
- 41.Therien HM, Mushynski WE. Isolation of synaptic junctional complexes of high structural integrity from rat brain. J Cell Biol. 1976;71:807–822. doi: 10.1083/jcb.71.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davis GA, Bloom FE. Isolation of synaptic junctional complexes from rat brain. Brain Res. 1973;62:135–153. doi: 10.1016/0006-8993(73)90624-0. [DOI] [PubMed] [Google Scholar]
- 43.Cotman CW, Taylor D. Isolation and structural studies on synaptic complexes from rat brain. J Cell Biol. 1972;55:696–711. doi: 10.1083/jcb.55.3.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsui CC, Pierchala BA. CD2AP and Cbl-3/Cbl-c constitute a critical checkpoint in the regulation of ret signal transduction. J Neurosci. 2008;28:8789–8800. doi: 10.1523/JNEUROSCI.2738-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]