Summary
The molecular mechanisms underlying folding of mammalian chromosomes remain poorly understood. The transcription factor CTCF is a candidate regulator of chromosomal structure. Using the auxin-inducible degron system in mouse embryonic stem cells, we show that CTCF is absolutely and dose-dependently required for looping between CTCF target sites and insulation of topologically associating domains (TADs). Restoring CTCF reinstates proper architecture on altered chromosomes, indicating a powerful instructive function for CTCF in chromatin folding. CTCF remains essential for TAD organization in non-dividing cells. Surprisingly, active and inactive genome compartments remain properly segregated upon CTCF depletion, revealing that compartmentalization of mammalian chromosomes emerges independently of proper insulation of TADs. Further, our data support that CTCF mediates transcriptional insulator function through enhancer-blocking but not as a direct barrier to heterochromatin spreading. Beyond defining the functions of CTCF in chromosome folding these results provide new fundamental insights into the rules governing mammalian genome organization.
Introduction
Chromosomes meet the dual challenge of packaging DNA into the nucleus and, at the same time, enabling access to genetic information. Decades of work on chromosome organization have tackled the link between chromosome structure and genetic functions (Belmont, 2014; Cremer et al., 2015). Patterns of genome folding have been scrutinized with ever-increasing precision, but the identity and roles of the underlying molecular actors are still poorly understood, limiting our functional understanding of chromosome architecture. While genome organization and molecular actors differ between distant species (Cubeñas-Potts et al., 2016; Dekker and Heard, 2015; Ea et al., 2015), here we focus on mammals.
Mammalian chromosomes are profoundly heterogeneous. Euchromatin comprises open chromatin fibers and gene-rich regions (Gilbert et al., 2004) while heterochromatin is condensed, gene-poor and transcriptionally dormant. This highlights the remarkable correlation between the cytological, biochemical and sequence organization of chromosomes. Chromosomes can be further segmented into domains belonging to two main types of spatial compartments, as revealed by high-throughput Chromosome Conformation Capture (3C), with chromatin contacts being more frequent between loci of the same compartment type, both within and between chromosomes (Lieberman-Aiden et al., 2009). When reported on linear genomic maps, the alternating pattern of compartment types forms a domain-wide arrangement that aligns strikingly with regional chromatin states (Bickmore and van Steensel, 2013; Bonev and Cavalli, 2016). The euchromatic A- compartment contains most actively transcribed regions, while the B-compartment corresponds to megabase-sized gene-poor Lamina-Associated Domains (LADs (Guelen et al., 2008; Kind et al., 2015)) which replicate late in S-phase (Ryba et al., 2010).
At a more local scale, chromosomes are partitioned into sub-megabase segments that tend to self-associate and thus are relatively insulated from neighboring domains forming Topologically Associating Domains (TADs, (Dixon et al., 2012; Nora et al., 2012)). The borders of TADs are frequently demarcated by the binding of the CCCTC-binding factor (CTCF) (Dixon et al., 2012; Phillips-Cremins et al., 2013), a broadly expressed zinc-finger nucleic acid binding protein initially involved in transcriptional insulation (Ghirlando and Felsenfeld, 2016; Merkenschlager and Nora, 2016). Ultra-high resolution Hi-C analyses demonstrated the existence of a peak of 3C signal between some CTCF-bound boundaries of a subset of TADs, referred to as contact domains at this scale – indicative of interaction through chromatin looping (Rao et al., 2014). Deleting such a TAD boundary, or even just the underlying CTCF site, can lead to loss of physical insulation and subsequent encapsulation of the two abutting TADs into a single domain (Lupiáñez et al., 2015; Narendra et al., 2015; Nora et al., 2012; Sanborn et al., 2015; Tsujimura et al., 2015). This highlights the crucial role of boundaries in mediating the physical insulation of neighboring chromosome domains, with important implications for disease-causing chromosomal rearrangements in humans (Flavahan et al., 2016; Franke et al., 2016; Hnisz et al., 2016).
Strikingly, in most of the cases, a pair of CTCF sites only engage in contact above local background if they are in a convergent linear orientation (Rao et al., 2014), creating an asymmetry in the insulation pattern (Vietri Rudan et al., 2015). This arrangement is important: inverting a single CTCF site can be enough to rewire the direction of looping and disrupt proper packaging of the underlying chromosomal segment into an insulated TAD (Guo et al., 2015; Lupiáñez et al., 2015; Sanborn et al., 2015; de Wit et al., 2015). Polymer modelling studies have proposed that CTCF mediates TAD insulation by acting as a polar blocking factor to Cohesin translocation along the DNA during the formation and expansion of chromatin loops (Fudenberg et al., 2016; Sanborn et al., 2015).
Locus-specific studies have implicated the CTCF protein itself in mediating chromosome folding (Splinter, 2006). Yet, genome-wide assays after RNAi revealed only very limited consequences, with CTCF depletion leading to slightly reduced intra-TAD chromosomal contacts, slightly increased inter-TAD contacts and modest transcriptional changes with no clear link to folding defects (Zuin et al., 2014). Genetic manipulation of CTCF has proven difficult as it is essential for development (Moore et al., 2012; Sleutels et al., 2012; Soshnikova et al., 2010; Wan et al., 2008) and proliferation of cultured cells (González-Buendía et al., 2014), hampering the understanding of the exact role of CTCF in mammalian chromosome folding and genome functions. It is currently unclear to what extent CTCF is actually required for chromatin architecture and which levels of genome organization this factor controls.
Here we used a conditional degradation strategy in mouse embryonic stem cells (mESCs), the Auxin-Inducible Degron (AID) system (Nishimura et al., 2009), to acutely and reversibly deplete CTCF below detectable levels. We demonstrate that CTCF is a major determinant of mammalian chromosome folding. Its role is however restricted to sub-megabase genome organization, with loss of CTCF leading dose-dependently to insulation defects at most TAD boundaries and abrogating the accumulation of chromatin loops between CTCF sites. A few boundaries (less than 20%) remain unaffected by CTCF depletion, highlighting that CTCF is a major driver of TAD insulation but that other processes also contribute. Importantly, CTCF depletion did not disrupt A/B compartments, revealing that local insulation and higher-order compartmentalization rely on distinct molecular determinants. CTCF depletion did not either alter how contact frequency scales overall with genomic distance, demonstrating that CTCF-mediated chromosomal interactions are not the ties that enable packaging of mammalian chromosomes. Beyond cementing the importance of CTCF in driving insulation between TADs, our observations also reveal an important activator effect of CTCF through direct promoter binding, support a role for CTCF as an enhancer blocker, and refute its proposed function as a direct barrier to H3K27me3 spreading.
Results
Acute CTCF Depletion with the Auxin-Inducible Degron System
To deplete endogenous CTCF in mESCs, we targeted the stop codon of both Ctcf alleles to introduce a 44–amino acid version of the AID tag (residues 71–114, (Morawska and Ulrich, 2013; Nishimura et al., 2009)) with an eGFP cassette (Figure 1A, Supplementary table1). We subsequently introduced a transgene encoding the Tir1 F-box protein from Oryza sativa (rice), which can bind to the AID in the presence of auxin, triggering proteasome-dependent degradation. The resulting cell line is referred to as CTCF-AID hereafter.
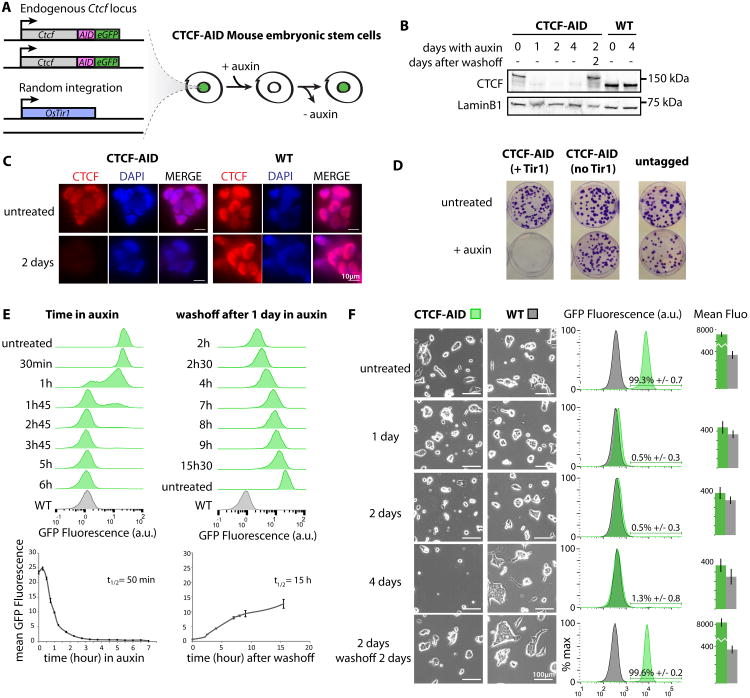
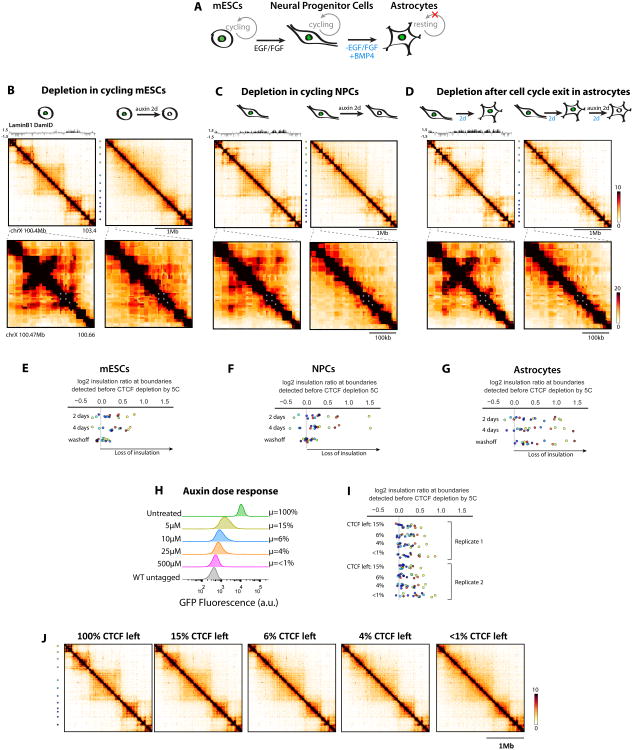
Figure 1. Acute and reversible depletion of CTCF with the auxin-inducible degron system in mESCs.
(A) Deploying the AID system at Ctcf in mESCs.
(B) Western-blot showing reversible loss of CTCF in CTCF-AID cells
(C) Immunofluorescence staining
(D) Long-term survival (12 days) is only compromised in CTCF-AID cells treated with auxin after introduction of the Tir1 transgene
(E) Time-course flow-cytometry
(F) Brightfield images of mESC colonies after auxin treatment indicating cells tolerate a 2-day depletion with no adverse effects on viability. See Figure S1
Adding auxin to the culture medium depleted CTCF to levels that could not be detected by Western blot, and washing out auxin allowed CTCF to accumulate back to initial levels (Figure 1B). Auxin in itself was neutral to untagged mESCs (Figure 1D and E), with no differential gene activity detected after up to 4 days of treatment (Supplementary tables 2 and 3). As reported previously the AID fusion led to slight constitutive destabilization (Morawska and Ulrich, 2013), so that basal CTCF levels were about 2–3-fold less in the AID-eGFP fusion line compared to the untagged parental line (Figure 1B and C). RNA-seq revealed 72 differentially expressed genes between the parental and untreated CTCF-AID lines (Supplementary table 3). Cells could nevertheless be expanded and subcloned normally (Figure 1D), indicating that the AID-eGFP fusion does not abrogate the essential functions of CTCF. In contrast, auxin-mediated degradation of CTCF prevented subcloning of CTCF-AID cells, recapitulating the full CTCF knockout phenotype in mESCs (Sleutels et al., 2012) (Figure 1D).
CTCF depletion was maximal as early as 3h45 after adding auxin (Figure 1E). Recovery initiated readily after washoff and was half complete by 15h (Figure 1E). Acute CTCF depletion was tolerated for 2 days without obvious cell death or differentiation (Figure 1F), but depleting for longer slowed cell proliferation dramatically (Figure 1F and S1A-B). Importantly, CTCF depletion in mESCs did not block cells in a specific phase of the cell cycle, and did not induce DNA damage or aneuploidy (Supplementary figures 1D-E). Cell death increased after 4 days of depletion (Figure S1F) but remained modest, unlike other cellular contexts (Soshnikova et al., 2010; Watson et al., 2014). Finally, expressing a stable doxycycline-inducible CTCF transgene at low levels largely rescued proliferation defects, demonstrating they are indeed due to acute depletion of endogenous CTCF (Figure S1G-J). Our system can therefore be used during at least two days after auxin addition (3-4 cell divisions) to study the immediate consequences of acute CTCF depletion without adverse effect on cell survival and proliferation.
Auxin Treatment Severely Depletes CTCF from Chromatin
CTCF binding patterns, as measured by ChIP-exo in untreated CTCF-AID mESCs, were highly similar to untreated or 2-day treated WT untagged cells, highlighting that auxin treatment in itself does not affect overall CTCF binding, nor does tagging with the AID-eGFP cassette (Figure S2A). Using ChIP-seq in CTCF-AID cells after 2 days of auxin we detected only 27% of the initial CTCF peaks. (Figure S1B-C and supplementary table 4). The enrichment level in persistent peaks was severely reduced (Figure S2D-E), indicating that CTCF occupancy is lost or considerably lower at all of its binding sites after depletion. ChIP-seq patterns from cells where auxin was washed off 2 days after a 2-day treatment was virtually identical to untreated cells, revealing that CTCF readily regains access to all of its cognate binding sites after transient depletion in mESCs (Figure S2B-E). Finally, depletion efficiency was equally efficient irrespective of local binding site density (Figure S2F-H).
CTCF is required for accumulating chromatin loops at CTCF sites
In order to measure changes in chromosome organization upon CTCF depletion we performed high-throughput 3C-based experiments. Current technologies require extremely deep sequencing to interrogate changes in contact frequencies between individual genomic loci below the megabase scale at the genome-wide level. Therefore, we first focused on the the X-inactivation centre locus (Xic) using 3C Carbon-Copy (5C, (Dostie et al., 2006)), with our male undifferentiated mESCs (which harbor a single active × chromosome). The Xic displays strong well-characterized CTCF-anchored interactions (Giorgetti et al., 2014; Nora et al., 2012) readily detected by 5C (Figure S2I and Supplementary Table 5). Chromosomal organization at the Xic in the untagged parental line was not perturbed by auxin and was identical in untreated CTCF-AID cells (Figure S2J). In contrast, auxin-mediated depletion of CTCF led to complete disappearance of these 5C peaks while auxin washoff restored them (Figure S2I).
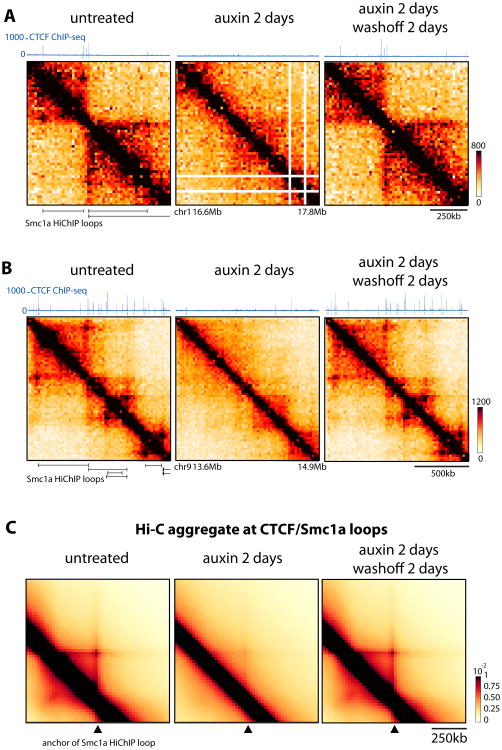
To extend these observations to the entire genome, we performed Hi-C in untreated, 2-day treated cells as well as after a 2-day washoff. Our 20kb resolution data (Figure 2A-B) did not allow us to perform robust de novo calling of loops. However, given that most CTCF binding events overlap with Cohesin enrichment by ChIP-seq (Parelho et al., 2008; Rubio et al., 2008; Wendt et al., 2008), we performed a meta-analysis by aggregating our Hi-C signal at CTCF/Cohesin bound loops, as previously detected by high-resolution HiChip for Smc1a in mESCs (Mumbach et al., 2016). This confirmed that CTCF is required for the interaction between CTCF/Cohesin bound loop-anchor loci genome-wide, and that bringing CTCF back is sufficient to restore these preferential contacts (Figure 2C).
Figure 2. CTCF is required for accumulating loops between CTCF/Cohesin binding sites.
