Abstract
Carbohydrates mediate their conversion to triglycerides in the liver by promoting both rapid posttranslational activation of rate-limiting glycolytic and lipogenic enzymes and transcriptional induction of the genes encoding many of these same enzymes. The mechanism by which elevated carbohydrate levels affect transcription of these genes remains unknown. Here we report the purification and identification of a transcription factor that recognizes the carbohydrate response element (ChRE) within the promoter of the L-type pyruvate kinase (LPK) gene. The DNA-binding activity of this ChRE-binding protein (ChREBP) in rat livers is specifically induced by a high carbohydrate diet. ChREBP's DNA-binding specificity in vitro precisely correlates with promoter activity in vivo. Furthermore, forced ChREBP overexpression in primary hepatocytes activates transcription from the L-type Pyruvate kinase promoter in response to high glucose levels. The DNA-binding activity of ChREBP can be modulated in vitro by means of changes in its phosphorylation state, suggesting a possible mode of glucose-responsive regulation. ChREBP is likely critical for the optimal long-term storage of excess carbohydrates as fats, and may contribute to the imbalance between nutrient utilization and storage characteristic of obesity.
Until recently, evolutionary pressures have favored the ability to efficiently store nutrients as fat during periods of abundant food supply as a safeguard against periodic famine (1). Coupled with dramatic changes in modern lifestyle and food consumption, these “thrifty genes” may now contribute to health defects suffered by as many as half of the American population presently overweight or obese (2). The liver is the principal organ responsible for the conversion of excess dietary carbohydrates to triglycerides. Within minutes, elevated glucose levels in the liver lead to posttranslational activation of several key enzymes of glycolysis and lipogenesis, including fructose-6-phosphate 2-kinase/fructose-2,6-bisphosphatase, fatty acid synthase, acetyl-CoA carboxylase, and L-type pyruvate kinase (LPK). A high-carbohydrate diet also induces transcription of many of the genes encoding these enzymes, thereby promoting long-term storage of sugars as triglycerides (3, 4). While the expression of some lipogenic enzymes is enhanced by insulin, which is secreted in response to high blood sugar levels, optimal transcription of most lipogenic genes requires elevated carbohydrate levels (reviewed in refs. 5 and 6). LPK gene transcription is stimulated by glucose, independent of insulin, in cultured hepatocytes expressing active glucokinase (7). The mechanism by which carbohydrates regulate transcription of these genes in the liver remains unknown. Previous studies have shown that many lipogenic genes contain carbohydrate response elements (ChREs) within their promoters that mediate glucose responsiveness (4, 8–11). These sequences have been used to characterize several ChRE-binding proteins (12–15). However, the identity of the carbohydrate-responsive transcription factor has remained elusive, as the expression/activity of these candidate proteins does not correlate with glucose-induced transcription regulated by the ChRE.
In this study, we report the purification and identification of a ChRE-binding protein, designated ChREBP. The tissue distribution, specificity, and carbohydrate responsiveness of ChREBP's DNA-binding activity are consistent with those of the putative glucose-responsive transcription factor. Furthermore, ChREBP contains several consensus phosphorylation sites that may regulate the activity of this protein. These data suggest that ChREBP is likely responsible for mediating carbohydrate induction of transcription of the LPK gene, and possibly other genes, in the liver and may influence long-term fat accumulation resulting from a high-carbohydrate diet.
Materials and Methods
Materials.
Okadaic acid was purchased from Boehringer Mannheim. The cAMP-dependent protein kinase (PKA) catalytic subunit and PKA inhibitor were purchased from New England Biolabs. The protein phosphatase 2A catalytic subunit was purchased from Bio-Rad. All other chemicals were reagent grade and obtained from commercial sources.
Preparation of Nuclear Extracts.
