Abstract
La Piedad Michoacán Mexico Virus (LPMV) is a member of the Rubulavirus genus within the Paramyxoviridae family. LPMV is the etiologic agent of “blue eye disease”, causing a significant disease burden in swine in Mexico with long-term implications for the agricultural industry. This virus mainly affects piglets and is characterized by meningoencephalitis and respiratory distress. It also affects adult pigs, causing reduced fertility and abortions in females, and orchitis and epididymitis in males.
Viruses of the Paramyxoviridae family evade the innate immune response by targeting components of the interferon (IFN) signaling pathway. The V protein, expressed by most paramyxoviruses, is a well-characterized IFN signaling antagonist. Until now, there were no reports on the role of the LPMV-V protein in inhibiting the IFN response. In this study we demonstrate that LPMV-V protein antagonizes type I but not type II IFN signaling by binding STAT2, a component of the type I IFN cascade. Our results indicate that the last 18 amino acids of LPMV-V protein are required for binding to STAT2 in human and swine cells. While LPMV-V protein does not affect the protein levels of STAT1 or STAT2, it does prevent the IFN-induced phosphorylation and nuclear translocation of STAT1 and STAT2 thereby inhibiting cellular responses to IFN α/β
Keywords: (LPMV) La Piedad Michoacan Mexico Virus, LPMV-V, STAT2, Interferon-Signaling antagonist
1. Introduction
La Piedad Michoacán Mexico Virus (LPMV) is a member of the Rubulavirus genus within the Paramyxoviridae family, in the order of Mononegavirales. The virus (LPMV) was first discovered in swine during the early 1980s in central-west Mexico, where the majority of the nation’s pig farms are located (Escobar-Lopez et al., 2012; Moreno-Lopez et al., 1986; Stephan et al., 1988). This swine virus is the causative agent of blue eye disease (BED) that appears with a varied symptomatology depending on the age of the animals. The piglets exhibit neurological and pulmonary symptoms characterized by encephalitis, corneal opacity and/or pneumonia (Mendoza-Magana et al., 2007; Rivera-Benitez et al., 2013; Rodriguez-Ropon et al., 2003; Wiman et al., 1998). In adults, the disease is characterized by symptoms that affect the reproductive system. Sows exhibit increased oestrus, and an increased incidence of stillbirth and mummified fetuses (Hernandez et al., 1998; Hernandez-Jauregui et al., 2004). In boars, the symptomatology is characterized by low sperm quality, orchitis and epididymitis that can lead to permanent sterility (Cuevas-Romero et al., 2014; Ramirez-Mendoza et al., 1997; Solis et al., 2007). BED is among the top four diseases that afflict the Mexican swine industry. The reduction of reproductive performance and the mortality associated with LMPV infections in pigs cause significant economic losses (Escobar-Lopez et al., 2012).
LPMV is an enveloped virus with a single-stranded negative-sense RNA genome of approximately15 Kb. The genome contains 6 genes encoding at least 9 proteins: the nucleoprotein (NP), matrix (M), fusion (F), hemagglutinin-neuraminidase (HN), large protein (L) and phosphoprotein (P) (Berg et al., 1997; Berg et al., 1991; Cuevas-Romero et al., 2013; Linne et al., 1992; Reyes-Leyva et al., 1999; Sanchez-Betancourt et al., 2012; Sundqvist et al., 1990; Svenda et al., 1997; Zenteno-Cuevas et al., 2007). The P gene of the paramyxoviruses encode several proteins by using different initiation codons and by a unique mechanism involving RNA editing that generates alternate mRNAs by site-specific co-transcriptional insertion of non-templated nucleotides (Hausmann et al., 1999a; Hausmann et al., 1999b; Iseni et al., 2002; Kolakofsky et al., 2005; Thomas et al., 1988). In particular the P gene of LPMV possesses several open reading frames (ORFs). The use of alternative initiation codons from the P gene mRNA results in the generation of a small 126 long amino acid protein called C as well as of P/V/W proteins LPMV V protein is 249 amino acids in length and its first 168 amino acids are identical to those of P and W proteins. Co-transcriptional editing at position corresponding to amino acid 168 of V results in non template addition of one or two G residues and in mRNAs encoding W (174 a.a.) and P (404) proteins respectively. The C-terminal region of the LPMV V protein (LPMV-V) is 60% similar to mumps virus V protein, 56% similar to parainfluenza virus type 5 (PIV5) V protein (PIV5-V), 51% similar to human parainfluenza virus type 4 V protein (hPIV4–V) and 49% similar to human parainfluenza virus type 2 V protein (hPIV2-V) at the amino acid level (Berg et al., 1992; Sundqvist et al., 1992).
The interferon (IFN) system plays a key role in the innate immune defense against viral infections. In most cell types, the production of type I IFN (IFN-α/β) is initiated immediately after viral infection leading to the subsequent activation of the IFN-α/β signaling cascade Type I IFN signaling is activated by the binding of IFN to its receptors (IFNAR1 and IFNAR2) resulting in heterodimerization of the IFNAR receptors and the resulting activation of the associated Janus kinases, Tyk2 and Jak1(Colamonici et al., 1994; Colamonici et al., 1995) 13). The activated Janus kinases phosphorylate STAT2 (Nadeau et al., 1999; Yan et al., 1996) on tyrosine 690 (Greenlund et al., 1995) and STAT1 (Gupta et al., 1996; Qureshi et al., 1995) 49) on tyrosine 701 (Shuai et al., 1993). Phosphorylated STAT1 and STAT2 heterodimerize and associate with interferon regulatory factor 9 (IRF9) to form IFN-stimulated gene factor 3 (ISGF3) (Fu et al., 1990), which translocate to the nucleus and binds the IFN-stimulated response elements (ISREs) within interferon-stimulated gene (ISG) promoters thereby inducing the expression of more than 100 ISGs to establish an antiviral state that limits viral replication and dissemination (Childs et al., 2012; Garcia-Sastre and Biron, 2006; Garcia-Sastre et al., 1998; Laurent-Rolle et al., 2014; Manicassamy et al., 2010; Morrison et al., 2013; Motz et al., 2013; Rajsbaum et al., 2014; Randall and Goodbourn, 2008). Type II IFN (IFN-γ) is produced by activated immune cells (Billiau and Matthys, 2009; Meyer, 2009) and leads to the production of a different subset of ISGs via a distinct signaling pathway. Type II IFN (IFNγ) binds to different receptor complex also formed of two subunits IFN-γR1, and IFN-γR2. Type II IFN signaling activates STAT1, by phosphorylation, which homodimerizes to form the interferon gamma factor (GAF), which translocates to the nucleus and binds to DNA at γ-activated sequence (GAS) elements.
Most Paramyxoviruses analyzed so far like Nipah virus, Hendra virus (Rodriguez et al., 2002; Rodriguez et al., 2003), PIV5 (Didcock et al., 1999a; Didcock et al., 1999b; Precious et al., 2005a), hPIV2 (Parisien et al., 2001), mumps virus (Kubota et al., 2005), canine distemper virus (CDV) (Rothlisberger et al., 2010) with the only exception of hPIV4 (Nishio et al., 2005b), are able to subvert both type I and type II IFN-mediated antiviral responses by targeting their signaling cascades.
Since Paramyxovirus IFN antagonistic activity is mainly linked to the V proteins, we examined whether the V protein of LPMV also had the ability to antagonize IFN signaling. In this study, we show that LPMV V evades type I IFN signaling by binding STAT2 and preventing phosphorylation of STAT2 and STAT1. As a result, the STAT proteins are retained in the cytoplasm, preventing IFN-induced STAT activation and nuclear translocation, thereby inhibiting cellular responses to IFN α/β. We also show that the last 18 amino acids of LPMV-V are required for inhibiting IFN signaling.
2. Material and Methods
2.1. Cells
293T, HeLa, PK13 and PK15 cells were grown in Dulbecco’s modified Eagle medium (Invitrogen) supplemented with 10% fetal calf serum (FCS), penicillin, and streptomycin. All cells were kept at 37°C in the presence of 5% CO2.
2.2. Plasmids
La Piedad Michoacán Mexico Virus V open reading frame (ORF), was amplified by reverse transcription PCR of RNA extracted from LPMV-infected cells with MGG55 strain. Primers are available upon request. PCR amplicons were cloned into pCAGGS (Niwa et al., 1991) and from here on are referred to as pCAGGS-LPMV-V. A HA-tagged version of the same amplicon was constructed by placing a hemagglutinin (HA) tag sequence encoding the amino acids MYPYDVPDYA downstream of the V protein, from here on referred to as pCAGGS-LPMV-V-HA. In order to determine the regions of interaction between LPMV-V and STAT proteins, four HA-tagged expression vectors were generated lacking the C-terminal 18, 25, 50 and 75 amino acids and from here on are referred to as V-ΔC18-HA, V-ΔC25-HA, V-ΔC50-HA, V-ΔC75-HA respectively.
2.3. Reporter assays
293T cells (1.5×106) were transfected in 6-well plates with 0.5 μg of a construct having an ISRE54 promoter driving the expression of a firefly reporter gene (pISRE54-firefly-luciferase), 0.1 μg of a constitutively expressing renilla luciferase reporter construct (pCAGGS-ren-luc), and 3 μg of the expression plasmids. Twenty-four hours post-transfection, cells were washed and treated with 1000U/mL IFN beta 1a (PBL 11410-2). Sixteen hours post-IFN treatment, cells were harvested using reporter lysis buffer (Promega, cat no E2810) and analyzed for luciferase activities normalized with Renilla activity. For IFNγ-dependent gene expression, a reporter having 3 copies of the γ activated sequence driving the expression of firefly luciferase (GAS-Luc) (0.5 μg) was transfected with 0.1 μg of a constitutively expressing Renilla-luciferase reporter construct (pCAGGS-ren-luc), and the indicated amounts of the expression plasmids. Twenty-four hours post-transfection, cells were washed and treated with IFN-γ (5 ng/ml) (PBL). Sixteen hours post-IFN treatment, cells were harvested then lysed, and the luciferase activity was measured by applying a dual-luciferase reporter (DLR) assay system (Promega) according to the manufacturer’s recommendation. The luminescence signals of the firefly and the renilla luciferase were measured with a TD-20/20 Luminometer (Promega), and their ratio was called relative luciferase activity, with the ratio over the empty vector pCAGGS set to 1 and analyzed using a DLR assay (Promega). The assays were performed in triplicate and p-values were calculated by a two-tailed Student’s t-test for unpaired samples using the software GraphPad Prism (GraphPad Software, Inc.).
