Abstract
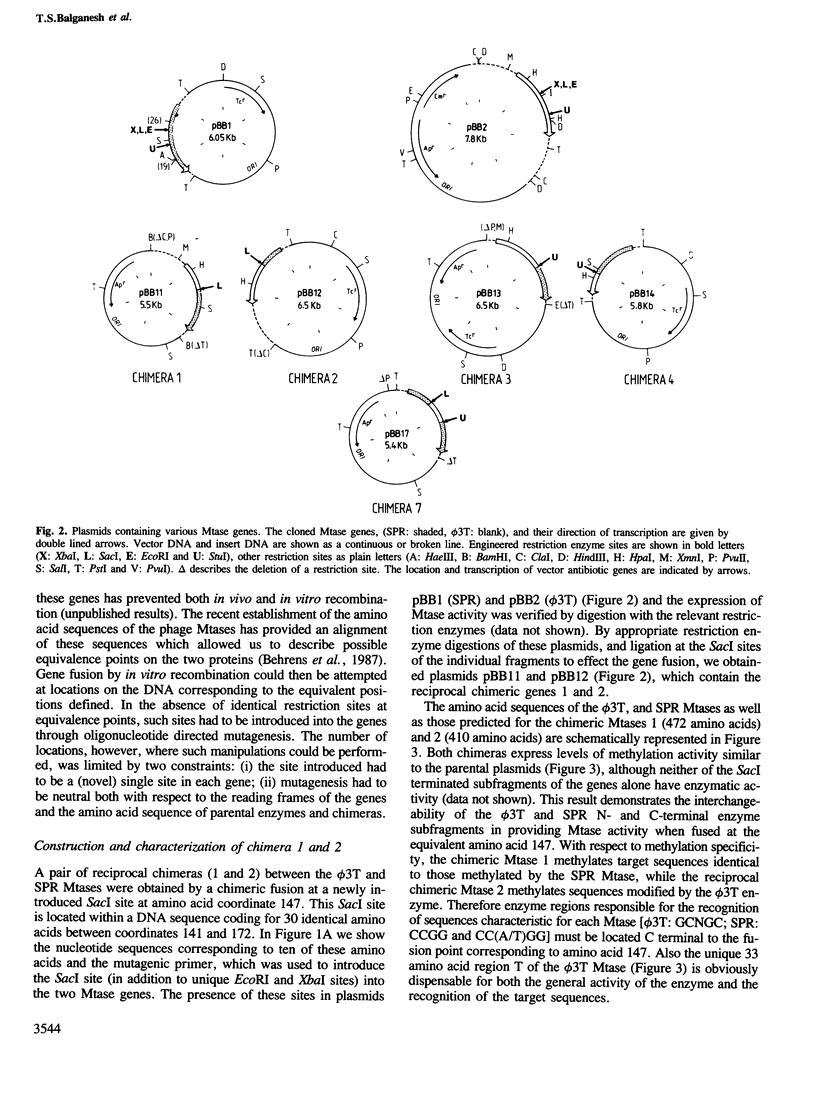
Multispecific DNA methyltransferases (Mtases) of temperate Bacillus subtilis phages SPR and phi 3T methylate the internal cytosine of the sequence GGCC. They differ in their capacity to methylate additional sequences. These are CCGG and CC(A/T)GG in SPR and GCNGC in phi 3T. Introducing unique restriction sites at equivalent locations within the two genes facilitated the construction of chimeric genes. These expressed Mtase activity at a level comparable to that of the parental genes. The methylation specificity of chimeric enzymes was correlated with the location of chimeric fusions. This analysis, which also included the use of mutant genes, showed that domains involved in the recognition of target sequences unique to each enzyme [CCGG, CC(A/T)GG or GCNGC] are represented by the central non-conserved parts of the proteins, whilst recognition of the sequence (GGCC), which is a target for both enzymes, is determined by an adjacent conserved region.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Behrens B., Noyer-Weidner M., Pawlek B., Lauster R., Balganesh T. S., Trautner T. A. Organization of multispecific DNA methyltransferases encoded by temperate Bacillus subtilis phages. EMBO J. 1987 Apr;6(4):1137–1142. doi: 10.1002/j.1460-2075.1987.tb04869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Buhk H. J., Behrens B., Tailor R., Wilke K., Prada J. J., Günthert U., Noyer-Weidner M., Jentsch S., Trautner T. A. Restriction and modification in Bacillus subtilis: nucleotide sequence, functional organization and product of the DNA methyltransferase gene of bacteriophage SPR. Gene. 1984 Jul-Aug;29(1-2):51–61. doi: 10.1016/0378-1119(84)90165-3. [DOI] [PubMed] [Google Scholar]
- Fuller-Pace F. V., Murray N. E. Two DNA recognition domains of the specificity polypeptides of a family of type I restriction enzymes. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9368–9372. doi: 10.1073/pnas.83.24.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J. A., Murray N. E. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983 May 5;166(1):1–19. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- Günthert U., Lauster R., Reiners L. Multispecific DNA methyltransferases from Bacillus subtilis phages. Properties of wild-type and various mutant enzymes with altered DNA affinity. Eur J Biochem. 1986 Sep 15;159(3):485–492. doi: 10.1111/j.1432-1033.1986.tb09912.x. [DOI] [PubMed] [Google Scholar]
- Günthert U., Reiners L. Bacillus subtilis phage SPR codes for a DNA methyltransferase with triple sequence specificity. Nucleic Acids Res. 1987 May 11;15(9):3689–3702. doi: 10.1093/nar/15.9.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günthert U., Reiners L., Lauster R. Cloning and expression of Bacillus subtilis phage DNA methyltransferase genes in Escherichia coli and B. subtilis. Gene. 1986;41(2-3):261–270. doi: 10.1016/0378-1119(86)90106-x. [DOI] [PubMed] [Google Scholar]