Rats were starved for 48 h and then fed 24 h with either the National Institutes of Health high-sucrose diet lab chow (by weight: 20% casein, 60.2% sucrose, 15% cellulose, 2.25% minerals, and 2.25% vitamins) (16), or a high-fat diet without starch (31% casein, 30.5% cellulose, 7% minerals, 1.5% vitamins, 1.5% corn oil, 1.5% peanut oil, and 27% lard) (17). Liver nuclear extracts were prepared according to Hattori et al. (18) with minor modifications (13). Fresh rat livers (≈10 g) were homogenized with a Potter–Elvehjem homogenizer in extraction buffer (10 mM Hepes, pH 7.9/10 mM KCl/0.1 mM EDTA/0.74 mM spermidine/1 mM DTT/0.5 mM PMSF) containing 0.3 M sucrose. The homogenate was mixed with 2 vol of cushion buffer (extraction buffer containing 2.2 M sucrose), layered on top of cushion buffer, and centrifuged at 75,000 × g for 30 min at 4°C. Pelleted nuclei were resuspended in 5 vol of nuclear lysis buffer [10% (vol/vol) glycerol/10 mM Hepes, pH 7.9/420 mM NaCl/0.1 mM EDTA/3 mM MgCl2/5 mM DTT/0.5 mM PMSF/0.5 μg/ml pepstatin A/1 mM benzamidine/0.2 μg/ml leupeptin) and homogenized with 10 strokes of a hand-held Dounce homogenizer. The nuclear extracts were centrifuged at 27,000 × g for 25 min at 4°C, and polyethylene glycol (PEG; Mr = 8,000) was added to the supernatant solution to 25% saturation. After centrifugation at 27,000 × g for 10 min, the pellet was dissolved in a buffer containing 20% glycerol, 20 mM Hepes (pH 7.9), 100 mM KCl, 0.2 mM EDTA, 5 mM DTT, and 0.5 mM PMSF and cleared by centrifugation at 27,000 × g for 5 min at 4°C. Protein concentration was determined by using the Bradford method (19).
Gel-Shift Assays.
Gel mobility-shift assays were performed as described by Liu et al. (8). Double-stranded oligonucleotides (Table 1) were prepared by mixing equal amounts of the complementary single-stranded DNAs in 50 mM NaCl, heating to 70°C for 15 min, and cooling to room temperature. The annealed oligonucleotides were labeled with 32P in the presence of [γ-32P]ATP (Amersham Pharmacia) and T4 polynucleotide kinase (New England Biolabs). Binding reactions were carried out in reaction mixture containing 20 mM Hepes (pH 7.9), 50 mM KCl, 5 mM DTT, 0.2 mM EDTA, 0.5 mM PMSF, 10% glycerol, 1 μg/μl poly(dI-dC) (Amersham Pharmacia), and 1 μg/μl nuclear extract. The reaction mixture was incubated at room temperature for 30 min, and DNA–protein complexes were separated by electrophoresis in a nondenaturing 4.5% polyacrylamide gel.
Table 1.
The sequences of the wild-type (WT) and mutated LPK ChREs
| Oligonucleotide | Sequence |
|---|---|
| WT | 5′-GGGCGCACGGGGCACTCCCGTGGTTCC-3′ |
| 3′-CCCGCGTGCCCCGTGAGGGCACCAAGG-5′ | |
| M1 | 5′-GGGCGaACGGGGCACTCCCGTtGTTCC-3′ |
| 3′-CCCGCtTGCCCCGTGAGGGCAaCAAGG-5′ | |
| M2 | 5′-GGGCGCcCGGGGCACTCCCGgGGTTCC-3′ |
| 3′-CCCGCGgGCCCCGTGAGGGCcCCAAGG-5′ | |
| M3 | 5′-GGGCGCAaGGGGCACTCCCtTGGTTCC-3′ |
| 3′-CCCGCGTtCCCCGTGAGGGaACCAAGG-5′ | |
| M4 | 5′-GGGCGCACtGGGCACTCCaGTGGTTCC-3′ |
| 3′-CCCGCGTGaCCCGTGAGGtCACCAAGG-5′ | |
| M5 | 5′-GGGCGCACGtGGCACTCaCGTGGTTCC-3′ |
| 3′-CCCGCGTGCaCCGTGAGtGCACCAAGG-5′ | |
| M6 | 5′-GGGCGCACGGtGCACTaCCGTGGTTCC-3′ |
| 3′-CCCGCGTGCCaCGTGAtGGCACCAAGG-5′ | |
| M3/5 | 5′-GGGCGCAtGcGGCACTCgCaTGGTTCC-3′ |
| 3′-CCCGCGTaCgCCGTGAGcGtACCAAGG-5′ | |
| S−1 | 5′-GGGCGCACGGGGCCTCCCGTGGTTCCT-3′ |
| 3′-CCCGCGTGCCCCGGAGGGCACCAAGGA-5′ | |
| S+1 | 5′-GGGCGCACGGGGCACTTCCCGTGGTTCC-3′ |
| 3′-CCCGCGTGCCCCGTGAAGGGCACCAAGG-5′ |
The E-boxes are in boldface type. Mutations within the E-box are indicated by lowercase letters. S−1 and S+1 indicate the deletion or addition, respectively, of one base between the two E-boxes.
Primary Hepatocytes Culture and Transfection.
The construction of luciferase reporter plasmids was described in ref. 13. The mouse ChREBP coding region (GenBank accession no. AF156604) was amplified by PCR and ligated into the pcDNA3.1/V5HisB expression vector (Invitrogen).