(A-B) Snapshots of 1.3 Mb of Hi-C data at 20kb resolution CTCF-AID mESCs aligned with CTCF ChIP-seq and the Smc1a HiChIP loops identified by Mumbach et al. 2016. Normalized Hi-C counts are multiplied by 105
(C) Genome-wide aggregation of normalized Hi-C signal anchored at Smc1a HiChip loops separated by 280 to 380kb (1196 loops). Similar results were obtained for smaller and larger loops. See Figure S2
CTCF Depletion Triggers Dramatic Loss of TAD Insulation
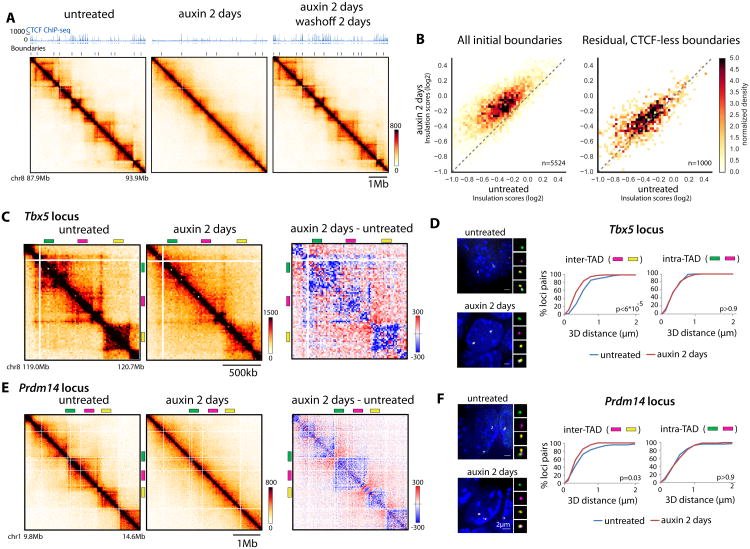
We next investigated the integrity of TAD folding upon CTCF depletion. Our Hi-C maps revealed extensive ectopic contacts across initial TAD boundaries, clearly visible by 5C as early as 24h after CTCF depletion (about two cell divisions; Figure 3A and S3A-B). These changes were again fully reversible after auxin washoff. Independently targeted CTCF-AID cell lines exhibited similar insulation defects (5/5 additional lines, Figure S3C). Ectopic CTCF expression from an inducible transgene prevented loss of insulation (Figure S3D-E) while auxin itself had no effect on WT untagged mESCs (Figure S3F), demonstrating that insulation defects upon CTCF depletion are specific and reproducible.
Figure 3. CTCF instructs insulation of TADs.
(A) Snapshots of 6Mb of Hi-C data at 20kb resolution from CTCF-AID mESCs aligned with CTCF ChIP-seq. Normalized Hi-C counts are multiplied by 105
(B) Left: CTCF depletion dampens insulation at TAD boundaries (higher insulation score over 100kb surrounding boundaries). Right: residual boundaries detected after CTCF depletion (and without persistent CTCF peaks, ∼20% of total boundaries) maintain insulation independently of CTCF. Note that lower score denotes higher insulation potential
(C) Snapshot of Hi-C data at the Tbx5 locus and differential contact map showing more inter-TAD (red) and fewer intra-TAD (blue) Hi-C signal after CTCF depletion
(D) 3D distance measurement from DNA FISH highlighting that CTCF depletion triggers inter-TAD compaction but does not affect intra-TAD packaging at the cytological level (E-F) same as C-D at the Prdm14 locus (n=90-100 alleles, Kolmogorov-Smirnov test). See Figure S3
To quantify this behavior genome-wide and identify loci that may deviate from it, we scored insulation potential across all chromosomes using our Hi-C data (Crane et al., 2015)(Supplementary Table 6 and Methods). Our resolution enabled calling 5524 boundaries for a median TAD size of 340 kb (mean of 450kb) in untreated cells. Loss of CTCF led to loss of insulation at most boundaries (>80%, Figure 3B). A subset of boundaries persisted after CTCF depletion. After removing those that displayed residual CTCF binding by ChIP-seq we identified 1000 persistent CTCF-less boundaries (18% of initial boundaries), where insulation was much less affected by CTCF depletion (Figure 3B).
To explore how changes measured by Hi-C translate at the cytological level, we used 3D DNA Fluorescent in situ Hybridization (FISH) with two probes in the same TAD and a third separated by one or more TAD boundaries (Figure 3C-F) – spanning a total of around 1.5Mb. For the two loci surveyed, loss of CTCF reduced inter-TAD 3D distances, which became equivalent to intra-TAD distances. This indicates that loss of insulation arises from compacting sequences initially in separate TADs. Intra-TAD FISH distances were unaffected by CTCF depletion, indicating that loss of CTCF does not trigger general chromatin compaction. In the absence of CTCF, linear genomic coordinates become a better predictor of 3D distances (Figure S3K) and, consistent with previous boundary-deletion experiments (Ji et al., 2016), TAD boundaries separate further apart in the three-dimensional space of the nucleus (Figure S3L).
In line with earlier less impactful (Figure S3M) RNAi-mediated CTCF depletion (Zuin et al., 2014) we detected fewer intra-TAD contacts upon loss of CTCF by Hi-C, while FISH did not detect changes in intra-TAD compaction (Figure 3C-F). This likely reflects the fact that total Hi-C read number is normalized between samples (so increased inter-TAD signal must be compensated by decreased signal elsewhere) while FISH distances are less resolutive but absolute – a limitation in comparing Hi-C and FISH (Dekker, 2016; Fudenberg and Imakaev, 2016; Giorgetti and Heard, 2016).
Disruption of Local Insulation Does Not Affect Higher-Order Chromosome Folding
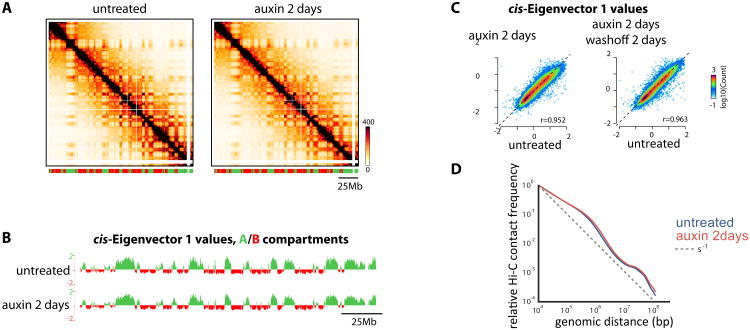
We next sought to investigate to what extent CTCF disruption affects higher-order segregation of active and inactive chromosome domains into A- and B- compartments (Gibcus and Dekker, 2013; Lieberman-Aiden et al., 2009). Contact maps (Figure 4A) as well as compartment signal (Imakaev et al., 2012) indicated that compartmentalization and genomic location of the transitions between A-and B- compartments are maintained after CTCF depletion (Figure 4B-C and S4A). We detected a minor but reproducible reduction (∼10%) in the strength of compartmentalization upon CTCF depletion (Figure S4B). Scaling of contact frequencies as a function of genomic separation did not change either (Figure 4D). Factors other than CTCF must therefore control the basal packaging regime of chromatin as well as its segregation in A- and B-compartments.
Figure 4. Proper insulation of TADs is not required for higher-order segregation of A and B compartments.
(A) Hi-C contact maps at 100kb resolution across entire chromosome 2. Bar denotes segments called as A (green) or B (red) compartment using 20kb-cis Eigenvector 1. Normalized Hi-C counts are multiplied by 105
(B) Distributions of cis Eigenvector 1 values across entire chromosome 2 are remarkably stable to depletion of CTCF
(C) cis Eigenvector 1 values are not affected genome-wide by CTCF depletion
(D) Overall scaling of Hi-C contact frequency as a function of genomic distance is not affect by the loss of CTCF, highlighting that CTCF does not affect general chromatin compaction. See Figure S4
We next explored if the residual TAD boundaries detected after CTCF depletion (18% of initial boundaries) could be explained by the maintenance of A/B compartmentalization. First, TAD boundaries in the A and B compartment both loose insulation potential upon CTCF depletion (Supplementary table S6). Second, out of the 1000 CTCF-less residual boundaries only 103 (10% - 3.1 fold enrichment over chance overlap, Figure S3E-F) were associated with a transition between A- and B- compartments. From these 1000 CTCF-less residual boundaries 609 (61%) had at least one CTCF ChIP-seq peak +/- 1 bin (20kb) prior to depletion, suggesting that CTCF binding at these sites is not what initially drove local insulation. Transcriptional activity (neighboring PolII ChIP-seq peak detected in untreated cells) was detected at 416 of the residual CTCF-less boundaries (41% - 2 fold enrichment over chance overlap). While this is compatible with compartment transition or transcription participating in the maintenance of CTCF-independent insulation, either of these features alone is not sufficient to drive CTCF-independent insulation since most boundaries associated with them are affected by CTCF depletion (Supplementary Table 6). Discrepancies with the reference genome may also account for some of the apparent retention of insulation.
Loss of CTCF also Triggers Misfolding in Non-Cycling Cells
To determine if insulation defects triggered by CTCF depletion require passage through DNA replication or mitosis, we differentiated our CTCF-AID mESCs stepwise into self-renewing Neural Precursor Cells (NPCs) and resting astrocytes (ACs; Figure 5A; Sofueva et al., 2013). 5C at the Xic revealed disrupted folding in cycling NPCs as well as resting ACs, whether CTCF was depleted before (Figure 5B-G) or after (Figure S5A-J) cell-cycle exit. Folding defects appeared somewhat less pronounced in differentiated cells, correlating with switching of a large portion of the region surveyed into a lamina associated domain (LAD) – and presumably B compartment (Figure 5A-D). Washing off auxin led to reformation of insulated TADs in mESCs and NPCs but not resting astrocytes. Passage through the cell cycle might therefore be required for restoring insulation or factors that cooperate with CTCF (e.g. Cohesin metabolism) may behave differently in terminally differentiated cells. Non-exclusively, loop formation or stabilization may not be a continuous process in these cells. Further experiments comparing different types of post-mitotic cells will clarify if this behavior is general to non-dividing cells.
Figure 5. CTCF remains essential for insulation of TADs in resting cells, and acts dose-dependently.
(A) mESCs can be converted into cycling NPCs and induced to exit cell cycle by terminal differentiation into astrocytes
(B-D) Extracts of restriction-fragment resolution interpolated 5C heatmaps at the Xic. LaminB1 DamID from(Peric-Hupkes et al., 2010). Color dots denote boundaries identified before CTCF depletion
(E-G) log2 ratio of 100kb insulation scores from depleted versus untreated cells at boundaries identified before depletion. Plots include boundaries probed beyond the region depicted in the heatmaps
(H) Titration of auxin leaves cells with intermediate CTCF levels. Percentages are relative to untreated CTCF-AID cells, where CTCF levels are 2-3 fold lower than parental untagged mESCs
(I) CTCF-dependent boundaries loose insulation as a function of leftover CTCF levels
(J) 5C heatmaps used to calculate insulation scores. See Figure S5.
CTCF depletion Needs to Be Near-Complete to Exhibit the Most Substantial Defects on TAD insulation
Previous studies with RNAi-mediated knock-down of CTCF in human HEK293 cells reported much milder folding defects than those we observed with CTCF-AID mESCs (Zuin et al., 2014). In order to address if differences are due to better depletion efficiency with the degron system than with RNAi, which leaves 10–15% CTCF (Zuin et al., 2014), we treated CTCF-AID mESCs with intermediate doses of auxin, and repeated 5C at the Xic in the context of various leftover amounts of CTCF, as quantified from fluorescence of the CTCF-AID-eGFP fusion (Figure 5H). Insulation defects scaled with the degree of CTCF depletion and samples with around 15% CTCF preserved more insulation than completely depleted cells (with some boundaries more sensitive than others, Figure 5I-J). This highlights that CTCF is very potent at mediating chromatin folding into TADs, acts in a dose-dependent fashion, and must therefore be very efficiently depleted to trigger major defects on chromosome organization.
CTCF and transcriptional regulation
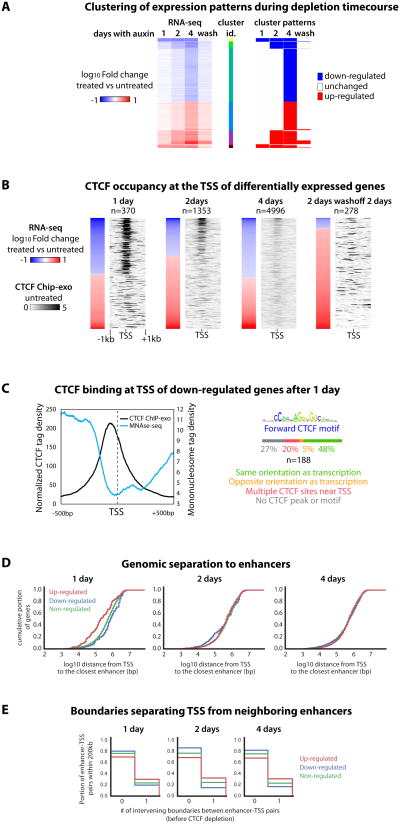
We then explored how the changes in local genome folding caused by acute CTCF depletion relate to transcriptional misregulation. We performed a time-course RNA-seq experiment in mESCs after 1, 2, or 4 days of auxin treatment (Figure 6A-B). The absolute number of differentially expressed genes increased over ten-fold between day 1 (370) and day 4 (4996) (Figure S6A-B), with around half of the dysregulated being down-regulated and half up-regulated at each time-point.
Figure 6. CTCF and transcriptional regulation.
(A) RNA-seq. fold change compared to untreated cells for genes differentially expressed at one or more time points. Wash denotes 2-day washoff after a 2-day treatment
(B) RNA-seq alignment with ChIP-exo (from untreated cells) for each time-point
(C) The CTCF site in the promoters of immediately down-regulated genes tends to be ∼60bp upstream of the TSS in direct orientation with transcription, and demarcates the beginning of the nucleosome-depleted region as previously measured by MNAse-seq (Teif et al., 2012)
(D) Immediately up-regulated genes tend to lie at shorter genomic distance to neighboring enhancers than down-or non-regulated genes. Trend is rapidly lost over time
(E) Enhancer-promoter pairs are more likely to be normally interrupted by a TAD boundary for genes that become up-regulated upon CTCF depletion. See Figure S6.
We first focused on down-regulated genes. Integration with CTCF Chip-exo data revealed that over 80% of the early down-regulated genes had CTCF bound within 1kb of the transcription start site (TSS) prior to depletion, as opposed to less than 20% of the up-regulated genes (Figure 6B). This trend is diluted with time as the number of differentially expressed genes rises. This indicates that the activity of a subset of CTCF-bound promoters (10% of all CTCF bound TSS) critically relies on CTCF, likely via direct binding. We explored if this activator role may be attributed to CTCF facilitating communication with distal regulatory elements. Out of the 188 genes down-regulated after 1 day of depletion only 53 (28%) overlap an anchor for SMC1a HiChIP loops (Mumbach et al., 2016) and 19 (10%) connect to an active regulatory region before treatment, based on H3K27Ac enrichment (Shen et al., 2012). Furthermore, down-regulated genes are not specifically positioned at TAD boundaries. Therefore down-regulation cannot be explained by loss of direct looping between promoters and enhancers. We noticed that at the promoter of the immediately down-regulated genes CTCF is bound slightly upstream of the TSS (around 60bp) and demarcates the beginning of the nucleosome-depleted region (Figure 6C). CTCF may therefore promote transcription by preventing promoter occlusion by nucleosomes. Strikingly, the orientation of the CTCF motif at these TSSs is almost systematically in direct orientation with the direction of transcription (90% of unequivocal sites, Figure 6C and Supplementary Table 7). This is reminiscent of the asymmetry of promoter positioning around CTCF ChIA-PET data in human cells (Tang et al., 2015). Given the implication of CTCF motif orientation in controlling long-range contacts it remains possible that CTCF depletion down-regulates the immediately responsive genes by disrupting tracking processes that are not associated with accumulation of chromatin loops as detected by a peak of Hi-C or HiChIP signal.
We then investigated up-regulated genes, and the possible effect of TAD dissolution on ectopic enhancer targeting. Previous studies have reported that CTCF is enriched around the TSS of both up-and down-regulated genes upon CTCF knock-down, but also noted that for up-regulated genes enrichment is shifted away from the promoter-proximal region (Zuin et al., 2014) – pointing to different mechanisms for up- and down-regulation upon CTCF depletion (Soshnikova et al., 2010). The fact that in our data CTCF does not bind the majority (80%) of TSS of genes up-regulated after 1 day suggests CTCF normally represses them indirectly. We find that immediately up-regulated genes tend to be located genomically closer to active enhancers (Figure 6D and S6B) than down- or non-regulated genes. However, a higher fraction of up-regulated genes normally have a TAD boundary separating them from neighboring (<200kb) enhancers, compared to down-regulated or non-regulated genes (Figure 6E and S6C). This suggests that CTCF depletion triggers up-regulation of a subset of genes formerly insulated from neighboring enhancers by a TAD boundary. This observation supports at the genome-wide level the notion that CTCF can mediate enhancer-blocking insulation through the specification of TAD boundaries, in line with previous locus-specific studies (Dowen et al., 2014; Doyle et al., 2014; Lupiáñez et al., 2015; Nora et al., 2012).
When focusing on TADs that that harbor multiple genes, 24% (24/99) have more than one up-regulated gene after one day of depletion. This indicates that up-regulated genes tend to localize in the same TAD more often than by chance (p=0.0042, and p=0.19 for down-regulated genes -Methods). However immediate up-regulation is not coordinated for all genes of the domain for all TADs. This argues against a simple model where upon losing a TAD boundary enhancers would immediately trigger up-regulation of all genes of the neighboring TADs homogeneously. It is possible we under-estimate transcriptional coordination because RNA-seq does not directly measure ongoing rates of transcription, and because our limited Hi-C resolution prevents us from robustly identifying small TADs. Taking advantage of the Smc1a HiChIP data we noticed that promoters of mis-regulated genes are more often close to loop anchors than promoters of non-regulated genes (Figure S6D), while the distribution is similar outside of the anchors. This indicates that promoters at loop anchors and TAD borders are more sensitive to CTCF disruption than genes away from boundaries. A function of TAD boundaries may therefore be to protect these promoters from the influence of neighboring enhancers.