Alternatively 293T cells were co-transfected with 3 μg of LPMV-V-HA, Nipah-V-HA and PIV5-V-HA plasmids encoding the respectively viral V proteins, 250 ng of the IFN-inducible chloramphenicol acetyltransferase (CAT) reporter (ISG54-CAT) and 50 ng of a plasmid constitutively expressing the firefly luciferase protein. 24 hours post transfection cells were treated with 1000 U/ml of human IFN-β (PBL). 24 hours post treatment; cells were lysed and CAT and Luciferase activity were measured. A Phosphoimager was used to quantify CAT activity and the value was normalized to firefly luciferase activity. The fold induction of each sample was then calculated as the CAT activity of the IFN-treated sample normalized to the firefly luciferase value of that sample. That value was then divided by the normalized value of the untreated empty vector transfected cells.
2.4. NDV-GFP bioassay
PK13 cells were transfected with either the empty pCAGGS plasmid or plasmids encoding various viral proteins as detailed in specific experiments. Empty pCAGGS was used as negative control for IFN antagonism, where PIV5-V-HA protein was included as positive control. At 24 hours post-transfection, cells were treated with 1000 U/ml of human IFN β (PBL). Following 24 hours of IFN β treatment, cells were infected with NDV-GFP as described previously (Park et al., 2003). Fluorescence images were obtained at 14 hours post-infection. In parallel experiments, VERO cells were first treated with 1000 U/ml of human IFN β (PBL) for 20 hours, then the cells were β transfected with either the empty pCAGGS plasmid or LPMV-V-HA plasmid. 24 hours post transfection the cells were infected with NDV-GFP described previously (Park et al., 2003). Fluorescence images were taken at 14 hours post-infection.
2.5. Real-time PCR
293T cells where transfected with our experimental and control plasmids and 24 hours later the cells where washed and treated with 1000 U/ml IFN β or with IFN-γ (5 ng/ml) for an additional 12 hours. Total cellular RNA was then extracted using Trizol (Invitrogen) and cDNA was generated by reverse transcription using random hexamers and oligo dT primers with Superscript III reverse transcriptase (Invitrogen). Real-time PCRs were carried out using SYBR green supermix (BioRad). MxA, ISG15, IP10 and IRF1 relative gene expression was calculated using the comparative threshold cycle method, employing 18S rRNA and GAPDH as references.
2.6. Western blot
For preparation of cell extracts, 293T and PK15 cells were cultured in six-well plates and transfected with our experimental and control plasmids. At 48 hours post-transfection, the cells were treated (or mock treated) with 1000 U/ml IFN-β or IFN-γ (5 ng/ml) for 30 minutes at 37°C and then washed with cold PBS and incubated on ice and subsequently lysed with 250 μl of whole-cell extract buffer (50 mM Tris [pH 8.0], 280 mM NaCl, 0.5% IGEPAL, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol, 1 mM dithiothreitol [DTT]) supplemented with protease inhibitor cocktail (Complete; Boehringer Mannheim) and sodium vanadate (0.1 mM). Lysates were incubated on ice for 10 minutes and centrifuged for 10 minutes at 20,000 g at 4°C. Supernatants were analyzed on sodium dodecyl sulfate (SDS) 4–15% polyacrylamide gels. Proteins were then transferred to nitrocellulose membranes and were probed with antibodies against STAT1 (Upstate Biotechnology, no. 06-501), STAT2 (Santa Cruz, no. sc-476), phosphotyrosine 701-STAT1 (Cell Signaling Technologies, no. 7649), Anti-phospho-STAT2 (Tyr689) (Millipore no: 07-224), anti-actin (Sigma–Aldrich # A 5060) anti β tubulin (Cell Signaling #2146) and anti-HA epitope tag (Sigma-Aldrich # H3663), and visualized by chemiluminescence (NEN Life Sciences).
2.7. Co-immunoprecipitation
293T cells were seeded in 6-well plates, and the next day cells were transfected with LPMV-V-HA, Nipah-V-HA and PIV5-V-HA plasmids. At 48 hours postransfection, the cells were treated (or left untreated) with 1000 U/ml IFN-β for 30 minutes at 37°C and then washed with ice-cold phosphate-buffered saline (PBS) and subsequently lysed with 250 μl of whole-cell extract buffer (50 mM Tris [pH 8.0], 280 mM NaCl, 0.5% IGEPAL, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol, 1 mM dithiothreitol [DTT]) supplemented with protease inhibitor cocktail (Complete; Boehringer Mannheim) and sodium vanadate (0.1 mM). After a 20-minute centrifugation at 20000g at 4°C, aliquots for total protein analyses were separated, and the remaining supernatants were incubated for 3 hours with the Red Anti-HA affinity gel antibody (Sigma-Aldrich Cat# E6779) or without antibody. Alternatively the supernatants were incubated for 3 hours with the Anti-STAT2 Antibody followed by the addition of protein G-Sepharose beads at 4°C overnight. After a 1-minute centrifugation at 5000g at 4°C, the pellets were washed six times with whole-cell extract buffer and finally dissolved directly in Laemmli sample buffer for western blotting performed as described above.
2.8. Indirect Immunofluorescence
For indirect immunofluorescence experiments, HeLa cells were grown to 70 to 80% confluence, on cover slides in 6-well plates, and 24 hours later were transfected with empty vector, LPMV-V-HA or NipahV-V-HA expression plasmids using lipofectamine 2000 (Invitrogen) following the manufacturer’s recommendations. Before fixation, cells were treated for 30 minutes with 1000 U/ml of IFN β. At 48 hours after transfection, the cells were, washed with PBS and fixed with 500 μl of ice-cold methanol-acetone solution (1:1, V/V) for 15 minutes. Following PBS washes cells were permeabilyzed with 0.5% IGEPAL for 10 minutes at room temperature. After 3 washes with PBS, blocking of nonspecific binding sites was performed in PBG (1X PBS, 5% BSA, 0.2% fish gelatin) for 30 minutes at room temperature. The incubation with primary antibodies diluted in PBG (anti-STAT1 and anti-STAT2 at 1:100 dilution, and anti-HA at 1:500 dilution) was performed for 1 hour at 37°C. Then the slides were washed in PBS and incubated for 1 hour at room temperature with the secondary antibody diluted in PBG Alexa Fluor 555 (Invitrogen) conjugated to mouse immunoglobulin G was used to visualize HA-tagged V proteins, Alexa Fluor 488 (Invitrogen) conjugate to rabbit immunoglobulin G was used to visualize either STAT1 or STAT2. Nuclear chromatin staining was performed by incubation in blocking solution containing 0.5mg/ml 4′, 6-diamidino-2-phenylindole, DAPI (Sigma-Aldrich). Cells were washed and coverslips mounted using Prolong antifade reagent (Invitrogen). Images were captured using a Leica SP5-DM confocal microscope at the Microscopy Shared Research Facility at Icahn School of Medicine at Mount Sinai.
3. RESULTS
3.1 LPMV-V protein antagonizes IFN α/β but not IFN γ signaling pathway
The IFN antagonist function of the V proteins of several paramyxoviruses is well characterized. In order to evaluate the potential of LPMV-V protein-mediated inhibition of IFN α/β - and IFN γ-mediated signaling, we first analyzed the expression and cellular localization of HA-tagged LPMV-V in 293T and HeLa cells, respectively. Western blot analysis showed the expression and expected molecular mass of approximately 42 kDa of LPMV-V, which is closer in size to PIV5 V protein than NipahV-V protein (Fig 1A). Indirect immunofluorescence analysis revealed both nuclear and cytoplasmic localization of LPMV-V protein, and solely cytoplasmic localization of the control protein NipahV-V (Fig 1B).
Fig. 1.
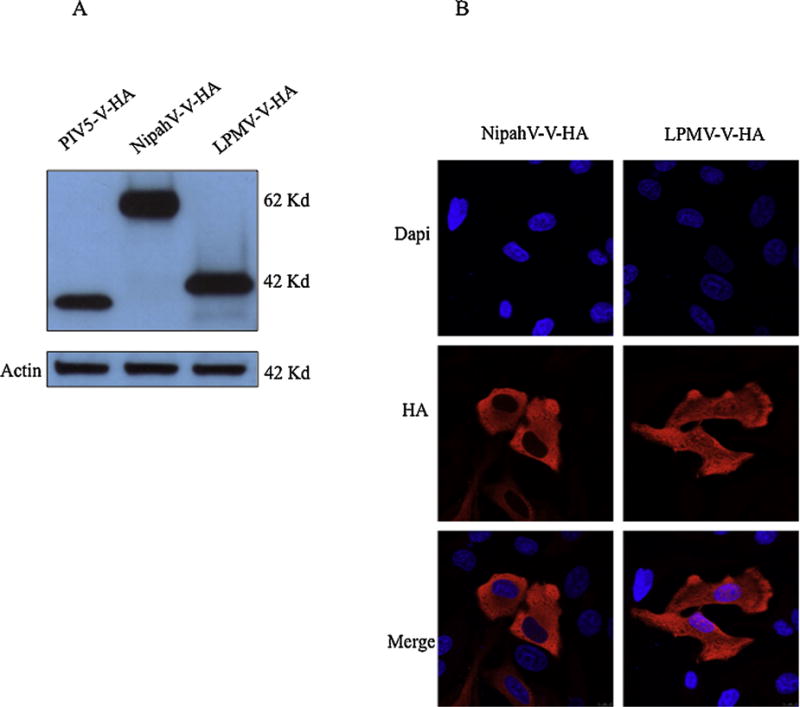
LPMV-V protein expression and cellular localization (A) 293T cells were transfected with expression vectors for HA-tagged PIV5-V, NipahV-V or LPMV-V, and lysates were separated by SDS-PAGE (12% polyacrylamide). V proteins were visualized by immunoblotting with antibodies to the HA epitope tag. Western blot analysis shows LPMV-V-HA expression protein, with the expected molecular mass about 42 kDa. (B) HeLa cells were transfected with expression vectors for HA-tagged NipahV-V or LPMV-V. Cells were fixed, permeabilized, and stained with antibodies to HA, as described in materials and methods. Nuclear DNA was stained with DAPI. Images were obtained using confocal microscopy.