Primary hepatocytes were prepared from male Sprague–Dawley rats by using the collagenase (Life Technologies, Grand Island, NY) perfusion method (20) and plated in collagen-coated six-well tissue culture plates (CMS/Fisher, Houston) at a density of 1 × 106 cells per well in glucose-free Dulbecco's modified Eagle's medium (DMEM; Life Technologies) supplemented with 100 nM dexamethasone, 10 nM insulin, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% dialyzed FBS (Life Technologies). After attachment, synthetic liposomes were used to transfect hepatocytes with 0.5 μg of the luciferase reporter plasmid and 0.1 μg of the internal control plasmid pRL-TK. After 14 h the medium was replaced by DMEM supplemented with 100 nM dexamethasone, 10 nM insulin, 10% FBS, and 10 mM lactate, 5.5 mM glucose, or 27.5 mM glucose. The cells were incubated for an additional 12 h before determination of luciferase activity by using the Dual-Luciferase Reporter Assay System (Promega).
Purification of ChREBP.
ChREBP DNA-binding activity was monitored by gel-shift analysis with 32P-labeled WT ChRE oligonucleotide (Table 1). Crude nuclear extract was prepared from the livers of 800 male Sprague–Dawley rats re-fed a high-carbohydrate diet for 24 h after starvation for 48 h. Livers were homogenized in 2 vol of buffer (300 mM sucrose/10 mM Hepes, pH 7.9/10 mM KCl/0.1 mM EDTA/0.74 mM spermidine/1 mM DTT/0.5 mM PMSF). The homogenate was mixed with 2 vol of buffer containing 2.2 M sucrose, layered on top of buffer containing 2.2 M sucrose, and centrifuged at 75,000 × g for 45 min at 4°C. Pelleted nuclei were homogenized in 2.5 vol of lysis buffer (10% glycerol/10 mM Hepes, pH 7.9/10 mM KCl/0.1 mM EDTA/3 mM MgCl2/5 mM DTT/0.5 mM PMSF/0.5 μg/ml pepstatin A/1 mM benzamidine/0.2 μg/ml leupeptin). After addition of 0.1 vol of 4.2 M NaCl, the homogenate was gently stirred for 1 h at 4°C and then centrifuged at 100,000 × g for 30 min at 4°C. The supernatant was diluted to 100 mM NaCl and cleared by centrifugation at 100,000 × g for 30 min. The nuclear extract was applied to a DE-52 DEAE-cellulose column equilibrated with buffer A (20 mM Hepes, pH 7.9/1 mM EDTA/10% glycerol/1 mM DTT) containing 0.1 M KCl. Proteins were eluted from the column with buffer A containing 0.2 M KCl. Active fractions were pooled and subsequently applied to a DNA-cellulose (native DNA) column equilibrated with buffer A containing 0.2 M KCl. The column was washed with buffer A containing 0.5 M KCl and eluted with buffer A containing 0.7 M KCl. The active fractions were pooled, and (NH4)2SO4 was added to a final concentration of 0.436 g/ml and stirred for 30 min at 4°C. After centrifugation at 100,000 × g for 20 min at 4°C, the precipitate was resuspended in buffer A containing 50 mM KCl and competitor DNAs [poly(dI-dC) and CATCAGGCCATCTGGCCCCTTGTTATTAA at 50 μg/ml and 10 μg/ml, respectively]. The sample was applied to a DNA-affinity column displaying double-stranded CATGGGCGCACGGGGCACTCCCGTGGTTCC DNA. The column was washed with buffer A containing 0.4 M KCl and nonspecific competitor DNAs and eluted with buffer A containing 0.8 M KCl. The DNA-affinity column step was repeated. The active fractions were pooled, resolved by SDS/PAGE, and visualized by silver staining. The 100-kDa band was excised from poly(vinylidene difluoride) (PVDF) membrane and digested, and individual peptides were sequenced as described in ref. 21. Four peptides were found to match human AF156603_1.
Results
A High-Carbohydrate Diet Induces ChREBP DNA-Binding Activity in Rat Livers.
