Abstract
We show that Mycobacterium smegmatis mutants disrupted in mscR, coding for a dual function S-nitrosomycothiol reductase and formaldehyde dehydrogenase, and mshC, coding for a mycothiol ligase and lacking mycothiol (MSH), are more susceptible to S-nitrosoglutathione (GSNO) and aldehydes than wild type. MSH is a cofactor for MscR, and both mshC and mscR are induced by GSNO and aldehydes. We also show that a mutant disrupted in egtA, coding for a γ-glutamyl cysteine synthetase and lacking in ergothioneine, is sensitive to nitrosative stress but not to aldehydes. In addition, we find that MSH and S-nitrosomycothiol reductase are required for normal biofilm formation in M. smegmatis, suggesting potential new therapeutic pathways to target to inhibit or disrupt biofilm formation.
Keywords: mycothiol, ergothioneine, formaldehyde dehydrogenase, S-nitrosothiol reductase, biofilms, mycobacteria, nitrosative stress
Introduction
Nitric oxide (NO) is produced by macrophages during the innate and adaptive immune responses to infection from a wide range of microbial pathogens (1,2). NO has diverse physiological functions, causing damage to both the pathogen and host cells by binding to metal and nitrosylating tyrosine and cysteine residues in proteins. It can also react with hydrogen peroxide to form peroxynitrite, causing oxidative damage. Paradoxically, nitrosylation is an important process in cell signaling as nitrosylation can activate or inhibit protein function (3). The functions of NO are not limited to its site of production, as NO is highly diffusible and low-molecular-weight (LMW) S-nitrosothiols can act as long-distance NO delivery vehicles. The LMW thiol, glutathione (GSH), reacts with NO to form nitrosoglutathione (GSNO), and GSNO can be catabolized to GSH and NO by S-nitrosoglutathione reductase (GSNOR), an enzyme common to most eukaryotes and prokaryotes (4,5). GSNOR is thus able to modulate effects of reactive oxygen and nitrogen species through catabolism of GSNO. Although its preferred substrates are GSNO and S-hydroxymethyl glutathione, an intermediate in formaldehyde metabolism, GSNOR has a dual NAD/H-dependent oxidoreductase activity toward a broad spectrum of aliphatic compounds including alcohols such as cinnamylalcohol, geraniol, ω-hydroxy fatty acids (6), and the aldehydes, glycolaldehyde and glyceraldehyde, which are byproducts of sugar metabolism. Indeed, a mutant of Haemophilus influenzae, disrupted in GSNOR, is sensitive to these toxic aldehydes (7).
Although GSH is the primary LMW thiol found in eukaryotes and Gram-negative bacteria, mycothiol (MSH) is the functional analog of GSH in actinobacteria, a phylum that includes a wide variety of bacteria, including the pathogen Mycobacterium tuberculosis (8). MSH contributes to cellular stress resistance against oxidants, antibiotics, toxins, and metals (9, 10). Additionally, a Mycobacterium smegmatis MSH-deficient mutant disrupted in mshA is sensitive to NO (11), and MSH acts as a cofactor for the MSH-dependent formaldehyde dehydrogenase/nitrosothiol reductase, MscR (12). Ergothioneine (ESH), another LMW thiol found in actinobacteria, is able to scavenge NO and oxidants in vitro (13) and in vivo (14).
Recently, a Nesseria meningitidis mutant lacking GSNOR demonstrated loss of viability in a biofilm (15). Whether MscR, MSH, or ESH play roles in biofilm formation in mycobacteria and other actinobacteria was not previously known. In this study, M. smegmatis mutants disrupted in mscR (SHP1), an MSH-deficient strain disrupted in mshC (S24) and an ESH-deficient strain disrupted in egtA (R119), were examined for their roles in detoxification of aldehydes and reactive nitrogen species. As biofilms produced during infection are highly resistant to broad-spectrum antibiotics and the host immune system and as the genus Mycobacterium includes pathogens such as M. tuberculosis and Mycobacterium avium, which are capable of forming biofilms (16), we also examined these mutants for biofilm formation in this study.
Materials and Methods
Culture Conditions
M. smegmatis mc2155 wild-type cultures were routinely started from glycerol stocks onto Middlebrook 7H10 solid medium with 0.5% glycerol supplemented with 1% glucose. Colonies were then transferred to Middlebrook 7H9 broth (Difco) with 0.05% Tween-80 supplemented with 1% glucose or 10% OADC (oleic acid, albumin, dextrose, and catalase). Antibiotics were added when appropriate (25 μg/mL kanamycin for S24 and R119, 75 μg/mL hygromycin for SHP1, and 25 μg/mL kanamycin and 75 μg/mL hygromycin for complemented strains, S24C and SHP1C). Cultures were incubated at 37 °C with shaking at 160 rpm.
Creation and Screening of a Transposon Mutant Library and Identification of the Disrupted Gene in S24 and R119
An M. smegmatis transposon mutant library (EZ::TN <kan-2> Tnp transposase and Tn5 kanamycin resistance marker) was screened for diamide-sensitive mutants as described by Rawat et al. (17). To identify the site of insertion, genomic DNA was digested with restriction enzymes Sa1 I and PstI followed by self-ligation and PCR amplification with primers complementary to the transposon. The sequence of the amplified PCR product was compared with the M. smegmatis genome sequence (17).
