Key Clinical Message
Maternal cell‐free DNA (cfDNA) results that are discordant with the diagnostic fetal karyotype should prompt further investigation. If deeper analysis of the cfDNA results demonstrates a “saw‐tooth” pattern characteristic of genome‐wide imbalance, maternal malignancy is suggested. Identifying the maternal malignancy can, however, be difficult.
Keywords: Cell‐free DNA, metastatic neoplasm, multiple monosomies, prenatal diagnosis
Introduction
Maternal plasma cell‐free DNA (cfDNA) analysis has become a preferred method chosen by patients to screen for common fetal trisomies. However, when the results are discordant with follow‐up diagnostic testing, there are limited follow‐up recommendations at present for practitioners and patients. Possible explanations for discordant results include confined placental mosaicism, maternal chromosomal mosaicism, co‐twin demise, DNA copy number variants in mother or fetus, maternal organ transplant from a male donor, and maternal malignancy 1. Here, we report a patient who had plasma cfDNA test results suggestive of full or partial monosomies for chromosomes 13, 18, 21, and X who was subsequently found to have hepatic lesions on magnetic resonance imaging (MRI). Postpartum the patient was diagnosed with stage IV colon cancer.
Case History
The patient was a 37‐year‐old G2P1001 woman who 2 years earlier had undergone in vitro fertilization and preimplantation genetic testing for cystic fibrosis as both she and her husband are carriers. This resulted in a full term, healthy female. During this first pregnancy, the patient had plasma cfDNA testing in that pregnancy that was reported as low risk for fetal aneuploidy.
In the current pregnancy, the couple used their remaining frozen embryos to conceive. Two embryos were transferred. A subsequent ultrasound scan demonstrated a single, viable intrauterine pregnancy. Maternal plasma cfDNA test results at 12 weeks suggested full or partial monosomies for chromosomes 13, 18, 21, and X. The patient then underwent diagnostic testing by amniocentesis at 18 weeks' gestation. The fetus had a 46, XX karyotype and a normal chromosomal microarray.
The concern for a maternal malignancy as an explanation for the discordant results between the cfDNA study and amniocentesis prompted a request for a deeper analysis of the whole‐genome sequencing results by the original testing laboratory (Fig. 1). This showed multiple areas of genome‐wide imbalance, suggestive of malignancy. The patient was subsequently referred to the cancer genetic counseling service for an oncologic evaluation at 21‐weeks' gestation. She was clinically asymptomatic. Her general physical examination was normal, and laboratory studies were unremarkable. Her family history was not suggestive of a hereditary cancer syndrome.
Figure 1.
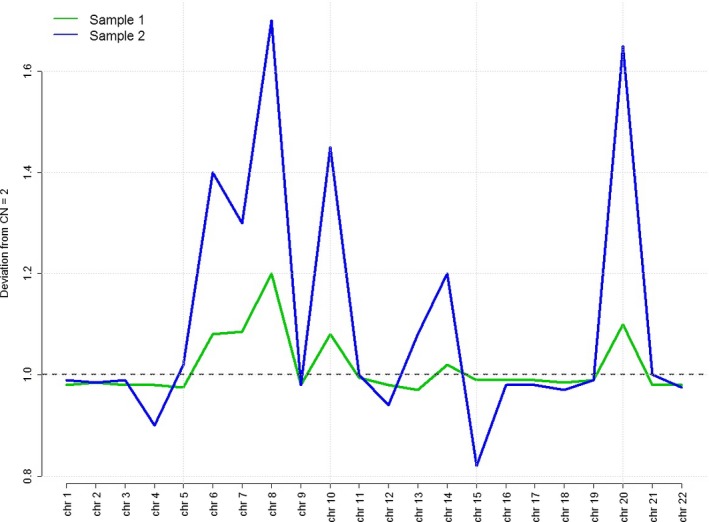
Genome‐wide data from NIPS performed at 12 weeks (green line) and then repeated early third trimester (blue line). Multiple copy number variants are seen across the genome leading to the saw‐tooth pattern seen above, with increasing copy numbers of mapped sequences in the repeat sample. Both test results reported full or partial monosomy for chromosomes 13, 18, 21, and X, which was the result of the excess amounts of DNA sequences in chromosomes 7, 8, 10, 14, and 20 causing the bioinformatics algorithm to interpret the abnormal ratios as monosomies of the test chromosomes.