Auxin washoff after a 2-day treatment did not completely restore the transcriptome, with most (252 out of 278, 90%) of the differentially expressed genes remaining up-regulated compared to untreated cells (Figure 6A-B). Transcript stability may to some extent account for persistent high mRNA levels. However, while some transcripts showed a trend downward their initial values while others kept rising (Supplementary Table 3), suggesting that for a small subset of genes transient loss of CTCF depletion can trigger transcriptional changes that become irreversible, indicating they are involved in a positive feedback mechanism.
CTCF Binding Is Not a Direct Impediment to H3K27me3 Spreading in mESCs
It has been proposed that CTCF may confer chromatin barrier activity by opposing the spreading of facultative heterochromatin, thereby demarcating active and inactive chromatin domains (Cuddapah et al., 2009; Dowen et al., 2014) and insulating against position effects (Essafi et al., 2011; Witcher and Emerson, 2009). This role has been debated (Bender et al., 2006; Huang et al., 2007; Recillas-Targa et al., 2002; Splinter, 2006).
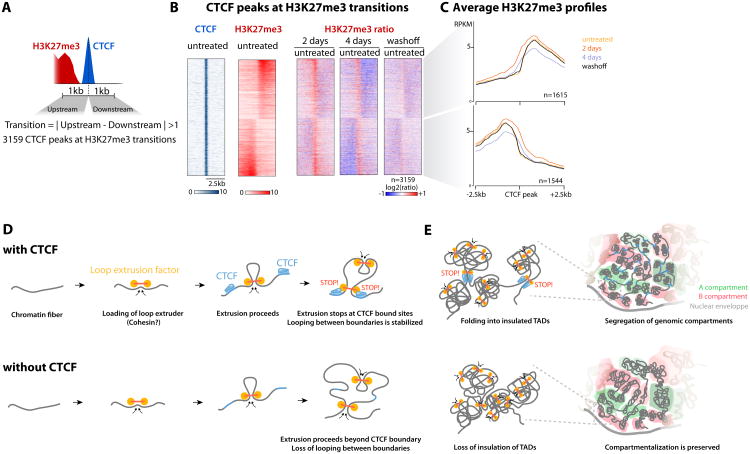
As reported in human cells (Cuddapah et al., 2009), we found that a subset of CTCF sites mark transitions in H3K27me3 enrichment in mESCs (∼7% of CTCF sites, Figure 7A). However CTCF depletion did not trigger spreading of H3K27me3 as measured by ChIP-seq (Figure 7B-C), even after 4 days (3-4 cell divisions Figure S1B). Changes were restricted to a very local gain of H3K27me3 signal at the initially bound CTCF site (Figure 7B and S7A), possibly due to nucleosomes becoming able to occupy the formerly bound CTCF site (Wiechens et al., 2016). On a more global scale, we observed a slight but significant decrease in overall H3K27me3 levels (Figure S7B). These changes are likely indirect effects as they are not restricted to the vicinity of CTCF sites, and may be accounted for by 2-fold transcriptional down-regulation of the essential PRC2 component EED (Supplementary Table 3).
Figure 7. CTCF does not constrain H3K27me3 spreading in mESCs and summary model.
(A) A subset of CTCF binding sites mark transitions in H3K27me3 patterns
(B-C) CTCF depletion does not trigger H3K27me3 spreading beyond the formerly bound CTCF site itself (center)
(D) Our observations are consistent with TAD formation by loop extrusion, and establish CTCF as the major factor defining domain boundaries genome-wide
(E) Statistical average cartoon representation of TAD disruption and compartment preservation upon loss of CTCF
See Figure S7.
Altogether our results demonstrate that the role of CTCF in genome organization is local, in controlling the accumulation of chromatin loops between TAD boundaries and physically insulating these domains from each other. In the absence of CTCF neighboring TADs merge, with consequences on transcriptional regulation. Overall chromosome compaction and organization are however not affected. Other factors than CTCF must therefore be responsible for general chromatin packaging and compartmentalization.
Discussion
Using a system enabling acute, reversible and near-complete loss of CTCF, we have elucidated the critical and dose-dependent roles of this enigmatic transcription factor in regulating 3D chromatin organization. Beyond establishing the central importance of CTCF for the insulation of TADs, this system has enabled addressing fundamental questions about the causal relationships between the different levels of genome organization, transcription, and large-scale chromatin states. Our findings indicate that spatial compartmentalization of mammalian genomes rely on molecular mechanisms that are distinct from those controlling the local insulation of chromosome neighborhoods. TADs and compartments therefore do not represent a hierarchy in the folding of mammalian chromosomes.
CTCF Is Necessary for TAD Insulation and Loops between Boundaries
CTCF depletion concomitantly disrupted loops between TAD boundaries and insulation of neighboring TADs. This substantiates the notion that these two aspects are molecularly coupled (Giorgetti et al., 2014). Our observations are compatible with mechanistic models where domain-wide enrichment of chromosomal contact is the result of a process that accumulates chromatin loops between CTCF-bound boundary elements (Fudenberg et al., 2016; Sanborn et al., 2015) (Figure 7D).
Pervasive Loss of Insulation upon CTCF Depletion Ascertains the Central Importance of Boundary elements
Our data support at the genome-wide level that CTCF binding confers the insulated nature of mammalian TADs, corroborating earlier boundary deletion experiments. This argues against models where segmental folding would arise from intrinsic interaction incompatibility between neighboring TADs (Chiariello et al., 2016). Block co-polymer incompatibility may be more relevant in other biological contexts where chromatin states are a better predictor of segmental packaging into TADs, such as in Drosophila (Jost et al., 2014).
Local Insulation and A/B Compartmentalization Are Molecularly Separable Principles of Mammalian Genome Folding
Long-range chromosome folding (above the megabase scale) is remarkably resistant to CTCF depletion, despite dramatic changes at the sub-megabase scale. We conclude that proper packaging of chromatin into TADs is not a prerequisite for the segregation of A and B compartments. It is possible that the precise boundaries of the chromosomal segments belonging to the same type of compartment are slightly altered at scales below what can be detected with our current 20-kb resolution.
This finding corroborates cases in which TAD folding and compartmentalization are uncoupled, such as the Drosophila polytene chromosomes that insulate TADs without compartmentalizing them (Eagen et al., 2015).
Our observations are consistent with the proposed mechanisms of TAD formation by intra-TAD loop extrusion and are in agreement with the idea that CTCF is a major blocking factor to the processivity of extrusion (Fudenberg et al., 2016; Sanborn et al., 2015). Notably, the extrusion model accurately describes mammalian chromosome folding at the sub-megabase scale but does not account for the segregation of genomic compartments, and the direct molecular drivers of CTCF-independent higher-order compartmentalization remain to be defined.
CTCF Does Not Directly Constrain the Spread of H3K27me3 but May Still Define Chromatin Domains
Our observation that H3K27me3 patterns remain largely unaltered challenges the notion that CTCF binding acts as a direct roadblock to heterochromatin spreading. This is consistent with the lack of H3K27me3 spreading after serial genetic deletions of the HoxD locus, removing large segments including CTCF sites (Schorderet et al., 2013). Our observations in undifferentiated ES cells do not, however, address the role of CTCF binding in defining the genomic segments that can undergo domain-wide chromatin state transitions during cell differentiation, which were initially found to align with TAD boundaries (Nora et al., 2012). Deleting single CTCF sites within the HoxA cluster enables ectopic developmental activation of genes across the former boundary, consistent with ectopic enhancer targeting, but again does not lead to H3K27me3 spreading (Narendra et al., 2015). Altogether current data support that CTCF mediates enhancer-blocker activity, through its ability to mediate insulation and segmental folding into TADs, but is not a direct impediment to heterochromatin spreading.
TAD Insulation and Transcriptional Regulation
The pervasiveness of the chromosome folding defects we observed upon CTCF depletion contrast with the rather limited immediate transcriptional defects measured by RNA-seq. It is difficult to interpret prolonged depletion, as secondary effects can rapidly become confounding and regulatory bleed-through is unlikely to be the only cause of transcriptional misregulation upon CTCF depletion. Our data highlight that exposure of a promoter to new enhancers has an initially mild and context-specific impact on transcriptional activity. This suggests that hijacking of cis-regulatory elements caused by altered insulation may require time to manifest pervasively, and that ectopic contact between enhancers and promoters is not in itself sufficient to predict the initial extent of transcriptional defects. Additional specificity or compatibility factors must contribute to how promoters respond after ectopic exposure to enhancers (van Arensbergen et al., 2014; Arnold et al., 2016).
Of note, we did not observe immediate coordinated TAD-wide transcriptional changes. This may appear at odds with previous reports of TAD-wide coordination of transcription dynamics upon deletion of a TAD boundary or during response to signaling (Le Dily et al., 2014; Narendra et al., 2015; Nora et al., 2012). The timing needed for transcriptional defects to accumulate may explain this apparent discrepancy, as boundary disruption experiments are typically analyzed long after the rearrangement has been induced, after cells have adapted. On the other hand acute degradation of CTCF provides the opportunity to monitor immediate effects, but is also expected to trigger a wide range of effects, where direct but slowly manifesting effect will be obscured by indirect but rapid secondary effects.
Finally, a parallel study employing near-complete removal of Cohesins from chromosomes reached a similar conclusion (Schwarzer et al., 2016). The consequences of losing CTCF or Cohesin on TAD folding are however nearly opposite, expanding on the observation that these two factors act different steps in edifying chromosome architecture (Zuin et al., 2014). The emerging model is that Cohesin packages the chromatin fiber while CTCF defines focal boundaries by constrains this packaging activity. This would explain why depleting CTCF does not affect how the frequency of chromosomal contacts scales overall with genomic distance, as opposed to altering factors that control Cohesin turnover on chromatin, such as Nipbl (Schwarzer et al., 2016). Understanding the molecular details of these processes and how they modulate transcriptional patterning as well as other nuclear processes is an exciting upcoming challenge.
Star Methods
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to the corresponding authors Benoit Bruneau (bbruneau@gladstone.ucsf.edu) and Elphège Nora (elphege.nora@gladstone.ucsf.edu).
Experimental Model and Subject Details
Mouse Embryonic Stem cells
E14Tg2a (karyotype 19, XY; 129/Ola isogenic background) and subclones were cultured in DMEM+Glutamax (ThermoFisher cat 10566-016) supplemented with 15% Fetal Bovine Serum (ThermoFisher SH30071.03), 550μM b-mercaptoethanol (ThermoFisher 21985-023), 1mM Sodium Pyruvate (ThermoFisher 11360-070), 1× non-essential amino-acids (ThermoFisher 11140-50) and 104U of Leukemia inhibitory factor (Millipore ESG1107). Cells were maintained at a density of 0.2-1.5×105 cells / cm² by passaging using TrypLE (12563011) every 24-48h on 0.1% gelatin-coated dishes (Millipore cat ES-006-B) at 37°C and 7% CO2. Medium was changed daily when cells were not passaged. Cells were checked for mycoplasma infection every 3-4 months and tested negative.
Neural progenitor cells and astrocytes
CTCF-AID mESCs were seeded at around 0.1 million cells in a 75cm² gelatinized dish in mESC medium. The following day cells were rinsed twice in 1X PBS and switched to NDiff227 differentiation medium (Stem Cells Inc.) and changed daily. After 7 days cells were detached using TryplE and seeded on non-gelatinized bacterial dishes for suspension culture at 3 million cells per 75cm² and cultured in NDiff227 containing 10ng/mL EGF and FGF (Peprotech). After 3 days floating aggregates were seeded on gelatinized dishes. After 2-4 days cells were dissociated using Accutase and passaged twice on gelatinized dishes in NDiff227+EGF+FGF. In order to overcome variable silencing of the Tir1 transgene the CTCF-AID NPCs were subcloned by limiting dilution and NPC colonies were manually picked after 10-15 days and expanded in NDiff227+EGF+FGF. For differentiation into quiescent astrocytes adherent NPC cultures were washed twice with NDiff227 and cultured for at least 48h with NDiff227+ 10ng/mL BMP4 (R&D Systems).
The Tir1 transgene variegated upon differentiation, which we overcame by first converting CTCF-AID mESCs into self-renewing Neural Precursor Cells (NPCs), subcloning NPCs and then selecting clonal lines that retained homogenous CTCF degradation upon auxin treatment. The CTCF-AID NPC subclones did not survive freeze and thawing.
For induction of the auxin-inducible degron indole-3-acetic acid (IAA, chemical analog of auxin) was added in the medium at 500μM from a 1000× stock diluted in sterile water. Stocks were kept at 4°C up to 4 weeks or -20°C for long term storage.
Method Details
Plasmid Construction
We used the smallest functional truncation of the AID tag (AID*, 44 amino-acids), initially developed in yeast (Morawska and Ulrich, 2013), shorter than the mini-AID (67 amino-acids (Kubota et al., 2013)). We observed equivalent CTCF depletion efficiency with the AID* as with the original full-length 231 amino-acid tag (Nishimura et al., 2009) (data not shown).
The CTCF-AID-EGFP targeting vector (pEN84) was assembled by serial modification of the base vector pFNF (Addgene #22687) using Gibson assembly with the following templates: the minimal functional AID tag (aa 71-114) described by (Morawska and Ulrich, 2013) was PCR amplified from pAID (Nishimura et al., 2009); homology arms to the last exon of Ctcf were PCR amplified from E14Tg2A genomic DNA (1kb each); the N-acteyl-transferase (PAC/PuroR) was PCR amplified from pLox-STOP-Lox TOPO (Addgene # 11854), the eGFP cDNA was PCR amplified from pTRE2-2A-eGFP (Kind gift from Kevin Monahan and Stavros Lomvardas). We also created a version of the plasmid conferring resistance to Blasticidin (pEN244).
The Tir1 expression vector (pEN113) for the cell line analyzed by Hi-C (#1) was assembled by serial modification of the base vector pFNF (Addgene #22687) using Gibson assembly with the following templates: CAGGS promoter was subcloned from pCAGEN (Addgene #11160), the Oryza Sativa Tir1 cDNA was PCR amplified from a synthetic mammalian codon-optimized vector (kind gift from Daphné Dambournet and David Drubin); homology arms to the Rosa26 locus were PCR amplified from E14Tg2A genomic DNA (1kb each). From this vector we created an alternative version of the vector with a puro selection cassette (pEN114). The Tir1 expression vectors for cell lines #4-6 (pEN396) contained a 2A-puro fusion and two 1kb homology arms surrounding the sgRNA target site at the Tigre acceptor locus (described below).
The BFP/mCherry FUCCI reporter (pEN435) was assembled by serial modification of the base vector vector pFNF (Addgene #22687) using Gibson assembly with the following templates: hGeminin and mCherry-Cdt1 were PCR amplified from pRetroX-S2G2M and pRetroX-G1-Red (Clonetech); tagBFP cDNA from pHR-Tet3G-2A-BFP (Kind gift from Stanley Qi); CAGGS promoter and puroR are of the same source as pEN113; homology arms to the Tigre locus (Zeng et al., 2008) were PCR amplified from E14Tg2A genomic DNA (1kb each).
The transgene for doxycycline-inducible CTCF expression (pEN366) was assembled by stitching an rtTA3G-encoding cassette (Clonetech) under a CAGGS promoter and a rabbit globin polyA termination sequence together with a TetO-3G element (Clonetech) and a bovine growth hormone polyA termination sequence. A cDNA encoding mouse CTCF (without UTRs; NCBI CCDS22606.1 sequence) was then produced by reverse-transcription of mESC cDNA (SuperscriptIII, ThermoFisher) using the following primers: tgctagcgcggccgcatcgatATGGAAGGTGAGGCGGTTGA and cacagtcgaggctatgtttaaacTCACCGGTCCATCATGCTGA (lower case = cloning adapters). An mRuby2 cassette was then introduced as a direct C-terminal fusion with the CTCF cDNA (LKGGAGG linker) and a 3×-FLAG tag in N-terminus (TG linker). The final targeting vector contained two 1kb homology arms surrounding the sgRNA target site of the Tigre locus described below, as well as an FRT-PGK-puro-FRT cassette for selection of stable integrants. The clone analyzed here was homozygous for the integration and the puro cassette was still present in the final cell line.
Maps of the targeting constructs in the Genbank format are available on Addgene and upon request. sgRNAs were cloned by annealing pairs of oligos either in pX330 (Addgene #42230) for single Cas9 nuclease or pX335 (Addgene #42335) for dual Cas9 nickase strategies, following the protocol described in (Cong et al., 2013). Ctcf-targeting sgRNAs were cloned in pX335 (dual nickase) by annealing oligos caccgATCACCGGTCCATCATGCTG and aaacCAGCATGATGGACCGGTGATc for the first sgRNA and caccgCTGGGGCCTTGCTCGGCACC and aaacGGTGCCGAGCAAGGCCCCAGc for the second sgRNA. Rosa26 sgRNAs were cloned in pX335 (dual nickase) by annealing oligos caccgTGGGCGGGAGTCTTCTGGGC and aaacGCCCAGAAGACTCCCGCCCAc for the first sgRNA and caccgACTGGAGTTGCAGATCACGA with aaacTCGTGATCTGCAACTCCAGTc for the second sgRNA. We noticed the dual nickase underperformed for Rosa26 and recommend using a single nuclease strategy approach with the first sgRNA only. The Tigre-targeting sgRNA was cloned into pX330 (single nuclease) by annealing caccgACTGCCATAACACCTAACTT and aaacAAGTTAGGTGTTATGGCAGTc.