- Kessler C., Höltke H. J. Specificity of restriction endonucleases and methylases--a review. Gene. 1986;47(1):1–153. doi: 10.1016/0378-1119(86)90245-3. [DOI] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Fritz H. J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984 Oct;38(3):879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- Kramer W., Drutsa V., Jansen H. W., Kramer B., Pflugfelder M., Fritz H. J. The gapped duplex DNA approach to oligonucleotide-directed mutation construction. Nucleic Acids Res. 1984 Dec 21;12(24):9441–9456. doi: 10.1093/nar/12.24.9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraja V., Shepherd J. C., Bickle T. A. A hybrid recognition sequence in a recombinant restriction enzyme and the evolution of DNA sequence specificity. Nature. 1985 Jul 25;316(6026):371–372. doi: 10.1038/316371a0. [DOI] [PubMed] [Google Scholar]
- Noyer-Weidner M., Diaz R., Reiners L. Cytosine-specific DNA modification interferes with plasmid establishment in Escherichia coli K12: involvement of rglB. Mol Gen Genet. 1986 Dec;205(3):469–475. doi: 10.1007/BF00338084. [DOI] [PubMed] [Google Scholar]
- Noyer-Weidner M., Jentsch S., Kupsch J., Bergbauer M., Trautner T. A. DNA methyltransferase genes of Bacillus subtilis phages: structural relatedness and gene expression. Gene. 1985;35(1-2):143–150. doi: 10.1016/0378-1119(85)90166-0. [DOI] [PubMed] [Google Scholar]
- Noyer-Weidner M., Jentsch S., Pawlek B., Günthert U., Trautner T. A. Restriction and modification in Bacillus subtilis: DNA methylation potential of the related bacteriophages Z, SPR, SP beta, phi 3T, and rho 11. J Virol. 1983 May;46(2):446–453. doi: 10.1128/jvi.46.2.446-453.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff S. W., Luh J., Ganesan A. T., Behrens B., Thompson R., Montenegro M. A., Morelli G., Trautner T. A. The genome of Bacillus subtilis phage SPP1: the arrangement of restriction endonuclease generated fragments. Mol Gen Genet. 1979 Jan 10;168(2):165–172. doi: 10.1007/BF00431442. [DOI] [PubMed] [Google Scholar]
- Soberon X., Covarrubias L., Bolivar F. Construction and characterization of new cloning vehicles. IV. Deletion derivatives of pBR322 and pBR325. Gene. 1980 May;9(3-4):287–305. doi: 10.1016/0378-1119(90)90328-o. [DOI] [PubMed] [Google Scholar]
- Tran-Betcke A., Behrens B., Noyer-Weidner M., Trautner T. A. DNA methyltransferase genes of Bacillus subtilis phages: comparison of their nucleotide sequences. Gene. 1986;42(1):89–96. doi: 10.1016/0378-1119(86)90153-8. [DOI] [PubMed] [Google Scholar]
- Trautner T. A., Pawlek B., Günthert U., Canosi U., Jentsch S., Freund M. Restriction and modification in Bacillus subtilis: identification of a gene in the temperate phage SP beta coding for a BsuR specific modification methyltransferase. Mol Gen Genet. 1980;180(2):361–367. doi: 10.1007/BF00425849. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]