Abstract
The n-3 PUFAs have many beneficial effects on human health, including roles in immunity, neurodevelopment, and preventing cardiovascular disease. In this study, we established reliable model fat-1 transgenic cattle using transgenic technology and performed a systematic investigation to examine the function of n-3 PUFAs. Our results showed that expression of the fat-1 gene improved several biochemical parameters related to liver function and to plasma glucose and plasma lipid metabolism. Results of global gene and plasma protein expression analysis showed that 310 genes and 13 plasma proteins differed significantly in the blood of fat-1 transgenic cattle compared with WT cattle, reflecting their regulatory roles in the immune and cardiovascular systems. Finally, changes in the gut microflora were also noted in the fat-1 transgenic cattle, suggesting novel roles for n-3 PUFAs in the metabolism of glucose and lipids, as well as anti-stress properties. To the best of our knowledge, this is the first report using multiple parallel analyses to investigate the role of n-3 PUFAs using models such as fat-1 transgenic cattle. This study provides novel insights into the regulatory mechanism of fat-1 in the immune and cardiovascular systems, as well as its anti-stress role.
Keywords: n-3 fatty acids, lipid metabolism, stress
The n-3 PUFAs are beneficial to human health. Previous studies have revealed that increased intake of n-3 PUFAs could reduce the risk of major human diseases, including cardiovascular disease, type 2 diabetes, and several types of cancer (1–4). Previous studies have also shown that n-3 PUFAs play a positive role in the immune system by mediating the improved inflammatory response (5–7). The n-3 PUFAs are a collection of PUFAs that include α-linolenic acid (ALA), DHA, and EPA. ALA is abundant in plant oils and can be obtained directly through the diet (8). Conversion of ALA in mammals can produce DHA and EPA through the desaturation-chain elongation pathway (9). Although a greater conversion capacity for ALA to DHA was found in women than men, the synthesis efficiency is limited (10). Therefore, DHA and EPA are also primarily obtained from the diet.
The fat-1 gene encodes n-3 PUFA desaturase, which can specifically convert n-6 PUFAs to n-3 PUFAs. Transgenic technology can be used to produce fat-1 transgenic domestic animals, which could then generate food to supply n-3 PUFAs for human consumption. In 2004, Kang et al. (11) reported the first fat-1 transgenic mice, which could synthesize n-3 PUFAs from n-6 PUFAs through constitutive expression of the fat-1 gene in vivo, showing that it would be feasible to obtain a rich supply of n-3 PUFAs from transgenic domestic animals. Subsequently, various fat-1 transgenic domestic animals have been generated using transgenic technology. These studies have focused more heavily on fat-1 transgenic pigs than either sheep or cattle. Lai et al. (12) generated the first fat-1 transgenic pigs in 2006 and demonstrated their elevated level of n-3 PUFAs, which were three times more abundant in the tail tissue of fat-1 transgenic pigs than in WT pigs. In 2010, Pan et al. (13) generated 21 piglets by transgenic somatic cell nuclear transfer; 15 of these piglets survived, and 13 of these were confirmed to positively express the fat-1 gene. The first fat-1 transgenic cattle were produced in our laboratory in 2009. The levels of four types of n-3 PUFA (18:3 n-3, 20:5 n-3, 22:6 n-3, and 22:5 n-3) in the ear tissues of the fat-1 transgenic cattle were significantly higher than those in the same tissues of WT cattle, whereas three types of n-6 PUFAs (18:2 n-6, 20:4 n-6, and 22:5 n-6) were significantly lower (14). Next, Zhang et al. (15) generated three fat-1 transgenic sheep in 2013. High levels of n-3 PUFAs, as well as a low ratio of n-6/n-3 PUFAs, were observed in the heart, liver, spleen, lung, kidney, brain, ear, tail, and muscle tissues. These results indicated the fat-1 genes had a physiological function converting n-6 PUFAs into n-3 PUFAs.
The mouse is a convenient model animal for experimental research. Therefore, fat-1 mice have been extensively employed to investigate the role of n-3 PUFAs in many diseases. Li et al. (10) used fat-1 mice rich in endogenous n-3 PUFAs to explore the protective effect of n-3 PUFAs in immune-mediated liver injury, showing that n-3 PUFAs limit concanavalin A-induced hepatitis via an autophagy-dependent mechanism. Additionally, an anti-tumor function of n-3 PUFAs in fat-1 mice was suggested by studies showing a reduction in colitis-associated colon cancer associated with a decreased inflammatory response (16). Recently, fat-1 mice have been used in several models of neurological disease, including Parkinson’s disease (17), Alzheimer’s disease (18), epilepsy (19), chronic inflammatory demyelinating disease of the central nervous system (20), and stroke-related brain injury (21). All fat-1 mice models were protected from neuronal damage when compared with their WT littermates (20).
In this study, we first confirmed the expression and target function of the fat-1 gene in transgenic cattle based on several parameters, including DNA, RNA, and protein and fatty acid properties. Furthermore, we used fat-1 cattle models to investigate the role of n-3 PUFAs using multiple methods, including measurement of blood biochemical parameters, levels of gene expression and plasma proteins in the blood; we also evaluated changes in the gut microflora. To the best of our knowledge, this is the first report using multiple parallel analyses to investigate the function of n-3 PUFAs using fat-1 transgenic cattle models. The present study provides a valuable reference as well as novel insights into the function of n-3 PUFAs.
MATERIALS AND METHODS
Ethics statement
All procedures performed for this study were consistent with the National Research Council Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of Inner Mongolia University.
Production and identification of transgenic cattle
Using the previously constructed fat-1 gene expression vector, PST200, bovine fetal fibroblasts were transfected, positive cells were pooled to produce transgenic embryos using somatic cell nuclear transfer, and the highest quality transgenic blastocysts were then selected for embryo transfer; all procedures were performed using previously described methods (14).
Ear tissue samples were taken from transgenic calves at 2 months of age. Total DNA and RNA were extracted from these tissues following previously described methods (14). The presence of the fat-1 gene in each DNA sample was confirmed using PCR. Expression of fat-1 mRNA was detected via RT-PCR. Primers specific for the fat-1 gene (5′-ATTGTCAGGGCGATGTAGGC-3′ and 5′-CGGCTATCTGGTGTGGAACA-3′) were used for PCR and RT-PCR. The amplification conditions for PCR and RT-PCR included 35 cycles of incubation at 94°C for 30 s, at 62°C for 30 s, and at 72°C for 40 s. The amplification products were subjected to electrophoresis on a 1.5% agarose gel. Subsequently, Western blotting was performed for detection of the fat-1-encoded protein based on a custom antibody (Genecreate, Wuhan, China).
Animal feeding
To obtain reliable data for this study, all cattle, including transgenic and WT cattle, were housed in a concrete-sided cowshed prior to sample collection, fed the same diet (commercial concentrated feed and wet corn silage), and monitored daily to ensure their health.
Tissue fatty acid analysis
Ear tissue samples were taken from six transgenic calves and six WT calves at 3 months of age to extract lipids, and PUFA extraction was performed using gas chromatography as previously described (14, 22).
Detection of blood biochemical parameters
Blood was collected from the jugular vein of three surviving transgenic calves and three WT calves at 6 months of age; these calves had been given limited feed for 24 h prior to sample collection. Each blood sample was placed into a tube containing 5% ethylene-diamine-tetraacetic acid. Plasma was separated by centrifugation at 3,500 g/min and 4°C for 15 min. Using the same methods, plasma was collected from the three surviving transgenic cattle and three WT cattle at 18 months and 4 years of age. Next, blood biochemical indices, including those for liver function [aspartate aminotransferase (AST), alanine aminotransferase (ALT), and lactate dehydrogenase (LDH)], renal function [creatinine (CRE)], plasma glucose (GLU), and plasma lipids [triglyceride (TG), total cholesterol (TC), high density lipoprotein cholesterol (HDL-C), and low density lipoprotein cholesterol (LDL-C)] were measured using the fully automatic biochemical analyzer, Glamour 3000 (Misiones Bernal, Buenos Aires, Argentina).