Gene targeting
For transfection plasmids were prepared using the Nucleobond Maxi kit (Macherey Nagel) followed by ethanol precipitation. Constructs were not linearized.
To knock in the AID-eGFP cassette at the N-terinus of CTCF E14Tg2a passage 19 were transfected by microporation using the Neon system (Thermofisher) using a 100μL tip with 1 million cells at 1400V, 10ms and 3 pulses. 2.5μg of each Ctcf-targeting sgRNA and 20μg of targeting construct (pEN84) was used. After electroporation cells were seeded in a 9cm² well and left to recover for 48h, at which stage around 10% of the cells show nuclear GFP fluorescence. Puromycine was then added to the media at 1μg/mL and cells were selected as a heterogenous pool of homozygous and heterozygous cells for around 10 days, at which stage over 95% of the cells showed nuclear GFP fluorescence. Cells were then transfected with the Neon system using a 10μL tip and 0.1 million cells with 250ng of a flippase-expressing plasmid (pCAGGS-FlpO-IRES-puro) in order to trigger FRT recombination and excision of the puromycine selection cassette. After electroporation cells were seeded in a 9cm² well and left to recover for 48h and transferred into a 78cm² petri dish from whish two serial 1:10 dilution were seeded in an additional two dishes. After 7-8 days of culture without antibiotic selection single colonies were manually picked, transferred into a 96-well plate, dissociated and re-plated. Clones were then genotyped by PCR for homozygous insertion of AID-eGFP and excision of the puro cassette. Over 95% of cells had one knock-in allele, of which 20% were homozygous. Half of the clones were found to have undergone FlpO-mediated recombination. When homozygous both alleles always underwent recombination.
To knock in the Tir1-expressing cassette one homozygous CTCF-AID-eGFP clone was transfected as described above using a 100μL tip format and pEN114 as the targeting construct. After a 48h recovery cells were subcloned and grown for 7 days in the presence of 200μg/μL Geneticin until single colonies could be picked. We noticed that only a handful of resistant clones were recovered, suggesting sub-optimal targeting – either because of the sgRNA or the targeting construct. Clonal lines were assessed for their ability to undergo auxin-mediated degradation of CTCF-AID-eGFP. We selected the clone with the fewest GFP-positive cells (<1%) after 24h of auxin treatment. This clone was then used for transient transfection of pCAGGS-FlpO-IRES-puro as described above to yield the CTCF-AID-eGFP, Tir1 line with which we conducted experiments presented in this manuscript (puromycine and neomycine sensitive). Rosa26 PCR genotyping revealed this clone had undergone random insertion of the Tir1 cassette. Unless stated this clone was used in all analyses (cell line #1) Robust expression of the Tir1 transgene was absolutely critical to mediate auxin responsiveness. Indeed our CTCF-AID lines down-regulated Tir1 during differentiation, even when targeted at Rosa26, leading to variegation of auxin response and limiting our analyses in committed cells that can be subcloned, such as neural progenitors. Further improvements in transgenesis will be necessary to enable reliable use of the AID system in both stem cells and their differentiated derivatives.
To create the additional cell lines #2 and #3 we used the intermediate CTCF-AID-eGFP clone (without Tir1), removed the FRT-puro-FRT selection cassette using transient transfection of pCAGGS-FlpO-IRES-puro and subcloning, and re-introduced the Tir1 expressing cassette at Rosa26 using pEN114 and puromycin selection and pX330-EN479 (Cas9 nuclease). Additional cell lines #4 and 5 were created from the same intermediate intermediate CTCF-AID-eGFP clone (without Tir1) but using the pEN396 to target a Tir1-2A-puro cassette at the Tigre locus. Cell line #6 was created by first targeting the Tir1-2A-puro cassette homozygously at the Tigre locus in WT E14Tg2a cells (with pEN396) and subsequently targeting AID-eGFP at CTCF, using a FRT-Blast-FRT selection cassette (pEN244) which was then removed by transient transfection of pCAGGS-FlpO-IRES-puro and subcloning.
We noticed that Tir1 targeting with the Tigre targeting vector was at least 5-fold more efficient than with our Rosa26 targeting vector. Basal CTCF-AID-eGFP levels were slightly lower (1.5-2 fold) than when Tir1 was inserted at Rosa26 or randomly (Figure S3C), suggesting that Tigre allows for higher expression or the Tir1 transgene, as reported previously (Madisen et al., 2015). We therefore recommend targeting Tigre instead of Rosa26 to drive Tir1 expression, unless basal expression level of the AID-fused protein is absolutely critical.
Crystal Violet staining
Limiting dilutions of mESCs were plated and grown for 14 days, after which they were rinsed with PBS and fixed/stained with 1% Formaldehyde 1% Methanol in PBS 0.05%w/v Crystal violet for 20 min. Plates were thoroughly rinsed with tap water and air dried.
Flow Cytometry
mESCs were dissociated with TryplE, resuspended in culture medium, spun and resuspended in 4% FBS-PBS before live flow cytometry on a MACSQuant instrument (Miltenyibiotec). Dissociation, wahs and Flow buffers were supplemented with auxin, when appropriate, to avoid re-expression of the CTCF-AID-eGFP fusion. Analysis was performed using the Flowjo sowftware.
CellTrace (CFSE) proliferation assay
Dissociated mESCs were labeled with CellTrace Violet dye (ThermoFisher) for 30min in PBS and washed following manufacturer's recommendations. Initial staining was measured by flow cytometry after 30 min, cells were plated and eventually treated with auxin. Remaining fluorescence was then measured daily for up to 4 days after cell dissociation by flow cytometry.
Western blots
mESCs were dissociated, resuspended in culture medium, pelleted, washed in PBS, pelleted again and kept at -80°C. 15-20 million cells were used to prepare nuclear extracts. Cell pellets were resuspended in 10mM Hepes pH 7.9, 2.5mM MgCl2, 0.25M sucrose, 0.1% NP40, 1mM DTT, 1× HALT protease inhibitors (ThermoFisher) and swell for 10 min on ice. After centrifugation at 500g nuclei were resuspended in on ice in (25mM Hepes pH 7.9, 1.5mM MgCl2, 700 mM NaCl, 0.5mM DTT, 0.1 mM EDTA, 20% glycerol, 1mM DTT, sonicated and centrifuged at 18,000g at 4°C for 10 minutes. Protein concentration from supernatants were measured using the Pierce Coomassie Plus assay kit (Thermofisher). For CTCF 10μg of nuclear extracts were loaded per lane while for histones 3μg were used. Samples were mixed with Laemmli buffer and 0.025% b-mercaptoethanol final, run on a 4-12% polyacrylamide TGX gel (Biorad). Transfer onto PVDF membranes was performed using the iBlot system (Thermofisher) Program 0 for 8 minutes. Membranes were incubated at least 30 minutes with Odyssey blocking buffer (Li-cor) prior to antibody incubation overnight at 4°C, following manufacturer's recommended dilutions and supplementing with 0.1% Tween-20 and 0.01%SDS. Membranes were washed five times 5minutes in PBS-0.1% Tween-20 at room temperature, incubated with secondaries antibodies (Goat Anti-Rabbit 680RD and Donkey Anti-Mouse 800CW (Li-cor), 1:10,000) in Odyssey blocking buffer with 0.1% Tween-20 and 0.01% SDS 1h at room temperature, washed 5 times and analyzed on a Li-cor imaging system. Pannels were mounted using imageJ preserving linearity.
Cell-cycle Analysis by propidium iodide staining
mESCs were dissociated, resuspended in culture medium, pelleted, washed in PBS, resuspended in ice-cold PBS at 2 million cells / mL. 9mL of 70% ethanol was then added drop-wise while mixing and cells were stored overnight at -20°C. Cells were pelleted at 200g 10 min at 4°C, washed with PBS, pelleted again and resuspended in 300μL of 0.1% Triton X-100 in PBS supplemented with 20μg/mL Propidium iodide and 0.2mg/mL RNAse A. After 30min incubation at 37°C cells were transferred on ice and used directly for flow cytometry.
Immunofluorescence
mESCs were grown on glass-coverslips, fixed with 3% formaldehyde in 1XPBS for 10′ at room temperature. Permeabilization was carried out in 0.5% Triton followed by blocking with 1% BSA diluted in 1X PBS (Gemini cat 700-110) for 15min at room temperature. Primary antibody (1/250) incubation was performed at room temperature for 45min, followed by three 5-minute washes in 1X PBS, secondary antibody (1/10.000) incubation, three 5-minute washes in 1X PBS, counter-staining with DAPI and mounting in 90% glycerol – 0.1× PB – 0.1% p-phenylenediamine pH9.
3D-DNA FISH
Procedure was carried out exactly as described in Nora et al. 2012. Probes were prepared by nick translation from following Bacterial Artificial Chromosomes obtained from CHORI/BACPAC: Tbx5 locus: RP24-164B17, RP24-267I14, RP23-469K13. Prdm14 locus: RP24-335O3, RP24-228J7, RP24-230I15.
Microscopy
Images were acquired on a DeltaVision widefield system (GE Healthcare) using a 100× objective and no binning. Images were deconvolved directly with the Softworks software.
ChIP-seq
For fixation mESCs were dissociated using TrypLE and resuspended in 10% FBS in PBS, counted and adjusted to 1 million cells per mL. Formaldehyde was then added to 1% final followed by 10 minute incubation at room temperature. Quenching was performed by adding 2.5M Glycine-PBS to 0.125M final followed by 5 min incubation at room temperature, 15 minute incubation at 4°C, centrifugation at 200g 5 minutes at 4°C, resuspended with 0.125M Glycine in PBS at 10 million cells per mL, aliquoted, spun at at 200g 5 minutes at 4°C and snap frozen on dry ice.
Fixed cells were thawed on ice, resuspended in ice cold 5mM PIPES pH 7.5, 85mM Kcl, 1% NP-40 and 1× HALT protease inhibitor, counted and readjusted to obtain 10 million cells total exactly, incubated on ice 15 min, centrifuged at 500g 5 min at 4°c, resuspended in 1mL 50mM Tris-HCl pH8, 10mM EDTA pH8, 1% SDS and 1× HALT protease inhibitor, transferred to a MilliTube (Covaris). Chromatin was sheared on a Covaris S2 sonicator for 7 minutes at 5% duty cycle, intensity 8, 200 cycles per burst in a waterbath maintained at 4°C, using 1 min sonication – 30 sec rest, resulting in 200-800bp fragments. Samples were clarified by centrifugation at 18,000g at 4°C for 10 min. Supernatents were transferred to 15mL conicals and 40ng of spike-in Drosophila chromatin (Active Motif) was added. 10% of the mixture was saved as input and the rest was diluted to 5mL with ice-cold 50mM Tris-Hcl pH 7.4, 150mM NacCl, 1% NP-40, 0.25% Sodium Deoxycholate, 1mM EDTA, 1× protease inhibitor. 10μg of anti-CTCF together with 4μg spike-in antibody (anti-H2Av, Active motif) or anti-H3K27me3 antibody together with 4μg spike-in antibody (Active motif) was added alongside with 40μL prewashed protein G Dynabeads (ThermoFisher) followed by overnight incubation at 4°C on a rotator. Beads were then collected on a magnetic rack and washed twice with 1mL cold 50mM Tris-Hcl pH 7.4, 150mM NacCl, 1% NP-40, 0.25% Sodium Deoxycholate, 1mM EDTA, twice with 1mL cold 100mM Tris-HCl pH9, 500mM LiCl, 1% NP-40, 1% Sodium deoxycholate and once with 1mL cold 100mM Tris-HCl pH9, 150mM Nacl, 500mM LiCl, 1% NP-40, 1% Sodium deoxycholate. Beads were then eluted with 100μL 50mM NaHCO3 1% SDS and heated at 65°C 30min with shaking. Input sample volumes were adjusted to 100μL with the same buffer. Eluates and inputs were supplemented with 10μg RNAse A and incubated 30 min at 30°C, then 20μg Proteinase K and 12μL of 5M NaCl were added followed by overnight incubation at 65°C. Samples were then purified using 1.8× Agencourt AMPure XP beads (Beckman-coulter) and eluted in 30μL Tris-HCl.
The entire Chip material or 50ng of the input DNA were used to construct Illumina sequencing libraries. End repair was performed in 100μL with 400μM dNTP, 15U T4 DNA polymerase (NEB), 5U Klenow large fragment DNA polymerase (NEB) and 50U T4 PNK (NEB) in 1× T4 ligase buffer (NEB), at room temperature 30min, followed by 1× AMPure purification. Entire eluate was used for A-tailing in a 50μL reaction with 1mM dATP and 15U Klenow 3′->5′ exo minus in 1X NEB buffer 2 followed by 1× AMPure purification. Entire eluate was used for adapter ligation in 50μL with 6,000U T4 ligase (NEB) and 20nM annealed and indexed adapters in 1× T4 ligase buffer (NEB) at room temperature for 2 hours, followed by 0.8× AMPure purification. Adapters were prepared by annealing following HPLC purified oligos: 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATC*T and 5′Phos-GATCGGAAGAGCACACGTCTGAACTCCAGTCACNNNNNNATCTCGTATGCCGTCTTCTGCTTG T where * represents a phosphothiorate bond and NNNNNN is a Truseq index sequence. The entire eluate was then used for PCR amplification in a 50μL reaction with 10μM primers 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGA and 5′-CAAGCAGAAGACGGCATACGAGAT and NEB Next high-fidelity 2× mix (NEB), using 98°C 30sec; 18 cycles of 98°C 10sec, 58°C 40sec, 72°C 30sec; 72°C 5min, followed by 0.9× AMPure purification. Entire eluate was then run on a 2% E-gel (ThermoFisher) and fragments 200pb-500bp were gel extracted. Library quality and quantity were estimated with Bioanalyzer and Qubit assays. Libraries were sequenced on a Next-seq 500 using 75bp single end.
ChIP-Exo
For fixation, 10 million adherent mESCs were incubated in 2% formaldehyde-10%FBS in PBS for 10 min at room temperature, quenched by adding glycine to 0.125M, washed with 0.125M glycine in PBS, scraped, pelleted, snap frozen on dry ice and stored at -80°C.
Procedure was based on (Luna-Zurita et al., 2016) with modification. Chip procedure was the same as for Chip-seq except that no spike-in antibody was used and washes consisted in 6 iterations of RIPA buffer (HEPES pH7.6 50mM, EDTA 1mM, Sodium Deoxycholate 0.7%, NP40 1% and LiCl 0.5M) followed by two iterations of Tris-HCl pH8. End repair was immediately followed by resuspending the DNA-antibody-bead matrix with 1mM ATP, 100μM dNTPs, 15U T4 DNA polymerase (NEB), 5U Klenow large fragment DNA polymerase (NEB) and 50U T4 PNK (NEB) in 1X NEB buffer 2 and incubating at 30°C for 30min. After two RIPA and two Tris-Cl pH8 washes ligation of p7 adapters was performed by resuspending the beads in 100μL of 1mM ATP, 150pmol p7 adapter and 2000U T4 DNA ligase (NEB) in 1X NEB buffer 2 and incubating at 25°C for 60min. p7 adapters were prepared by mixing the following HPLC purified oligos at 10μM final in 10M Tris-Hcl pH8, 50m NaCl, 1M EDTA: 5′Phos-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATC 3′ and 5′-GATCGGAAGAGCACACGTCT. After two RIPA and two Tris-Cl pH8 washes Nick repair was performed by resuspending the beads in 100μL of 150μM dNTPs, 15U Phi29 polymerase (NEB) in 1× Phi29 polymerase buffer (NEB) and incubating at 30°C for 20min. After two RIPA and two Tris-Cl pH8 washes lambda exonuclease digestion was performed by resuspending the beads in 100μL 1× Lambda exonuclease buffer supplemented with 10U lambda exonuclease (NEB) and incubating at 37°C for 30min. After two RIPA and two Tris-Cl pH8 washes RecJf exonuclease digestion was performed by resuspending the beads in 100μL 1× RecJf exonuclease buffer supplemented with 30U lambda exonuclease (NEB) and incubating at 37°C for 30min. After two RIPA and two Tris-Cl pH8 washes DNA was finally eluted by adding 100μL of 50mM NaHCO3, 1%SDS and incubating at 65°C for 30min. Supernatent was collected and supplemented with 1μL of 10mg/mL RNAse A, incubated at 37°C for 30min. 1μL of 20mg/mL Proteinase K and 12μL of 5M NaCl was then added and samples were reverse-crosslinked by incubation at 65°C overnight.