Next we examined the effects of LPMV-V protein on type I IFN- or type II IFN-mediated gene expression using luciferase reporter genes under the control of IFN-responsive promoters. 293T cells were co-transfected with pCAGGS plasmids encoding NipahV-V, PIV5-V and LPMV-V proteins, a reporter plasmid pISRE54-CAT), a reporter plasmid pGAS-firefly-luciferase and a plasmid driving the constitutive expression of renilla-luciferase used as internal control. Twenty-four hours post transfection the cells were treated with 1000 U/ml of IFNβor IFN–γ and 24 hours later cells were lysed and analyzed for luciferase activity. Our results show that cells transfected with empty plasmid and treated with IFN showed a significant increase in luciferase expression, as expected. The cells expressing LPMV-V, showed significantly reduced luciferase expression driven by the ISRE promoter, which was comparable to that seen with the positive controls NipahV-V and PIV5-V. In contrast, LPMV-V did not reduce luciferase gene expression driven by the GAS promoter, while both NipahV-V and PIV5-V, as expected, were able to do so. (Fig 2A, 2B). These results indicate that LPMV-V protein specifically evades type I but not type II IFN signaling.
Fig. 2.
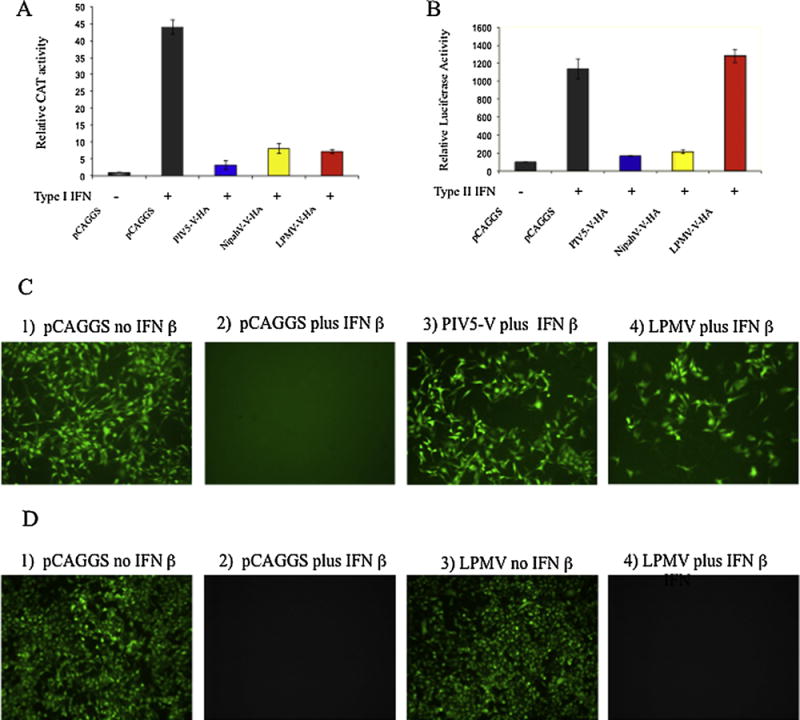
LPMV V inhibits Type I IFN signaling but not Type II IFN signaling. (A) 293T Cells were co-transfected with pCAGGS plasmids encoding NipahV-V-HA, PIV5-V-HA and LPMV-V-HA proteins, plus an empty vector (pCAGGS), a plasmid encoding an ISRE-54-CAT reporter and with a plasmid constitutively expressing firefly-luciferase used as control. At 24 hours post transfection, cells were treated with IFN-β (1000 U/ml) for 24 hours prior to assaying for CAT activity. Induction of CAT activity was normalized to firefly luciferase activity. (B) 293T Cells were co-transfected with pCAGGS plasmids encoding NipahV-V, PIV5-V and LPMV-V proteins, plus an empty vector (pCAGGS), GAS-luciferase plasmid and a plasmid driving the constitutive expression of renilla-luciferase used as internal control. 24 hours post transfection the cells were treated with IFN-γ (5 ng/ml) (PBL) and 24 hours later cells were lysed and analyzed for GAS-luciferase activity normalized to renilla luciferase activity.
(C) PK13 cells were transfected with either the empty pCAGGS plasmid or plasmids encoding PIV5-V-HA and LPMV-V-HA viral proteins. Empty pCAGGS was used as negative control for IFN antagonism, whereas the PIV5-V-HA protein was included as positive control. At 24 hours post-transfection, cells were treated with 1000 U/ml of human IFN β (PBL). Following 24 hours of IFN β treatment, cells were infected with NDV-GFP as described previously (Park et al., 2003). Fluorescence images were obtained at 14 hours post-infection. (D). VERO cells were first treated with 1000 U/ml of human IFN β (PBL) for 20 hours, than were transfected with either the empty pCAGGS plasmid or LPMV-V-HA plasmid. 24 hours post transfection the cells were infected with NDV-GFP as described previously (Park et al., 2003). Fluorescence images were taken at 14 hours post-infection.
To examine the biological relevance of LPMV-V protein in IFN antagonism, we analyzed the effect of LPMV-V on replication of an IFN-sensitive Newcastle disease virus that expresses GFP (NDV-GFP) in the presence of type I IFN (Park et al., 2003). PK13 cells were transfected with plasmids encoding LMPV-V and PIV5-V. Twenty-four hours post transfection, cells were either mock treated or treated overnight with 1000 U/mL of type I IFN. The following day the cells were infected with NDV-GFP. As expected, in cells transfected with empty plasmid and treated with IFN-β, NDV was unable to replicate as evidenced by lack of GFP expression (Fig 2C panels 1 and 2). However, in cells expressing LPMV-V, NDV-GFP replication was enhanced, similar to both of the positive control PIV5-V confirming that LPMV-V blocks type I IFN mediated antiviral activity (Fig. 2C panels 3 and 4). However, when cells were transfected with LMPV-V-HA expression plasmid 20 hours after type I IFN treatment, LMPV-V-HA was unable to rescue NDV-GFP infectivity, indicating that while LMPV-V protein inhibits type I IFN signaling, LMPV-V protein does not inhibit an already establish type I IFN-induced antiviral state (Fig. 2D).
3.2. LPMV-V protein dampens the induction of Type I interferon-stimulated genes ISG15 and MXA
To determine if LPMV-V inhibits ISG expression at the RNA level, quantitative RT-PCR was used to measure ISG15 and MXA gene expression in 293T cells that had been stimulated with IFN β for 12 hours. Results showed that the cells expressing LPMV-V protein had 25- and 325-fold lower ISG15 and MXA transcript levels, respectively, than cells transfected with an empty plasmid, comparable to the reduction showed by the control proteins NipahV-V and PIV5-V (Fig. 3A and 3B).
Fig 3.
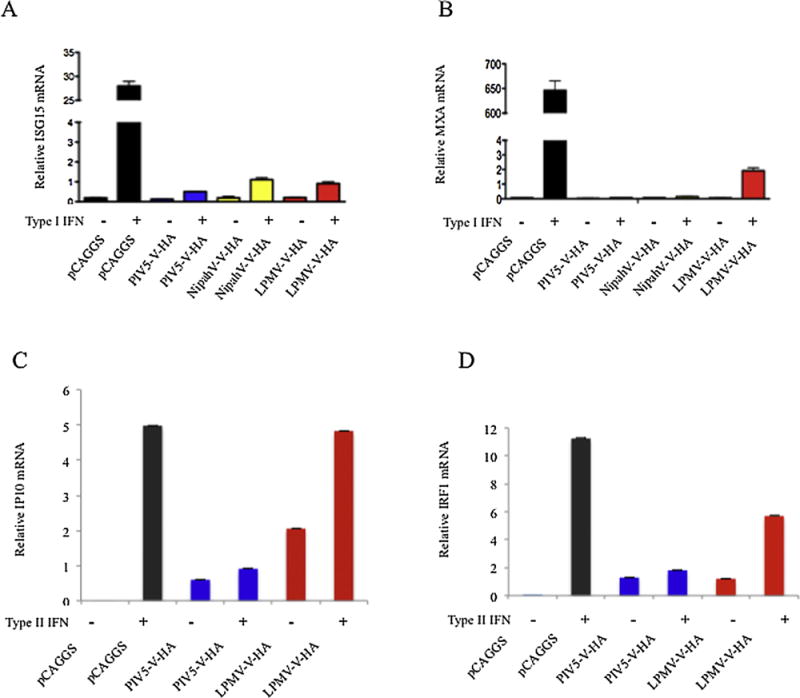
(A and B). 293T cells were transfected with our experimental and control plasmids and 24 hours later the cells were washed and treated with IFNβ at 1000 U/ml for an additional 12 h. Total cellular RNA was then extracted and cDNA was generated by reverse transcription. Real-time PCRs were carried out using SYBR green supermix. MxA hand ISG15 relative gene expression was calculated using the comparative threshold cycle method, employing 18S rRNA and GAPDH as references.