Minimum Inhibitory Concentrations
Starter cultures were used to inoculate 7H9 medium containing 1% glucose supplemented with 0.05% Tween-80. These cultures were grown to mid-log phase and then diluted to an OD600 of 0.2. Formaldehyde, methylglyoxal, glycolaldehyde, glyceraldehyde, or GSNO were serially diluted twofold in 100 μL and added to 100 μL of culture to obtain a final OD600 of 0.1 in ELISA plates. These cultures were incubated for 48 h, and then the minimum inhibitory concentration (MIC) was determined. Assays were performed in quadruplicate and repeated at least three times.
Growth Curves
Cultures were grown to mid-log phase and then diluted to an OD600 of 0.1. Ten milliliters of cultures in 50-mL tubes was treated with 0.75 mM formaldehyde, 0.75 mM methylglyoxal, 2.25 mM glycolaldehyde, 9 mM glyceraldehyde, 0.75 mM NaNO2, or 0.75 mM GSNO, concentrations that were 75–90% of the determined MIC. Time points for growth curves were taken until stationary phase, and the growth curves were performed three times in triplicate or more. To analyze the effects of oxygen tension on growth, the headspace–volume ratio was adjusted to 4.0 (high oxygen tension), 1.5 (moderate oxygen tension), or 0.66 (low oxygen tension) in 50-mL tubes (18).
Creation of mscR Mutant, SH1, and Complementation of Mutant with M. Tuberculosis mscR Gene
The mscR (MSMEG_4340) gene and flanking regions were amplified using primers 5′-CTCGAGTCACAACATCACCAC-3′ and 5′-GAATTCATGAGTCAGACGGTG-3′. The resulting PCR amplicon was purified and subsequently ligated into the TA cloning vector, pCR2.1, containing a kanamycin resistance cassette to obtain pSH1. A hygromycin resistance cassette was excised from pHINT using the restriction enzymes PstI and SmaI, blunted with the Klenow fragment, and then inserted into the StuI site in the mscR gene. The resulting vector, pSH2, was then electroporated into competent M. smegmatis mc2155, and electroporated cells were plated on selective media with hygromycin, followed by replica plating of mutant colonies on media with kanamycin to rule out nonhomologous recombination events. Confirmation of mutation was performed using PCR with the above primers and Southern blot hybridization. One confirmed colony, SHP1, was further characterized.
To validate the phenotype of the mscR mutant, the native gene from M. tuberculosis, Rv2259, was amplified with fdhco5, 5′-CTCGAGTTCGCTCTGCATCGTCAC-3′, and fdhco3, 5′-GGTACCGACCAGCCAGATGTTGTT-3′, primers with XhoI and KpnI restriction sites engineered, respectively, into the primer sequence. The 1.5-kb gene fragment containing the gene and 345 bp upstream of the start site was purified from a 1% agarose gel and TA cloned into pCR2.1, and the resulting plasmid, pSH3, was then subjected to digestion with XhoI and KpnI for cloning into the integrative vector, pAINT that had been similarly restriction digested. Selection of this new plasmid, pSH4, was performed on LB media supplemented with kanamycin followed by transformation into SHP1 competent cells and plating on 7H10 supplemented with 10% OADC and hygromycin and kanamycin. Confirmation of an integrated copy of the M. tuberculosis mscR was carried out by PCR using primers fdhco5 and fdhco3. The M. smegmatis strain SHP1C contained a mutated copy of mscR and a copy of the M. tuberculosis mscR.
RNA Extraction and qPCR
To analyze gene expression, M. smegmatis mc2155 was grown in Middlebrook 7H9 media with 1% glucose at 37 °C with shaking to an OD600 of 0.5. Aliquots of 10 mL of culture were treated with 0.75 mM formaldehyde, 0.75 mM methylglyoxal, 2.25 mM glycolaldehyde, 9 mM glyceraldehyde, or 0.75 mM GSNO in quadruplicates. After 1-h incubation, the aliquots were centrifuged at 4,000 rpm at 4 °C for 10 min, and pelleted cells were flash frozen and stored at −80 °C. RNA was extracted using a Qiagen Rneasy Mini Kit with the optional on-column DNase treatment (Qiagen) as specified in the manufacturer’s protocol, except cells were lysed with glass beads using a ribolyzer (three 45-sec cycles at maximum speed). The RNA extraction was followed by an additional Turbo DNase treatment (Life Technologies). Samples were then checked for DNA contamination using PCR. One microgram of RNA and 0.5 μg of random primers were heated for 5 min at 70 °C, cooled on ice for 5 min, and then reverse transcribed using a Promega M-MLV Reverse Transcriptase Kit according to the manufacturer’s instructions. After confirmation of synthesis of cDNA, qPCR reactions were performed using Thermo Scientific ABsolute SYBR Green ROX Mix with 50 ng RNA and 200 nM primers in a reaction volume of 25 μL. The qPCR conditions were an initial 50 °C cycle for 15 min, followed by a 95 °C cycle for 15 min, 40 cycles of 95 °C for 30 sec, 55 °C for 30 sec, and 72 °C for 1 min, and a final elongation step of 72 °C for 4 min. Primer efficiency was calculated by generating a standard curve and by analysis of the melting curve. Real-time quantitative PCR (qPCR) was performed in an Eppendorf Realplex2 Mastercycler with primers (Table 1). Relative gene expression was calculated using the −2ΔΔCT method (ΔCt – Ct sample − Ct control) and reported as fold change in gene expression of each sample normalized to the mysA relative to the untreated control. Each experiment was performed in triplicate.