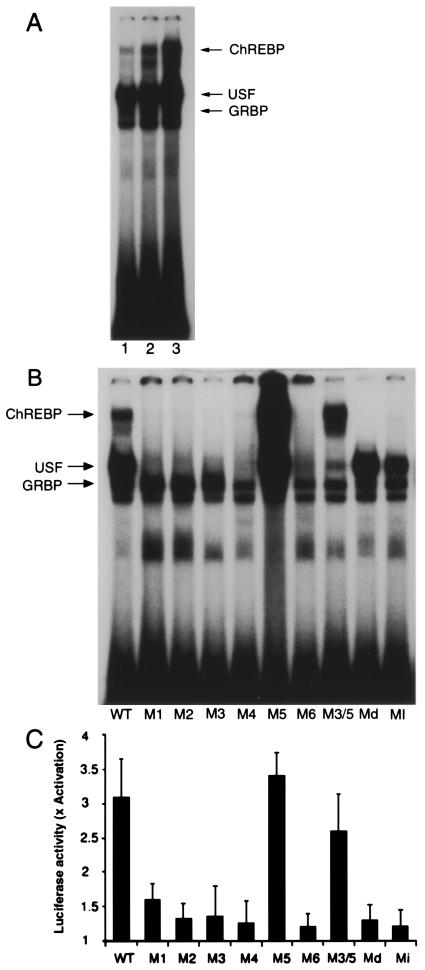
It has been shown that glucose-induced transcription of the LPK gene is mediated by a ChRE located within its promoter (4, 8–11). The ChRE identified within the promoter of the LPK gene contains two imperfect E-box motifs in a head-to-tail orientation separated by 5 bp (WT; Table 1). This sequence was used to examine DNA-binding activity in liver nuclear extracts prepared from rats fed either a high-carbohydrate or high-fat diet for 24 h after starvation. One of the DNA-binding complexes observed in nuclear extracts, designated ChREBP, was enriched 2–3-fold only in rats fed the high-carbohydrate diet (Fig. 1A, lanes 2 and 3). Conversely, formation of the previously described DNA-binding complex containing the upstream stimulatory factor (USF) (12) did not vary as a function of diet (Fig. 1A).
Figure 1.
(A) Gel-shift analysis of hepatic nuclear factors that recognize the WT LPK ChRE. Rats were starved for 48 h (lane 1) and subsequently re-fed a high-fat diet for 24 h (lane 2) or a high-carbohydrate diet for 24 h (lane 3). Nuclear extracts were prepared from the rat livers and incubated with a radiolabeled oligonucleotide corresponding to the WT LPK ChRE. Lanes 2 and 3 contain equal protein loads. Arrows indicate the positions of the DNA-binding complexes containing USF or the glucose response element binding protein (GRBP) as determined by supershift of the DNA-binding complex with antibodies specific for each protein. (B) Mutations to the LPK ChRE differentially affect the formation of the various DNA-binding complexes. The seuences of the mutated oligonucleotides are shown in Table 1 (Md and Mi correspond to S−1 and S+1, respectively). Liver nuclear extracts from rats r-fed a high-carbohydrate diet for 24 h after 48-h starvation were incubated with the WT and the mutated oligonucleotides and examined by gel-shift analysis. (C) Mutations to the LPK ChRE affect glucose-responsive transcription of a downstream luciferase reporter gene. Construction of luciferase reporters driven by the WT and variant LPK promoter sequences (Table 1) was described in ref. 13. Primary hepatocytes were transfected with the reporter constructs and the fold activation of luciferase expression in response to high glucose (27.5 mM) was determined. Values represent the mean ± standard error of four experiments.
ChREBP's DNA-Binding Specificity in Vitro Correlates with Promoter Activity in Vivo.
The relative affinities of the observed DNA-binding proteins for the native LPK ChRE sequence and mutated variants (Table 1) are shown in Fig. 1B. Mutation of the modified E-box motifs to perfect palindromes (sequence M5) enhanced formation of the ChREBP DNA-binding complex, whereas further deviation from the canonical E-box motif significantly impeded complex formation (sequences M1–4, M6). Unlike formation of USF and GRBP complexes (13), formation of the ChREBP complex was particularly sensitive to changes in the spacing between the two E-box motifs (sequences S−1 and S+1). Importantly, only complex formation between the variant sequences and ChREBP correlated precisely with the ability of the ChRE variants to mediate glucose-induced transcription of a luciferase reporter in primary hepatocytes (Fig. 1C). The agreement between ChREBP's DNA-binding specificity and the relative ability of the various DNA sequences to confer carbohydrate-responsive transcriptional activation suggests that this complex might contain the long-sought glucose-regulated transcription factor.
ChREBP Is a Member of the Basic Helix–Loop–Helix Leucine Zipper (bHLH-ZIP) Family of Transcription Factors Observed Only in the Liver.
ChREBP was purified to homogeneity from the livers of rats fed a high-carbohydrate diet and shown to consist of a single polypeptide with an apparent molecular mass of 100 kDa. The amino acid sequences of four peptides generated by tryptic digestion were determined. Each peptide was found to match the Williams–Beuren syndrome critical region 14 protein (WBSCR14), encoded by one of the 17 genes heterozygously deleted from patients suffering from the neurodevelopmental disorder Williams–Beuren syndrome (22). The contributions of most of the deleted genes, including WBSCR14, to the WBS phenotypes are not known.