(C and D). 293T cells were transfected with our experimental and control plasmids and 24 hours later the cells were washed and treated with IFN-γ (5 ng/ml) for 12 hours. Total cellular RNA was then extracted and cDNA was generated by reverse transcription. Real-time PCRs were carried out using SYBR green supermix. IP10 and IRF1 relative gene expression was calculated using the comparative threshold cycle method, employing 18S rRNA and GAPDH as references
3.3. LPMV-V protein does not reduce the induction of Type II interferon-stimulated genes IP10, IRF1
A quantitative RT-PCR was also used to measure IP10 and IRF1 gene expression in 293T cells that had been stimulated with IFN γ for 12 hours. Results showed that expression of LMPV-V had no effect on IP10 and RF1 transcript levels (Fig 3C and 3D).
3.4. LPMV-V reduces Type I IFN-dependent phosphorylation of STAT2 and STAT1 but not Type II IFN-dependent phosphorylation of STAT1
STAT1 and STAT2 are the targets of several Paramyxovirus V proteins (Ramachandran and Horvath, 2009). To determine the mechanism of LPMV-V protein inhibition of IFN signaling, we analyzed the effect of LPMV-V protein expression on the total levels of cellular STAT1 and STAT2 and the type I and type II IFN-induced phosphorylation of STAT1 and STAT2. Western blot analysis showed that the total STAT1 and STAT2 levels remained constant in cells expressing LPMV-V, comparable to cells expressing an empty plasmid. However, expression of PIV5-V protein resulted in a loss of total STAT1, which is consistent with published reports (Didcock et al., 1999b) (Fig. 4A and 4B). Examination of LPMV-V protein effect on the type I IFN-induced phosphorylation of STAT1 and STAT2 by western blot analysis showed that 293T cells expressing LPMV-V protein, similarly to those expressing NipahV-V and PIV5-V proteins, had a reduced level of IFN-induced phosphorylation of STAT1-Y701 and STAT2-Y690. The transfection efficiency (close to 90%) in this case could explain the residual, phosho STAT1 and STAT2 activity in cells that express LPMV-V, NipahV-V, and PIV5-V proteins. This is in contrast to what was observed in cells transfected with an empty vector (Fig. 4 A). The results indicate that LPMV protein had no effect on the total cellular levels of STAT1 and STAT2 but instead inhibited the type I IFN-activated phosphorylation of STAT1 and STAT2. Different results were obtained when we analyzed the role of LPMV-V protein on the type II IFN-induced phosphorylation of STAT1. Our data suggest that LPMV-V protein still does not inhibit efficiently the induction the STAT1 phosphorylation under gamma interferon stimulation (Fig 4B).
Fig. 4.
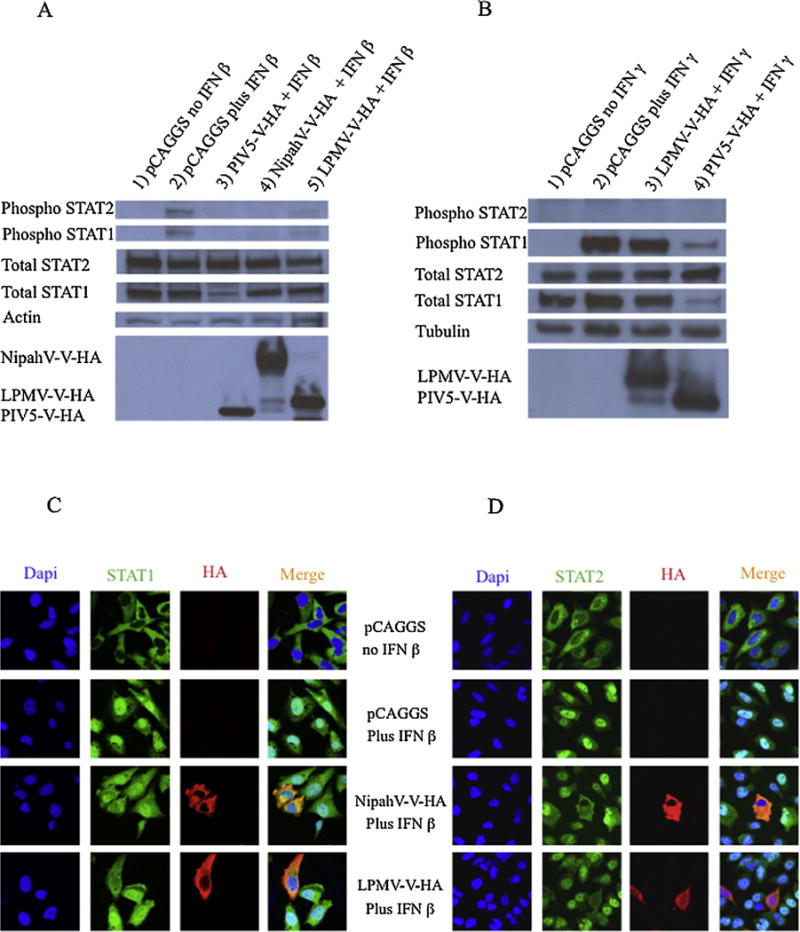
LPMV-V protein inhibits Type I IFN induced phosphorylation of STAT1 and STAT2 and prevents STAT1 and STAT2 nuclear translocation. (A) 293T cells were transfected with empty vector, LPMV-V-HA, NipahV-V-HA or PIV5-V-HA expression plasmids. Cells were treated for 30 min l with 1,000 U/ml of IFN β prior to lysis. Lysates were separated by SDS-PAGE (12 % polyacrylamide), transferred to nitrocellulose, and visualized by immunoblotting with antisera to STAT1 STAT2 phosphotyrosine 701-STAT1 phosphotyrosine 690-STAT2, anti-actin and anti-HA epitope tag as describe in Materials and Methods.
(B) 293T cells were transfected with empty vector, LPMV-V-HA, or PIV5-V-HA expression plasmids. Cells were treated with 1,000 U/ml of IFN β per 30 min prior to lysis. Lysates were separated by SDS-PAGE (12 % polyacrylamide), transferred to nitrocellulose, and visualized by immunoblotting with antisera to STAT1 STAT2 phosphotyrosine 701-STAT1, phosphotyrosine 690-STAT2, anti-tubulin and anti-HA epitope tag as describe in Materials and Methods.
(C) HeLa cells were transfected with empty vector, LPMV-V-HA or NipahV-V-HA expression plasmids. Before fixation, cells were treated for 30 minutes with 1000 U/ml of IFN β. Cells were fixed, permeabilized, and stained with antibodies against STAT1 and HA as described in Materials and Methods.
(D) HeLa cells were transfected with empty vector, LPMV-V-HA or NipahV-V-HA expression plasmids. Before fixation, cells were treated for 30 minutes with 1000 U/ml of IFN β. Cells were fixed, permeabilized, and stained with antibodies to STAT2 and HA, as described in Materials and Methods. Nuclear DNA was stained with DAPI. Images were obtained using confocal microscopy.
3.5. LPMV-V protein prevents Type I IFN-induced STAT1 and STAT2 nuclear translocation
Since we had observed that the LPMV-V protein did not affect the total level of STAT1 and STAT2 but reduced the type I IFN-induced phosphorylation of STAT1 and STAT2, we hypothesized that the STATs protein would be retained in the cytoplasm in the presence of LPMV-V protein. Using confocal laser scanning microscopy, we showed that in cells transfected with an empty plasmid then treated with type I IFN, STAT1 and STAT2 translocate to the nucleus. However, the IFN-induced translocation of STAT1 and STAT2 was inhibited in cells expressing LPMV-V protein similar to the positive control NipahV-V protein (Fig 4C and Fig 4D). Thus LPMV-V prevents the type I IFN-induced phosphorylation and nuclear translocation of STAT1 and STAT2, thus preventing formation of the ISGF3 complex and the subsequent binding at the ISRE of ISGs.
3.6. LPMV-V protein binds STAT2 protein
Since the V protein of several paramyxoviruses binds either STAT1 and STAT2 or both, we examined whether LPMV-V protein has the ability to bind STAT1 and STAT2 using co-immunoprecipitation assays. 293T cells were transfected with LPMV-V HA, NipahV-V-HA, PIV5-V HA plasmids and empty vector HA tagged. Cell extracts were immunoprecipitated taking advantage of the HA epitope, and immune complexes were processed for immunoblotting with anti-STAT1 and anti-STAT2 antibodies. Only STAT2 protein was found in the LPMV-V immune complexes, whereas NipahV-V protein, as predicted, pulled down both STAT1 and STAT2 proteins as have been previously reported (Fig 5A).
Fig. 5.