Detection of the blood gene expression profile
Total RNA was extracted from the blood of three transgenic cattle and three WT cattle using the Trizol extraction protocol and purified using an RNeasy mini kit (Qiagen, Germany), following the manufacturer’s protocol. For quality control, total RNA was quantified using a NanoDrop ND-2000 spectrophotometer (Thermo Scientific), and RNA integrity was assessed using an Agilent Bioanalyzer 2100 (Agilent Technologies). After completion of quality control procedures, total RNA was reverse transcribed into double-stranded cDNA; next, cRNA was synthesized, labeled with cyanine-3-CTP, purified, and hybridized to the bovine gene expression microarray (4 × 44K, Design ID: 023647). After elution onto hybridized arrays, the arrays were scanned at 5 μm using the Agilent scanner G2505C (Agilent Technologies). Feature Extraction software (version 10.7.1.1, Agilent Technologies) was used to analyze the array images and obtain raw data, whereas Gene Spring software was employed to perform general analysis. The raw data were normalized using the quantile algorithm. Gene expression data were deposited into the NCBI Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) and can be accessed via accession number GSE66651. Differentially expressed genes were then identified based on fold changes. P values were calculated using the t-test, and the false discovery rate (FDR) was calculated to correct the P values using the R statistical package. The threshold for differentially expressed genes was set as a fold change >2.0, P < 0.01, and FDR value <0.05. Hierarchical clustering was performed to visualize gene expression patterns among the samples. GO analysis was used to determine the roles of these differentially expressed mRNAs, and biological functions with a P < 0.05 were considered statistically significant.
Detection and analysis of differential plasma proteins
Blood was collected in heparinized tubes from three transgenic cattle and three WT cattle. All six cattle were allowed limited feed for 24 h prior to sample collection. Whole blood was centrifuged at 7,000 g/min and 4°C for 10 min to obtain plasma, which was stored at −80°C until two-dimensional gel electrophoresis (2-DE) was performed. Equal amounts of plasma from three WT cattle were pooled to obtain the control group sample. The total plasma protein concentration was measured via the Bradford method using a protein assay reagent (Bio-Rad, Hercules, CA). The 2-DE was carried out based on the methods used in previous reports (23). After 2-DE was performed, differentially expressed protein spots were screened based on the threshold of >1.5-fold change and a P value <0.05. These differentially expressed protein spots were identified using mass spectrometry, as previously described (23). The annotation of the identified differential proteins was performed using the online tool, STRING (http://string-db.org/). Interactions among these differentially expressed proteins were also predicted using this tool. Common interaction proteins identified from the three transgenic cattle were extracted. These common interaction proteins were subjected to GO and KEGG analyses to gain insight into the effects on the three fat-1 transgenic cattle at the plasma protein level.
Detection and analysis of gut microbes
Fecal samples were obtained from three transgenic cattle and three WT cattle by rectal palpation using sterile technique on the same morning. During collection, the outside air temperature was 15–18°C, and each sample was transferred into a separate sterilized container and immediately stored at 4°C, followed by long-term storage at −80°C until DNA was extracted. None of the cattle had received antibiotics within the past 3 months, and none had experienced gastrointestinal or acute disease. All cattle were housed in a concrete-sided cattle shed for 1 month prior to sample collection, were fed the same diet (commercial concentrate feed and wet corn silage), and were monitored every day to ensure their health. Total genomic DNA was extracted from fecal samples using a QIAamp stool DNA mini kit, in accordance with the manufacturer’s protocol (Qiagen; 51504). Pyrobest DNA polymerase (Ta-KaRa, DR500A) was used for amplification of the V4 hypervariable region of the 16S rRNA gene from microbial genomic DNA. Next, the V4 amplicons were sequenced using the paired-end method on an Illumina MiSeq sequencer with a six-cycle index read. The raw sequence data were deposited into the NCBI database (https://www.ncbi.nlm.nih.gov/home/submit.shtml) and can be accessed via accession number SUB2038674. Sequence reads were trimmed so that the average Phred quality score for each read was greater than 30 and the read was longer than 50 bp; after trimming, these reads were assembled using Flash software (http://ccb.jhu.edu/software/FLASH/), and reads that could not be assembled were discarded. Only reads with series of consecutive identical bases shorter than 6 bp and without ambiguous bases were used for further analysis. Sequence clustering was performed using UCLUST (QIIME) with a similarity cutoff of 99% to form operational taxonomic units (OTUs). Next, the number and abundance of OTUs were determined for all samples. The species diversity (Chao1, ACE, Simpson, Shannon, and Coverage) of gut microbes was analyzed using the single summary command in MOTHUR software (http://www.mothur.org/). Differences in species and their distribution in the gut microbiota were analyzed based on the abundance profiles in transgenic and WT cattle.
Statistical analysis
All data are expressed as mean values ± SD. The results were analyzed using Student’s t-test, and differences were considered significant at either P < 0.05 or P < 0.01.
RESULTS
Generation of transgenic cattle expressing the fat-1 gene
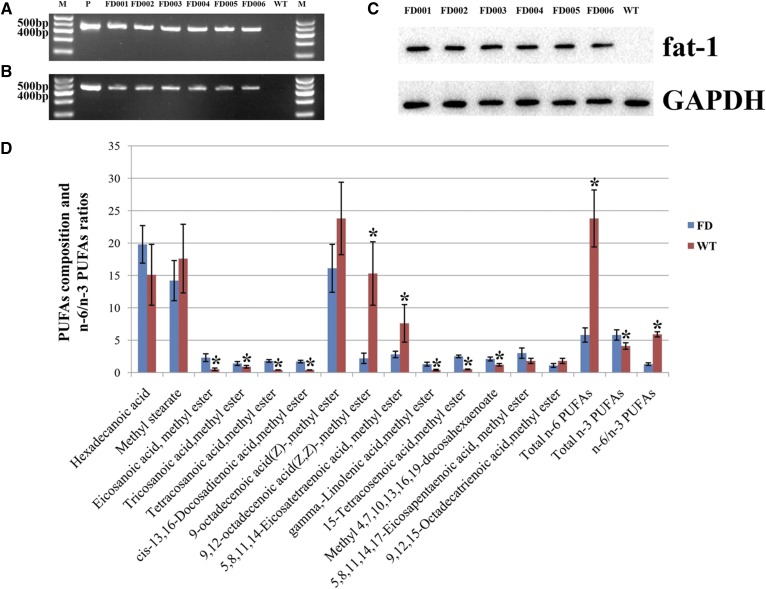
In total, 1,156 reconstructed oocytes were produced after transferring fat-1 transgenic cells into enucleated oocyte cytoplasts. The cleavage and blastocyst rates were 80% and 32%, respectively. In total, 157 blastocysts were transferred to 106 synchronized recipient cattle. Pregnancy rates were 37.7% (40/106), 28.3% (30/106), and 18.8% (20/106) at 60, 90, and 210 days, respectively. In total, nine female calves were delivered naturally 280–286 days after transfer (Table 1). Of these nine calves, six fat-1 transgenic cattle were identified based on DNA, RNA, and protein analyses, and they were designated FD001, FD002, FD003, FD004, FD005, and FD006, respectively (Fig. 1A–C). At the present time, three of the fat-1 transgenic cattle (FD002, FD005, and FD006) are still alive and healthy. The survival rate of transgenic cattle was 33.3%.
TABLE 1.
Development of bovine transgenic cloned embryos in vitro
| Item | Transferred Blastocysts (n) | Recipient Cattle (n) | 60-day Pregnancy (% Recipients) | 120-Day Pregnancy (% Recipients) | 210-Day Pregnancy (% Recipients) | Number Birth (% Recipients) | Number Survived (% Births) |
| Total | 157 | 106 | 40 (37.7) | 30 (28.3) | 20 (18.8) | 9 (8.5) | 3 (33.3) |
Fig. 1.