Following a discussion with multiple medical specialists, a full body MRI scan without contrast was performed at 23 weeks' gestation to search for a possible malignancy (Fig. 2). The imaging identified multiple T2 hyperintense and T1 hypointense lesions in the liver: the largest measured 5.5 × 4.3 × 6.6 cm. The differential diagnosis included hepatic adenomas, primary hepatocellular carcinoma, or metastatic lesions. The patient was further counseled regarding these new findings. The decision was made not to perform a liver biopsy.
Figure 2.
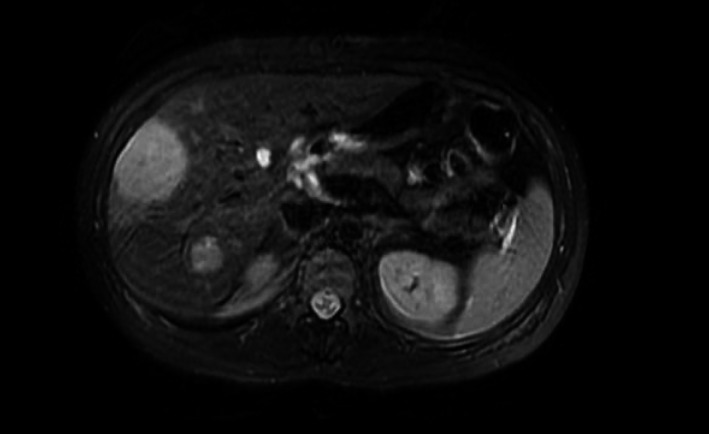
Abdominal MRI at 23‐weeks' gestation identified multiple T2 hyperintense and T1 hypointense lesions in the liver.
The patient underwent another MRI scan at 27 weeks' gestation. This demonstrated that the hepatic lesions had increased in size with the largest one measuring 9.9 × 5.4 × 8.8 cm. Due to the concern that the lesions could become hemorrhagic, at 28 weeks' gestation the patient underwent an invasive radiology‐guided embolization procedure. A repeat maternal plasma cfDNA analysis continued to show multiple monosomies. Evaluation of the whole‐genome sequencing results showed a similar but more exaggerated pattern of genome‐wide imbalance compared to the previous test (Fig. 1). Because of a dropping hematocrit and increasing right upper quadrant pain, a third MRI scan was performed, which demonstrated that the largest lesion measured 10.5 × 5.4 × 9.7 cm and the smaller lesions were reduced in size. Her liver enzyme values remained normal.
The patient underwent a planned cesarean delivery at 32‐weeks' gestation to facilitate her medical management. At the time of her surgery, fine needle biopsies of four liver lesions were performed. Three lesions demonstrated necrotic type material consistent with the patient's recent embolization. One biopsy demonstrated poorly differentiated adenocarcinoma. Postpartum, she had a CT scan that demonstrated cecal thickening. Subsequent colonoscopy revealed a circumferential mass involving the cecum and proximal ascending colon. Multiple biopsies were taken but did not reveal any evidence of malignancy. The patient underwent a positron emission tomography (PET) scan that demonstrated a fluorodeoxyglucose(FDG)‐avid cecal mass consistent with colon cancer along with FDG‐avid pericecal lymph nodes consistent with metastasis. In addition, there were FDG‐avid right lobe hepatic lesions, which were consistent with metastases. The patient had a repeat biopsy of the hepatic lesion that demonstrated metastatic poorly differentiated adenocarcinoma. The diagnosis was stage IV colon cancer, and systemic chemotherapy was initiated. There was no response, so she underwent a right colectomy and partial hepatectomy. She then had a second round of chemotherapy but did not respond and died approximately 10‐months postpartum. The infant is alive and well.