DNA was then purified using AMPure XP beads at a ratio 1.8× to sample and eluted in 20μL Tris-HClpH8. DNA was then denatured by incubation at 95°C for 5min and immediate transfer on ice. Second strand was then synthesized by adding 5pmol of P7 primer (5′-GACTGGAGTTCAGACGTGTGCT) in50μL total of 1× Phi29 buffer (NEB) and incubating at 65°C for 5 min then 30°C for 2min, followed by addition of 10U Phi29 polymerase and 1μL of 10M dNTPs and incubation at 30°C for 20min and 65°C for 10 minutes. Following AMPure XP purification (1.8×) and elution in 20μL ligation of p5 adapter was performed by incubation with 15pmol p5 adapter, 2000U T4 ligase in 1× T4 ligase buffer (NEB) in 50μL total at 25°C for 60min then 65°C for 10min. p5 adapters were prepared by mixing the following HPLC purified oligos at 10μM final in 10M Tris-Hcl pH8, 50m NaCl, 1M EDTA: 5′-AGATCGGAAGAGCG and 5′-TACACTCTTTCCCTACACGACGCTCTTCCGATCT. Following AMPure XP purification (1.8×) and elution in 20μL PCR amplification with indexed primers was performed using the NEB Next high-fidelity 2× PCR Master Mix with 25μM primers in 50μL and using 98°C for 30sec, 18 cycles of 98°C for10sec, 65°C for 30sec and 72°C for 30sec, followed by 72°C for 5min. PCR primer sequences are 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACG*A and 5′-CAAGCAGAAGACGGCATACGAGATNNNNNNGTGACTGGAGTTCAGACGTGTGC*T where * represents a phosphothiorate bond and NNNNNN is a Truseq index sequence. Following AMPure XP purification (0.9×) and elution in 20μL libraries were loaded on a 2% E-gel (ThermoFisher) and fragments 200pb-500bp were gel extracted. Library quality and quantity were estimated with Bioanalyzer and Qubit assays. Libraries were sequenced on a Hi-seq2000 or 4000 using 75bp single end.
RNA-Seq
Total RNA was prepared by Ethanol precipitation as described in Jay & Ciaudo 2013. Six to ten million adherent mESCs where washed with PBS and lysed directly with Trizol (Thermofisher), transferred into a 15mL conical tube, vortexed, supplemented with 1.6mL Chloroform, vortexed again and centrifuged at 3200g at 4°c for 15 min. Upper phase was mixed with and equal volume of isopropanol and spun at 3200g 4°C 30 min. Pellet was washed with 70% ethanol, air dried and resuspended in 100μL water. 10μg total RNA was used with the DNAse turbo kit (Ambion) in 50μL with 1μL DNAse. To purify polyA+ species 10μg DNAse treated RNA was heated at 65°C 5 min, transferred on ice, mixed with 20μL oligodT(25) magnetic beads (ThermoFisher) prewashed and resuspended in 45μL binding buffer and incubated 15min at room temperature. After two 200μL washes beads were resuspended in 10μL Tris pH 7.5, heated at 75°C for 2 min and eluate was immediately subjected to a second round of purification using 10μL beads per sample and eluting in 20μL – resulting in 30-100ng RNA. RNA-seq library were constructed using the NEBNext ultra (non-directional) RNA library kit for Illumina using 10ng polyA+ RNA as input and 12-15 PCR cycles. Library concentrations were estimated using Bioanalyzer (Agilent) and Qubit (ThermoFisher) assays, pooled and sequenced on a Next-seq instrument (Illumina) using 1.8pM, 75bp paired-end.
Chromosome Conformation Capture Carbon-Copy (5C)
We made substantial improvements over previously published protocols (Dostie et al. 2006), incorporating in situ (in nuclei) ligation (Rao et al. 2014), circumventing the need for phenol-chloroform purification and adopting a single-PCR strategy to construct 5C-sequencing libraries from the 3C template. These changes enable proceeding through the 3C protocol in a single tube per sample, allow handling of over 20 samples in parallel, reduce the amount of cells needed by a factor 5 to 10 and cut down the time needed to complete the protocol from 8 days (Nora et al. 2012) to 4 days.
10 million adherent mESCs were fixed as described for Chip-exo except that 2% formaldehyde was used. For 3C, 5 million cells were Lysed in 1mL 10mM Tris-HCl pH8.0, 10mM Nacl 0.2% NP40 for 15 min, pelleted at 4°C and washed twice with 1mL ice cold 1X NEB buffer 2. Cells were then resuspended in a 1.5mL tube in 400L 0.1% SDS in 1X NEB buffer 2 at room temperature, incubated at 65°C for 10min, cooled, supplemented with 44μL 10% Triton X-100, incubated at 37°C for 15 min. 1000U of HindIII (high-concentration, NEB) was then added for overnight incubation in a thermomixer at 800rpm. Cells were then incubated at 65°C 20min, cooled at room temperature and supplemented with 800μL of 50μM Tris-HCl pH7.5, 10mM MgCl2, 10mM DTT, 1% Triton X-100, 0.1mg/mL BSA, 1mM ATP and 10U T4 ligase (ThermoFisher cat 15224017). After 4h incubation at 25°c in a thermomixer at 800rpm cells were centrifuged at 1000rpm, resuspended in 500μL of 1% SDS with 1μg Proteinase K in 1× TE buffer, incubated at 55°C for 30min, supplemented with 50μL of 5M NaCl and incubated at 65°C overnight. DNA was then purified by adding 500μL isopropanol and incubating at -80°c for 30min following by centrifugation at 18,000g at 4°C, one 70% Ethanol wash, air drying and resuspension in 50μL 1× TE buffer, followed by incubation with 10μg RNAse at 37°C.
For 5C-sequencing we used the set of oligonucleotides described in Nora et al. 2012 that we pooled omitting the ones that were previously found to produce aspecific ligation (Supplementary table 5). 3C template were quantified using gel electrophoresis or the PicoGreen assay (ThermoFisher). Two to four 20μL 5C annealing reactions were assembled in parallel, each using 1μg 3C template, 1μg Salmon Sperm (ThermoFisher), 10fmol of each 5C oligonucleotide in 1X NEB buffer4. For neural progenitor cells and astrocytes 4μg of 3C template was used per 20μL annealing reaction. Samples were denatured at 95°C for 5 minutes and incubated at 48°C for 12-16h. 20μL of 1× Taq ligase buffer with 5U Taq ligase were added to each annealing reaction followed by incubation at 48°C 1h and 65°C 10 min. Negative controls (no ligase, no template, no 5C oligonucleotide) were included during each experiments to ensure the absence of contamination.
To fuse Illumina-compatible sequences 5C libraries were directly PCR amplified with primers annealing to the universal T3/T7 portion of the 5C oligonucleotides (underlined) and harboring 5′ tails containing Illumina sequences (italic):
5C-PCR_FOR: 5′AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTATTAACCCTCACTAAAGGGA
5C-PCR_REV: 5′CAAGCAGAAGACGGCATACGAGATnnnnnnGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTAATACGACTCACTATAGCC
Where nnnnnn denotes a 6-bp Truseq index sequence (Illumina) for multiplexing.
For this each 5C ligation reaction was used to template two parallel PCRs (so 4-8 PCRs total), using per reaction 6μL of 5C ligation with 1.125U Amplitaq gold (ThermoFisher) in 1× PCR buffer II, 1.8mM MgCl2, 0.2 dNTPs, 1.25μM 5C-PCR_FOR and 5C-PCR_REV primers in 25μL total. Cycling conditions were 95°C 9 min, 25 cycles of 95°C 30sec, 60°C 30sec, 72°C 30 sec followed by 72°C 8min. PCR products from the same 3C sample were pooled and purified using the PCR purification MinElute kit (Qiagen) and run on a 2.5% agarose electrophoresis. 5C libraries (231bp) were then excised and purified with the Gel extraction MinElute kit (Qiagen). Library concentrations were estimated using Bioanalyzer (Agilent) and Qubit (ThermoFisher) assays, pooled and sequenced on a Next-seq instrument (Illumina) using 1.2 to 1.5pM and 20-40% PhiX, 92bp single end.
Hi-C
Hi-C was performed as described (Lieberman-Aiden et al., 2009; Naumova et al., 2013). 25 million 2% formaldehyde cross-linked cells were incubated in 1000 μl of cold lysis buffer (10 mM Tris-HCl pH8.0, 10 mM NaCl, 0.2% (v/v) Igepal CA630, mixed with 10 μl protease inhibitors (Thermofisher 78438) immediately before use) on ice for 15 minutes. Next, cells were lysed with a Dounce homogenizer and pestle A (KIMBLE Kontes # 885303-0002) by moving the pestle slowly up and down 30 times, incubating on ice for one minute followed by 30 more strokes with the pestle. The suspension was centrifuged for 5 minutes at 2,000 g at RT using a table top centrifuge (Centrifuge 5810R, (Eppendorf). The supernatant was discarded and the pellet was washed twice with ice cold 500 μl 1X NEBuffer 2.1 (NEB). After the second wash, the pellet was resuspended in 1X NEBuffer 2 in a total volume of 250 μl and split into five 50 μl aliquots. Next, 312 μl 1X NEBuffer 2 was added to each aliquot. Chromatin was solubilized by addition of 38 μl 1% SDS per tube and the mixture was resuspended and incubated at 65°C for 10 minutes. Tubes were put on ice and 44 μl 10% Triton X-100 was added. Chromatin was subsequently digested by adding 400 Units HindIII (NEB) at 37°C for overnight digestion with alternating rocking. Digested chromatin solutions were spun shortly and transferred to ice. One tube was kept separate and used for generating a 3C control library as described (Naumova et al., 2013). The chromatin samples in the remaining four tubes were used for generating Hi-C libraries and were treated as follows: The HindIII DNA ends were filled in and marked with biotin by adding 60 μl fill-in mix [1.5 μl 10 mM dATP, 1.5 μl 10 mM dGTP, 1.5 μl 10 mM dTTP, 37.5 μl 0.4 mM biotin-14-dCTP (ThermoFisher #19518-018), 6 μl 10× NEBuffer 2.1, 2 μl water and 10 μl 5U/μl Klenow polymerase (NEB M0210L)] followed by incubation at 37°C for 80 minutes in a thermomixer. Klenow polymerase was inactivated by adding 96 μl 10% SDS followed by incubation at 65°C for 30 minutes. Tubes were then placed on ice immediately afterwards. The content of each of the tubes was transferred to 15 ml conical tube containing 7.58 ml ligation mix [820 μl 10% Triton X-100, 758 μl 10× ligation buffer (500 mM Tris-HCl pH7.5, 100 mM MgCl2, 100 mM DTT), 82 μl 10 mg/ml BSA, 82 μl 100 mM ATP and 5.84 ml water]. 50 μl 1U/μl T4 DNA ligase (Invitrogen #15224) was added and ligation was performed at 16°C for 4 hours. DNA was then purified as follows. 50 μl 10 mg/ml Proteinase K (ThermoFisher # 25530-031) was added to each tube and samples were incubated at 65°C for 4 hours followed by a second addition of 50 μl 10 mg/ml Proteinase K solution, followed by overnight incubation at 65°C. Tubes were cooled to RT and transferred to 50 ml conical tubes. The DNA was extracted by adding an equal volume of phenol pH8.0:chloroform (1:1) (Fisher BP1750I-400), vortexing for 3 minutes and spinning for 10 minutes at 4,000 rpm in a table top centrifuge (centrifuge 5810R, Eppendorf). The supernatants were transferred to new 50 ml conical tubes. Another extraction was performed with an equal volume of phenol pH8.0:chloroform (1:1). After vortexing and centrifugation for 10 minutes at 4,000 rpm, all four supernatants of the Hi-C samples were pooled into a single 250ml centrifuge tube and the volume was brought to 40 ml with 1× TE buffer (10 mM Tris pH8.0, 1 mM EDTA). To precipitate the DNA, 4 ml 3M Na-acetate pH5.0 was added, mixed well and then 100 ml of ice cold 100% ethanol was added. The volume of the 3C control sample was brought to 10 ml with TE. DNA precipitation was done by addition of 1 ml of 3M Na-acetate and 25 ml ice-cold 100% ethanol in a 35 mL centrifuge tube. Tubes were inverted slowly several times to mix the contents and then were incubated at least one hour at -80°C. Next, the tubes were spun at 4°C for 30 minutes at 16,000g AvantiTM J-25 Centrifuge (Beckman). The supernatants were discarded and DNA pellets were dissolved in 500 μl 1× TE buffer and transferred to a 0.5 mL AMICON® Ultra Centrifugal Filter Unit – 0.5 ml 30K (UFC5030BK EMD Millipore) for desalting. Columns were spun at 14,000g for 10min, in a microfuge. The flow throughs were discarded. Columns were washed three times with 450 μl TE. After the final wash, the 3C library was dissolved in 25 μl TE; the Hi-C library was dissolved in 100 μl TE. Any RNA was degraded by incubation with 1 μl of 10 mg/ml RNAse A at 37°C for 30 minutes. The quality and quantity of 3C and Hi-C libraries were checked by running aliquots on a 0.8% agarose gel along with a 1 kb ladder (NEB #N3232S). Libraries should run as a rather discrete band with a molecular weight that is larger than 10 kb. With a successful biotin fill-in and marking of DNA ends, HindIII (AAGCTT) restriction sites get converted into NheI sites (GCTAGC). To test the efficiency of this process we used PCR to amplify a ligation product formed by two nearby restriction fragments followed by digestion with HindIII, NheI and by a double digestion with HindIII+NheI restriction enzymes. The relative efficiency of Hi-C ligation product formation and biotin fill-in was defined as the proportion of ligation product digested with NheI and varied from 50 to 80% in different Hi-C libraries. The following two pairs of primers were used: mGAPDH_1 and mGAPDH2.
mGAPDH_1 ATGGAGACCTGCCGCCGGCTCATCA
mGAPDH_2 CGTGCTGTGACTTCGCACTTTTCTGA
Next, Hi-C libraries were treated with T4 DNA polymerase to remove biotinylated ends that did not ligate (dangling ends). Eight reactions were assembled as follows: 5 μg of Hi-C library, 5 μl 10× NEBuffer 2.1, 0.5 μl 2.5 mM dATP, 0.5 μl 2.5 mM dGTP and 5 Units T4 DNA polymerase (NEB # M0203L) in a total volume of 50 μl. Reactions were incubated at 20°C for 4 hours. The reaction was stopped by incubating 20min at 75°C. To desalt and concentrate the DNA, the reactions were pooled together and added on 0.5 mL AMICON® Ultra Centrifugal Filter Unit – 0.5 ml 30K (UFC5030BK EMD Millipore). Columns were spun at 14,000g for 10min, in a microfuge. The flow through was discarded. Columns were washed twice with 450 μl TE. After the final wash, the Hi-C libraries was dissolved in 120 μl TE. The DNA was sheared to a size of 100-400 bp (with the majority of molecules around 200 bp) using a Covaris S2 instrument (Covaris, Woburn, MA). The settings were as follows: Duty cycle 10%, Intensity 5, Cycles per burst 200, Set mode - Frequency sweeping, Process time 60 sec per process, Cycles number 3. To enrich for DNA fragments of 100-300 bp an Ampure XP fractionation was performed (Beckman Coulter, A63881) and the DNA was eluted with 50 μL of water. The size range of the DNA fragments after fractionation was determined by running an aliquot on an agarose gel. The sheared DNA ends were repaired by addition of 7 μl 10× ligation buffer (NEB # B0202S), 7 μl 2.5 mM dNTP mix, 2.5 μl T4 DNA polymerase (NEB # M0203L), 2.5 μl T4 polynucleotide kinase (NEB #M0201S), 0.5 μl Klenow DNA polymerase (NEB #M0210S) and 5.5 μl water to the 45uL of DNA. The DNA was purified using Ampure beads (Beckman Coulter, A63881) and eluted in 32uL of TLE (10 mM Tris pH8.0, 0.1 mM EDTA (TLE buffer). Next, an ‘A’ was added to the 3′ ends of the end-repaired DNA by addition of 5 μl 10× NEBuffer2, 10 μl 1 mM dATP, 3 μl Klenow (exo-) (NEB #M0212L) and 16 μl water. The reaction was incubated at 37°C for 30 min followed by incubation at 65°C for 20 minutes to inactivate Klenow polymerase. The reactions were cooled on ice. All subsequent steps were performed in DNA LoBind tubes (Eppendorf #22431021) and each step was performed in a fresh tube. 50 μl of streptavidin Dynabeads (MyOne Streptavin C1 Beads, ThermoFisher #650-01) were washed twice with 400 μl Tween Wash Buffer (TWB) (5 mM Tris-HCl pH8.0, 0.5 mM EDTA, 1 M NaCl, 0.05% Tween20) by incubating for 3 minutes at RT with rotation, reclaiming against a magnetic separation rack for 1 minute and removing all supernatant. Next, reclaimed beads were resuspended in 400 μl 2× Binding Buffer (BB) (10 mM Tris-HCl pH8.0, 1 mM EDTA, 2 M NaCl) and combined with 400 μl Hi-C DNA from the previous step. The mixture was incubated at RT for 15 minutes with rotation. The supernatant was removed and the DNA-bound Streptavidin beads were washed once with 400 μl 1× BB. The beads were then washed with 100 μl 1× ligation buffer (Invitrogen 5× buffer), and then resuspended in 19 μl of 1× ligation buffer (NEB quick ligase, M2200S). Ligation reaction was set-up as follows: 19 μl Hi-C library on beads, 6 μl Illumina paired end adapters (Illumina), 10 μl 2× quick ligation buffer (NEB), 1 μl quick DNA ligase (NEB quick ligase, M2200S). The reaction was incubated at RT for 15min. The beads with bound ligated Hi-C DNA were collected by holding against a magnetic separation rack and were then washed twice with 400 μl 1× TWB, once with 200 μl 1xBB and once with 200 μl 1X NEBuffer2 to remove non-ligated Paired End adapters. The beads were resuspended in 20 μl 1X NEBuffer 2. Next, test PCR reactions were performed to determine the optimal number of PCR cycles needed to generate enough Hi-C library for sequencing. Four trial PCR reactions were set up, each containing 0.9 μl Dynabead-bound Hi-C library, Illumina PE1.0 and PE2.0 PCR primers (0.21 μl of each; 25 μM), 0.12 μl 25mM dNTPs, 0.3 μl Pfu Ultra II Fusion DNA polymerase (Agilent #600670), 1.5 μl 10× Pfu Ultra buffer and 11.76 μl water. The temperature profile during the PCR amplification was 30 seconds at 98°C followed by 5, 7, 9 or 11 cycles of 10 seconds at 98°C, 45 seconds at 65°C, 30 seconds at 72°C and a final 7-minute extension at 72°C. The PCR reactions were run on a 2% agarose gel and the minimal cycle number was determined that yielded sufficient DNA for sequencing. Typically, 6 cycles were chosen for amplification of Hi-C libraries. PCR was then performed in nine reactions with the remaining Dynabead-bound Hi-C library. The PCR product was run on a 2% agarose gel and smear 200-400bp to assess the DNA concentration. A final quality control was performed by NheI digestion of an aliquot of the final Hi-C library. Without NheI digestion, the DNA sizes of the libraries ranged from 300-400bp. After NheI digestion, the DNA sizes of the libraries shifted and ranged from 100-350bp. It indicated that the majority of the ligation products have been digested by NheI and validated that the libraries were mainly constituted of true ligation products. The libraries were sequenced using 50 bp paired end reads with a HiSeq2000 machine and HiSeq4000.