TABLE 1.
Primers for quantitative real-time PCR
| Name | Primer sequence |
|---|---|
| mscR-Fwd | CCT GGA AGC AGG CCT TCT A |
| mscR-Rev | GCA GGT ACA GCG AGA TCA G |
| mysA-Fwd | ATC GAC GAG CCG TCC GAG AA |
| mysA-Rev | CCT TGC CGA TCT GCT TGA GG |
| egtA-Fwd | GGT GAT GCT GGT GAA CTC T |
| egtA-Rev | CGT CGT CAG GTG ATA GTC C |
| mshC-Fwd | ATC GCC GAG GTC GTC GAG AT |
| mshC-Rev | GGC GAA CAA CGT GAG CAT GG |
Mycobacterium Smegmatis Biofilm Assay
M. smegmatis wild type and mutant strains were incubated on Trypticase Soy Agar (TSA) plates containing antibiotics if necessary at 37 °C for 48 hours. A single colony from each strain was inoculated in 4 ml of Trypticase Soy Broth (TSB) with 0.05% Tween 80 with antibiotics if needed. The cultures were incubated at 37 °C at 225 rpm for 72 hours. When the OD600 reached 2.0, the cells were diluted 1:1000 and incubated in TSB media supplemented with 1% Procalamine (B. Braun Medical Inc.), which enhances biofilm formation in M. smegmatis, and incubated at 30 °C for 72 hours with no disturbance. Four replicates for each strain were seeded for biofilm formation. After 72 hours, the biofilms, visible on the liquid-air interface were documented. The thickness of the biofilms was quantitated by measuring absorbance at OD630 in a BioTek EON plate reader after removal of the media from each well.
Results and Discussion
Mutants Disrupted in mscR, mshC but not egtA Are Sensitive to Nitrosative Stress
We had previously reported that MSH was required for growth of M. smegmatis when cultures were exposed to gaseous NO; here we confirm that this is also true with GSNO (11). To determine if mutants deficient in mscR, MSH, or ESH are sensitive to aldehyde and nitrosative stress, MICs were determined and growth curves were performed using sub-lethal concentrations of stressors. The transposon mutant, S24, disrupted in mshC, which codes for MSH ligase, the fourth step of a five step MSH biosynthetic pathway, was most sensitive to killing by GSNO (Fig. 1j), followed by SHP1 (Fig. 1e), a deletion mutant of mscR, and then the transposon mutant, R119, disrupted in egtA, which codes for a γ-glutamate-cysteine ligase that provides sulfur for the synthesis of ESH (Fig. 1o), when compared with wild type. ESH thus appears to play a smaller role in nitrosative stress resistance than MSH in M. smegmatis despite the fact that ESH is an excellent scavenger of peroxynitrite (19) and GSNO (13).
FIG 1.
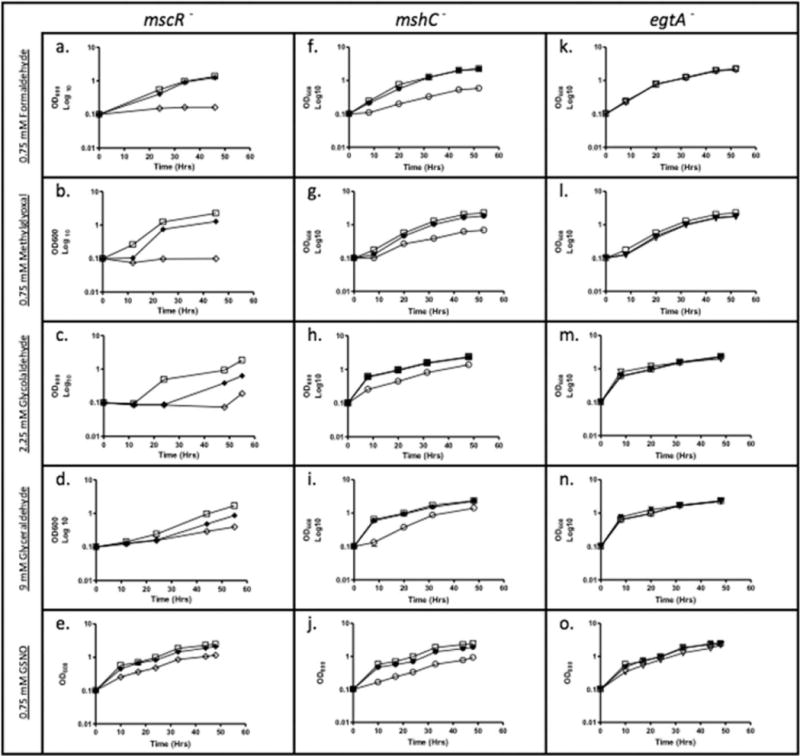
Growth of M. smegmatis in 7H9 media treated with 0.75 mM formaldehyde, 0.75 mM methylglyoxal, 2.25 mM glycolaldehyde, 9 mM glyceraldehyde, or 0.75 mM GSNO; wild type (□), SHP1 (mscR−) (◊), SHP1C (◆), S24 (mshC−) (○), S24C (•), R119 (egtA−) (∇), and R119C (▼).