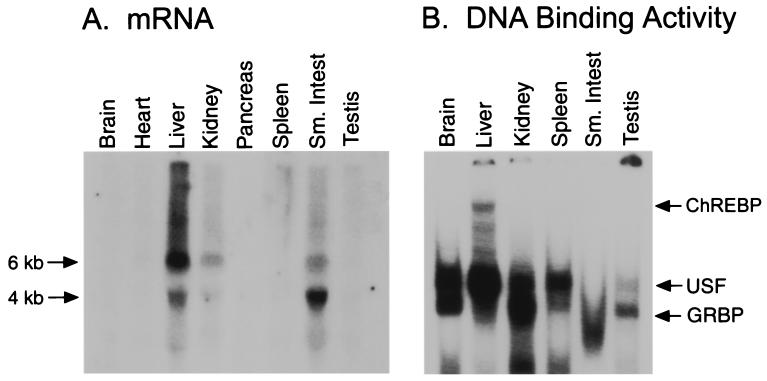
ChREBP is a member of the basic helix–loop–helix leucine zipper (bHLH-ZIP) family of transcription factors known to recognize E-box motifs within their target promoters. Northern analysis identified two ChREBP isoforms (6 kb and 4 kb) present in the liver, kidney, and small intestine (Fig. 2A). These results are consistent with the ChREBP expression pattern observed in the mouse (22). However, gel-shift analysis of nuclear extracts prepared from various tissue sources demonstrated that ChREBP DNA-binding activity is observable only in liver nuclear extracts (Fig. 2B). ChREBP's limited expression profile is consistent with previous studies demonstrating that LPK is expressed exclusively in the liver (23) and β cells of the pancreas (24). The failure to detect the ChREBP mRNA in the pancreas is likely due to the fact that β cells represent only 1% of the cells in this organ. Conversely, the USF-containing complex was observed in many tissues that do not express LPK, as expected from USF's ubiquitous expression pattern (25). Although several other unidentified complexes are observed, only the ChREBP-containing complex is induced by a high-carbohydrate diet and is specific to the site of LPK expression.
Figure 2.
(A) Tissue distribution of ChREBP as determined by Northern blot analysis. Message levels were visualized from total RNA (25 μg) with a 32P-radiolabeled probe derived from the ChREBP cDNA. (B) ChREBP activity is detected only in liver nuclear extracts. Nuclear extracts were prepared from rats re-fed a high carbohydrate diet for 24 h after 48-h starvation and incubated with the 32P-radiolabeled WT oligonucleotide (Table 1) followed by gel-shift analysis. Arrows indicate the positions of the DNA-binding complexes containing USF or GRBP as determined by supershift of the DNA-binding complex with antibodies specific for each protein.
ChREBP Activates Transcription from the LPK Promoter in Response to Glucose in Hepatocytes.
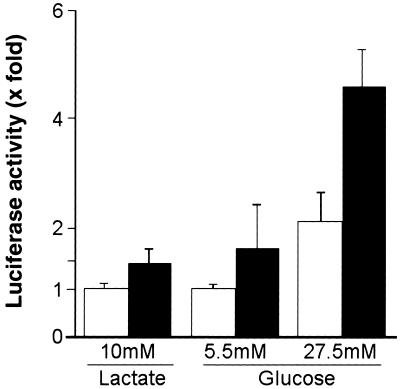
To determine whether ChREBP can mediate glucose-activated transcription in cells, primary hepatocytes were cotransfected with the luciferase reporter gene driven by a 200-bp fragment of the LPK promoter containing the ChRE (13) and an expression vector featuring ChREBP under the control of the constitutively active cytomegalovirus (CMV) promoter. When transfected cells were cultured in the absence of glucose (10 mM lactate) or in the presence of a low concentration of glucose (5.5 mM), expression of the ChRE-driven reporter remained low and was unresponsive to forced overexpression of ChREBP (Fig. 3, dark bars). Supplementation of the culture medium with 27.5 mM glucose did lead to a 2-fold induction of reporter expression in cells transfected with empty vector (Fig. 3, open bars). This reporter induction reflects the presence of the endogenous glucose-responsive transcription factor that recognizes the LPK ChRE (Fig. 1C). Forced expression of ChREBP was able to further induce expression of the reporter an additional 2-fold above the endogenous levels. The modest level of reporter induction, above endogenous ChREBP activity, in response to glucose is typical of the glucose-responsive transcriptional induction of many glycolytic enzymes in vivo. These results indicate that ChREBP may function as a glucose-responsive transcription factor in vivo.
Figure 3.
Forced expression of ChREBP activates expression from the LPK promoter in response to glucose in vivo. Primary hepatocytes were cotransfected with 0.5 μg of the luciferase reporter construct driven by the LPK promoter and 1.5 μg of the ChREBP cDNA under the control of the constitutively active cytomegalovirus (CMV) promoter (filled bars) or with the empty expression vector (open bars). After transfection, the cells were incubated for 12 h in medium containing 10 mM lactate, 5.5 mM glucose, or 27.5 mM glucose. Values represent the mean ± standard error of five experiments.