Identification for the fat-1 transgenic cattle. A: Detection of fat-1 gene (450 bp) delivered into transgenic cattle by PCR. B: Detection of fat-1 gene (450 bp) expression in transgenic cattle by RT-PCR. The fat-1 expression vector was used as the template for the positive control (P) and WT cattle were used as the negative control. C: Detection of the protein expression of fat-1 gene in transgenic cattle by Western blot. D: Analysis of the fatty acid composition in the tissues of fat-1 transgenic cattle. Fatty acids were measured as methyl esters by gas chromatography. Bars represent the mean ± SD from six ear samples in each group (six transgenic calves (FD) and six age-matched WT calves) with two independent measurements for each sample. Significant differences of fatty acid composition in the tissues were found between the WT cattle and transgenic cattle (*P < 0.05).
Tissue fatty acid composition of fat-1 transgenic cattle
To assess the activity of the fat-1 gene in transgenic cattle, fatty acids in the ear tissues were analyzed. As shown in Fig. 1D, various changes were found in the 14 types of fatty acid. Among the saturated fatty acids, eicosanoic acid, tricosanoic acid, and tetracosanoic acid increased significantly compared with the levels in the WT cattle used as the control group. The concentrations of two n-3 PUFAs, 5,8,11,14,17-EPA and 4,7,10,13,16,19-DHA were elevated in transgenic cattle compared with WT cattle, and the difference between the two groups for DHA was significant. On the other hand, the concentration of 9,12,15-octadecatrienoic acid (ALA) was lower in transgenic cattle than in WT cattle, but this difference was not significant. Levels of the n-6 PUFAs, 9,12-octadecenoic acid and 5,8,11,14-eicosatetraenoic acid, were significantly reduced in transgenic cattle compared with WT cattle, whereas both γ-linolenic acid and 15-tetracosenoic acid were increased significantly. Total n-6 PUFAs were significantly reduced and total n-3 PUFAs were significantly elevated in the transgenic cattle. However, the degree of reduction of n-6 PUFAs was greater than the degree of increase of n-3 PUFAs in the transgenic cattle. The n-6/n-3 PUFA ratio, which is approximately 1:1, was significantly reduced in the transgenic cattle. These results indicated that the expected function of the delivered fat-1 gene was successfully established using transgenic cattle.
Blood biochemical parameters of fat-1 transgenic cattle
The results of blood biochemical levels for both fat-1 transgenic and WT cattle are shown in Table 2, which shows that only ALT, the biochemical parameter representing liver function, was significantly lower in transgenic cattle at 6 months than in WT cattle of the same age. When these cattle reached 18 months of age, AST, GLU, TC, and LDL-L were significantly reduced in the transgenic cattle relative to the WT cattle. Only GLU was significantly lower in the transgenic cattle than in the WT cattle when the cattle were 4 years old.
TABLE 2.
Comparison of the blood biochemical parameters between the fat-1 transgenic cattle and WT cattle
| Item | Calf (6 months old) | Adult Cattle (18 months old) | Adult Cattle (4 years old) | |||
| FD | WT | FD | WT | FD | WT | |
| AST (U/l) | 63.34 ± 23.51 | 69.20 ± 2.61 | 77.98 ± 13.45a | 91.02 ± 3.72 | 83.35 ± 9.84 | 92.01 ± 2.74 |
| ALT (U/l) | 13.90 ± 1.91a | 16.63 ± 0.75 | 35.90 ± 1.35 | 38.27 ± 0.35 | 36.08 ± 0.38 | 37.46 ± 2.59 |
| LDH (U/l) | 693.33 ± 105.08 | 785.33 ± 70.00 | 871.7 ± 79.31 | 993.37 ± 133.55 | 894.16 ± 82.17 | 969.41 ± 102.49 |
| CRE (μmol/l) | 55.33 ± 31.82 | 48.67 ± 9.61 | 89.67 ± 15.57 | 66.67 ± 13.20 | 88.56 ± 13.69 | 68.11 ± 10.84 |
| GLU (mmol/l) | 4.73 ± 0.25 | 4.80 ± 0.95 | 3.53 ± 0.12a | 4.13 ± 0.21 | 3.63 ± 0.24a | 4.16 ± 0.16 |
| TG (mmol/l) | 0.10 ± 0.00 | 0.17 ± 0.06 | 0.23 ± 0.12 | 0.23 ± 0.06 | 0.22 ± 0.05 | 0.27 ± 0.03 |
| TC (mmol/l) | 2.10 ± 0.36 | 2.30 ± 1.23 | 2.03 ± 0.06a | 2.90 ± 0.05 | 2.14 ± 0.05 | 2.66 ± 0.48 |
| HDL-C (mmol/l) | 1.82 ± 0.12 | 1.88 ± 1.03 | 1.85 ± 0.07 | 2.45 ± 0.48 | 2.00 ± 0.10 | 2.41 ± 0.34 |
| LDL-C (mmol/l) | 0.14 ± 0.09 | 0.17 ± 0.05 | 0.19 ± 0.03a | 0.31 ± 0.08 | 0.24 ± 0.03 | 0.29 ± 0.05 |
Plasma content of AST, ALT, LDH, CRE, GLU, TG, TC, HDL-C, and LDL-C in the WT cattle and transgenic cattle.
P < 0.05, WT cattle compared with the transgenic cattle.
Analysis of integration site and copy number in fat-1 transgenic cattle
In the present study, only the FD006 fat-1 transgenic animal was used for identification of the integration site and copy number using high-throughput sequencing (see supplemental Materials and Methods). Through a BLAST search of these sequencing reads, we obtained four bridging paired-end reads in which one end mapped to chromosome 16 of the bovine genome and the other end mapped to the fat-1 vector region. The four bridging paired-end reads were split for further analysis of the specific integration break points. The right boundary was located between position 15726078 of chromosome 16 and position 6475 of the inserted fat-1 vector (supplemental Fig. S1). However, we did not obtain bridging paired-end reads for the left boundary between chromosome 16 and the fat-1 vector region. More interestingly, introduction of a 10-nucleotide portion of the bovine genome at insertion site 15726078 of chromosome 16 was also observed, which is a characteristic signature of transgene integration (supplemental Fig. S1). To verify the transgene integration site, event-specific PCR was performed on FD006 DNA samples (supplemental Fig. S2). The sequencing depths for insertion site 15726078 of chromosome 16 and insertion site 6475 of fat-1 vector were 6× and 11×, respectively (supplemental Fig. S3). Therefore, we speculate that the transgene is a single copy. This result represents preliminary work to identify the transgene integration site and copy number in FD006. Identification of the integration site and copy number for two other transgenic cattle (FD002 and FD005) and analysis of genetic stability in these cattle will be performed in subsequent studies.
Global gene expression in fat-1 transgenic cattle
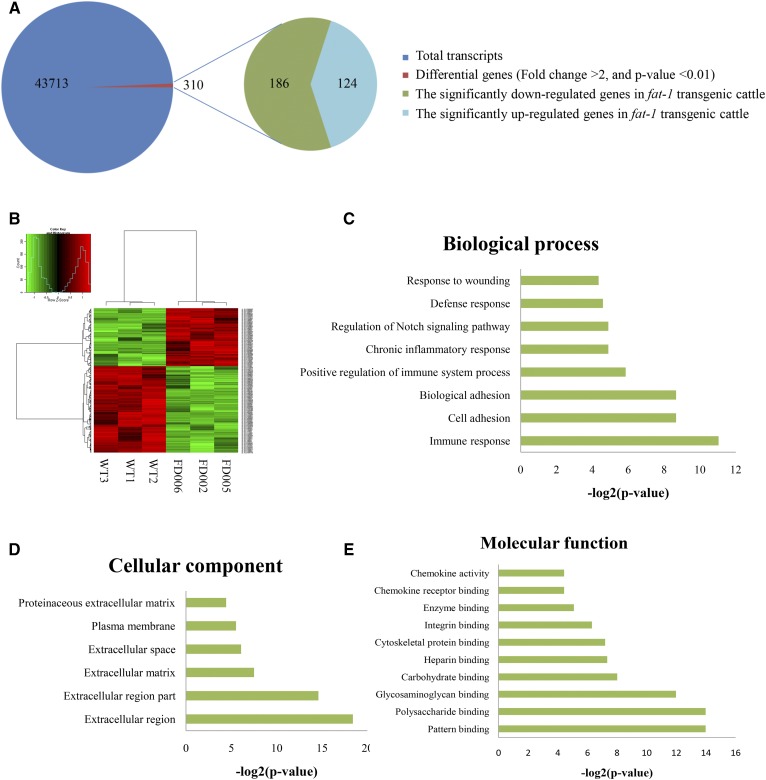
Next, the gene expression patterns of the fat-1 transgenic and WT cattle were compared using an expression profile microarray. In total, 43,711 transcript sequences were used as probes. The results showed that 310 transcripts differed significantly in expression (P < 0.01, FDR < 0.05), reflecting up to a 2-fold change between the fat-1 transgenic and WT cattle (Fig. 2A). Of these 310 transcripts, 124 were significantly upregulated in fat-1 transgenic cattle compared with WT cattle (Fig. 2A). After hierarchical clustering analysis of the 310 genes, all three transgenic cattle showed consistent genetic backgrounds, which differed from the expression pattern of the WT cattle (Fig. 2B).