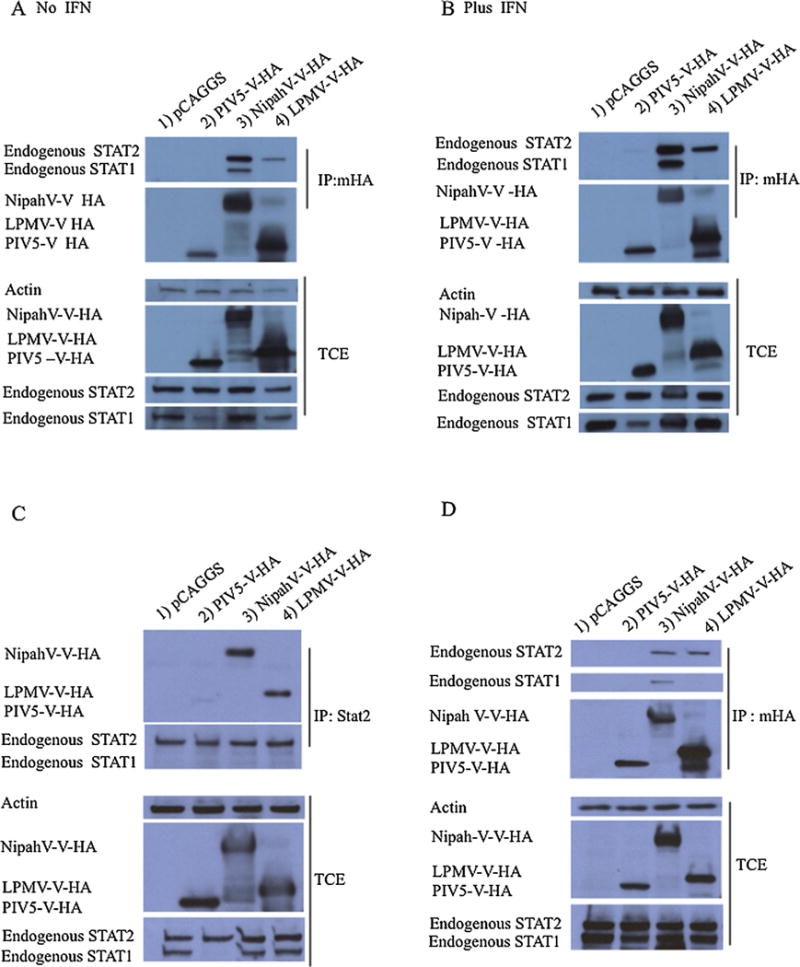
LPMV-V protein interacts with human and porcine STAT2. (A and B) LPMV-V-HA, NipahV-V-Ha Ha, PIV5-V-Ha plasmids and empty vector were transfected in 293T cells. At 28 hours postransfection, the cells were treated (or left untreated) with 1000 U/ml IFN-β for 24 hours at 37°C. 48 hours after transfection cells were lysate. Aliquots for total protein analyses were separated, and the remaining supernatants were incubated for 3 hours with the Red Anti-HA affinity gel antibody or without antibody. The pellets were washed six times with whole-cell extract buffer and finally dissolved directly in Laemmli sample buffer. The samples were analyzed on sodium dodecyl sulfate (SDS) 4–15% polyacrylamide gels. Proteins were then transferred to nitrocellulose membranes and were probed with antibodies against STAT1, STAT2, HA and Actin. (C) Reciprocal co-immunoprecipitation in 293t cells using an anti-STAT2 antibody was performed. (D) Co-immunoprecipitation assay was performed in porcine cells PK-15.
To rule out the possibility that low levels of STAT1 was responsible for the phenotype of specific binding with STAT2, we stimulated cells with IFN for 24 hours to increase the levels of STAT1, which is an ISG. We then performed a co-immunoprecipitation assay to evaluate whether LPMV-V protein would be able to precipitate STAT1 protein when higher levels of STAT1 are present. The result was similar to what we observed performing the co-immunoprecipitation in the absence of IFN β no binding with STAT1 was detected (Fig 5B). We then proceeded to confirm our results by performing a reciprocal co-immunoprecipitation. 293T cells were mock transfected or transfected with plasmids of interest. The cell extracts were immunoprecipitated with STAT2 antibody, and the immune complexes were probed for with anti-HA antibody. The results confirmed that STAT2 binds LPMV-V protein (Fig 5 C). Since LPMV affects pigs, we also examined whether LPMV V protein binds porcine STAT2. Using co-immunoprecipitation assays in PK13 cells, a cell line originated from swine, we show that LPMV-V protein binds porcine STAT2 and not STAT1 (Fig 5D).
In order to determine the region of LPMV-V protein that is required for its interaction with STAT2, we decided to focus our attention on the C-terminal region that has a main role for IFN antagonism in the Paramyxoviridae family (Ramachandran and Horvath, 2009). Four HA-tagged expression vectors were engineered (Fig. 6A). One construct with a deletion of the last 18 amino acids (V-ΔC18-HA), a second construct with a deletion of the last 25 amino acids (V-ΔC25-HA), a third construct with a deletion of the last 50 amino acids (V-ΔC50-HA) and a fourth construct with a deletion of the last 75 amino acids (V-ΔC75-HA) were engineered and tested in a co-immunoprecipitation assay in PK13 cells as in Figure 6 panel B. The construct that had a deletion of the last 18 amino acids did not bind STAT2 protein indicating that the C-terminus of LPMV-V protein is critical for its binding to STAT2 (Fig 6 B). Next we analyzed the ability of the mutant proteins to inhibit the type I IFN signaling cascade using a reporter gene assay. Our results show that cells expressing the LPMV-V protein lacking the last 18 amino acids were unable to inhibit IFN signaling similar to mock-transfected cells that were treated with IFN (Fig 6C). These results are consistent with those obtained in the co-immunoprecipitation experiment with the LPMV-V proteins deletion mutant, confirming that the last 18 amino acids are critical for the binding of LPMV-V protein to STAT2 and for LPMV V protein type I IFN antagonist activity.
Fig. 6.
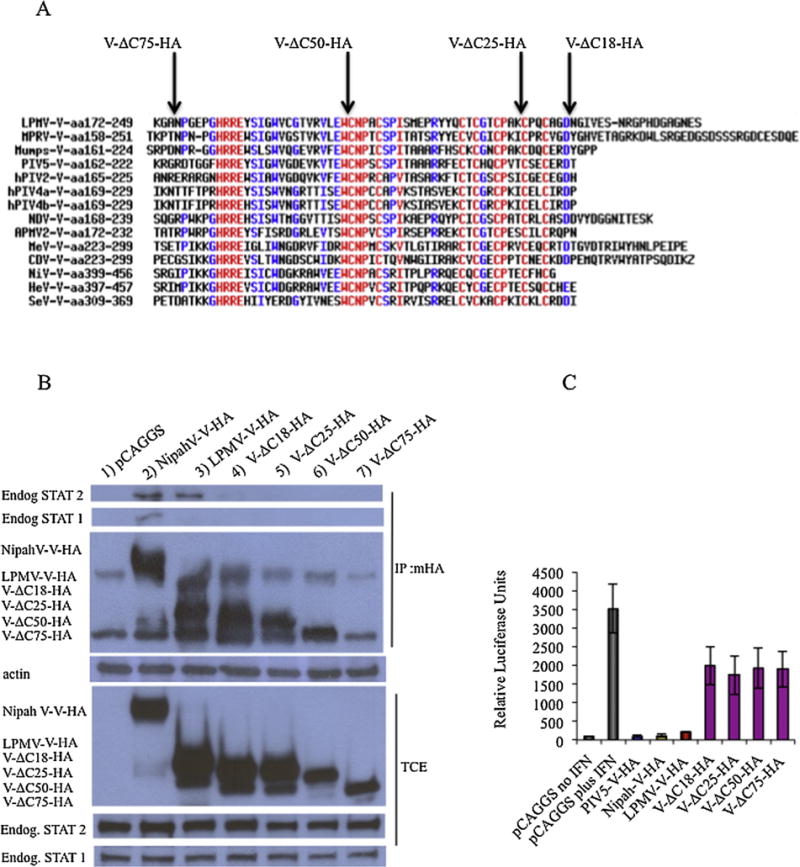
The last 18 amino acids of LPMV-V are necessary for binding STAT2. (A) Alignment of V proteins from different virus in the Paramyxovirus family and schematic representation LPMV-V-HA C-Terminal deletions mutants (V-ΔC18-HA, V-ΔC25-HA, V-ΔC50-HA, V-ΔC75-HA). (B) Co-immunoprecipitation assay of 293T cells transfected with LPMV-V HA deletions mutants (V-ΔC18-HA, V-ΔC25-HA, V-ΔC50-HA, V-ΔC75-HA) plasmids and empty vector were transfected in 293T cells. At 48 hours postransfection, the cells were lysate. Aliquots for total protein analyses were separated, and the remaining supernatants were incubated for 3 hours with the Red Anti-HA affinity gel antibody or without antibody. The pellets were washed six times with whole-cell extract buffer and dissolved directly in Laemmli sample buffer. The samples were analyzed on sodium dodecyl sulfate (SDS) 4–15% polyacrylamide gels. Proteins were then transferred to nitrocellulose membranes and were probed with antibodies against STAT1, STAT2, HA and Actin.
(C) 293T cells were co-transfected with pCAGGS plasmids encoding NipahV-V, PIV5-V and LPMV-V-Ha, V-ΔC18-HA, V-ΔC25-HA, V-ΔC50-HA, V-ΔC75-HA proteins, a reporter plasmid pISRE54-firefly-luciferase and a plasmid driving the constitutive expression of renilla-luciferase used as internal control. 24 hours post transfection the cells were treated with 1000 U/ml of IFNβ and 24 hours later cells were lysed and analyzed for ISRE54 luciferase activity, normalized with renilla luciferase activity.
4. Discussion
The ability of paramyxoviruses to subvert the IFN response is well conserved, with the only exception being HuPIV4 (Goodbourn and Randall, 2009; Nishio et al., 2005b; Ramachandran and Horvath, 2009). This characteristic is due mainly to the V proteins. In the Henipavirus genus, the Nipah virus and Hendra virus V proteins, for example, inhibit IFN responses by forming high-molecular-weight complexes with both STAT1 and STAT2 and preventing STATs phosphorylation and translocation (Rodriguez et al., 2002; Rodriguez et al., 2003). The V protein of measles virus, of the Morbillivirus genus, blocks IFN-induced STAT1 and STAT2 nuclear import without degrading STATs and preventing STATs phosphorylation (Takeuchi et al., 2003). STAT polyubiquitination and proteasome-dependent degradation is the mechanism by which the V proteins of Rubulavirus members, like human parainfluenza virus 5, human parainfluenza virus 2 and mumps virus inhibit IFN signaling (Andrejeva et al., 2002; Didcock et al., 1999b; Kubota et al., 2005; Nishio et al., 2002; Nishio et al., 2005a; Parisien et al., 2001; Parisien et al., 2002; Ulane and Horvath, 2002) with the exception of Maupera virus that blocks IFN signaling without STATs degradation (Hagmaier et al., 2007).