Quantification and Statistical Analysis
ChIP-seq Analysis
Fastq files were trimmed using the fastq-mcf program, aligned to the mm9 reference genome with bowtie2 (Langmead and Salzberg, 2012). Reads with a mapq score of 30 or greater were retained, using Samtools. Data used to generate the heatmaps presented in the manuscript were obtained by downsampling the number of reads to match the most shallow sample (for CTCF and H3K27me3 separately) and pooling the reads of each biological replicates. Heatmap visualization and integration with RNA-seq was performed using Easeq version 1.03 (Lerdrup et al., 2016). The Euler diagram was drawn using eulerAPE (Micallef and Rodgers, 2014). Chip-seq peaks were called on each replicate individually using all available reads. For peak calling we followed the guidelines described in (Thomas et al., 2016). For CTCF, which display focal enrichment, we used the Genome-wide Event finding and Motif discovery (GEM) method (Guo et al., 2012). For H3K27me3, which marks broad domains, we used the Baysian Change Point (BCP) method (Xing et al., 2012). The consensus peak list was obtained by retaining peaks that overlapped for at least 1bp between biological replicates. For exemple loci in figures S2 and S7 read depth-normalized tag densities were generated directly by the Easeq software using the “filled track” tool. The normalized tag density bigwig tracks used for visualization with the UCSC genome browser were generated by dividing into 20bp bins and a normalized tag density was calculated for each bin as follow:
ChIP-Exo Analysis
Analysis and footprint identification was carried out as described in (Luna-Zurita et al., 2016). The 5′-most position of reads that mapped to the reference strand and the 3′-most position of reads that mapped to the non-reference strand were identified for each read as the actual edges of each exonuclease-treated fragment. To identify broad regions of binding, bins with tag densities of greater than 100 were merged to generate a peak list for each sample. Within 1kb of each region, strand-specific single-base-resolution tag densities were calculated for each dataset by dividing each region into 1bp bins, then counting the number of tags within 5bp of each bin. For each region of binding, the footprint for each bound region was defined as the span from the peak position of ‘+’ strand binding to the peak position of ‘-’ strand binding as seen from the high-resolution tag densities.
RNA-seq Analysis
Alignment and differential expression was performed on the BaseSpace environment version 1.0.0 (Illumina). Alignment was produced using STAR version 2.5.0a (Dobin et al., 2013) with default parameters except that novel transcript assembly was not performed. mm9 RefSeq was used as reference gene set and adapters were trimmed, Cufflinks version 2.2.1 (Trapnell et al., 2010)was used with fragment bias and multi-read correction with Bedtools version 2.17.0 (Quinlan and Hall, 2010). Differential expression analysis was analyzed using Cuffdiff (Trapnell et al., 2013) with default parameters within BaseSpace. Genes with an FPKM below 1.1 in all conditions were not considered in the differential expression analysis. Heatmap visualization and integration with Chip-seq and Chip-exo was performed using Easeq version 1.03 (Lerdrup et al., 2016). For integration with enhancer positions we took the enhancer list assembled by (Chen et al., 2012) with the same probability threshold (0.8). The super-enhancer list was retrieved from (Hnisz et al., 2013). FPKM provided in Supplementary Table 3 are means from 3 independent biological replicates.
To determine the significance of co-localization of TSS of differentially expressed genes with HiChIP loop anchors (figure S6D) we used the exact Fisher test. In the test we used a 2×2 contingency table containing the numbers of DE genes with the TSS co-localized or not in the same 5kb bin with a HiChIP loop anchor and the numbers of HiChIP loop anchors co-localized or not with a DE gene
CTCF Motif orientation analysis
First, we established a consensus list of CTCF ChIP-seq peak from the CTCF-AID line by retaining only the peaks identified in both replicates (overlap of at least 1bp between replicates). We then retrieved the DNA sequence from each peak using the TableBrowser tool of the UCSC genome browser, using the mm9 assembly. Each of these sequences were then searched for CTCF motifs using FIMO (Grant et al., 2011) with the CTCF position frequency matrix obtained from the JASPAR database, motif MA0139.1 and default parameters. Promoters of affected genes were not specifically enriched for tandem CTCF sites compared to their occurrence in CTCF ChIP-seq peaks genome-wide (around 1/3 of peaks have multiple CTCF motifs (Pugacheva et al., 2015)).
Hi-C analysis
Mapping, filtering, and normalization of Hi-C data
We mapped the sequence of Hi-C molecules to reference mouse genome assembly mm9 using Bowtie 2.2.8 and the iterative mapping strategy, as described in (Imakaev et al., 2012; Lajoie et al., 2015). Upon filtering PCR duplicates and reads mapped to multiple or zero locations, we aggregated the reads pairs into 20kb and 100kb genomic bins to produce Hi-C contact matrices. For downstream analyses, data from biological replicates were pooled. Low-coverage bins were then excluded from further analysis using the MAD-max (maximum allowed median absolute deviation) filter on genomic coverage, set to 4.5 median absolute deviations from the median (corresponding to three standard deviations in the case of a normal distribution). To remove the short-range Hi-C artifacts - unligated and self-ligated Hi-C molecules - we ignored the contacts mapping to the same or adjacent genomic bins in all downstream analyses. The filtered 20kb and 100kb contacts matrices were then normalized using the iterative correction procedure (IC), such that the genome-wide sum of contact probability for each row/column equals 1.0. Observed/expected contact maps were obtained by dividing each diagonal of a contact map by its chromosome-wide average value over non-filtered genomic bins. The compartment structure of Hi-C maps was detected using a modified procedure from (Imakaev et al., 2012). Compartments were quantified as the dominant eigenvector of the observed/expected 20kb and 100kb cis contacts maps upon subtraction of 1.0, as implemented in hiclib. Segmentation of eigenvectors into regions corresponding to active (A) and inactive (B) compartments was performed using a 2-state HMM model. The code for mapping, filtering and normalization analysis of Hi-C data is available at https://github.com/dekkerlab/cworld-dekker (lab of Job Dekker) and https://bitbucket.org/mirnylab/hiclib (lab of Leonid Mirny).
Insulation scores from Hi-C data
To local contact insulation analysis was based on the algorithm described in (Crane et al., 2015). For every 20kb bin, the insulation score was calculated as the total number of normalized and filtered contacts formed across that bin by pairs of loci located on the either side, up to 100kb away. The score was normalized by its genome-wide median. To find insulating boundaries, we detected peaks in log2-transformed insulation score track using the peakdet algorithm [Billauer E (2012). peakdet: Peak detection using MATLAB, http://billauer.co.il/peakdet.html]. Briefly, this algorithm seeks a sequence of local maxima and minima whose values differ by more than a pre-specified threshold (i.e. peak prominence). The detected minima in the insulation score correspond to a local depletion of contacts across the genomic bin, are then called as insulating boundaries. To find the optimal threshold for peak calling, we varied the peak calling threshold, and for each value compared the called boundaries with the loop anchoring regions detected in (Mumbach et al., 2016). This comparison revealed that at high threshold values, corresponding to stricter boundary selection, up to 62% of detected boundaries co-aligned with the previously detected loop anchors within +-1 20kb bin precision. As we lowered the boundary detection threshold, fewer added boundaries co-aligned with the loops; this analysis suggested the optimal threshold of 0.3, where the specificity of loop anchor recall dropped 3-fold. Finally, we selected only the boundaries that had zero or one filtered 20kb bin in a 100kb range, since the presence of filtered bins affects insulation. We then used the same approach to call boundaries in all Hi-C samples. The boundaries detected in the auxin-treated sample within +-1 20kb bin from a position of a boundary in the untreated sample were called “residual” boundaries. To correlate the presence of boundaries with presence of active promoters or compartment transition we found all boundaries that had a PolII peak or a transition between A and B HMM compartment assignment, correspondingly; to account for the inaccuracy in boundary calls we allowed +/- 1 20kb bin mismatch.
Chromosome Conformation Capture Carbon-Copy (5C) analysis
Mapping and insulation scores from 5C matrices
Adapters were trimmed and aligment was performed using Bowtie2 against a pseudo-genome composed of all possible Forward-Reverse pairs of 5C oligonucleotides (Nora et al. 2012). Results were then transformed into a matrix table. Primers giving artefactual signal were removed using the code deposited by the lab of Job Dekker on Github https://github.com/dekkerlab/cworld-dekker. Heatmaps were generated using the my5C tools (Lajoie et al., 2009). For the insulation score analysis, the primer-based heatmaps were aggregated at 20kb resolution by calculating the median interaction frequency between primers belonging to all pairs of 20kb genomic bins. The aggregated maps were then filtered by removing the contacts in the first two diagonals and normalized using IC. The insulation score and boundary detection was then performed using the same method and parameters as described above for Hi-C maps.
Display of restriction fragment level 5C heatmaps
5C primers matrices were filtered as previously described methods (Sanyal, Dekker 2012). We detected and flagged all outlier (anchor) row/cols that are defined as having a having an aggregate (row/col) signal greater than or less than 1.5 * IQR (of the distribution of all row/col signals). We then took the union of all flagged (anchor) row/col outliers across all the 5C matrices, and removed these (anchor) row/cols from all datasets. 68 anchors were removed. Then, the matrices were balanced according to the ICE method developed for Hi-C (Imakaev, Mirny 2012. 5C cannot interrogate contacts between two restriction fragments harboring a forward oligonucleotide or a reverse oligonucleotide. In order to display intelligible heatmaps we interpolated the uninterrogated forward-forward and reverse-reverse pixels by the median of the eight pixels surrounding it, producing smoothed matrices using the my5C tools (Lajoie et al., 2009).
3D-DNA FISH analysis
3D distance measurement was performed on ImageJ using the scripts described in (Nora et al., 2012).
Data and software accessibility
Software from this study has been previously published as detailed under “QUANTIFICATION AND STATISTICAL ANALYSIS.”
Data Resources
Raw and processed sequencing data reported in this paper have been submitted to GEO, accession number pending.
Plasmids can be requested through Addgene.
Supplementary Material
(A) Principle of the CellTrace dye dilution assay for proliferation
(B) Flow cytometry of dilution kinetics of the CellTrace dye indicates that auxin-treated CTCF-AID mESCs keep proliferating after two days of CTCF depletion and slow down afterwards. Auxin does not trigger any proliferation defect in CTCF-AID mESCs lacking the Tir1 F-box protein transgene.
(C) Propidium iodide staining indicates CTCF depleted mESCs are not blocked in any specific stage of the cell cycle and do not become aneuploidy.
(D) A tagBFP2/mCherry FUCCI cassette was created and knocked in in CTCF-AID or WT mESCs. Auxin treatment only leads to a slight increase of the G1 FUCCI-signal after 4 days of CTCF depletion, confirming that loss of CTCF does not block cell cycle progression overall.
(E) CTCF depleted did not show increased DNA damage as monitored by western blot, and displayed overall constant bulk H3K27me3 levels. LaminB1 used as loading control
(F) CTCF depletion did not lead to massive apoptosis, although number of dying cells increase after long depletion.
(G) Strategy for introducing dox-inducible CTCF transgenes in CTCF-AID cells
(H) Flow cytometry confirms that most auxin+dox-treated cells loose endogenous CTCF (>99%) and express transgenic CTCF (>95%) after 4 days of auxin+dox treatment
(I) Western blot using a CTCF antibody indicates that the dox-inducible transgene can be readily detected but drives lower expression than normal endogenous CTCF levels
(J) Inducing CTCF expression from the transgene largely alleviates the proliferation defects caused by from depleting of endogenous CTCF.
Supplementary Table 3 RNA-seq FPKM values, Related to figure 6
Supplementary Table 5 - 5C oligonucleotides, Related to figure 5
Supplementary Table 7 - CTCF motif orientation inside ChIP-seq Peaks found in untreated CTCF-AID mESCs Related to figure 6
(A) 5C at the Xic confirms that chromatin loops do not accumulate at CTCF peaks after CTCF depletion and are reaquired upon CTCF resoration.
(B) Auxin treatment of WT cells has no effect on chromatin folding
(C) CTCF ChIP-exo signal at CTCF ChIP-seq peaks detected in untreated CTCF-AID cells. Auxin treatment of WT cells has no effect on CTCF binding. Tagging with the CTCF-AID-eGFP does not disrupt CTCF binding pattern.
(D) Auxin treatment of CTCF-AID cells dramatically reduces CTCF enrichment at peaks detected in untreated cells and is fully reversible after washoff
(E) Easeq Genome browser visualization of an example locus. A subset of CTCF ChIP-seq peaks are still detected, but of low intensity, after depletion and are restored in strength after washoff
(F) Loss of ChIP-seq signal upon CTCF depletion is equivalent in the A and B genomic compartment as defined by Hi-C.
(G) A compartment tends to have stronger CTCF ChIP-seq peaks than B compartment
(H) CTCF binding is 5-fold denser in the A compartment than in B.
(I) Restriction-fragment level interpolated visualization of 5C around the Linx-Chic1-Xite loops. CTCF depletion disrupts CTCF binding and underlying loops while CTCF recovery re-stablishes binding and chromatin contacts.
(J) Auxin treatment in itself does not perturb the accumulation of chromatin loops in WT untagged mESCs, as exemplified at the Linx/Chic1/Xist 300kb TAD within the 4.5Mb segment covered by our 5C assay.
(A) Restriction-fragment level interpolated visualization of 5C at the Xic. Color dots denote TAD boundaries.
(B) Insulation score ratio between treated and untreated cells at boundaries detected in untreated cells, highlighting that a subset of TAD boundaries rapidly loose insulation upon CTCF depletion
(C) Insulation score analysis using 5C on independently generated cell lines (see Methods for details). Lines #2-5 were created by re-introducing Tir1 transgenes in the intermediate CTCF-AID-eGFP (no Tir1) clone used to generate the cell line used for the other analyses (#1), at the Rosa26 or Tigre acceptor loci. Cell line #6 was created by first introducing a Tir1 transgene at Tigre in WT cells and then re-creating the CTCF-AID-eGFP allele homozygously.
(D) 5C in the CTCF-AID line (#1) complemented with CTCF transgene indicating
(E) Insulation score analysis indicating that expression of the CTCF transgene mitigate the insulation defects caused by the loss of endogenous CTCF. Note that transgene expression is not as high as endogenous CTCF (Figure S1I)
(F-G) Auxin treatment has no effect on TAD insulation in WT untagged and CTCF-AID (no Tir1) cells
(H) Probability of calling a TAD boundary at Smc1a HiChIP loop as a function of the local prominence of the insulation score calculated at 100kb with our Hi-C. We chose the threshold (0.3) below which improvement in retrieving Sm1a HiChIP loop is below 50% (see methods).
(I) Hi-C snapshot illustrating that a subset of boundaries resist CTCF depletion Shown is an example region harboring boundaries that resist CTCF depletion. The one is associated with a strong promoter and the other one with a A/B compartment transition.
(J) Hi-C snapshot illustrating that a small subset of boundaries retain strong insulation after depletion without being associated with transcription or compartment transition
(K) Replot of the DNA FISH data presented in figure 3D illustrating that after CTCF depletion inter-TAD 3D distances becomes equivalent to intra-TAD when probe pairs are equally spaced on the chromosome and not overlapping boundaries.
(L) Replot of the DNA FISH data presented in figure 3D illustrating that probe pairs partially overlapping boundaries (green and yellow) become more separated after CTCF depletion
(M) Scaling of ICE normalized Hi-C contacts between loop anchors or matched random loci pairs, comparing CTCF depletion by RNAi in human HEK293T cells (Zuin et al., 2014) to the CTCF-AID mESCs. Loop anchors are from GM12878 (human) and CH12-LX cells (mouse), respectively (Rao et al., 2014). Thick line is the median, shaded area highlights 25-75 percentile and dotted lines are landmarks for visual comparison.