Inside macrophages, NO produced by the inducible NO synthase (iNOS) system can combine with GSNO; both NO and GSNO are known to be inhibitory to bacteria such as M. tuberculosis (20). In bacteria with GSH as the major LMW thiol, such as Escherichia coli, or in bacteria that import GSH from the environment, such as Haemophilus influenzae (21) and Streptococcus pneumoniae (22), S-nitrosothiol reductases that catabolize GSO have been found to be protective against nitrosative stress. Indeed, in S. pneumoniae, nitrosothiol reductase is required for survival in blood after an intraperitoneal challenge experiment, but does not affect the adherence of pneumococci to the nasopharynx in vivo (22). The situation may be different in MSH-containing bacteria, where GSH and GSNO are known to control the growth of M. tuberculosis within macrophages (23). GSNO produced by macrophages could be catabolized to NO and GSH, either chemically or enzymatically through the actions of carbonyl reductase 1, the thioredoxin system, or Cu–Zn superoxide dismutase (24), and the NO released could be used to form MSNO, the substrate for MscR. Interestingly, a mutant disrupted in the M. tuberculosis mscR gene has not been obtained, implying that this gene is essential (25), whereas a Corynebacterium glutamicum mscR mutant is viable and is able to grow as well as the wild type. In addition, a C. glutamicum double mutant disrupted in a gene encoding an acetaldehyde dehydrogenase, ald, and mscR and another double mutant disrupted in disrupted in ald and mshC were severely impaired in their ability to oxidize formaldehyde (26).
Mutants Disrupted in mscR and mshC but Not egtA Are Sensitive to Aldehyde Stress
Endogenous sources of reactive aldehydes include sugars, such as fructose whose metabolism results in glyceraldehyde and glycolaldehyde intermediates, the latter being further oxidized to glyoxal (27) and glucose, whose metabolism can result in the formation of methylglyoxal (28). Both SHP1 (Figs. 1b–1d) and S24 (Figs. 1g–1i) were sensitive to killing by various short-chain aliphatic aldehydes, formaldehyde, methylglyoxal, glycolaldehyde, and glyceraldehyde. SHP1 and S24 were also sensitive to formaldehyde, a reactive byproduct of methanol metabolism, as well as demethylation reactions in the cell (Figs. 1a and 1f, respectively). R119 showed no significant sensitivity to aldehydes, indicating that ESH is not involved in the protection against these stresses (Figs. 1k–1n). Taken together, these data indicate that both MscR and MSH also play roles in the detoxification of short-chain aldehydes, which are formed as metabolic byproducts and are toxic to cells.
Oxygen Tension Increases the Stress of Aldehydes
Growth curves were set up with tubes having varying head space–volume ratio to determine if oxygen tension has an effect on killing in wild type, SHP1, and SHP1C. There was no obvious difference in growth when the cultures were treated with NaNO2 (data not shown), which is not surprising as most bacteria can generate NO either aerobically by NO synthases or anaerobically from nitrite. Under aldehyde stress, SHP1 was more sensitive than wild type to killing by aldehydes, and the complemented strain (SHP1C) had intermediate sensitivity under all headspace–volume ratios (Fig. 2). However, SHP1 was substantially more sensitive to formaldehyde or glycolaldehyde when grown under high oxygen tension (Figs. 2i and 2k) when compared with lower oxygen tension (Figs. 2a and 2c). Short-chain aldehydes can oxidize to form dicarbonyl species in the presence of oxygen, and these highly reactive dicarbonyl species may be one reason for the increased sensitivity of SHP1 to formaldehyde and glycolaldehyde under aerobic conditions (29).
FIG 2.