In this study we show that LPMV-V protein evades type I interferon signaling but not type II (Fig 2A and 2B). A drastic reduction of ISG15 and MXA transcript levels was found in cells with LPMV-V protein expression stimulated with type I IFN, compared to the mock-transfected cells (Fig 3a and 3B) which is consistent with published reports (Flores-Ocelotl Mdel et al., 2011). In contrast, cells expressing LPMV-V protein, type II IFN stimulation does not reduce IP10 and IRF1 mRNA levels (Fig 3C and 3D). The biological impact of LPMV-V protein in type I IFN antagonism was confirmed by its capacity to rescue NDV-GFP replication in the presence of IFN (Fig 2C panel 4). We show that LPMV-V protein does not affect the total level of STAT1 and STAT2, but inhibits the type I IFN-induced phosphorylation and nuclear translocation of both STAT1 and STAT2. However, LMPV-V does not inhibit the type II IFN-induced phosphorylation of STAT1 (Fig 4A, 4B, 4C, and 4D). Furthermore, we show that LPMV-V protein binds selectively to STAT2 in human and porcine cells (Fig 5A, 5B, 5C and 5D) and that the last 18 amino acids of LPMV V are required for binding STAT2. (Fig 6B and 6C)
Taken together these data show that the LPMV-V protein antagonizes the type I IFN response but not the type II without targeting STAT1 and STAT2 proteins for degradation. This contrasts with V proteins from other members of the Rubulavirus genus, such as PIV5, mumps or HuPI2, which antagonize type I IFN signaling by inducing degradation of the STAT1 and STAT2 proteins (Didcock et al., 1999a; Didcock et al., 1999b; Kubota et al., 2005; Parisien et al., 2001; Parisien et al., 2002; Precious et al., 2005a; Precious et al., 2005b; Ulane and Horvath, 2002). LPMV-V protein IFN evasion strategy partially resembles that of Henipavirus V protein, with its selective binding with STAT2 protein, though not with STAT1, and with the inhibition of STAT phosphorylation and subsequent nuclear translocation (Rodriguez et al., 2002; Rodriguez et al., 2003). However, LPMV-V protein does not affect the type II interferon signaling pathway despite its inhibition of type I interferon-mediated STAT1 phosphorylation. The IFN signaling antagonism strategy used by the V protein of measles, a Morbillivirus genus member, is most similar to that used by LPMV-V protein. In fact measles V protein inhibits type I IFN but not type II by inhibiting type I IFN-mediated STAT1 and STAT2 phosphorylation. (Takeuchi et al., 2003). These data suggest that by targeting STAT2, LPMV-V protein inhibits both STAT2 and STAT1 phosphorylation in response to type I IFN signaling, resulting in inhibition of STATs protein nuclear translocation. These data are consistent with what is described in the literature on the primary role that STAT2 protein has in the type I signaling activation (Leung et al., 1995; Li et al., 1997; Qureshi et al., 1996). Similar to the strategy used by LPMV-V protein to evade the IFN signaling cascade in the Rubulavirus genus, is the one adopted by Maupera virus V protein. Both inhibit IFN signaling without degrading STATs. While Maupera virus V protein inhibits IFN signaling by binding both STAT1 and STAT2 and preventing their nuclear translocation, without affecting their phosphorylation (Hagmaier et al., 2007), LPMV-V protein antagonizes type I IFN signaling cascade by binding specifically to STAT2, preventing STATs nuclear translocation and affecting their phosphorylation.
Across the Paramyxovirus family, V proteins evade the IFN signaling pathway using a variety of strategies (Andrejeva et al., 2004; Didcock et al., 1999a; Didcock et al., 1999b; Kubota et al., 2005; Motz et al., 2013; Nishio et al., 2002; Nishio et al., 2005a; Parisien et al., 2001; Parisien et al., 2002; Precious et al., 2005b; Rodriguez et al., 2002; Rodriguez et al., 2003). Our study has found that LPMV-V protein’s IFN signaling evasion strategy differs from the proteasome-mediated degradation strategy that is normally used by V proteins from members of its genus, Rubulavirus. This is despite the fact that the C-terminal domain of LPMV-V protein is similar to that of the other V proteins in the Rubulavirus genus and that it has eight cysteine residues, of which seven are conserved in all V proteins (Berg et al., 1992) (Fig. 5A). The similar C-terminus region of the LPMV-V protein with the C-terminus of other paramyxovirus V proteins is likely to be related to their essential role in interacting with MDA-5, (Davis et al., 2014; Motz et al., 2013; Rodriguez and Horvath, 2014) however, the last 21 amino acids of LPMV-V protein is unique, and this could have a key role in the IFN signaling evasion mediated by STAT2 inactivation. This is likely the case as we found that the last 18 amino acids of LPMV-V are critical for STAT2 binding and type I IFN signaling inhibition. Although our results still need to be confirmed in the context of LPMV infection, this information could be of use in the future when designing LPMV vaccine candidates or antiviral therapeutics against LPMV.
Highlights.
La Piedad Michoacán Mexico Virus (LPMV) V protein antagonizes type I but not type II IFN signaling.
Type I IFN interferon-signaling inhibition is due to the binding with LPMV-V protein with STAT2 protein, a fundamental component of type I IFN cascade.
LPMV-V protein does prevent the IFN-induced phosphorylation and nuclear translocation of STAT1 and STAT2 thereby inhibiting cellular responses to IFN α/β.
The last 18 amino acids are necessary for binding to STAT2 in human and swine origin cells
Acknowledgments
These studies were partly supported by NIAID grant U19AI083025 (to AG-S) and by a National Institute of Health fellowship, (to M.L.-R.). Confocal laser scanning microscopy was performed at ISMMS-Microscopy Shared Resource facility.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(49):17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrejeva J, Young DF, Goodbourn S, Randall RE. Degradation of STAT1 and STAT2 by the V proteins of simian virus 5 and human parainfluenza virus type 2, respectively: consequences for virus replication in the presence of alpha/beta and gamma interferons. Journal of virology. 2002;76(5):2159–2167. doi: 10.1128/jvi.76.5.2159-2167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg M, Bergvall AC, Svenda M, Sundqvist A, Moreno-Lopez J, Linne T. Analysis of the fusion protein gene of the porcine rubulavirus LPMV: comparative analysis of paramyxovirus F proteins. Virus genes. 1997;14(1):55–61. doi: 10.1023/a:1007987407250. [DOI] [PubMed] [Google Scholar]
- Berg M, Hjertner B, Moreno-Lopez J, Linne T. The P gene of the porcine paramyxovirus LPMV encodes three possible polypeptides P, V and C: the P protein mRNA is edited. J Gen Virol. 1992;73(Pt 5):1195–1200. doi: 10.1099/0022-1317-73-5-1195. [DOI] [PubMed] [Google Scholar]
- Berg M, Sundqvist A, Moreno-Lopez J, Linne T. Identification of the porcine paramyxovirus LPMV matrix protein gene: comparative sequence analysis with other paramyxoviruses. J Gen Virol. 1991;72(Pt 5):1045–1050. doi: 10.1099/0022-1317-72-5-1045. [DOI] [PubMed] [Google Scholar]
- Billiau A, Matthys P. Interferon-gamma: a historical perspective. Cytokine & growth factor reviews. 2009;20(2):97–113. doi: 10.1016/j.cytogfr.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Childs K, Randall R, Goodbourn S. Paramyxovirus V proteins interact with the RNA Helicase LGP2 to inhibit RIG-I-dependent interferon induction. Journal of virology. 2012;86(7):3411–3421. doi: 10.1128/JVI.06405-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colamonici O, Yan H, Domanski P, Handa R, Smalley D, Mullersman J, Witte M, Krishnan K, Krolewski J. Direct binding to and tyrosine phosphorylation of the alpha subunit of the type I interferon receptor by p135tyk2 tyrosine kinase. Mol Cell Biol. 1994;14(12):8133–8142. doi: 10.1128/mcb.14.12.8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colamonici OR, Platanias LC, Domanski P, Handa R, Gilmour KC, Diaz MO, Reich N, Pitha-Rowe P. Transmembrane signaling by the alpha subunit of the type I interferon receptor is essential for activation of the JAK kinases and the transcriptional factor ISGF3. The Journal of biological chemistry. 1995;270(14):8188–8193. doi: 10.1074/jbc.270.14.8188. [DOI] [PubMed] [Google Scholar]
- Cuevas-Romero JS, Hernandez-Baumgarten E, Kennedy S, Hernandez-Jauregui P, Berg M, Moreno-Lopez J. Long-term RNA persistence of Porcine rubulavirus (PorPV-LPMV) after an outbreak of a natural infection: The detection of viral mRNA in sentinel pigs suggests viral transmission. Virus research. 2014 doi: 10.1016/j.virusres.2014.04.012. [DOI] [PubMed] [Google Scholar]