ChREBP's DNA-Binding Activity Is Inhibited by Phosphorylation in Vitro.
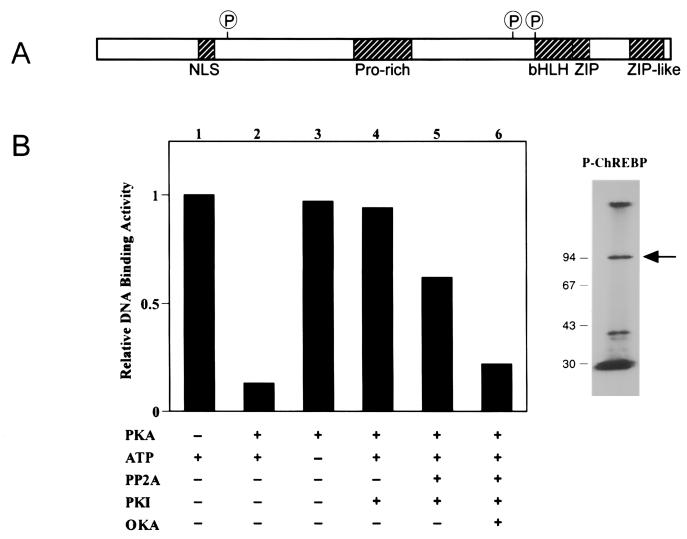
The mechanism by which excess glucose regulates the DNA-binding activity of ChREBP remains unclear. However, cAMP is known to inhibit glucose-responsive signaling pathways (6, 26, 27). Interestingly, the repressive effects of cAMP on LPK gene transcription have been reported to be localized to the ChRE despite the complete lack of similarity to known cAMP-regulated promoter elements (6, 9). ChREBP contains three consensus PKA phosphorylation sites, including one located within the basic region of the putative DNA-binding domain (Fig. 4A). When circulating carbohydrate levels are low, elevated glucagon levels promote increased cAMP production, which activates PKA and may result in phosphorylation of ChREBP. Addition of a negatively charged phosphoryl group to the basic region would be expected to disrupt DNA-binding activity, thereby inactivating the transcription factor. Conversely, excess carbohydrates would lower cAMP levels and could also lead to the activation of a protein phosphatase that hydrolyzes the phosphoryl group from inactive ChREBP. Dephosphorylation of inactivated ChREBP would restore DNA-binding activity and induce transcription of genes required for carbohydrate metabolism. To test this model, purified ChREBP was incubated with the catalytic subunit of PKA in the presence of ATP, resulting in phosphorylation of ChREBP and a 90% reduction in its DNA-binding activity (Fig. 4B). ChREBP inactivation required ATP and was inhibited by addition of a protein kinase inhibitor, PKI (Fig. 4B). DNA-binding activity of inactivated ChREBP was restored by the addition of the catalytic subunit of protein phosphatase 2A (PP2A). Reactivation of phosphorylated ChREBP by PP2A is blocked by the phosphatase inhibitor okadaic acid (Fig. 4B). These data are consistent with the model that ChREBP activity is regulated by changes in its phosphorylation state.
Figure 4.
(A) ChREBP contains several recognizable protein motifs, including a bipartite nuclear localization signal (NLS), a basic helix–loop–helix leucine zipper (bHLH-ZIP) motif, and three consensus PKA phosphorylation sites (P) including one (sequence RRIT) within the bHLH region. (B) (Left) DNA-binding activity of ChREBP can be regulated by phosphorylation in vitro. Incubation of active ChREBP (lane 1) for 20 min with both ATP (1 mM) and PKA (0.1 unit/μl) abolished DNA-binding activity (lane 2) as measured by gel-shift analysis with the 32P-radiolabeled WT oligonucleotide. Omission of ATP from the reaction mixture prevents ChREBP inactivation (lane 3). PKA-dependent ChREBP inactivation is blocked by 0.2 unit/μl protein kinase inhibitor (PKI) (lane 4). After PKI addition, DNA-binding activity of phosphorylated ChREBP can be restored by addition of 0.025 unit/μl protein phosphatase 2A (PP2A) (lane 5). Addition of the PP2A inhibitor okadaic acid (OKA) (10 nM) prevents reactivation of the ChREBP DNA-binding activity. (Right) Incubation of the protein with [γ-32P]ATP in the presence of PKA leads to radiolabeling of ChREBP observed after SDS/PAGE.