Fig. 2.
Analysis of gene expression in the fat-1 transgenic cattle. A: Schematic represents the differentially expressed genes between fat-1 transgenic cattle and WT cattle. B: The analysis on hierarchical clustering of the expressed genes in fat-1 transgenic cattle and WT cattle. C: Classification of microarray biological process. D: Classification of microarray cellular components; E: Classification of microarray molecular function. The x axis indicates the likelihood [−log2(P-value)] in a category and the y axis indicates the different subcategories of biological process.
To gain further insight into the potential influence of these 310 differentially expressed genes, GO enrichment analysis was performed. Twenty-four GO terms were significantly enriched, including eight GO terms representing biological processes, six for cellular components, and 10 for molecular functions (Fig. 2C–E). Among the GO terms for biological processes, the most highly enriched terms were associated with immune and inflammatory responses, including “immune response,” “positive regulation of immune system process,” “chronic inflammatory response,” and “defense response” (Fig. 2C). Among the GO terms for molecular functions, many binding processes, especially those related to immune functions, were significantly enriched, including “glycosaminoglycan binding,” “polysaccharide binding,” “integrin binding,” and “chemokine receptor binding” (Fig. 2E). These results further confirmed the possibility of immune regulation by the differentially expressed genes in fat-1 transgenic cattle. Finally, “extracellular region,” “extracellular region part,” “extracellular matrix,” “extracellular space,” and “plasma membrane” accounted for most of the terms in the cellular components category (Fig. 2D). The cell membrane system, including the extracellular region, plasma membrane, and other components, indicated an important role in the processes of cell signal propagation, suggesting that the cell membrane system’s important role in immune regulation is mediated by the fat-1 gene. Taken together, these results showed that insertion of the fat-1 gene into the cattle genome resulted in changes in gene expression related to the regulation of biological processes in both the immune system and the inflammatory response.
Plasma protein expression in fat-1 transgenic cattle
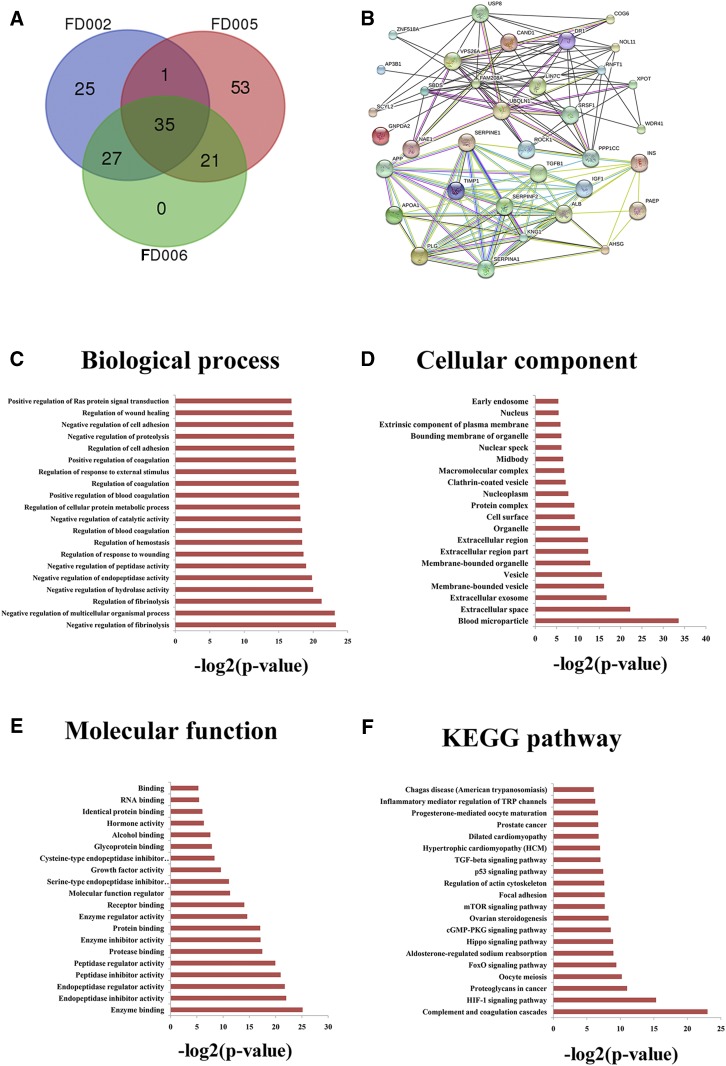
Differentially expressed plasma proteins were identified in three transgenic cattle using 2-DE and mass spectrometry. In total, six, eight, and four differentially expressed proteins were found in the FD002, FD005, and FD006 transgenic cattle, respectively (Table 3). To further investigate the functions of significantly differentially expressed proteins, potential interaction proteins were predicted for each transgenic animal. In total, 162 interaction proteins were found in the three transgenic cattle, including 35 common to all three transgenic cattle (Fig. 3A, B). GO and KEGG enrichment analyses of the 35 common interaction proteins resulted in the top 20 terms listed in Fig. 3C–F. Most of the GO terms were closely associated with blood coagulation, such as “negative regulation of fibrinolysis,” “regulation of fibrinolysis,” “regulation of blood coagulation,” “positive regulation of blood coagulation,” and “regulation of coagulation” (Fig. 3A). The “blood microparticle,” “extracellular exosome,” “membrane-bounded vesicle,” and “vesicle” terms were enriched in the cellular component (Fig. 3D), and the “complement and coagulation cascades” pathway was significantly enriched among KEGG terms (Fig. 3F). The “complement and coagulation cascades” pathway plays a positive role in innate immunity, acting as a nonspecific defense mechanism against pathogens and blood coagulation. These results indicate that the fat-1 gene potentially mediates regulation of thrombosis and the immune system.
TABLE 3.
Identified plasma differential proteins between transgenic and WT cattle
| Item | Protein Name |
| FD002 versus WT | IGJ, ALB, AZGP1, APOA4 ,Fam208A, CTAGE5 |
| FD005 versus WT | CLU, KNG1, AHSG, LOC511240, ORM1, ENSBTAG00000046739, Fam208A, Serpin A3-5 |
| FD006 versus WT | LOC511240, ALB, Fam208A, APOA4 |
Fig. 3.
Analysis of plasma proteins in the fat-1 transgenic cattle. A: Venn diagram of the three comparisons (WT/FD002, WT/FD005, and WT/FD006) based on the interacting proteins lists from the significantly differential plasma proteins in each group. The number of interacting proteins is indicated in the respective segments. B: Schematic of regulation network for the 35 common interacting proteins from the significantly differential plasma proteins of three groups (WT/FD002, WT/FD005, and WT/FD006). C–F: Function analysis of 35 common interacting proteins between the fat-1 transgenic cattle and WT cattle. C: Biological process classification. D: Cellular component classification. E: Molecular function classification. F: KEGG pathway classification. The x axis indicates the likelihood [−log2(P-value)] in a category and the y axis indicates the different subcategories of biological process.