- Cuevas-Romero S, Blomstrom AL, Alvarado A, Hernandez-Jauregui P, Rivera-Benitez F, Ramirez-Mendoza H, Berg M. Development of a real-time RT-PCR method for detection of porcine rubulavirus (PoRV-LPMV) Journal of virological methods. 2013;189(1):1–6. doi: 10.1016/j.jviromet.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis ME, Wang MK, Rennick LJ, Full F, Gableske S, Mesman AW, Gringhuis SI, Geijtenbeek TB, Duprex WP, Gack MU. Antagonism of the phosphatase PP1 by the measles virus V protein is required for innate immune escape of MDA5. Cell host & microbe. 2014;16(1):19–30. doi: 10.1016/j.chom.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didcock L, Young DF, Goodbourn S, Randall RE. Sendai virus and simian virus 5 block activation of interferon-responsive genes: importance for virus pathogenesis. Journal of virology. 1999a;73(4):3125–3133. doi: 10.1128/jvi.73.4.3125-3133.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didcock L, Young DF, Goodbourn S, Randall RE. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. Journal of virology. 1999b;73(12):9928–9933. doi: 10.1128/jvi.73.12.9928-9933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Lopez AC, Rivera-Benitez JF, Castillo-Juarez H, Ramirez-Mendoza H, Trujillo-Ortega ME, Sanchez-Betancourt JI. Identification of Antigenic Variants of the Porcine Rubulavirus in Sera of Field Swine and their Seroprevalence. Transbound Emerg Dis. 2012;59(5):416–420. doi: 10.1111/j.1865-1682.2011.01286.x. [DOI] [PubMed] [Google Scholar]
- Flores-Ocelotl Mdel R, Rosas-Murrieta NH, Vallejo-Ruiz V, Reyes-Leyva J, Herrera-Camacho I, Santos-Lopez G. Transcription of interferon stimulated genes in response to Porcine rubulavirus infection in vitro. Brazilian journal of microbiology: [publication of the Brazilian Society for Microbiology] 2011;42(3):1167–1175. doi: 10.1590/S1517-838220110003000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XY, Kessler DS, Veals SA, Levy DE, Darnell JE., Jr ISGF3, the transcriptional activator induced by interferon alpha, consists of multiple interacting polypeptide chains. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(21):8555–8559. doi: 10.1073/pnas.87.21.8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sastre A, Biron CA. Type 1 interferons and the virus-host relationship: a lesson in detente. Science. 2006;312(5775):879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252(2):324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- Goodbourn S, Randall RE. The regulation of type I interferon production by paramyxoviruses. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2009;29(9):539–547. doi: 10.1089/jir.2009.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenlund AC, Morales MO, Viviano BL, Yan H, Krolewski J, Schreiber RD. Stat recruitment by tyrosine-phosphorylated cytokine receptors: an ordered reversible affinity-driven process. Immunity. 1995;2(6):677–687. doi: 10.1016/1074-7613(95)90012-8. [DOI] [PubMed] [Google Scholar]
- Gupta S, Yan H, Wong LH, Ralph S, Krolewski J, Schindler C. The SH2 domains of Stat1 and Stat2 mediate multiple interactions in the transduction of IFN-alpha signals. The EMBO journal. 1996;15(5):1075–1084. [PMC free article] [PubMed] [Google Scholar]
- Hagmaier K, Stock N, Precious B, Childs K, Wang LF, Goodbourn S, Randall RE. Mapuera virus, a rubulavirus that inhibits interferon signalling in a wide variety of mammalian cells without degrading STATs. J Gen Virol. 2007;88(Pt 3):956–966. doi: 10.1099/vir.0.82579-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann S, Garcin D, Delenda C, Kolakofsky D. The versatility of paramyxovirus RNA polymerase stuttering. Journal of virology. 1999a;73(7):5568–5576. doi: 10.1128/jvi.73.7.5568-5576.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann S, Garcin D, Morel AS, Kolakofsky D. Two nucleotides immediately upstream of the essential A6G3 slippery sequence modulate the pattern of G insertions during Sendai virus mRNA editing. Journal of virology. 1999b;73(1):343–351. doi: 10.1128/jvi.73.1.343-351.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez J, Reyes-Leyva J, Zenteno R, Ramirez H, Hernandez-Jauregui P, Zenteno E. Immunity to porcine rubulavirus infection in adult swine. Veterinary immunology and immunopathology. 1998;64(4):367–381. doi: 10.1016/s0165-2427(98)00169-x. [DOI] [PubMed] [Google Scholar]
- Hernandez-Jauregui P, Ramirez Mendoza H, Mercado Garcia C, Moreno-Lopez J, Kennedy S. Experimental porcine rubulavirus (La Piedad-Michoacan virus) infection in pregnant gilts. Journal of comparative pathology. 2004;130(1):1–6. doi: 10.1016/s0021-9975(03)00058-6. [DOI] [PubMed] [Google Scholar]
- Iseni F, Baudin F, Garcin D, Marq JB, Ruigrok RW, Kolakofsky D. Chemical modification of nucleotide bases and mRNA editing depend on hexamer or nucleoprotein phase in Sendai virus nucleocapsids. Rna. 2002;8(8):1056–1067. doi: 10.1017/s1355838202029977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolakofsky D, Roux L, Garcin D, Ruigrok RW. Paramyxovirus mRNA editing, the “rule of six” and error catastrophe: a hypothesis. J Gen Virol. 2005;86(Pt 7):1869–1877. doi: 10.1099/vir.0.80986-0. [DOI] [PubMed] [Google Scholar]
- Kubota T, Yokosawa N, Yokota S, Fujii N, Tashiro M, Kato A. Mumps virus V protein antagonizes interferon without the complete degradation of STAT1. Journal of virology. 2005;79(7):4451–4459. doi: 10.1128/JVI.79.7.4451-4459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent-Rolle M, Morrison J, Rajsbaum R, Macleod JM, Pisanelli G, Pham A, Ayllon J, Miorin L, Martinez-Romero C, tenOever BR, Garcia-Sastre A. The interferon signaling antagonist function of yellow fever virus NS5 protein is activated by type I interferon. Cell host & microbe. 2014;16(3):314–327. doi: 10.1016/j.chom.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung S, Qureshi SA, Kerr IM, Darnell JE, Jr, Stark GR. Role of STAT2 in the alpha interferon signaling pathway. Mol Cell Biol. 1995;15(3):1312–1317. doi: 10.1128/mcb.15.3.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Leung S, Kerr IM, Stark GR. Functional subdomains of STAT2 required for preassociation with the alpha interferon receptor and for signaling. Mol Cell Biol. 1997;17(4):2048–2056. doi: 10.1128/mcb.17.4.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linne T, Berg M, Bergvall AC, Hjertner B, Moreno-Lopez J. The molecular biology of the porcine paramyxovirus LPMV. Vet Microbiol. 1992;33(1–4):263–273. doi: 10.1016/0378-1135(92)90054-w. [DOI] [PubMed] [Google Scholar]
- Manicassamy B, Manicassamy S, Belicha-Villanueva A, Pisanelli G, Pulendran B, Garcia-Sastre A. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(25):11531–11536. doi: 10.1073/pnas.0914994107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Magana ML, Godoy-Martinez DV, Guerrero-Cazares H, Rodriguez-Peredo A, Duenas-Jimenez JM, Duenas-Jimenez SH, Ramirez-Herrera MA. Blue eye disease porcine rubulavirus (PoRv) infects pig neurons and glial cells using sialo-glycoprotein as receptor. Veterinary journal. 2007;173(2):428–436. doi: 10.1016/j.tvjl.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Meyer O. Interferons and autoimmune disorders. Joint, bone, spine : revue du rhumatisme. 2009;76(5):464–473. doi: 10.1016/j.jbspin.2009.03.012. [DOI] [PubMed] [Google Scholar]
- Moreno-Lopez J, Correa-Giron P, Martinez A, Ericsson A. Characterization of a paramyxovirus isolated from the brain of a piglet in Mexico. Archives of virology. 1986;91(3–4):221–231. doi: 10.1007/BF01314282. [DOI] [PubMed] [Google Scholar]
- Morrison J, Laurent-Rolle M, Maestre AM, Rajsbaum R, Pisanelli G, Simon V, Mulder LC, Fernandez-Sesma A, Garcia-Sastre A. Dengue virus co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling. PLoS pathogens. 2013;9(3):e1003265. doi: 10.1371/journal.ppat.1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motz C, Schuhmann KM, Kirchhofer A, Moldt M, Witte G, Conzelmann KK, Hopfner KP. Paramyxovirus V proteins disrupt the fold of the RNA sensor MDA5 to inhibit antiviral signaling. Science. 2013;339(6120):690–693. doi: 10.1126/science.1230949. [DOI] [PubMed] [Google Scholar]
- Nadeau OW, Domanski P, Usacheva A, Uddin S, Platanias LC, Pitha P, Raz R, Levy D, Majchrzak B, Fish E, Colamonici OR. The proximal tyrosines of the cytoplasmic domain of the beta chain of the type I interferon receptor are essential for signal transducer and activator of transcription (Stat) 2 activation. Evidence that two Stat2 sites are required to reach a threshold of interferon alpha-induced Stat2 tyrosine phosphorylation that allows normal formation of interferon-stimulated gene factor 3. The Journal of biological chemistry. 1999;274(7):4045–4052. doi: 10.1074/jbc.274.7.4045. [DOI] [PubMed] [Google Scholar]
- Nishio M, Garcin D, Simonet V, Kolakofsky D. The carboxyl segment of the mumps virus V protein associates with Stat proteins in vitro via a tryptophan-rich motif. Virology. 2002;300(1):92–99. doi: 10.1006/viro.2002.1509. [DOI] [PubMed] [Google Scholar]