(A) cis-Eigenvector 1 values in 100kb genomic bins are ranked and pairwise enrichment of Hi-C contacts between each of the 50 ranks are calculated (pooled replicates). Genomic regions with similar ranks of Eigenvector 1 values display more Hi-C contact while regions of opposite ranks are depleted (see methods). This trend is conserved overall after CTCF depletion or restoration.
(B)Compartmentalization strength is only mildly affected by CTCF depletion
(C) Scatter plot of the insulation score of genomic elements that are transitions between A/B compartments and TAD boundaries before CTCF depletion (left) or after (right), highlighting that insulation is also weaker at these compartments transition after loss of CTCF. Note that strong boundaries have the lowest insulation scores.
(A-G) Restriction-fragment level 5C interpolated heatmaps highlighting that auxin treatment of CTCF-AID NPCs and astrocytes disrupts TAD insulation, irrespective of the timing of CTCF depletion to cell-cycle exit. Blue = time after adding BMP4 to convert NPCs into astrocytes.
(H) Summary of all experiments with NPCs and astrocytes.
(I) Quantification of insulation loss from the 5C in all NPC and astrocyte samples.
(J) Comparative levels of CTCF-AID-eGFP in mESCs, NPCs and astrocytes measured by flow cytometry. mESCs display a broader fluorescence distribution as they are mostly in S/G2, while NPCs display both G1 and S/G2 cells and astrocytes are in G0.
(A) Scatter plot of the fold change in treated versus untreated cells as a function of the expression level in untreated cells. Up and down-regulation are observed for genes with a wide-range of initial expression levels. Misregulation is not restricted to lowly-expressed genes.
(B-C) Same analysis as in 6D-E but focused on super-enhancers active in mESCs.
(D) Gene misregulated upon CTCF depletion are more often found close to Smc1a HiChIP loop anchors than expected by chance. See methods for statistical details.
(A) CTCF and H3K27me3 ChIP-seq centered at all CTCF peaks detected in untreated cells. A small subset is embedded in large H3K27me3 regions with a dip at the CTCF site. This dip disappears upon CTCF depletion and reappears after CTCF restoration. This suggests that nucleosomes become able to cover the previously occupied CTCF site when CTCF binding is lost.
(B) Overall H3K27me3 levels at H3K27me3 ChIP-seq peaks are unaffected after two day, become slightly lower after 4 days of depletion and are readjusted upon CTCF restoration.
(C-D) Easeq Genome browser visualization of an example locus illustrating H3K27me3 does not spread beyond flanking CTCF sites upon CTCF depletion in mESCs.
Supplementary Table 1 sgRNA sequences, Related to Figure 1
Key resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-CTCF antibody, rabbit polyclonal | Active Motif | Cat #61311 |
| Anti-H3S10Ph, rabbit polyclonal | Millipore | Cat #06-570 |
| Anti-LaminB1, mouse monoclonal | Abcam | Cat #Ab8982 |
| Anti-γH2AX mouse monoclonal | Millipore | Cat #05-636 |
| Anti-H3K27me3, rabbit monoclonal | Cell Signaling Technology | Cat #9733 |
| Anti-Cleaved Caspase3 (Asp175) | Cell Signaling Technology | Cat #9664 |
| Spike-in Antibody | Active Motif | Cat #61686 |
| Spike-in Chromatin | Active Motif | Cat #53083 |
| Chemicals, Peptides and Recombinant Proteins | ||
| Indole-3-acetic acid sodium salt (auxin analog) | Sigma-Aldrich | Cat #I5148-2G |
| EGF | Peprotech | Cat #AF-100-15 |
| FGF basic | Peprotech | Cat #100-18B |
| BMP4 | R&D Systems | Cat #314-BP-010 |
| Critical commercial assays | ||
| Celltrace Proliferation kit | Thermofisher | Cat #C34564 |
| Neon™ Transfection system | Thermofisher | Cat #MPK10025 and Cat #MPK1025 |
| NEBNext ultra RNA library kit for illumina | NEB | Cat #E7530L |
| Experimental models: Cell lines | ||
| E14TG2a | Hooper et al., 1987 | |
| Recombinant DNA | ||
| pX330-U6-Chimeric_BB-CBh-hSpCas9 | Addgene | Cat #42230 |
| pX335-U6-Chimeric_BB-CBh-hSpCas9n(D10A) | Addgene | Cat #42335 |
| pEN84 - CTCF-AID[71-114]-eGFP-FRT-PuroR-FRT | This study | Deposited to Addgene Cat #86230 |
| pEN244 - CTCF-AID[71-114]-eGFP-FRT-Blast-FRT | This study | Deposited to Addgene Cat #92140 |
| pEN113 - pCAGGS-Tir1-V5-BpA-Frt-PGK-EM7-NeoR-bpA-Frt-Rosa26 | This study | Deposited to Addgene Cat #86233 |
| pEN114 - pCAGGS-Tir1-V5-BpA-Frt-PGK-EM7-PuroR-bpA-Frt-Rosa26.ape | This study | Deposited to Addgene Cat #92143 |
| pEN396 - pCAGGS-Tir1-V5-2A-PuroR Tigre donor | This study | Deposited to Addgene Cat #92142 |
| pEN435 - pCAGGS-TagBFP-hGeminin-2A-mCherry-hCdt1-rbgpA-Frt-PGK-EM7-PuroR-bpA-Frt Tigre targeting | This study | Deposited to Addgene Cat #92139 |
| pX335-EN475 (spCas9nickase with CTCF sgRNA1) | This study | Deposited to Addgene Cat #86231 |
| pX335-EN477 (spCas9nickase with CTCF sgRNA2) | This study | Deposited to Addgene Cat #86232 |
| pX330-EN479 (spCas9nuclease with Rosa26 sgRNA) | This study | Deposited to Addgene Cat #86234 |
| pX330-EN1201 (spCas9nuclease with Tigre sgRNA) | This study | Deposited to Addgene Cat # 92144 |
| pCAGGS-FlpO-IRES-puro | Kranz et al. 2010 | |
| BAC #RP24-335O3 | CHORI/BACPAC | |
| BAC #RP24-228J7 | CHORI/BACPAC | |
| BAC #RP24-230I15 | CHORI/BACPAC | |
| BAC #RP24-164B17 | CHORI/BACPAC | |
| BAC #RP24-267I14 | CHORI/BACPAC | |
| BAC #RP23-469K13 | CHORI/BACPAC | |
| Software and algorithms | ||
| FlowJo | FlowJo LLC | https://www.flowjo.com/ |
| imageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Easeq | Lerdrup et al., 2016 | http://easeq.net/ |
| my5C | Lajoie et al., 2009 | http://my5c.umassmed.edu/ |
| R | R Core Team, 2014 | http://www.R-project.org/ |
| Basespace environment | Illumina | https://basespace.illumina.com/ |
| C-world (Hi-C analysis software) | Job Dekker lab | https://github.com/dekkerlab/cworld-dekker |
| Python | Python Software Foundation | https://www.python.org/ |
| Hiclib (Hi-C analysis software) | Leonid Mirny lab | https://bitbucket.org/mirnylab/hiclib/ |
| FIMO | Grant et al., 2011 | http://meme-suite.org/tools/fimo |
| Datasets reanalyzed | ||
| Smc1a HiCHIP | GSE80820 | Mumbach et al. 2016 |
| LaminB1 DamID | GSE40112 | Peric-Hupkes et al. 2010 |
| MNAse-seq | GSE40896 | Teif et al, 2009 |
| Pol2 Chip-seq | GSM918749 | ENCODE Mar 2012 Freeze |
| H3K36me3 Chip-seq | GSM1000109 | ENCODE Mar 2012 Freeze |
| CTCF RNAi and mock Hi-C | GSE44267 | Zuin et al. 2014 |
| in situ Hi-C | GSE63525 | Rao et al. 2014 |
Acknowledgments
We apologize for not citing numerous relevant studies due to space constraints. We thank Daphné Dambournet, Baptiste Roelens and Matthias Merkenschlager for discussions; Casey Gifford for help with sequencing; Elizabeth Blackburn for access to microscopy resources; Hakan Ozadam for bioinformatics support during revisions; Sean Thomas, Alex Williams and the Gladstone Bioinformatics core; Gary Howard for editorial assistance; Edith Heard and Geoffrey Fudenberg for critical comments on the manuscript. This work was supported by the EMBO (ALTF523-2013) and HSFP (E.P.N.); the National Institutes of Health/National Heart, Lung, and Blood Institute (Bench to Bassinet Program U01HL098179), the Gladstone Institutes, and William H. Younger, Jr. (B.G.B); the UCSF-Gladstone Institute of Virology & Immunology Center for AIDS Research (CFAR) NIH P30 AI027763 (Gladstone flow cytometry core); the National Human Genome Research Institute (R01 HG003143, U54 HG007010, U01 HG007910), the National Cancer Institute (U54 CA193419), the NIH Common Fund (U54 DK107980, U01 DA 040588), the National Institute of General Medical Sciences (R01 GM 112720), and the National Institute of Allergy and Infectious Diseases (U01 R01 AI 117839). J.D. is an investigator of the Howard Hughes Medical Institute. B.G.B. is a co-founder of Tenaya Therapeutics.
Footnotes
Author Contributions: E.P.N. conceived and designed the study with input from B.G.B; E.P.N. engineered and cultured cell lines, performed 5C, ChIP-exo, ChIP-seq, RNA-seq and FISH with help from A.U., and analyzed data. A.-L.V. and J.H.G. performed Hi-C in the lab of J.D and pre-processed Hi-C and 5C data. A.G. and N.A. performed computational analyses of Hi-C data in the lab of L.A.M. E.P.N. wrote the manuscript with B.G.B and with input from all authors.
References
- van Arensbergen J, van Steensel B, Bussemaker HJ. In search of the determinants of enhancer-promoter interaction specificity. Trends Cell Biol. 2014;24:695–702. doi: 10.1016/j.tcb.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold CD, Zabidi MA, Pagani M, Rath M, Schernhuber K, Kazmar T, Stark A. Genome-wide assessment of sequence-intrinsic enhancer responsiveness at single-base-pair resolution. Nat Biotechnol. 2016 doi: 10.1038/nbt.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont AS. Large-scale chromatin organization: the good, the surprising, and the still perplexing. Curr Opin Cell Biol. 2014;26:69–78. doi: 10.1016/j.ceb.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender MA, Byron R, Ragoczy T, Telling A, Bulger M, Groudine M. Flanking HS-62.5 and 3′ HS1, and regions upstream of the LCR, are not required for β-globin transcription. Blood. 2006;108:1395–1401. doi: 10.1182/blood-2006-04-014431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickmore WA, van Steensel B. Genome Architecture: Domain Organization of Interphase Chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Bonev B, Cavalli G. Organization and function of the 3D genome. Nat Rev Genet. 2016;17:661–678. doi: 10.1038/nrg.2016.112. [DOI] [PubMed] [Google Scholar]
- Chiariello AM, Annunziatella C, Bianco S, Esposito A, Nicodemi M. Polymer physics of chromosome large-scale 3D organisation. Sci Rep. 2016;6:29775. doi: 10.1038/srep29775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, Meyer BJ. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015;523:240–244. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer M, Hübner B, Strickfaden H, Smeets D, Popken J, Sterr M, Markaki Y, Rippe K, Cremer C. The 4D nucleome: Evidence for a dynamic nuclear landscape based on co-aligned active and inactive nuclear compartments. FEBS Lett. 2015;589:2931–2943. doi: 10.1016/j.febslet.2015.05.037. [DOI] [PubMed] [Google Scholar]
- Cubeñas-Potts C, Rowley MJ, Lyu X, Li G, Lei EP, Corces VG. Different enhancer classes in Drosophila bind distinct architectural proteins and mediate unique chromatin interactions and 3D architecture. Nucleic Acids Res. 2016:gkw1114. doi: 10.1093/nar/gkw1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J. Mapping the 3D genome: Aiming for consilience. Nat Rev Mol Cell Biol. 2016;17:741–742. doi: 10.1038/nrm.2016.151. [DOI] [PubMed] [Google Scholar]
- Dekker J, Heard E. Structural and functional diversity of Topologically Associating Domains. FEBS Lett. 2015;589:2877–2884. doi: 10.1016/j.febslet.2015.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012 doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, et al. Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, et al. Control of Cell Identity Genes Occurs in Insulated Neighborhoods in Mammalian Chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle B, Fudenberg G, Imakaev M, Mirny LA. Chromatin Loops as Allosteric Modulators of Enhancer-Promoter Interactions. PLoS Comput Biol. 2014;10:e1003867. doi: 10.1371/journal.pcbi.1003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ea V, Sexton T, Gostan T, Herviou L, Baudement MO, Zhang Y, Berlivet S, Le Lay-Taha MN, Cathala G, Lesne A, et al. Distinct polymer physics principles govern chromatin dynamics in mouse and Drosophila topological domains. BMC Genomics. 2015;16:607. doi: 10.1186/s12864-015-1786-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagen KP, Hartl TA, Kornberg RD. Stable Chromosome Condensation Revealed by Chromosome Conformation Capture. Cell. 2015;163:934–946. doi: 10.1016/j.cell.2015.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essafi A, Webb A, Berry RL, Slight J, Burn SF, Spraggon L, Velecela V, Martinez-Estrada OM, Wiltshire JH, Roberts SGE, et al. A Wt1-Controlled Chromatin Switching Mechanism Underpins Tissue-Specific Wnt4 Activation and Repression. Dev Cell. 2011;21:559–574. doi: 10.1016/j.devcel.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suvà ML, Bernstein BE. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke M, Ibrahim DM, Andrey G, Schwarzer W, Heinrich V, Schöpflin R, Kraft K, Kempfer R, Jerković I, Chan WL, et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature. 2016;538:265–269. doi: 10.1038/nature19800. [DOI] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M. FISH-ing for captured contacts: towards reconciling FISH and 3C. 2016 doi: 10.1038/nmeth.4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirlando R, Felsenfeld G. CTCF: making the right connections. Genes Dev. 2016;30:881–891. doi: 10.1101/gad.277863.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Dekker J. The Hierarchy of the 3D Genome. Mol Cell. 2013;49:773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA. Chromatin Architecture of the Human Genome∷ Gene-Rich Domains Are Enriched in Open Chromatin Fibers. Cell. 2004;118:555–566. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Giorgetti L, Heard E. Closing the loop: 3C versus DNA FISH. Genome Biol. 2016;17:215. doi: 10.1186/s13059-016-1081-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti L, Galupa R, Nora EP, Piolot T, Lam F, Dekker J, Tiana G, Heard E. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell. 2014;157:950–963. doi: 10.1016/j.cell.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Buendía E, Pérez-Molina R, Ayala-Ortega E, Guerrero G, Recillas-Targa F. Experimental Strategies to Manipulate the Cellular Levels of the Multifunctional Factor CTCF. In: Robles-Flores M, editor. Cancer Cell Signaling. Springer; New York: 2014. pp. 53–69. [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016:aad9024. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Li X, Yusufzai TM, Qiu Y, Felsenfeld G. USF1 recruits histone modification complexes and is critical for maintenance of a chromatin barrier. Mol Cell Biol. 2007;27:7991–8002. doi: 10.1128/MCB.01326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imakaev M, Fudenberg G, McCord RP, Naumova N, Goloborodko A, Lajoie BR, Dekker J, Mirny LA. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Dadon DB, Powell BE, Fan ZP, Borges-Rivera D, Shachar S, Weintraub AS, Hnisz D, Pegoraro G, Lee TI, et al. 3D Chromosome Regulatory Landscape of Human Pluripotent Cells. Cell Stem Cell. 2016;18:262–275. doi: 10.1016/j.stem.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost D, Carrivain P, Cavalli G, Vaillant C. Modeling epigenome folding: formation and dynamics of topologically associated chromatin domains. Nucleic Acids Res. 2014;42:9553–9561. doi: 10.1093/nar/gku698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Pagie L, de Vries SS, Nahidiazar L, Dey SS, Bienko M, Zhan Y, Lajoie B, de Graaf CA, Amendola M, et al. Genome-wide maps of nuclear lamina interactions in single human cells. Cell. 2015;163:134–147. doi: 10.1016/j.cell.2015.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Dily F, Baù D, Pohl A, Vicent GP, Serra F, Soronellas D, Castellano G, Wright RHG, Ballare C, Filion G, et al. Distinct structural transitions of chromatin topological domains correlate with coordinated hormone-induced gene regulation. Genes Dev. 2014;28:2151–2162. doi: 10.1101/gad.241422.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M, Nora EP. CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annu Rev Genomics Hum Genet. 2016;17:17–43. doi: 10.1146/annurev-genom-083115-022339. [DOI] [PubMed] [Google Scholar]
- Moore JM, Rabaia NA, Smith LE, Fagerlie S, Gurley K, Loukinov D, Disteche CM, Collins SJ, Kemp CJ, Lobanenkov VV, et al. Loss of Maternal CTCF Is Associated with Peri-Implantation Lethality of Ctcf Null Embryos. PLOS ONE. 2012;7:e34915. doi: 10.1371/journal.pone.0034915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska M, Ulrich HD. An expanded tool kit for the auxin-inducible degron system in budding yeast: A tool kit for the AID system. Yeast. 2013;30:341–351. doi: 10.1002/yea.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumbach MR, Rubin AJ, Flynn RA, Dai C, Khavari PA, Greenleaf WJ, Chang HY. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat Methods. 2016 doi: 10.1038/nmeth.3999. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra V, Rocha PP, An D, Raviram R, Skok JA, Mazzoni EO, Reinberg D. Transcription. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015;347:1017–1021. doi: 10.1126/science.1262088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. Cohesins Functionally Associate with CTCF on Mammalian Chromosome Arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SWM, Solovei I, Brugman W, Gräf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. Molecular Maps of the Reorganization of Genome-Nuclear Lamina Interactions during Differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria MEG, Sanyal A, Gerasimova TI, Lajoie BR, Bell JSK, Ong CT, Hookway TA, Guo C, Sun Y, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recillas-Targa F, Pikaart MJ, Burgess-Beusse B, Bell AC, Litt MD, West AG, Gaszner M, Felsenfeld G. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc Natl Acad Sci U S A. 2002;99:6883–6888. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20:761–770. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn AL, Rao SSP, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci. 2015;112:E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorderet P, Lonfat N, Darbellay F, Tschopp P, Gitto S, Soshnikova N, Duboule D. A Genetic Approach to the Recruitment of PRC2 at the HoxD Locus. PLoS Genet. 2013;9:e1003951. doi: 10.1371/journal.pgen.1003951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, Fonseca NA, Huber W, Haering C, Mirny L, et al. Two independent modes of chromosome organization are revealed by cohesin removal. bioRxiv 094185. 2016 doi: 10.1038/nature24281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F, Soochit W, Bartkuhn M, Heath H, Dienstbach S, Bergmaier P, Franke V, Rosa-Garrido M, van de Nobelen S, Caesar L, et al. The male germ cell gene regulator CTCFL is functionally different from CTCF and binds CTCF-like consensus sites in a nucleosome composition-dependent manner. Epigenetics Chromatin. 2012;5:8. doi: 10.1186/1756-8935-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofueva S, Yaffe E, Chan WC, Georgopoulou D, Vietri Rudan M, Mira-Bontenbal H, Pollard SM, Schroth GP, Tanay A, Hadjur S. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 2013;32:3119–3129. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnikova N, Montavon T, Leleu M, Galjart N, Duboule D. Functional Analysis of CTCF During Mammalian Limb Development. Dev Cell. 2010;19:819–830. doi: 10.1016/j.devcel.2010.11.009. [DOI] [PubMed] [Google Scholar]