Gut microbial community in fat-1 transgenic cattle
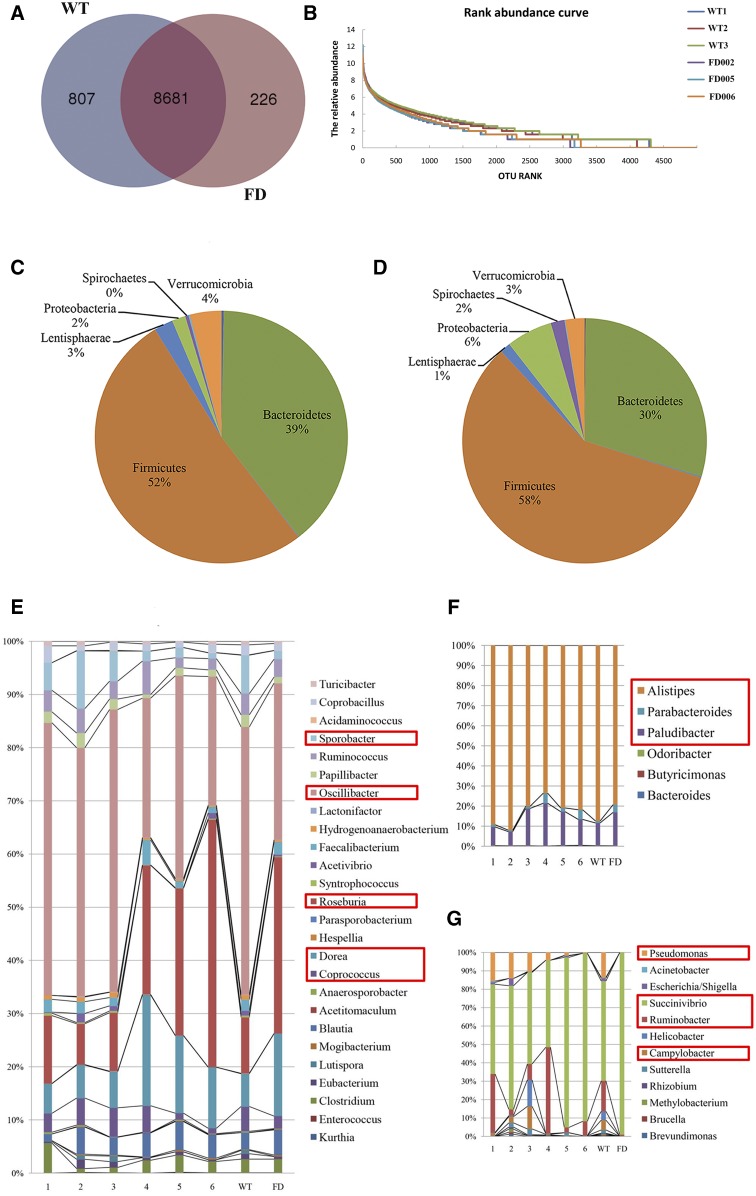
Fewer OTUs (8,907) were detected in the fat-1 transgenic cattle than in the WT cattle (9,488). There were 8,681 common OTUs found in both the fat-1 transgenic and WT cattle, 807 OTUs were unique to the WT cattle, and only 226 were unique to the fat-1 transgenic cattle (Fig. 4A). Species diversity indices of gut microbes, including Chao1 and Shannon, were significantly lower in the fat-1 transgenic cattle than in WT cattle (Table 4). Observation of the total abundance of gut microbes led to detection of a low abundance in fat-1 transgenic cattle (Fig. 4B).
Fig. 4.
Analysis of gut microbial community in the fat-1 transgenic cattle. A: Venn diagram of the comparison between the WT cattle and fat-1 transgenic cattle (FD) based on the OTU lists at 99% similarity. The number of OTUs is indicated in the respective segments. B: Distribution curve of species abundance for the WT cattle (WT1, WT2, WT3) and fat-1 transgenic cattle (FD002, FD005, FD006). Distribution of the gut microbiota composition at phylum level for WT (C) cattle and fat-1 transgenic (D) cattle. The genus distributions levels for Firmicutes (E), Bacteroidetes (F), and Proteobacteria (G), respectively. The numbers 1, 2, and 3 represent the distribution levels for each WT cow. The numbers 4, 5, and 6 represent the distributions level for FD002, FD005, and FD006 transgenic cattle. WT represents the mean distribution level for three WT cattle; FD represents the mean distribution level for three fat-1 transgenic cattle.
TABLE 4.
Analysis of species diversity at the level of 99% similarity
| Item | Chao1 | Ace | Simpson | Shannon | Coverage |
| WT | 11,257.7 ± 321.9 | 13,739.7 ± 471.3 | 0.0041 ± 0.001 | 7.031 ± 0.075 | 0.967 ± 0.002 |
| FD | 10,177.3 ± 156.2 | 13,759.3 ± 246.9 | 0.0062 ± 0.002 | 6.638 ± 0.158 | 0.962 ± 0.003 |
| P | 0.00638 | 0.952 | 0.184 | 0.018 | 0.055 |
P < 0.05 indicates significant differences in the WT cattle compared with the transgenic cattle.
Analysis of these microbes at the phylum and genus levels showed that Proteobacteria and Spirochaetes were significantly more abundant in fat-1 transgenic cattle, whereas Euryarchaeota were significantly less common (Table 5). Additionally, four genera (Odoribacter, Methanobrevibacter, Lutispora, and Spirochaetes) were significantly reduced in the fat-1 transgenic cattle, whereas five genera (Bacteroides, Treponema, Dorea, Roseburia, and Acetitomaculum) increased significantly (Table 6). Of these nine genera, five belonged to the Firmicutes, indicating that Firmicutes could be the phylum most influenced by the fat-1 gene.
TABLE 5.
Comparison of species abundance at the phylum levels between the fat-1 transgenic cattle and WT cattle
| Taxon | WT | FD | WT/FD | P |
| Euryarchaeota | 214.49 | 45.49 | 2.24 | 0.0076 |
| Proteobacteria | 1,270.39 | 4,715.62 | −1.89 | 0.0319 |
| Spirochaetes | 303.86 | 1,401.20 | −2.21 | 0.0139 |
P < 0.05 indicates significant differences and P < 0.01 indicates extremely significant differences between the fat-1 transgenic cattle and WT cattle.
TABLE 6.
Comparison of the species abundance at the genus levels between the fat-1 transgenic cattle and WT cattle
| Taxon | WT | FD | WT/FD | P | |
| Bacteroidetes | Odoribacter | 9.12 | 0.01 | 9.83 | 0.00080 |
| Bacteroides | 4.39 | 11.48 | −1.39 | 0.0027 | |
| Euryarchaeota | Methanobrevibacter | 100.55 | 36.99 | 1.44 | 0.0327 |
| Spirochaetes | Treponema | 303.86 | 1,401.20 | −2.21 | 0.0221 |
| Firmicutes | Lutispora | 13.39 | 0.84 | 3.99 | 0.0458 |
| Dorea | 130.67 | 509.45 | −1.96 | 0.0320 | |
| Roseburia | 223.24 | 1,093.53 | −2.29 | 0.0023 | |
| Sporobacter | 153.13 | 52.94 | 1.53 | 0.0385 | |
| Acetitomaculum | 0.78 | 3.18 | −2.02 | 0.0411 | |
P < 0.05 indicates significant differences and P < 0.01 indicates extremely significant differences between the fat-1 transgenic cattle and WT cattle.