- Nishio M, Tsurudome M, Ito M, Garcin D, Kolakofsky D, Ito Y. Identification of paramyxovirus V protein residues essential for STAT protein degradation and promotion of virus replication. Journal of virology. 2005a;79(13):8591–8601. doi: 10.1128/JVI.79.13.8591-8601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio M, Tsurudome M, Ito M, Ito Y. Human parainfluenza virus type 4 is incapable of evading the interferon-induced antiviral effect. Journal of virology. 2005b;79(23):14756–14768. doi: 10.1128/JVI.79.23.14756-14768.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108(2):193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Parisien JP, Lau JF, Rodriguez JJ, Sullivan BM, Moscona A, Parks GD, Lamb RA, Horvath CM. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology. 2001;283(2):230–239. doi: 10.1006/viro.2001.0856. [DOI] [PubMed] [Google Scholar]
- Parisien JP, Lau JF, Rodriguez JJ, Ulane CM, Horvath CM. Selective STAT protein degradation induced by paramyxoviruses requires both STAT1 and STAT2 but is independent of alpha/beta interferon signal transduction. Journal of virology. 2002;76(9):4190–4198. doi: 10.1128/JVI.76.9.4190-4198.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MS, Shaw ML, Munoz-Jordan J, Cros JF, Nakaya T, Bouvier N, Palese P, Garcia-Sastre A, Basler CF. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. Journal of virology. 2003;77(2):1501–1511. doi: 10.1128/JVI.77.2.1501-1511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Precious B, Childs K, Fitzpatrick-Swallow V, Goodbourn S, Randall RE. Simian virus 5 V protein acts as an adaptor, linking DDB1 to STAT2, to facilitate the ubiquitination of STAT1. Journal of virology. 2005a;79(21):13434–13441. doi: 10.1128/JVI.79.21.13434-13441.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Precious B, Young DF, Andrejeva L, Goodbourn S, Randall RE. In vitro and in vivo specificity of ubiquitination and degradation of STAT1 and STAT2 by the V proteins of the paramyxoviruses simian virus 5 and human parainfluenza virus type 2. J Gen Virol. 2005b;86(Pt 1):151–158. doi: 10.1099/vir.0.80263-0. [DOI] [PubMed] [Google Scholar]
- Qureshi SA, Leung S, Kerr IM, Stark GR, Darnell JE., Jr Function of Stat2 protein in transcriptional activation by alpha interferon. Mol Cell Biol. 1996;16(1):288–293. doi: 10.1128/mcb.16.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi SA, Salditt-Georgieff M, Darnell JE., Jr Tyrosine-phosphorylated Stat1 and Stat2 plus a 48-kDa protein all contact DNA in forming interferon-stimulated-gene factor 3. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(9):3829–3833. doi: 10.1073/pnas.92.9.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajsbaum R, Versteeg GA, Schmid S, Maestre AM, Belicha-Villanueva A, Martinez-Romero C, Patel JR, Morrison J, Pisanelli G, Miorin L, Laurent-Rolle M, Moulton HM, Stein DA, Fernandez-Sesma A, tenOever BR, Garcia-Sastre A. Unanchored K48-linked polyubiquitin synthesized by the E3-ubiquitin ligase TRIM6 stimulates the interferon-IKKepsilon kinase-mediated antiviral response. Immunity. 2014;40(6):880–895. doi: 10.1016/j.immuni.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran A, Horvath CM. Paramyxovirus disruption of interferon signal transduction: STATus report. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2009;29(9):531–537. doi: 10.1089/jir.2009.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Mendoza H, Hernandez-Jauregui P, Reyes-Leyva J, Zenteno E, Moreno-Lopez J, Kennedy S. Lesions in the reproductive tract of boars experimentally infected with porcine rubulavirus. Journal of comparative pathology. 1997;117(3):237–252. doi: 10.1016/s0021-9975(97)80018-7. [DOI] [PubMed] [Google Scholar]
- Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89(Pt 1):1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Reyes-Leyva J, Espinosa B, Santos G, Zenteno R, Hernandez J, Vallejo V, Zenteno E. Purification and characterization of the hemagglutinin-neuraminidase of Porcine rubulavirus LPMV. Glycoconjugate journal. 1999;16(9):517–522. doi: 10.1023/a:1007022021301. [DOI] [PubMed] [Google Scholar]
- Rivera-Benitez JF, Cuevas-Romero S, Perez-Torres A, Reyes-Leyva J, Hernandez J, Ramirez-Mendoza H. Respiratory disease in growing pigs after Porcine rubulavirus experimental infection. Virus research. 2013;176(1–2):137–143. doi: 10.1016/j.virusres.2013.05.017. [DOI] [PubMed] [Google Scholar]
- Rodriguez JJ, Parisien JP, Horvath CM. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. Journal of virology. 2002;76(22):11476–11483. doi: 10.1128/JVI.76.22.11476-11483.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JJ, Wang LF, Horvath CM. Hendra virus V protein inhibits interferon signaling by preventing STAT1 and STAT2 nuclear accumulation. Journal of virology. 2003;77(21):11842–11845. doi: 10.1128/JVI.77.21.11842-11845.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez KR, Horvath CM. Paramyxovirus V protein interaction with the antiviral sensor LGP2 disrupts MDA5 signaling enhancement but is not relevant to LGP2-mediated RLR signaling inhibition. Journal of virology. 2014;88(14):8180–8188. doi: 10.1128/JVI.00737-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Ropon A, Hernandez-Jauregui P, Sanchez-Torres L, Favila-Castillo L, Estrada-Parra S, Moreno-Lopez J, Kennedy S. Apoptosis in lymph nodes and changes in lymphocyte subpopulations in peripheral blood of pigs infected with porcine rubulavirus. Journal of comparative pathology. 2003;128(1):1–8. doi: 10.1053/jcpa.2002.0598. [DOI] [PubMed] [Google Scholar]
- Rothlisberger A, Wiener D, Schweizer M, Peterhans E, Zurbriggen A, Plattet P. Two domains of the V protein of virulent canine distemper virus selectively inhibit STAT1 and STAT2 nuclear import. Journal of virology. 2010;84(13):6328–6343. doi: 10.1128/JVI.01878-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Betancourt JI, Trujillo ME, Mendoza SE, Reyes-Leyva J, Alonso RA. Genetic and antigenic changes in porcine rubulavirus. Canadian journal of veterinary research = Revue canadienne de recherche veterinaire. 2012;76(1):33–37. [PMC free article] [PubMed] [Google Scholar]
- Shuai K, Stark GR, Kerr IM, Darnell JE., Jr A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science. 1993;261(5129):1744–1746. doi: 10.1126/science.7690989. [DOI] [PubMed] [Google Scholar]
- Solis M, Ramirez-Mendoza H, Mercado C, Espinosa S, Vallejo V, Reyes-Leyva J, Hernandez J. Semen alterations in porcine rubulavirus-infected boars are related to viral excretion and have implications for artificial insemination. Research in veterinary science. 2007;83(3):403–409. doi: 10.1016/j.rvsc.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Stephan HA, Gay GM, Ramirez TC. Encephalomyelitis, reproductive failure and corneal opacity (blue eye) in pigs, associated with a paramyxovirus infection. Vet Rec. 1988;122(1):6–10. doi: 10.1136/vr.122.1.6. [DOI] [PubMed] [Google Scholar]
- Sundqvist A, Berg M, Hernandez-Jauregui P, Linne T, Moreno-Lopez J. The structural proteins of a porcine paramyxovirus (LPMV) J Gen Virol. 1990;71(Pt 3):609–613. doi: 10.1099/0022-1317-71-3-609. [DOI] [PubMed] [Google Scholar]
- Sundqvist A, Berg M, Moreno-Lopez J, Linne T. The haemagglutinin-neuraminidase glycoprotein of the porcine paramyxovirus LPMV: comparison with other paramyxoviruses revealed the closest relationship to simian virus 5 and mumps virus. Archives of virology. 1992;122(3–4):331–340. doi: 10.1007/BF01317194. [DOI] [PubMed] [Google Scholar]
- Svenda M, Berg M, Moreno-Lopez J, Linne T. Analysis of the large (L) protein gene of the porcine rubulavirus LPMV: identification of possible functional domains. Virus research. 1997;48(1):57–70. doi: 10.1016/s0168-1702(96)01426-8. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Kadota SI, Takeda M, Miyajima N, Nagata K. Measles virus V protein blocks interferon (IFN)-alpha/beta but not IFN-gamma signaling by inhibiting STAT1 and STAT2 phosphorylation. FEBS letters. 2003;545(2–3):177–182. doi: 10.1016/s0014-5793(03)00528-3. [DOI] [PubMed] [Google Scholar]
- Thomas SM, Lamb RA, Paterson RG. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell. 1988;54(6):891–902. doi: 10.1016/S0092-8674(88)91285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulane CM, Horvath CM. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology. 2002;304(2):160–166. doi: 10.1006/viro.2002.1773. [DOI] [PubMed] [Google Scholar]
- Wiman AC, Hjertner B, Linne T, Herron B, Allan G, McNeilly F, Adair B, Moreno-Lopez J, Berg M. Porcine rubulavirus LPMV RNA persists in the central nervous system of pigs after recovery from acute infection. Journal of neurovirology. 1998;4(5):545–552. doi: 10.3109/13550289809113499. [DOI] [PubMed] [Google Scholar]
- Yan H, Krishnan K, Greenlund AC, Gupta S, Lim JT, Schreiber RD, Schindler CW, Krolewski JJ. Phosphorylated interferon-alpha receptor 1 subunit (IFNaR1) acts as a docking site for the latent form of the 113 kDa STAT2 protein. The EMBO journal. 1996;15(5):1064–1074. [PMC free article] [PubMed] [Google Scholar]
- Zenteno-Cuevas R, Huerta-Yepez S, Reyes-Leyva J, Hernandez-Jauregui P, Gonzalez-Bonilla C, Ramirez-Mendoza H, Agundis C, Zenteno E. Identification of potential B cell epitope determinants by computer techniques, in hemagglutinin-neuraminidase from the porcine rubulavirus La Piedad Michoacan. Viral immunology. 2007;20(2):250–260. doi: 10.1089/vim.2006.0066. [DOI] [PubMed] [Google Scholar]