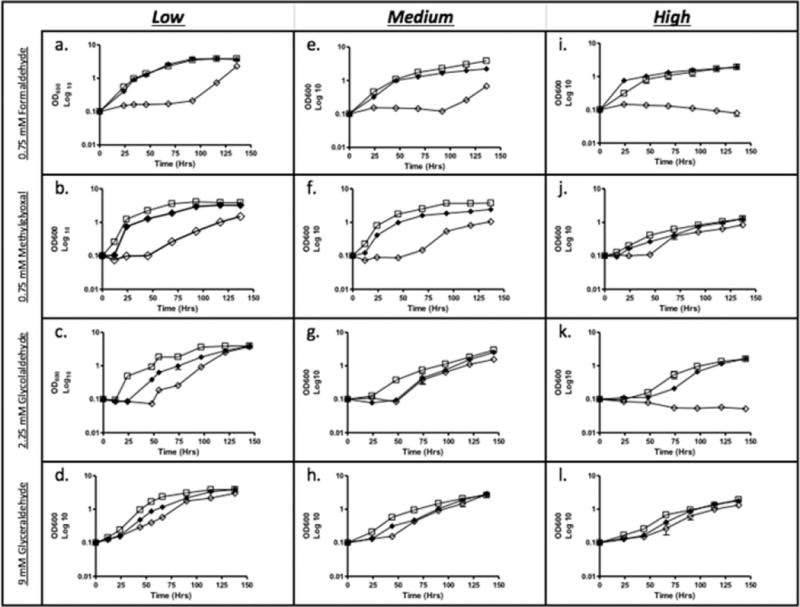
Growth of M. smegmatis in 7H9 media treated with 0.75 mM formaldehyde, 0.75 mM methylglyoxal, 2.25 mM glycolaldehyde, and 9 mM glyceraldehyde. Cultures were grown in tubes with a headspace–volume ratio of 4.0 (a–d), 1.5 (e–h), and 0.66 (i–l); wild type (□), SHP1 (mscR−) (◊), and SHP1C (◆).
mscR and mshC expression increases under aldehyde stress and. mscR, mshC, and egtA Expression Increases under Nitrosative Stress
Induction of bacterial S-nitrosthiol reductases has been studied in several organisms and seems to be triggered by distinct stimuli. The GSNOR gene of H. influenzae is induced by formaldehyde but not GSNO (21), whereas the S. pneumoniae gene is induced in response to GSNO (22). An increase in mscR expression in response to formaldehyde has been documented in Rhodococcus erythropolis, a methylotrophic bacterium (30), and the expression of both adhCs (nitrosothiol reductases) present in Acinetobacter baumannii is significantly enhanced by the presence of subinhibitory concentrations of formaldehyde in the culture media (31). To determine how the expression of mscR, mshC, and egtA changes in response to nitrosative stress and aldehyde stress in M. smegmatis, qPCR was performed to monitor gene expression. A fivefold to 10-fold increase in the expression of mscR and mshC was detected in response to aldehyde and nitrosative stress. In contrast, expression of egtA did not significantly change on exposure to aldehyde stress; however, there was a significant increase in the expression of egtA in response to GSNO, consistent with the finding that the R119 mutant is sensitive to GSNO but not aldehyde stress (Fig. 3).
FIG 3.
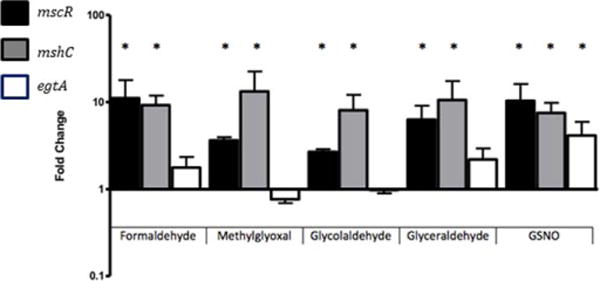
Expression of mscR (black), mshC (gray), and egtA (white) in M. smegmatis mc2155 after 1-h incubation in 0.75 mM formaldehyde, 0.75 methylglyoxal, 2.25 mM glycolaldehyde, 9 mM glyceraldehyde, or 0.75 mM GSNO.
Mutants Disrupted in mscR and mshC Have Decreased Biofilm Formation
M. smegmatis forms biofilms that appear as surface pellicles at the air–liquid interface due to the presence of large amounts of free mycolic acids in its extracellular matrix, instead of polysaccharides commonly found in the matrix of other biofilms (32). The biofilm appears as a complex network of ridges with a bulk fluid sequestered underneath the surface pellicle (32). Examination of biofilms produced by S24 and SHP1 revealed a striking difference in the appearance of these biofilms when compared with the wild type; both mutants displayed thin biofilms with less ridges, and this phenotype was partially reversed in the complemented strains (Fig. 4 and Table 2). MSH and ESH can chemically react with NO directly or in a transnitrosylation reaction with GSNO, and thus, the lack of either thiol should lead to increased levels of NO and GSNO, as would the disruption of mscR, resulting in increased protein nitrosylation and presumably inactivation and/or alteration of cell signaling involving NO. The role of NO signaling during the biofilm lifecycle has been examined in P. aeruginosa, where NO was discovered to be produced at the same time as cell death and dispersal (33), and thus, NO may also play important functional roles throughout the developmental stages of the mycobacteria biofilm lifecycle.
FIG 4.