Discussion
The identity of the glucose-responsive ChRE-binding activity has until now remained elusive. Although the USF1/USF2 heterodimer has been shown to bind the ChRE in vitro, the expression, regulation, and selectivity of USF do not correlate with the glucose response (12). The same is true for GRBP (13), which shares mobility properties with the ChRE-binding protein purified by Yamada et al. (14) and the ChRE-binding protein reported by Koo and Towle (15), which does not vary with diet and was not regulated by phosphorylation. Alternatively, the expression, regulation, and selectivity of ChREBP are consistent with its assignment as a carbohydrate-responsive transcription factor. ChREBP activity is induced by consumption of a high-carbohydrate diet, but not a high-fat diet, after starvation (Fig. 1A). ChREBP's DNA-binding specificity precisely correlates with promoter activity in vivo (Fig. 1 B and C). Gel-shift analysis using oligonucleotides corresponding to the related ChREs from other carbohydrate-responsive genes such as S14 (28) and fatty acid synthase (29) also resulted in the formation of a complex with mobility properties identical to the ChREBP–LPK ChRE complex (data not shown). ChREBP's DNA-binding activity was observable only in nuclear extracts prepared in the liver (Fig. 2). This expression pattern is consistent with the expression pattern of the LPK target gene. Significantly, forced ChREBP expression in liver hepatocytes activates transcription from the LPK promoter specifically in response to high glucose levels but not low glucose levels or lactate (Fig. 3).
Analysis of the primary sequence of the ChREBP protein reveals the presence of several putative domains typical of transcription factors (Fig. 4A). The ChREBP carboxyl terminus contains a basic helix–loop–helix motif that likely mediates binding of the factor to the E-box motifs within the ChRE. In addition, there are regions of homology to proline-rich domains, leucine zipper domains, and leucine zipper-like domains (22) that could mediate additional protein–protein interactions, including dimerization. The amino terminus features a bipartite nuclear localization signal that likely mediates nuclear import. The role of these domains in mediating the function of this factor requires further investigation.
Elucidation of the mechanism by which elevated carbohydrate levels in the liver induce transcription by means of ChREBP continues to be the subject of much study. Glucose must first be metabolized in the liver to promote carbohydrate-responsive transcription of the LPK gene (reviewed in ref. 6). However, the identity of important metabolic intermediates and the pathways by which they regulate transcription remain unknown. Identification of ChREBP as a critical downstream component of this pathway should provide new insights into these regulatory mechanisms. For example, the presence of several potential phosphorylation sites suggests that the activity of this transcription factor could be regulated by changes in its phosphorylation state, particularly within the DNA-binding domain. As we show here, phosphorylation of ChREBP by PKA inactivates DNA-binding activity in vitro, consistent with a model in which cAMP serves an intracellular mediator to inactivate this transcription factor in vivo. In addition, this model predicts that a protein phosphatase could activate ChREBP, providing an additional point of regulation that could be mediated by a glucose metabolite. If the model proposed in this study is correct, identification of the particular protein phosphatase(s) responsible for ChREBP activation in vivo should provide additional insight into the nature of a long-sought glucose metabolite responsible for the glucose signaling and the molecular mechanism by which it induces ChREBP activity. Putative phosphorylation sites also reside near the nuclear localization signal (NLS) and may hint at other layers of regulation. We close by hypothesizing that ChREBP-dependent induction of genes involved in carbohydrate metabolism and lipogenesis is likely required for efficient, long-term storage of excess dietary carbohydrates. Inhibition of ChREBP activation would be expected to attenuate excess fat accumulation resulting from a high-carbohydrate diet and provide novel opportunities to address the health consequences stemming from obesity and diabetes.
Acknowledgments
We thank Dr. Steven L. McKnight and Dr. Bonnie C. Miller for helpful advice. R.K.B. is supported by a National Research Service Award from the National Institutes of Health.