Given the abnormal cfDNA test results seen in the second pregnancy, the genome‐wide tracings from the first pregnancy were retrospectively reviewed and were still considered to be unremarkable.
Discussion
Fetal cfDNA is detectable in maternal serum as early as 5–7 weeks of gestation 2. In the first trimester, approximately 10% of cfDNA is fetal in origin and is almost entirely derived from placental trophoblast cells. Several different techniques exist to analyze cfDNA 2. The technique used to analyze the DNA in the patient's sample was massively parallel shotgun sequencing (MPSS). MPSS involves identifying and counting DNA fragments. Both maternal and fetal DNA segments are sequenced simultaneously. The segments are sequenced, aligned, and uniquely mapped to sites from a reference human genome. Each individual laboratory employs its own statistical method to determine when to call a sample monosomic or trisomic for a specific chromosome. The test utilized here incorporated a software program called bowtie to align the sequences to the 19th reference version of the human genome sequence map 3. The clinical laboratory's proprietary software then evaluated the target chromosomes (13, 18, 21, X, and Y) by calculating a ratio between the normalized coverage on each target chromosome to the sum of normalized coverage on a respective set of reference chromosomes (typically two to six chromosomes). The software has upper and lower limits that it applies to the test results in order to generate an aneuploidy classification status for chromosomes 13, 18, and 21. These include aneuploidy detected, suspected, or no aneuploidy. For sex chromosomes it includes aneuploidy detected or not. The excess amount of circulating DNA sequences from the reference chromosomes, particularly the chromosomes with peak sequences above the horizontal line in Figure 1, resulted in abnormal ratios, thus generating the test results of monosomies for 13, 18, 21, and X. The screening result of multiple monosomies is caused by a bioinformatics artifact.
Because the patient's sample was analyzed by MPSS, the genome‐wide data were available and could be re‐analyzed. These demonstrated an abnormal pattern of multiple chromosomes across the genome that led to a “saw‐tooth” pattern (Fig. 1). Given the multiple abnormalities across the genome, this pattern was suspicious for a malignancy. However, genome‐wide aberrations have also been reported for benign, neoplastic lesions in pregnancy, such as uterine leiomyomas 4. If the patient's sample had been tested using the targeted sequencing method that does not use ratios, the results from chromosomes 13, 18, and 21 would likely have been normal and the suspicion for cancer may not have been raised.
CfDNA levels are frequently elevated in patients with cancer 5, 6, 7, 8. Ongoing research is addressing whether the increased cfDNA levels can be used for different purposes in cancer screening and monitoring response to treatment 6. Several studies have demonstrated that plasma cfDNA is increased in metastatic colon cancer 8. Other, nonratio approaches use detection and monitoring of a tumor‐specific oncogene such as KRAS. This was not done here, and in fact, would require a separate test from the MPSS counting approach.
In 2013, the first case of a pregnant patient with discordant results subsequently being diagnosed with metastatic cancer was published 5. The patient was a 37‐year‐old G2P1 woman with cfDNA test results that demonstrated fetal aneuploidy for chromosomes 13 and 18. At two‐week postpartum, the patient was diagnosed with metastatic small cell carcinoma of vaginal origin. In June 2015, three more patients diagnosed with cancer (ovarian carcinoma, follicular lymphoma, and Hodgkin lymphoma) after discordant cfDNA results were reported 6. In July 2015, an additional ten pregnant patients were reported to have a malignancy after undergoing cfDNA testing (neuroendocrine of unknown origin, non‐Hodgkin lymphoma in three patients, colorectal, Hodgkin lymphoma, acute T‐cell lymphoblastic leukemia, and two patients critically ill with type of cancer not reported) 7.