Analysis of microbial community composition indicated that six phyla dominated the gut microbiota in both transgenic and WT cattle (Fig. 4C, D). Of these six phyla, Firmicutes (58% vs. 52%) and Proteobacteria (6% vs. 2%) were more abundant in the fat-1 transgenic cattle than in the WT cattle, whereas the Bacteroidetes (30% vs. 39%) had lower numbers in the fat-1 transgenic cattle than in the WT. Further analysis of the microbes in the Firmicutes revealed that five genera, Roseburia, Dorea, Oscillibacter, Sporobacter, and Coprococcus, changed markedly (Fig. 4E). Roseburia and Dorea abundances were higher in the fat-1 transgenic cattle than in the WT cattle, whereas Oscillibacter and Sporobacter were lower in the fat-1 transgenic cattle than in the WT cattle. Coprococcus exhibited differences among individual cattle, with lower numbers in the FD005 and FD006 transgenic cattle than in the WT cattle, but similar distribution levels in FD002 and the WT cattle. In the Bacteroidetes, Alistipes was a dominant genus in both the transgenic and WT cattle, but they had lower frequency in the fat-1 transgenic cattle than the WT. On the other hand, Parabacteroides and Paludibacter were more abundant in fat-1 transgenic cattle than in the WT cattle (Fig. 4F). Finally, for genera within the Proteobacteria, Succinivibrio was significantly elevated in fat-1 transgenic cattle, whereas Pseudomonas, Ruminobacter and Campylobacter were significantly decreased (Fig. 4G).
DISCUSSION
Consistent with previous reports involving other fat-1 transgenic animals (11, 12, 15), we confirmed that the fat-1 gene could elevate the level of n-3 PUFAs and reduce that of n-6 PUFAs in transgenic cattle. ALT and AST are the two key indices for the assessment of liver function. If the liver was impaired, ALT and AST levels would be beyond the normal physiological range. Kim et al. (24) reported that mild symptoms were observed in fat-1 transgenic mice when a high-fat diet induced nonalcoholic fatty liver disease. Because of the endogenous synthesis of n-3 PUFAs from n-6 PUFAs, hepatocyte steatosis weakened, and hepatocyte ballooning and fibrosis were ameliorated. More importantly, the ALT and AST levels were within their normal ranges, which suggested that n-3 PUFAs improve liver function (24). Blood glucose provides nutrition to humans and animals as the immediate source of energy. It has been reported that when n-3 PUFAs were lacking in the diet, the risk of type 2 diabetes increased (25). The pathogenesis of type II diabetes is associated with insulin resistance, and n-3 PUFAs may improve insulin sensitivity, promote insulin signal transduction, and reduce plasma triglyceride levels, thereby decreasing morbidity due to type 2 diabetes (26). Additionally, n-3 PUFAs increase the utilization of ambient blood glucose such that the more unsaturated n-3 PUFAs are present, the more insulin secretion is stimulated (27). This finding suggests that n-3 PUFAs may reduce blood glucose levels. Studies have also shown that n-3 PUFAs inhibited the biosynthesis of TC by decreasing the enzymatic activity of HMG-CoA and increasing the enzymatic activity of ACAT and promoted the metabolism of TC into bile, leading to reduction of the total serum TC level (28, 29). A high level of LDL-C, resulting from a sudden blood clot in an artery narrowed by atherosclerosis, is regarded as a significant risk factor for a heart attack. The results presented above include detection of a low ALT level in the 6-month-old transgenic calves; low AST, GLU, TC, and LDL-C levels in the 18-month-old transgenic cattle; and a low GLU level in these cattle at 4 years of age, consistent with the findings of previous studies (24–29). Therefore, our results indicate that fat-1 gene expression may have protective effects on the liver and cardiovascular system.
It has been reported that n-3 PUFAs can modulate the host’s inflammatory and immune responses through several potential mechanisms (5–7). Increased n-3 PUFA levels reduced T cell proliferative capacity, leading to responses to both mitogenic and antigenic stimulation and to decreased pro-inflammatory responses (30). The n-3 PUFAs may directly interact with the NF-kB transcription factor to reduce the expression of inflammatory cytokines resulting in a decrease in pro-inflammatory responses (31). In human clinical studies, n-3 PUFAs have been used to treat autoimmune diseases, such as interleukin-1 (IL-1)-induced coronary heart disease, caducity, depression, cancers, IL-1- and leukotriene-B4-induced arthritis, regional enteritis, ulcerative colitis, and lupus erythematosus (32–37). Recently, Delpech et al. (7) reported that an inflammatory episode was induced in fat-1 and WT mice by injecting bacterial endotoxin (lipopolysaccharide), which resulted in different phenotypes of microglia and the expression of cytokines and chemokines. The abundant n-3 PUFAs in fat-1 mice modulated the brain’s innate immune system activity and improved the protection of animals against lipopolysaccharide-induced pro-inflammatory cytokine production and subsequent spatial memory alteration (7). In the present study, 310 significantly differentially expressed genes were detected in the fat-1 transgenic cattle, and these genes were closely associated with many biological processes associated with immune responses. Some biological processes involved in cytomembrane components or reactions were also detected, suggesting a potential immune response regulatory mechanism modulated by the fat-1 gene’s cytomembrane system reactions. The present results not only confirmed the immune-modulatory properties of n-3 PUFAs but also identified a potential regulatory mechanism for immune responses.
Because fish oil is rich in n-3 PUFAs, especially EPA and DHA, the American Heart Association recommends daily intake of fish oil to reduce the incidence of coronary artery disease (38). Liu et al. (1) confirmed that the intake of EPA and DHA affect the polymorphism of FADS1 rs174547, thereby reducing the incidence of coronary artery disease. Thrombosis may be induced by the common effects of blood coagulation, platelet aggregation, and fibrinolysis. It has been reported that thromboxane A2 could promote platelet aggregation and vasoconstriction, inducing thrombus formation, but n-3 PUFAs could promote production of prostaglandin, which then decreases the synthesis of thromboxane A2, leading to an anti-thrombosis effect (39). Another study showed that high levels of plasminogen activator inhibitor-1 (PAI-1) and tissue plasminogen activator (t-PA) can increase the risk of platelet aggregation, but n-3 PUFAs can reduce the levels of PAI-1 and t-PA (40). In the present study, GO and KEGG enrichment analysis showed that the biological processes associated with thrombus formation, such as fibrinolysis and blood coagulation, were significantly enriched in the fat-1 transgenic and WT cattle. The results corresponded to low levels of TC and LDL-C, which further proved that n-3 PUFAs play important protective roles in the cardiovascular system.
Previous studies have reported no significant changes in the gut microbiota in transgenic pigs, cattle, and sheep (13–15). However, our study presents evidence that the fat-1 gene caused changes in the gut microbiota of transgenic cattle. The most significantly affected bacterial genera are those with metabolic pathways associated with glucose and lipids. For example, Dorea and Roseburia, belonging to the Firmicutes, were highly abundant in the fat-1 transgenic cattle. Dorea is reported to be associated with serum TC and LDL-C levels (41). Roseburia can induce the formation of butyrate from various dietary polysaccharide substrates, is involved in the regulation of glucose metabolism, and may increase the anti-inflammatory properties of the host (42, 43). Among Proteobacteria, a high level of Succinivibrio, which ferments cellulose and starch, was detected in the transgenic cattle, indicating that Succinivibrio played an important role in carbohydrate metabolism (44–46). Alistipes (Bacteroidetes) can improve glucose homeostasis in diabetes by negative regulation of the vitamin D receptor (47). In this study, both GLU and Alistipes were detected at low levels in the fat-1 transgenic cattle; these results were inconsistent with those of a previous study (47), implying that Alistipes might not have a decisive effect of reducing the level of plasma GLU. To our surprise, a significantly lower abundance (more than 9-fold) of Odoribacter was observed in the fat-1 transgenic cattle, indicating that Odoribacter might be the bacterial genus most significantly affected by the inserted fat-1 gene. In mice, Odoribacter was closely associated with stress (48), and the intake of n-3 PUFAs reduced stress responses by changing the composition of the gut microbiota in a rat study (49). Therefore, we speculate that the reduction of Odoribacter probably had an anti-stress effect on the fat-1 transgenic cattle.