Abbreviations
- ChRE
carbohydrate response element
- ChREBP
ChRE-binding protein
- LPK
L-type pyruvate kinase
- PKA
cAMP-dependent protein kinase A
- WT
wild type
- USF
upstream stimulatory factor
- GRBP
glucose response element binding protein
References
- 1.Neel J N. Nutr Rev. 1999;57:S2–S9. doi: 10.1111/j.1753-4887.1999.tb01782.x. [DOI] [PubMed] [Google Scholar]
- 2.Troiano R P, Flegal K M. Int J Obes Relat Metab Disord. 1999;23:S22–S27. doi: 10.1038/sj.ijo.0800855. [DOI] [PubMed] [Google Scholar]
- 3.Goodridge A G. Annu Rev Nutr. 1987;7:157–185. doi: 10.1146/annurev.nu.07.070187.001105. [DOI] [PubMed] [Google Scholar]
- 4.Granner D, Pilkis S. J Biol Chem. 1990;265:10173–10176. [PubMed] [Google Scholar]
- 5.Girard J, Ferré P, Foufelle F. Annu Rev Nutr. 1997;17:325–352. doi: 10.1146/annurev.nutr.17.1.325. [DOI] [PubMed] [Google Scholar]
- 6.Towle H C, Kaytor E N, Shih H-M. Annu Rev Nutr. 1997;17:405–433. doi: 10.1146/annurev.nutr.17.1.405. [DOI] [PubMed] [Google Scholar]
- 7.Doiron B, Cuif M-H, Kahn A, Diaz-Guerra M-J. J Biol Chem. 1994;269:10213–10216. [PubMed] [Google Scholar]
- 8.Liu Z, Thompson K S, Towle H C. J Biol Chem. 1993;268:12787–12795. [PubMed] [Google Scholar]
- 9.Bergot M O, Diaz-Guerra M J, Puzenat N, Raymondjean M, Kahn A. Nucleic Acids Res. 1992;20:1871–1877. doi: 10.1093/nar/20.8.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuif M H, Porteu A, Kahn A, Vaulont S. J Biol Chem. 1993;268:13769–13772. [PubMed] [Google Scholar]
- 11.Thompson K S, Towle H C. J Biol Chem. 1991;266:8679–8682. [PubMed] [Google Scholar]
- 12.Kaytor E N, Shih H-M, Towle H C. J Biol Chem. 1997;272:7525–7531. doi: 10.1074/jbc.272.11.7525. [DOI] [PubMed] [Google Scholar]
- 13.Hasegawa J, Osatomi K, Wu R F, Uyeda K. J Biol Chem. 1999;274:1100–1107. doi: 10.1074/jbc.274.2.1100. [DOI] [PubMed] [Google Scholar]
- 14.Yamada K, Tanaka T, Noguchi T. Biochem Biophys Res Commun. 1999;257:44–49. doi: 10.1006/bbrc.1999.0410. [DOI] [PubMed] [Google Scholar]
- 15.Koo S-H, Towle H C. J Biol Chem. 2000;275:5200–5207. doi: 10.1074/jbc.275.7.5200. [DOI] [PubMed] [Google Scholar]
- 16.Casazza J P, Veech R L. Biochem J. 1986;236:635–641. doi: 10.1042/bj2360635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francone O L, Griffaton G, Kalopissis A-D. Am J Physiol. 1992;263:E615–E623. doi: 10.1152/ajpendo.1992.263.4.E615. [DOI] [PubMed] [Google Scholar]
- 18.Hattori M, Tugores A, Veloz L, Karin M, Brenner D A. DNA Cell Biol. 1990;9:777–781. doi: 10.1089/dna.1990.9.777. [DOI] [PubMed] [Google Scholar]
- 19.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 20.Berry M N, Friend D S. J Cell Biol. 1969;43:506–520. doi: 10.1083/jcb.43.3.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K H, Brose K, Arnot D, Kidd T, Goodman C S, Henzel W, Tessier-Lavigne M. Cell. 1999;96:771–784. doi: 10.1016/s0092-8674(00)80588-7. [DOI] [PubMed] [Google Scholar]
- 22.de Luis O, Carmen Valero M, Pérez Jurado L A. Eur J Hum Genet. 2000;8:215–222. doi: 10.1038/sj.ejhg.5200435. [DOI] [PubMed] [Google Scholar]
- 23.Imamura K, Tanaka T. J Biochem. 1972;71:1043–1051. doi: 10.1093/oxfordjournals.jbchem.a129852. [DOI] [PubMed] [Google Scholar]
- 24.Noguchi T, Yamada K, Yamagata K, Takenaka M, Nakajima H, Imai E, Wang Z, Tanaka T. Biochem Biophys Res Commun. 1991;181:259–264. doi: 10.1016/s0006-291x(05)81411-1. [DOI] [PubMed] [Google Scholar]
- 25.Sirito M, Lin Q, Maity T, Sawadogo M. Nucleic Acids Res. 1994;22:427–433. doi: 10.1093/nar/22.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Viollet B, Kahn A, Raymondjean M. Mol Cell Biol. 1997;17:4208–4219. doi: 10.1128/mcb.17.8.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gourdon L, Lou D Q, Raymondjean M, Vasser-Cognet M, Kahn A. FEBS Lett. 1999;459:9–14. doi: 10.1016/s0014-5793(99)01203-x. [DOI] [PubMed] [Google Scholar]
- 28.Shih H M, Liu A, Towle H C. J Biol Chem. 1995;270:21991–21997. doi: 10.1074/jbc.270.37.21991. [DOI] [PubMed] [Google Scholar]
- 29.Foufelle F, Lepetit N, Bosc D, Delzenne N, Morin J, Raymondjean M, Ferre P. Biochem J. 1995;308:521–527. doi: 10.1042/bj3080521. [DOI] [PMC free article] [PubMed] [Google Scholar]