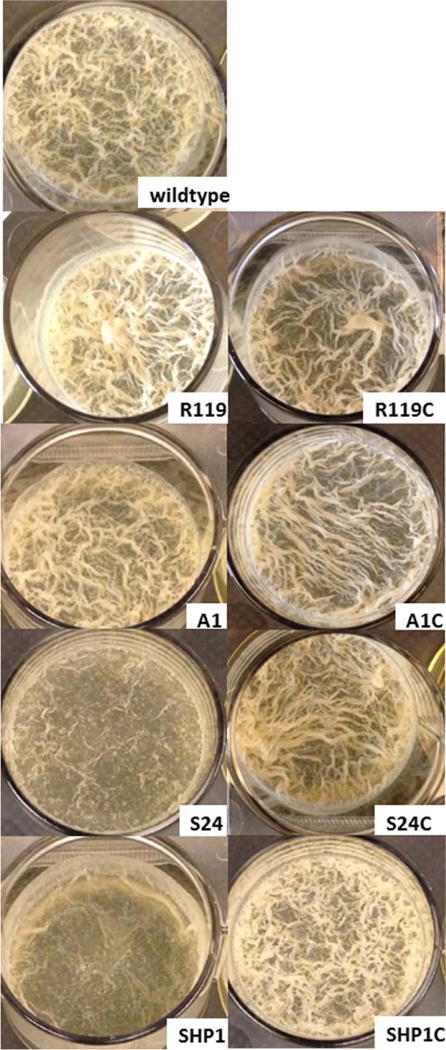
M. smegmatis biofilms formed in TSB media supplemented with 1% procalamine and incubated at 30 °C for 72 h with no disturbance. Wild-type strain compared with SHP1 (mscR−), SHP1C (complemented SHP1), S24 (mshC−), S24C (complemented S24), R119 (egtA−), and R119C (complemented R119).
TABLE 2.
Biofilm formation in M. smegmatis strains
| Strain | OD630 | P-value* |
|---|---|---|
| Wild type | 1.153 ± 0.012 | |
| A1 (mshA−) | 1.057 ± 0.028 | 0.01514 |
| A1C | 1.116 ± 0.020 | 0.0180** |
| S24 (mshC)− | 0.852 ± 0.132 | 0.04119 |
| S24C | 1.159 ± 0.034 | 0.88105 |
| R119 (egtA)− | 1.188 ± 0.003 | 0.02736 |
| R119C | 1.054 ± 0.030 | 0.01847 |
| SHP1 (mscR)− | 0.713 ± 0.101 | 0.00249 |
| SHP1C | 1.120 ± 0.009 | 0.03352 |
Biofilms were assayed at OD630 for four replicates.
P-value when compared with wild type calculated using a paired Student’s two-tailed t-test.
Partial complementation due to inducible nature of the acetamidase promoter controlling the mshA expression in pALACE vector (22).
Mutants, disrupted in mshC, such as S24, are sensitive to oxidative stress (34), which may hinder biofilm formation due to the accumulation of reactive oxygen species. Interestingly, A1, disrupted in mshA, which codes for a glycosyl transferase catalyzing the first step in the MSH biosynthetic pathway (35) also had reduced biofilm formation albeit not as severely reduced as S24 and SHP1 (Table 2), and this was partially reversed in the complemented strain. R119 had slightly improved biofilm formation when compared with wild type, and this was reversed in the complemented strain R119C. The less attenuated phenotype of A1 may be due to the overproduction of Ohr, an organic hydroperoxide resistance protein, and increased levels of ESH, which compensate for the lack of MSH and reduce oxidative stress in this mutant (36). Recently, Trivedi et al. reported that M. tuberculosis exposed to DTT, which causes an increase in reductive stress, produced more biofilm. Genes involved in protection against oxidative stress, such as katG, ahpC, and ahpD, were upregulated on DTT exposure (37). It is also possible that in A1 and R119, an enzyme that neutralizes NO and/or GSNO maybe upregulated in a similar manner as Ohr. Mutants disrupted in other genes involved in oxidative stress protection, such as the superoxide dismutase mutant in Listeria monocytogenes (38) and a mutant disrupted in glutathione reductase in N. gonorrhoeae (39), were previously found to be impaired in biofilm formation. There was no difference in biofilm formation in Pseudomonas aeruginosa catalase mutants (40), and an alkyl hydroperoxide reductase mutant of Campylobacter jejuni (41) had increased biofilm formation.
As the formation of a biofilm often leads to persistent and chronic infections, which increase morbidity and mortality, our results suggest that novel strategies to inhibit and/or disrupt biofilm formation and possibly to alter biofilm dispersal (inducing or preventing it) may exist through targeting LMW thiols and S-nitrosothiol reductases in bacteria.
Acknowledgments
This study was supported by the National Institutes of Health (NIH) grant R00AI100896 to Clarissa J. Nobile and NIH grant 5SC3GM100855-03 to Mamta Rawat. The authors thank Glenda Polack, Ruzan Orkusyan, and Pisiwat Kongsomboonvech for technical assistance.