Conclusion
Management of the pregnant woman with discordant cfDNA results remains a clinical dilemma, particularly when genome‐wide sequencing results suggest malignancy. The most common cancers that have been diagnosed in pregnant patients include breast, cervical, Hodgkin lymphoma, leukemias, and malignant melanoma. These are also the most common types of cancers seen in women of reproductive age 9. As more information becomes available, specific cfDNA test result patterns may be helpful in guiding the subsequent evaluation. The extent of the diagnostic work up may be limited by the pregnancy itself. Standard serologic tumor markers are unreliable in a pregnant woman.
The current recommendations for evaluation of malignancy in the setting of discordant cfDNA results are only based on expert opinions; these include obtaining a complete blood count, chemistry panel, whole‐body MRI scan without contrast 6, and referral to medical oncology. Similarly for women who are not pregnant, for whom there is a suspicion of malignancy, there are no standard evaluations for cancer of unknown primary cell type 10. Patients diagnosed with cancer typically present with signs or symptoms that together with focused diagnostic testing lead to an eventual diagnosis. Most of the pregnant women identified to date because of abnormal cfDNA test results have been initially asymptomatic. A systematic multidisciplinary approach to cataloging additional cases of discordant cfDNA results, and their associated diagnoses, is needed in order to better define patient‐specific risks and consistent recommendations for diagnosis and treatment.
Conflict of interest
No conflicts of interest.
Authorship
JS: wrote the first draft, performed the literature search, and edited the manuscript. VK: edited the manuscript and obtained patient permission for publication. DWB: edited and critically reviewed the manuscript. GF: edited and critically reviewed the manuscript. NP: edited and critically reviewed the manuscript. MS: edited and critically reviewed the manuscript. BG: edited and critically reviewed the manuscript.
Acknowledgment
We would like to thank Dr. Catalin Barbacioru for preparation of Figure 1.
References
- 1. Bianchi, D. W. 2015. Prepare for unexpected prenatal test results. Nature 522:29–30. [DOI] [PubMed] [Google Scholar]
- 2. Rink, B. D. , and Norton M. E.. 2015. Screening for fetal aneuploidy. Semin. Perinatol. 40:35–43. [DOI] [PubMed] [Google Scholar]
- 3. Langmead, B. 2010. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics; Chapter 11:Unit 11.7. https://doi.org/10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dharajiya, N. G. , Namba A., Horiuchi I., Miyai S., Farkas D., Almasri E., et al. 2015. Uterine leiomyoma confounding a noninvasive prenatal test result. Prenat. Diagn. 35:990–993. [DOI] [PubMed] [Google Scholar]
- 5. Osborne, C. M. , Hardisty E., Devers P., Kaiser‐Rogers K., Hayden M., Goodnight W., and Vora N.. 2013. Discordant noninvasive prenatal testing results in a patient subsequently diagnosed with metastatic disease. Prenat. Diagn. 33:609–611. [DOI] [PubMed] [Google Scholar]
- 6. Amant, F. , Verheecke M., Wlodarska I., Dehaspe L., Brady P., Brison N., et al. 2015. Presymptomatic identification of cancers in pregnant women during noninvasive prenatal testing. JAMA Oncol. 1:814–819. [DOI] [PubMed] [Google Scholar]
- 7. Bianchi, D. W. , Chudova D., Sehnert A. J., Bhatt S., Murray K., Prosen T. L., et al. 2015. Noninvasive prenatal testing and incidental detection of occult maternal malignancies. JAMA 314:162–169. [DOI] [PubMed] [Google Scholar]
- 8. Lecomte, T. , Ceze N., Dorval E., and Laurent‐Puig P.. 2010. Circulating free tumor DNA and colorectal cancer. Gastroenterol. Clin. Biol. 34:662–681. [DOI] [PubMed] [Google Scholar]
- 9. Pavlidis, N. A. 2002. Coexistence of pregnancy and malignancy. Oncologist 7:279–287. [PubMed] [Google Scholar]
- 10. Varadhachary, G. R. 2011. Carcinoma of unknown primary: focused evaluation. J. Natl. Compr. Canc. Netw. 9:1406–1412. [DOI] [PubMed] [Google Scholar]