CONCLUSIONS
In conclusion, the fat-1 gene promoted an increase in n-3 PUFAs and reduced the ratio of n-6/n-3 PUFAs, functions that are consistent with the expected role of the fat-1 gene. Accordingly, through analysis of blood biochemical indexes, differential gene expression, differential plasma protein expression, and changes in the gut microbiota, we found that elevated n-3 PUFAs improved several biochemical parameters related to liver function, plasma glucose, and plasma lipid metabolism in the fat-1 transgenic cattle. We also found that elevated n-3 PUFAs exerted a positive effect on the fat-1 transgenic cattle’s immune system and cardiovascular system as well as showed anti-stress properties. Furthermore, based on the present results, we speculate that changes in plasma proteins and gut microbiota might originate from differentially expressed genes, but the underlying mechanism remains unknown. Therefore, further investigation into the causes of the changes in gene expression when the fat-1 gene is inserted into the bovine genome remains necessary.
Supplementary Material
Acknowledgments
The authors thank the Tianjin CKRG Biological Engineering Technology & Services Co., Ltd. and the Shanghai Personal Biotechnology Co., Ltd. for providing technical assistance in bioinformatics analysis.
Footnotes
Abbreviations:
- ALA
- α-linolenic acid
- AST
- aspartate aminotransferase
- ALT
- alanine aminotransferase
- CRE
- creatinine
- 2-DE
- two-dimensional gel electrophoresis
- FDR
- false discovery rate
- HDL-C
- high density lipoprotein cholesterol
- LDH
- lactate dehydrogenase
- LDL-C
- low density lipoprotein cholesterol
- OTU
- operational taxonomic unit
- TC
- total cholesterol
- TG
- triglyceride
This work was supported by National Transgenic Animal Program of China Grant 2013ZX08007-002/2014ZX08007-002.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Liu F., Li Z., Lv X., and Ma J.. 2015. Dietary n-3 polyunsaturated fatty acid intakes modify the effect of genetic variation in fatty acid desaturase 1 on coronary artery disease. PLoS One. 10: e0121255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Psota T. L., Gebauer S. K., and Kris-Etherton P.. 2006. Dietary omega-3 fatty acid intake and cardiovascular risk. Am. J. Cardiol. 98: 3i–18i. [DOI] [PubMed] [Google Scholar]
- 3.White P. J., Arita M., Taguchi R., Kang J. X., and Marette A.. 2010. Transgenic restoration of long-chain n-3 fatty acids in insulin target tissues improves resolution capacity and alleviates obesity-linked inflammation and insulin resistance in high-fat-fed mice. Diabetes. 59: 3066–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Algamas-Dimantov A., Yehuda-Shnaidman E., Hertz R., Peri I., Bar-Tana J., and Schwartz B.. 2014. Prevention of diabetes-promoted colorectal cancer by (n-3) polyunsaturated fatty acids and (n-3) PUFA mimetic. Oncotarget. 5: 9851–9863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harbige L. S. 2003. Fatty acids, the immune response, and autoimmunity: a question of n-6 essentiality and the balance between n-6 and n-3. Lipids. 38: 323–341. [DOI] [PubMed] [Google Scholar]
- 6.Gravaghi C., La Perle K. M., Ogrodwski P., Kang J. X., Quimby F., Lipkin M., and Lamprecht S. A.. 2011. Cox-2 expression, PGE(2) and cytokines production are inhibited by endogenously synthesized n-3 PUFAs in inflamed colon of fat-1 mice. J. Nutr. Biochem. 22: 360–365. [DOI] [PubMed] [Google Scholar]
- 7.Delpech J. C., Madore C., Joffre C., Aubert A., Kang J. X., Nadjar A., and Layé S.. 2015. Transgenic increase in n-3/n-6 fatty acid ratio protects against cognitive deficits induced by an immune challenge through decrease of neuroinflammation. Neuropsychopharmacology. 40: 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swanson D., Block R., and Mousa S. A.. 2012. Omega-3 fatty acids EPA and DHA: health benefits throughout life. Adv. Nutr. 3: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X., Pang D., Yuan T., Li Z., Li Z., Zhang M., Ren W., Ouyang H., and Tang X.. 2016. N-3 polyunsaturated fatty acids attenuates triglyceride and inflammatory factors level in hfat-1 transgenic pigs. Lipids Health Dis. 15: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y., Tang Y., Wang S., Zhou J., Zhou J., Lu X., Bai X., Wang X. Y., Chen Z., and Zuo D.. 2016. Endogenous n-3 polyunsaturated fatty acids attenuate T cell-mediated hepatitis via autophagy activation. Front. Immunol. 7: 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang J. X., Wang J., Wu L., and Kang Z. B.. 2004. Transgenic mice: fat-1 mice convert n-6 to n-3 fatty acids. Nature. 427: 504. [DOI] [PubMed] [Google Scholar]
- 12.Lai L., Kang J. X., Li R., Wang J., Witt W. T., Yong H. Y., Hao Y., Wax D. M., Murphy C. N., Rieke A., et al. . 2006. Generation of cloned transgenic pigs rich in omega-3 fatty acids. Nat. Biotechnol. 24: 435–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan D., Zhang L., Zhou Y., Feng C., Long C., Liu X., Wan R., Zhang J., Lin A., Dong E., et al. . 2010. Efficient production of omega-3 fatty acid desaturase (sFat-1)-transgenic pigs by somatic cell nuclear transfer. Sci. China Life Sci. 53: 517–523. [DOI] [PubMed] [Google Scholar]
- 14.Wu X., Ouyang H., Duan B., Pang D., Zhang L., Yuan T., Xue L., Ni D., Cheng L., Dong S., et al. . 2012. Production of cloned transgenic cow expressing omega-3 fatty acids. Transgenic Res. 21: 537–543. [DOI] [PubMed] [Google Scholar]
- 15.Zhang P., Liu P., Dou H., Chen L., Chen L., Lin L., Tan P., Vajta G., Gao J., Du Y., et al. . 2013. Handmade cloned transgenic sheep rich in omega-3 fatty acids. PLoS One. 8: e55941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia Q., Lupton J. R., Smith R., Weeks B. R., Callaway E., Davidson L. A., Kim W., Fan Y. Y., Yang P. Y., Newman R. A., et al. . 2008. Reduced colitis-associated colon cancer in fat-1 (n-3 fatty acid desaturase) transgenic mice. Cancer Res. 68: 3985–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bousquet M., Saint-Pierre M., Julien C., Salem N., Cicchetti F., and Calon F.. 2008. Beneficial effects of dietary omega-3 polyunsaturated fatty acid on toxin-induced neuronal degeneration in an animal model of Parkinson’s disease. FASEB J. 22: 1213–1225. [DOI] [PubMed] [Google Scholar]
- 18.Lebbadi M., Julien C., Phivilay A., Tremblay C., Emond V., Kang J. X., and Calon F.. 2011. Endogenous conversion of omega-6 into omega-3 fatty acids improves neuropathology in an animal model of Alzheimer’s disease. J. Alzheimers Dis. 27: 853–869. [DOI] [PubMed] [Google Scholar]
- 19.Taha A. Y., Huot P. S., Reza-López S., Prayitno N. R., Kang J. X., Burnham W. M., and Ma D. W.. 2008. Seizure resistance in fat-1 transgenic mice endogenously synthesizing high levels of omega-3 polyunsaturated fatty acids. J. Neurochem. 105: 380–388. [DOI] [PubMed] [Google Scholar]
- 20.Siegert E., Paul F., Rothe M., and Weylandt K. H.. 2017. The effect of omega-3 fatty acids on central nervous system remyelination in fat-1 mice. BMC Neurosci. 18: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu X., Zhang F., Leak R. K., Zhang W., Iwai M., Stetler R. A., Dai Y., Zhao A., Gao Y., and Chen J.. 2013. Transgenic overproduction of mega-3 polyunsaturated fatty acids provides neuroprotection and enhances endogenous neurogenesis after stroke. Curr. Mol. Med. 13: 1465–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu Y., Nie D., Witt W. T., Chen Q., Shen M., Xie H., Lai L., Dai Y., and Zhang J.. 2008. Expression of the fat-1 gene diminishes prostate cancer growth in vivo through enhancing apoptosis and inhibiting GSK-3 beta phosphorylation. Mol. Cancer Ther. 7: 3203–3211. [DOI] [PubMed] [Google Scholar]