References
- 1.Bogdan C. Nitric oxide and the immune response. Nat Immunol. 2001;2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 2.Chan ED, Chan J, Schluger NW. What is the role of nitric oxide in murine and human host defense against tuberculosis? Am J Respir Cell Mol Biol. 2001;25:606–612. doi: 10.1165/ajrcmb.25.5.4487. [DOI] [PubMed] [Google Scholar]
- 3.Seth D, Stamler JS. The SNO-proteome: causation and classifications. Curr Opin Chem Biol. 2011;15:129–136. doi: 10.1016/j.cbpa.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu L, Hausladen A, Zeng M, Que L, Heitman J, et al. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–494. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- 5.Staab CA, Hellgren M, Höög JO. Medium- and short-chain dehydrogenase/reductase gene and protein families. Cell Mol Life Sci. 2008;65:3950–3960. doi: 10.1007/s00018-008-8592-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kubienova L, Kopečný D, Tylichovà M, Briozzo P, Skopalovà J, et al. Structural and functional characterization of a plant S-nitrosoglutathione reductase from Solanum lycopersicum. Biochimie. 2013;95:889–902. doi: 10.1016/j.biochi.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 7.Kidd SP, Jiang D, Tikhomirova A, Jennings MP, McEwan AG. A glutathione-based system for defense against carbonyl stress in Haemophilus influenzae. BMC Microbiol. 2012;12:159. doi: 10.1186/1471-2180-12-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newton GL, Buchmeier N, Fahey RC. Biosynthesis and functions of mycothiol, the unique protective thiol of Actinobacteria. Microbiol Mol Biol Rev. 2008;72:471–494. doi: 10.1128/MMBR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guerra-Lopez D, Daniels L, Rawat M. Mycobacterium smegmatis mc2 155 fbiC and MSMEG_2392 are involved in triphenylmethane dye decolorization and coenzyme F420 biosynthesis. Microbiology. 2007;153:2724–2732. doi: 10.1099/mic.0.2006/009241-0. [DOI] [PubMed] [Google Scholar]
- 10.Liu YB, Long MX, Yin YJ, Si MR, Zhang L, et al. Physiological roles of mycothiol in detoxification and tolerance to multiple poisonous chemicals in Corynebacterium glutamicum. Arch Microbiol. 2013;195:419–429. doi: 10.1007/s00203-013-0889-3. [DOI] [PubMed] [Google Scholar]
- 11.Kaakoush N, Sterzenbach T, Miller W, Suerbaum S, Mendz G. Identification of disulfide reductases in Campylobacterales: a bioinformatics investigation. Antonie Van Leeuwenhoek. 2007;92:429–441. doi: 10.1007/s10482-007-9171-5. [DOI] [PubMed] [Google Scholar]
- 12.Vogt RN, Steenkamp DJ, Zheng R, Blanchard JS. The metabolism of nitrosothiols in the mycobacteria: identification and characterization of S-nitrosomycothiol reductase. Biochem J. 2003;374:657–666. doi: 10.1042/BJ20030642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Misiti F, Castagnola M, Zuppi C, Giardina B, Messana I. Role of ergothioneine on S-nitrosoglutathione catabolism. Biochem J. 2001;356:799–804. doi: 10.1042/0264-6021:3560799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paul BD, Snyder SH. The unusual amino acid l-ergothioneine is a physiologic cytoprotectant. Cell Death Differ. 2010;17:1134–1140. doi: 10.1038/cdd.2009.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen NH, Couñago RM, Djoko KY, Jennings MP, Apicella MA, et al. A glutathione-dependent detoxification system is required for formaldehyde resistance and optimal survival of Neisseria meningitidis in biofilms. Antioxidants Redox Signaling. 2013;18:743–755. doi: 10.1089/ars.2012.4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards JP, Ojha AK. Mycobacterial biofilms. Microbiol Spectr. 2014;2:1–13. doi: 10.1128/microbiolspec.MGM2-0004-2013. [DOI] [PubMed] [Google Scholar]
- 17.Rawat M, Heys J, Av-Gay Y. Identification and characterization of a diamide sensitive mutant of Mycobacterium smegmatis. FEMS Microbiol. 2002;220:161–169. doi: 10.1016/S0378-1097(03)00127-7. [DOI] [PubMed] [Google Scholar]
- 18.Seebeck FP. In vitro reconstitution of mycobacterial ergothioneine biosynthesis. J Am Chem Soc. 2010;132:6632–6633. doi: 10.1021/ja101721e. [DOI] [PubMed] [Google Scholar]
- 19.Franzoni F, Colognato R, Galetta F, Laurenza I, Barsotti M, et al. An in vitro study on the free radical scavenging capacity of ergothioneine: comparison with reduced glutathione, uric acid and trolox. Biomed Pharmacother. 2006;60:453–457. doi: 10.1016/j.biopha.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 20.MacMicking J, Xie Q, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 21.Kidd SP, Jiang D, Jennings MP, McEwan AG. Glutathione-dependent alcohol dehydrogenase AdhC is required for defense against nitrosative stress in Haemophilus influenzae. Infect Immun. 2007;75:4506–4513. doi: 10.1128/IAI.00487-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stroeher UH, Kidd SP, Stafford SL, Jennings MP, Paton JC, et al. A pneumococcal MerR-like regulator and S-nitrosoglutathione reductase are required for systemic virulence. J Infect Dis. 2007;196:1820–1826. doi: 10.1086/523107. [DOI] [PubMed] [Google Scholar]