- Splinter E. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, Szalaj P, Trzaskoma P, Magalska A, Wlodarczyk J, Ruszczycki B, et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell. 2015;163:1611–1627. doi: 10.1016/j.cell.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teif VB, Vainshtein Y, Caudron-Herger M, Mallm JP, Marth C, Höfer T, Rippe K. Genome-wide nucleosome positioning during embryonic stem cell development. Nat Struct Mol Biol. 2012;19:1185–1192. doi: 10.1038/nsmb.2419. [DOI] [PubMed] [Google Scholar]
- Tsujimura T, Klein FA, Langenfeld K, Glaser J, Huber W, Spitz F. A Discrete Transition Zone Organizes the Topological and Regulatory Autonomy of the Adjacent Tfap2c and Bmp7 Genes. PLoS Genet. 2015;11:e1004897. doi: 10.1371/journal.pgen.1004897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri Rudan M, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, Hadjur S. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015;10:1297–1309. doi: 10.1016/j.celrep.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan LB, Pan H, Hannenhalli S, Cheng Y, Ma J, Fedoriw A, Lobanenkov V, Latham KE, Schultz RM, Bartolomei MS. Maternal depletion of CTCF reveals multiple functions during oocyte and preimplantation embryo development. Dev Camb Engl. 2008;135:2729–2738. doi: 10.1242/dev.024539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson LA, Wang X, Elbert A, Kernohan KD, Galjart N, Bérubé NG. Dual Effect of CTCF Loss on Neuroprogenitor Differentiation and Survival. J Neurosci. 2014;34:2860–2870. doi: 10.1523/JNEUROSCI.3769-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Wiechens N, Singh V, Gkikopoulos T, Schofield P, Rocha S, Owen-Hughes T. The Chromatin Remodelling Enzymes SNF2H and SNF2L Position Nucleosomes adjacent to CTCF and Other Transcription Factors. PLOS Genet. 2016;12:e1005940. doi: 10.1371/journal.pgen.1005940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Vos ESM, Holwerda SJB, Valdes-Quezada C, Verstegen MJAM, Teunissen H, Splinter E, Wijchers PJ, Krijger PHL, de Laat W. CTCF Binding Polarity Determines Chromatin Looping. Mol Cell. 2015;60:676–684. doi: 10.1016/j.molcel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- Witcher M, Emerson BM. Epigenetic Silencing of the p16INK4a Tumor Suppressor Is Associated with Loss of CTCF Binding and a Chromatin Boundary. Mol Cell. 2009;34:271–284. doi: 10.1016/j.molcel.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin J, Dixon JR, Reijden MIJA van der, Ye Z, Kolovos P, Brouwer RWW, Corput MPC van de, Werken HJG van de, Knoch TA, IJcken WFJ van, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for the Methods Sectons
- Chen C, Morris Q, Mitchell JA. Enhancer identification in mouse embryonic stem cells using integrative modeling of chromatin and genomic features. BMC Genomics. 2012;13:152. doi: 10.1186/1471-2164-13-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013 doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, Meyer BJ. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015;523:240–244. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinforma Oxf Engl. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. FIMO: Scanning for occurrences of a given motif. Bioinformatics btr064. 2011 doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Mahony S, Gifford DK. High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLoS Comput Biol. 2012;8:e1002638. doi: 10.1371/journal.pcbi.1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. Super-Enhancers in the Control of Cell Identity and Disease. Cell. 2013:1–14. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper M, Hardy K, Handyside A, Hunter S, Monk M. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. 1987;326:292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- Imakaev M, Fudenberg G, McCord RP, Naumova N, Goloborodko A, Lajoie BR, Dekker J, Mirny LA. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T, Nishimura K, Kanemaki MT, Donaldson AD. The Elg1 Replication Factor C-like Complex Functions in PCNA Unloading during DNA Replication. Mol Cell. 2013;50:273–280. doi: 10.1016/j.molcel.2013.02.012. [DOI] [PubMed] [Google Scholar]
- Lajoie BR, van Berkum NL, Sanyal A, Dekker J. My5C: web tools for chromosome conformation capture studies. Nat Meth. 2009;6:690–691. doi: 10.1038/nmeth1009-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie BR, Dekker J, Kaplan N. The Hitchhiker's guide to Hi-C analysis: practical guidelines. Methods San Diego Calif. 2015;72:65–75. doi: 10.1016/j.ymeth.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerdrup M, Johansen JV, Agrawal-Singh S, Hansen K. An interactive environment for agile analysis and visualization of ChIP-sequencing data. Nat Struct Mol Biol. 2016;23:349–357. doi: 10.1038/nsmb.3180. [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna-Zurita L, Stirnimann CU, Glatt S, Kaynak BL, Thomas S, Baudin F, Samee MAH, He D, Small EM, Mileikovsky M, et al. Complex Interdependence Regulates Heterotypic Transcription Factor Distribution and Coordinates Cardiogenesis. Cell. 2016;164:999–1014. doi: 10.1016/j.cell.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Garner AR, Shimaoka D, Chuong AS, Klapoetke NC, Li L, van der Bourg A, Niino Y, Egolf L, Monetti C, et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron. 2015;85:942–958. doi: 10.1016/j.neuron.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micallef L, Rodgers P. eulerAPE: drawing area-proportional 3-Venn diagrams using ellipses. PloS One. 2014;9:e101717. doi: 10.1371/journal.pone.0101717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska M, Ulrich HD. An expanded tool kit for the auxin-inducible degron system in budding yeast: A tool kit for the AID system. Yeast. 2013;30:341–351. doi: 10.1002/yea.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumbach MR, Rubin AJ, Flynn RA, Dai C, Khavari PA, Greenleaf WJ, Chang HY. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat Methods. 2016 doi: 10.1038/nmeth.3999. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J. Organization of the mitotic chromosome. Science. 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva EM, Rivero-Hinojosa S, Espinoza CA, Méndez-Catalá CF, Kang S, Suzuki T, Kosaka-Suzuki N, Robinson S, Nagarajan V, Ye Z, et al. Comparative analyses of CTCF and BORIS occupancies uncover two distinct classes of CTCF binding genomic regions. Genome Biol. 2015;16:161. doi: 10.1186/s13059-015-0736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R, Thomas S, Holloway AK, Pollard KS. Features that define the best ChIP-seq peak calling algorithms. Brief Bioinform. 2016 doi: 10.1093/bib/bbw035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H, Mo Y, Liao W, Zhang MQ. Genome-wide localization of protein-DNA binding and histone modification by a Bayesian change-point method with ChIP-seq data. PLoS Comput Biol. 2012;8:e1002613. doi: 10.1371/journal.pcbi.1002613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Horie K, Madisen L, Pavlova MN, Gragerova G, Rohde AD, Schimpf BA, Liang Y, Ojala E, Kramer F, et al. An inducible and reversible mouse genetic rescue system. PLoS Genet. 2008;4:e1000069. doi: 10.1371/journal.pgen.1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Principle of the CellTrace dye dilution assay for proliferation
(B) Flow cytometry of dilution kinetics of the CellTrace dye indicates that auxin-treated CTCF-AID mESCs keep proliferating after two days of CTCF depletion and slow down afterwards. Auxin does not trigger any proliferation defect in CTCF-AID mESCs lacking the Tir1 F-box protein transgene.
(C) Propidium iodide staining indicates CTCF depleted mESCs are not blocked in any specific stage of the cell cycle and do not become aneuploidy.
(D) A tagBFP2/mCherry FUCCI cassette was created and knocked in in CTCF-AID or WT mESCs. Auxin treatment only leads to a slight increase of the G1 FUCCI-signal after 4 days of CTCF depletion, confirming that loss of CTCF does not block cell cycle progression overall.
(E) CTCF depleted did not show increased DNA damage as monitored by western blot, and displayed overall constant bulk H3K27me3 levels. LaminB1 used as loading control
(F) CTCF depletion did not lead to massive apoptosis, although number of dying cells increase after long depletion.
(G) Strategy for introducing dox-inducible CTCF transgenes in CTCF-AID cells
(H) Flow cytometry confirms that most auxin+dox-treated cells loose endogenous CTCF (>99%) and express transgenic CTCF (>95%) after 4 days of auxin+dox treatment
(I) Western blot using a CTCF antibody indicates that the dox-inducible transgene can be readily detected but drives lower expression than normal endogenous CTCF levels
(J) Inducing CTCF expression from the transgene largely alleviates the proliferation defects caused by from depleting of endogenous CTCF.
Supplementary Table 3 RNA-seq FPKM values, Related to figure 6
Supplementary Table 5 - 5C oligonucleotides, Related to figure 5
Supplementary Table 7 - CTCF motif orientation inside ChIP-seq Peaks found in untreated CTCF-AID mESCs Related to figure 6
(A) 5C at the Xic confirms that chromatin loops do not accumulate at CTCF peaks after CTCF depletion and are reaquired upon CTCF resoration.
(B) Auxin treatment of WT cells has no effect on chromatin folding
(C) CTCF ChIP-exo signal at CTCF ChIP-seq peaks detected in untreated CTCF-AID cells. Auxin treatment of WT cells has no effect on CTCF binding. Tagging with the CTCF-AID-eGFP does not disrupt CTCF binding pattern.
(D) Auxin treatment of CTCF-AID cells dramatically reduces CTCF enrichment at peaks detected in untreated cells and is fully reversible after washoff
(E) Easeq Genome browser visualization of an example locus. A subset of CTCF ChIP-seq peaks are still detected, but of low intensity, after depletion and are restored in strength after washoff
(F) Loss of ChIP-seq signal upon CTCF depletion is equivalent in the A and B genomic compartment as defined by Hi-C.
(G) A compartment tends to have stronger CTCF ChIP-seq peaks than B compartment
(H) CTCF binding is 5-fold denser in the A compartment than in B.
(I) Restriction-fragment level interpolated visualization of 5C around the Linx-Chic1-Xite loops. CTCF depletion disrupts CTCF binding and underlying loops while CTCF recovery re-stablishes binding and chromatin contacts.
(J) Auxin treatment in itself does not perturb the accumulation of chromatin loops in WT untagged mESCs, as exemplified at the Linx/Chic1/Xist 300kb TAD within the 4.5Mb segment covered by our 5C assay.
(A) Restriction-fragment level interpolated visualization of 5C at the Xic. Color dots denote TAD boundaries.
(B) Insulation score ratio between treated and untreated cells at boundaries detected in untreated cells, highlighting that a subset of TAD boundaries rapidly loose insulation upon CTCF depletion
(C) Insulation score analysis using 5C on independently generated cell lines (see Methods for details). Lines #2-5 were created by re-introducing Tir1 transgenes in the intermediate CTCF-AID-eGFP (no Tir1) clone used to generate the cell line used for the other analyses (#1), at the Rosa26 or Tigre acceptor loci. Cell line #6 was created by first introducing a Tir1 transgene at Tigre in WT cells and then re-creating the CTCF-AID-eGFP allele homozygously.
(D) 5C in the CTCF-AID line (#1) complemented with CTCF transgene indicating
(E) Insulation score analysis indicating that expression of the CTCF transgene mitigate the insulation defects caused by the loss of endogenous CTCF. Note that transgene expression is not as high as endogenous CTCF (Figure S1I)
(F-G) Auxin treatment has no effect on TAD insulation in WT untagged and CTCF-AID (no Tir1) cells
(H) Probability of calling a TAD boundary at Smc1a HiChIP loop as a function of the local prominence of the insulation score calculated at 100kb with our Hi-C. We chose the threshold (0.3) below which improvement in retrieving Sm1a HiChIP loop is below 50% (see methods).
(I) Hi-C snapshot illustrating that a subset of boundaries resist CTCF depletion Shown is an example region harboring boundaries that resist CTCF depletion. The one is associated with a strong promoter and the other one with a A/B compartment transition.
(J) Hi-C snapshot illustrating that a small subset of boundaries retain strong insulation after depletion without being associated with transcription or compartment transition
(K) Replot of the DNA FISH data presented in figure 3D illustrating that after CTCF depletion inter-TAD 3D distances becomes equivalent to intra-TAD when probe pairs are equally spaced on the chromosome and not overlapping boundaries.
(L) Replot of the DNA FISH data presented in figure 3D illustrating that probe pairs partially overlapping boundaries (green and yellow) become more separated after CTCF depletion
(M) Scaling of ICE normalized Hi-C contacts between loop anchors or matched random loci pairs, comparing CTCF depletion by RNAi in human HEK293T cells (Zuin et al., 2014) to the CTCF-AID mESCs. Loop anchors are from GM12878 (human) and CH12-LX cells (mouse), respectively (Rao et al., 2014). Thick line is the median, shaded area highlights 25-75 percentile and dotted lines are landmarks for visual comparison.
(A) cis-Eigenvector 1 values in 100kb genomic bins are ranked and pairwise enrichment of Hi-C contacts between each of the 50 ranks are calculated (pooled replicates). Genomic regions with similar ranks of Eigenvector 1 values display more Hi-C contact while regions of opposite ranks are depleted (see methods). This trend is conserved overall after CTCF depletion or restoration.
(B)Compartmentalization strength is only mildly affected by CTCF depletion
(C) Scatter plot of the insulation score of genomic elements that are transitions between A/B compartments and TAD boundaries before CTCF depletion (left) or after (right), highlighting that insulation is also weaker at these compartments transition after loss of CTCF. Note that strong boundaries have the lowest insulation scores.
(A-G) Restriction-fragment level 5C interpolated heatmaps highlighting that auxin treatment of CTCF-AID NPCs and astrocytes disrupts TAD insulation, irrespective of the timing of CTCF depletion to cell-cycle exit. Blue = time after adding BMP4 to convert NPCs into astrocytes.
(H) Summary of all experiments with NPCs and astrocytes.
(I) Quantification of insulation loss from the 5C in all NPC and astrocyte samples.
(J) Comparative levels of CTCF-AID-eGFP in mESCs, NPCs and astrocytes measured by flow cytometry. mESCs display a broader fluorescence distribution as they are mostly in S/G2, while NPCs display both G1 and S/G2 cells and astrocytes are in G0.
(A) Scatter plot of the fold change in treated versus untreated cells as a function of the expression level in untreated cells. Up and down-regulation are observed for genes with a wide-range of initial expression levels. Misregulation is not restricted to lowly-expressed genes.
(B-C) Same analysis as in 6D-E but focused on super-enhancers active in mESCs.
(D) Gene misregulated upon CTCF depletion are more often found close to Smc1a HiChIP loop anchors than expected by chance. See methods for statistical details.
(A) CTCF and H3K27me3 ChIP-seq centered at all CTCF peaks detected in untreated cells. A small subset is embedded in large H3K27me3 regions with a dip at the CTCF site. This dip disappears upon CTCF depletion and reappears after CTCF restoration. This suggests that nucleosomes become able to cover the previously occupied CTCF site when CTCF binding is lost.
(B) Overall H3K27me3 levels at H3K27me3 ChIP-seq peaks are unaffected after two day, become slightly lower after 4 days of depletion and are readjusted upon CTCF restoration.
(C-D) Easeq Genome browser visualization of an example locus illustrating H3K27me3 does not spread beyond flanking CTCF sites upon CTCF depletion in mESCs.
Supplementary Table 1 sgRNA sequences, Related to Figure 1