- 23.Ding J., Berryman D. E., and Kopchick J. J.. 2011. Plasma proteomic profiles of bovine growth hormone transgenic mice as they age. Transgenic Res. 20: 1305–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim E. H., Bae J. S., Hahm K. B., and Cha J. Y.. 2012. Endogenously synthesized n-3 polyunsaturated fatty acids in fat-1 mice ameliorate high-fat diet-induced non-alcoholic fatty liver disease. Biochem. Pharmacol. 84: 1359–1365. [DOI] [PubMed] [Google Scholar]
- 25.Raheja B. S., Sadikot S. M., Phatak R. B., and Rao M. B.. 1993. Significance of the n-6/n-3 ratio for insulin action in diabetes. Ann. N. Y. Acad. Sci. 683: 258–271. [DOI] [PubMed] [Google Scholar]
- 26.Romanatto T., Fiamoncini J., Wang B., Curi R., and Kang J. X.. 2014. Elevated tissue omega-3 fatty acid status prevents age-related glucose intolerance in fat-1 transgenic mice. Biochim. Biophys. Acta. 1842: 186–191. [DOI] [PubMed] [Google Scholar]
- 27.Opara E. C., Garfinkel M., Hubbard V. S., Burch W. M., and Akwari O. E.. 1994. Effect of fatty acids on insulin release: role of chain length and degree of unsaturation. Am. J. Physiol. 266: E635–E639. [DOI] [PubMed] [Google Scholar]
- 28.Ikeda I., Wakamatsu K., Inayoshi A., Imaizumi K., Sugano M., and Yazawa K.. 1994. alpha-Linolenic, eicosapentaenoic and docosahexaenoic acids affect lipid metabolism differently in rats. J. Nutr. 124: 1898–1906. [DOI] [PubMed] [Google Scholar]
- 29.Conquer J. A., and Holub B. J.. 1998. Effect of supplementation with different doses of DHA on the levels of circulating DHA as non-esterified fatty acid in subjects of Asian Indian background. J. Lipid Res. 39: 286–292. [PubMed] [Google Scholar]
- 30.Fowler K. H., Chapkin R. S., and McMurray D. N.. 1993. Effects of purified dietary n-3 ethyl esters on murine T lymphocyte function. J. Immunol. 151: 5186–5197. [PubMed] [Google Scholar]
- 31.Tomio K., Kawana K., Taguchi A., Isobe Y., Iwamoto R., Yamashita A., Kojima S., Mori M., Nagamatsu T., Arimoto T., et al. . 2013. Omega-3 polyunsaturated fatty acids suppress the cystic lesion formation of peritoneal endometriosis in transgenic mouse models. PLoS One. 8: e73085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris M. C., Evans D. A., Bienias J. L., Tangney C. C., Bennett D. A., Wilson R. S., Aggarwal N., and Schneider J.. 2003. Consumption of fish and n-3 fatty acids and risk of incident Alzheimer disease. Arch. Neurol. 60: 940–946. [DOI] [PubMed] [Google Scholar]
- 33.Nemets B., Stahl Z., and Belmaker R. H.. 2002. Addition of omega-3 fatty acid to maintenance medication treatment for recurrent unipolar depressive disorder. Am. J. Psychiatry. 159: 477–479. [DOI] [PubMed] [Google Scholar]
- 34.James M. J., and Cleland L. G.. 1997. Dietary n-3 fatty acids and therapy for rheumatoid arthritis. Semin. Arthritis Rheum. 27: 85–97. [DOI] [PubMed] [Google Scholar]
- 35.Shoda R., Matsueda K., Yamato S., and Umeda N.. 1995. Therapeutic efficacy of n-3 polyunsaturated fatty acid in experimental Crohn’s disease. J. Gastroenterol. 30 (Suppl. 8): 98–101. [PubMed] [Google Scholar]
- 36.Aslan A., and Triadafilopoulos G.. 1992. Fish oil fatty acid supplementation in active ulcerative colitis: a double-blind, placebo-controlled, crossover study. Am. J. Gastroenterol. 87: 432–437. [PubMed] [Google Scholar]
- 37.Clark W. F., Parbtani A., Huff M. W., Reid B., Holub B. J., and Falardeau P.. 1989. Omega-3 fatty acid dietary supplementation in systemic lupus erythematosus. Kidney Int. 36: 653–660. [DOI] [PubMed] [Google Scholar]
- 38.Kris-Etherton P. M., Harris W. S., and Appel L. J.; American Heart Association Nutrition Committee. 2002. Fish consumption, fish oil, omega-3 fatty acids, and cardiovascular disease. Circulation. 106: 2747–2757. [DOI] [PubMed] [Google Scholar]
- 39.de Lorgeril M., Salen P., Martin J. L., Monjaud I., Delaye J., and Mamelle N.. 1999. Mediterranean diet, traditional risk factors, and the rate of cardiovascular complications after myocardial infarction: final report of the Lyon Diet Heart Study. Circulation. 99: 779–785. [DOI] [PubMed] [Google Scholar]
- 40.Montegaard C., Tulk H. M., Lauritzen L., Tholstrup T., and Robinson L. E.. 2010. Acute ingestion of long-chain (n-3) polyunsaturated fatty acids decreases fibrinolysis in men with metabolic syndrome. J. Nutr. 140: 38–43. [DOI] [PubMed] [Google Scholar]
- 41.Chen D., Yang Z., Chen X., Huang Y., Yin B., Guo F., Zhao H., Zhao T., Qu H., Huang J., et al. . 2014. The effect of Lactobacillus rhamnosus hsryfm 1301 on the intestinal microbiota of a hyperlipidemic rat model. BMC Complement. Altern. Med. 14: 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scott K. P., Martin J. C., Chassard C., Clerget M., Potrykus J., Campbell G., Mayer C. D., Young P., Rucklidge G., Ramsay A. G., et al. . 2011. Substrate-driven gene expression in Roseburia inulinivorans: importance of inducible enzymes in the utilization of inulin and starch. Proc. Natl. Acad. Sci. USA. 108 (Suppl. 1): 4672–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vital M., Penton C. R., Wang Q., Young V. B., Antonopoulos D. A., Sogin M. L., Morrison H. G., Raffals L., Chang E. B., Huffnagle G. B., et al. . 2013. A gene-targeted approach to investigate the intestinal butyrate-producing bacterial community. Microbiome. 1: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Herrin S. M., and Kenealy W. R.. 1993. Glucose and carbon dioxide metabolism by Succinivibrio dextrinosolvens. Appl. Environ. Microbiol. 59: 748–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van den Abbeele P., Van de Wiele T., Verstraete W., and Possemiers S.. 2011. The host selects mucosal and luminal associations of coevolved gut microorganisms: a novel concept. FEMS Microbiol. Rev. 35: 681–704. [DOI] [PubMed] [Google Scholar]
- 46.Li Z., Zhang Z., Xu C., Zhao J., Liu H., Fan Z., Yang F., Wright A. D., and Li G.. 2014. Bacteria and methanogens differ along the gastrointestinal tract of Chinese roe deer (Capreolus pygargus). PLoS One. 9: e114513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin D., Wu S., Zhang Y. G., Lu R., Xia Y., Dong H., and Sun J.. 2015. Lack of vitamin D receptor causes dysbiosis and changes the functions of the murine intestinal microbiome. Clin. Ther. 37: 996–1009.e7. [DOI] [PubMed] [Google Scholar]
- 48.Bangsgaard Bendtsen K. M., Krych L., Sorensen D. B., Pang W., Nielsen D. S., Josefsen K., Hansen L. H., Sorensen S. J., and Hansen A. K.. 2012. Gut microbiota composition is correlated to grid floor induced stress and behavior in the BALB/c mouse. PLoS One. 7: e46231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pusceddu M. M., El Aidy S., Crispie F., O’Sullivan O., Cotter P., Stanton C., Kelly P., Cryan J. F., and Dinan T. G.. 2015. N-3 polyunsaturated fatty acids (PUFAs) reverse the impact of early-life stress on the gut microbiota. PLoS One. 10: e0139721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.