- 23.Venketaraman V, Dayaram YK, Talaue MT, Connell ND. Glutathione and nitrosoglutathione in macrophage defense against Mycobacterium tuberculosis. Infect Immun. 2005;73:1886–1889. doi: 10.1128/IAI.73.3.1886-1889.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broniowska KA, Diers AR, Hogg N. S-Nitrosoglutathione. Biochim Biophys Acta. 2013;1830:3173–3181. doi: 10.1016/j.bbagen.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sassetti CM, Boyd DH, Rubin EJ. Comprehensive identification of conditionally essential genes in mycobacteria. Proc Natl Acad Sci USA. 2001;98:12712–12717. doi: 10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Witthoff S, Mühlroth A, Marienhagen J, Bott M. C1 metabolism in Corynebacterium glutamicum: an endogenous pathway for oxidation of methanol to carbon dioxide. Appl Environ Microbiol. 2013;79:6974–6983. doi: 10.1128/AEM.02705-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isobe K, Kataoka M, Ogawa J, Hasegawa J, Shimizu S. Microbial oxidases catalyzing conversion of glycolaldehyde into glyoxal. New Biotechnol. 2012;29:177–182. doi: 10.1016/j.nbt.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Ferguson GP, Totemeyer S, Maclean MJ, Booth IR. Methylglyoxal production in bacteria: suicide or survival? Arch Microbiol. 1998;170:209–219. doi: 10.1007/s002030050635. [DOI] [PubMed] [Google Scholar]
- 29.Yang K, Feng C, Lip H, Bruce WR, O’Brien PJ. Cytotoxic molecular mechanisms and cytoprotection by enzymic metabolism or autoxidation for glyceraldehyde, hydroxypyruvate and glycolaldehyde. Chem Biol Interact. 2011;191:315–321. doi: 10.1016/j.cbi.2011.02.027. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida N, Hayasaki T, Takagi H. Gene expression analysis of methylotrophic oxidoreductases involved in the oligotrophic growth of Rhodococcus erythropolis N9T-4. Biosci Biotechnol, Biochem. 2011;75:123–127. doi: 10.1271/bbb.100700. [DOI] [PubMed] [Google Scholar]
- 31.Echenique JR, Dorsey CW, Patrito LC, Petroni A, Tolmasky ME, et al. Acinetobacter baumannii has two genes encoding glutathione-dependent formaldehyde dehydrogenase: evidence for differential regulation in response to iron. Microbiology. 2001;147:2805–2815. doi: 10.1099/00221287-147-10-2805. [DOI] [PubMed] [Google Scholar]
- 32.Ojha AK, Baughn AD, Sambandan D, Hsu T, Trivelli X, et al. Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol Microbiol. 2008;69:164–174. doi: 10.1111/j.1365-2958.2008.06274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barraud N, Kelso MJ, Rice SA, Kjelleberg S. Nitric oxide: a key mediator of biofilm dispersal with applications in infectious diseases. Curr Pharm Des. 2015;21:31–42. doi: 10.2174/1381612820666140905112822. [DOI] [PubMed] [Google Scholar]
- 34.Rawat M, Newton GL, Ko M, Martinez GJ, Fahey RC, et al. Mycothiol-deficient Mycobacterium smegmatis mutants are hypersensitive to alkylating agents, free radicals, and antibiotics. Antimicrob Agents Chromother. 2002;46:3348–3355. doi: 10.1128/AAC.46.11.3348-3355.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newton GL, Koledin T, Gorovitz B, Rawat M, Fahey RC, et al. The glycosyltransferase gene encoding the enzyme catalyzing the first step of mycothiol biosynthesis (mshA) J Bacteriol. 2003;185:3476–3479. doi: 10.1128/JB.185.11.3476-3479.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ta P, Buchmeier N, Newton GL, Rawat M, Fahey RC. Organic hydroperoxide resistance protein and ergothioneine compensate for loss of mycothiol in Mycobacterium smegmatis mutants. J Bacteriol. 2011;193:1981–1990. doi: 10.1128/JB.01402-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trivedi A, Mavi PS, Bhatt D, Kumar A. Thiol reductive stress induces cellulose-anchored biofilm formation in Mycobacterium tuberculosis. Nat Commun. 2016;7:11392. doi: 10.1038/ncomms11392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suo Y, Huang Y, Liu Y, Shi C, Shi X. The expression of superoxide dismutase (SOD) and a putative ABC transporter permease is inversely correlated during biofilm formation in Listeria monocytogenes 4b G. PLoS One. 2012;7:e48467. doi: 10.1371/journal.pone.0048467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seib KL, Wu HJ, Kidd SP, Apicella MA, Jennings MP, et al. Defenses against oxidative stress in Neisseria gonorrhoeae: a system tailored for a challenging environment. Microbiol Mol Biol Rev. 2006;70:344–361. doi: 10.1128/MMBR.00044-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elkins JG, Hassett DJ, Stewart PS, Schweizer HP, McDermott TR. Protective role of catalase in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide. Appl Environ Microbiol. 1999;65:4594–4600. doi: 10.1128/aem.65.10.4594-4600.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oh E, Jeon B. Role of alkyl hydroperoxide reductase (AhpC) in the biofilm formation of Campylobacter jejuni. PLoS One. 2014;9:e87312. doi: 10.1371/journal.pone.0087312. [DOI] [PMC free article] [PubMed] [Google Scholar]


